We narrowed to 10,309 results for: yeast
-
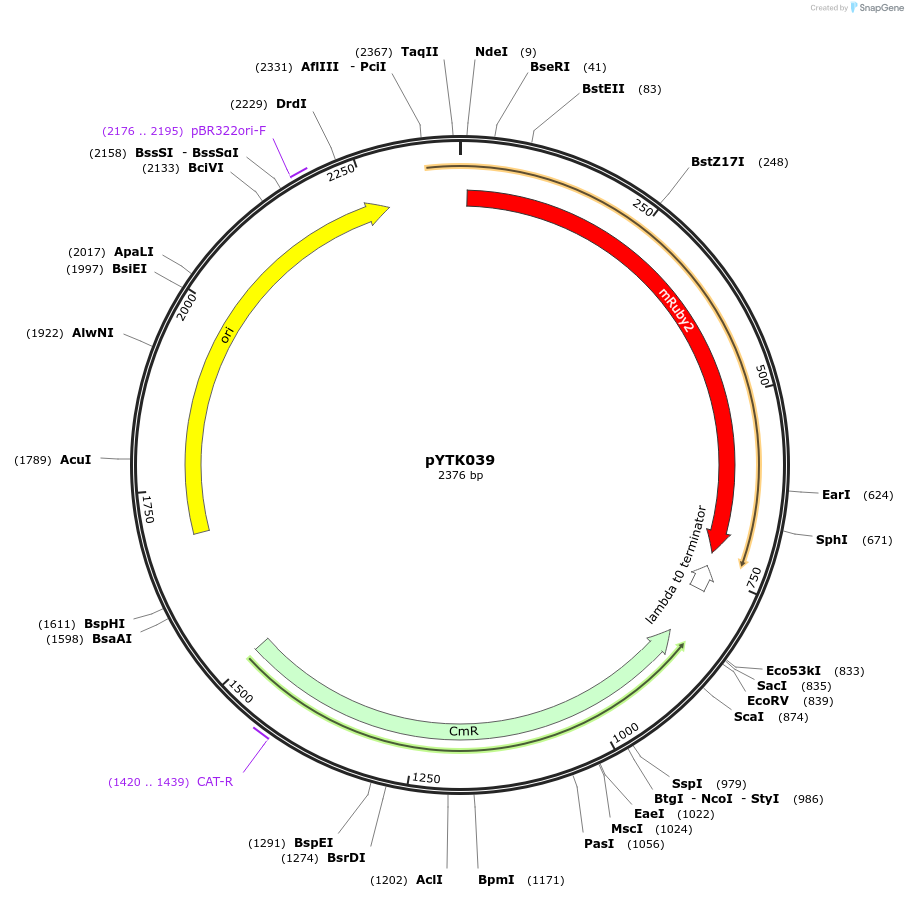
Plasmid#65146PurposeEncodes mRuby2 as a Type 3a part to be used in the Dueber YTK systemDepositorInsertmRuby2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
pYTK040
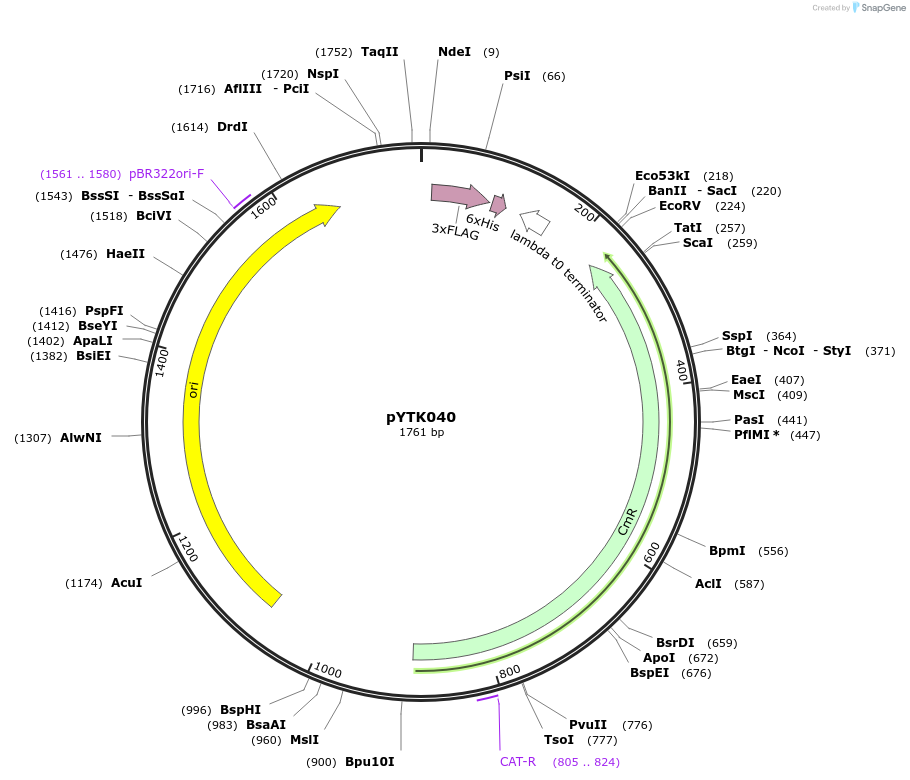
Plasmid#65147PurposeEncodes 6XHis_3XFlag as a Type 3a part to be used in the Dueber YTK systemDepositorInsert6XHis_3XFlag
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
pYTK044
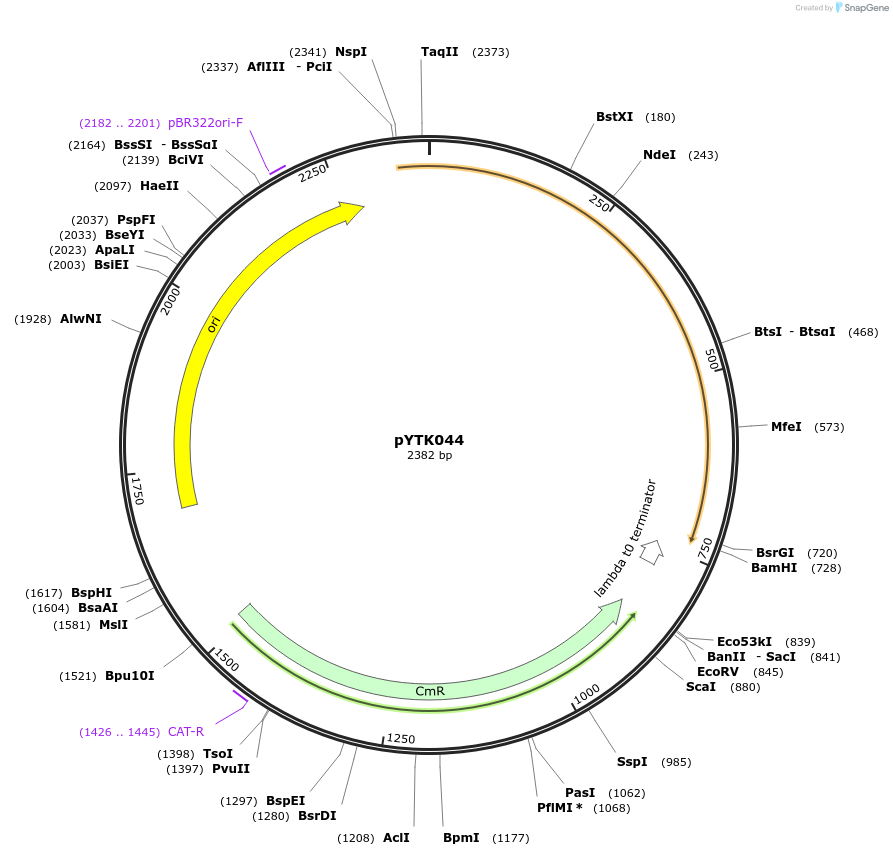
Plasmid#65151PurposeEncodes mTurquoise2 as a Type 3b part to be used in the Dueber YTK systemDepositorInsertmTurquoise2
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
pYTK045
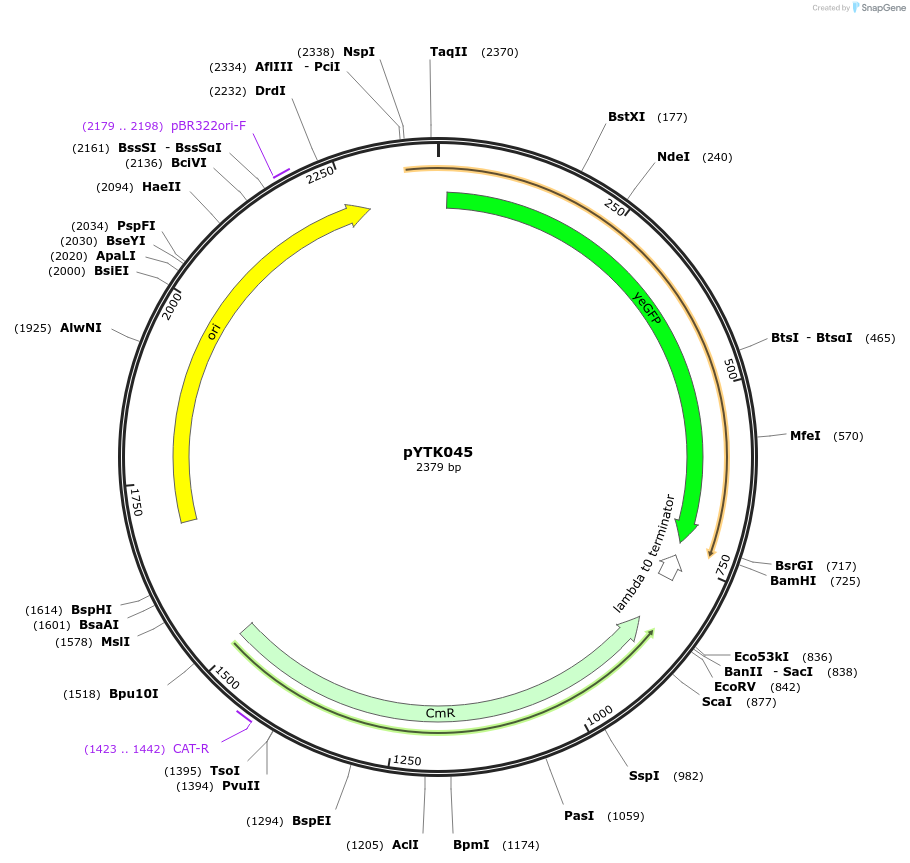
Plasmid#65152PurposeEncodes Venus as a Type 3b part to be used in the Dueber YTK systemDepositorInsertVenus
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
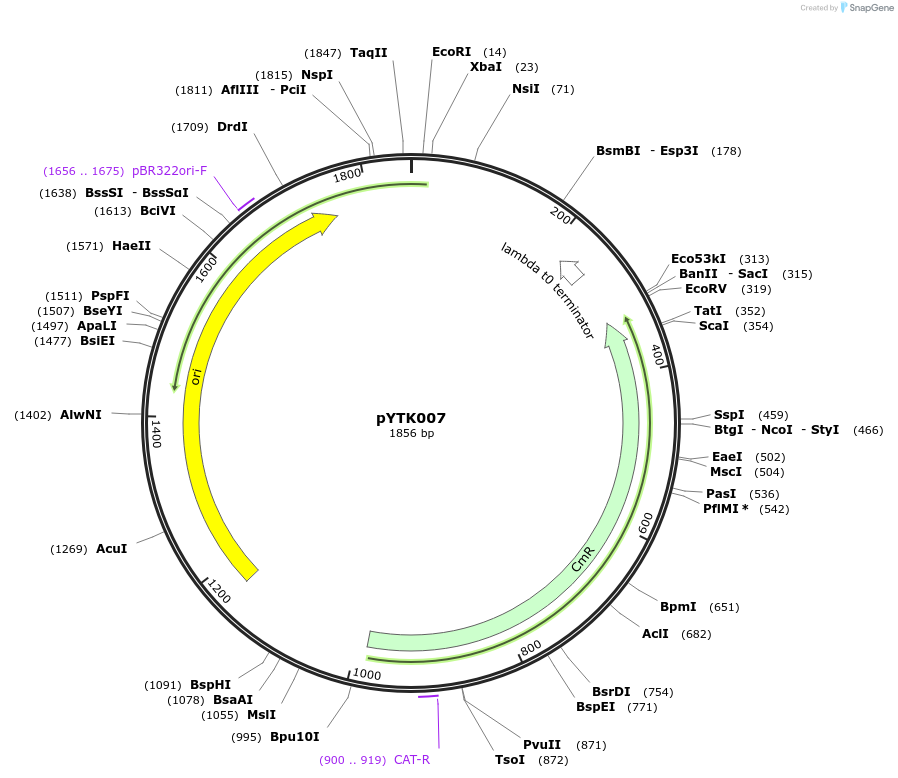
pYTK007
Plasmid#65114PurposeEncodes ConL5 as a Type 1 part to be used in the Dueber YTK systemDepositorInsertConL5
ExpressionBacterialAvailable SinceJune 16, 2015AvailabilityAcademic Institutions and Nonprofits only -
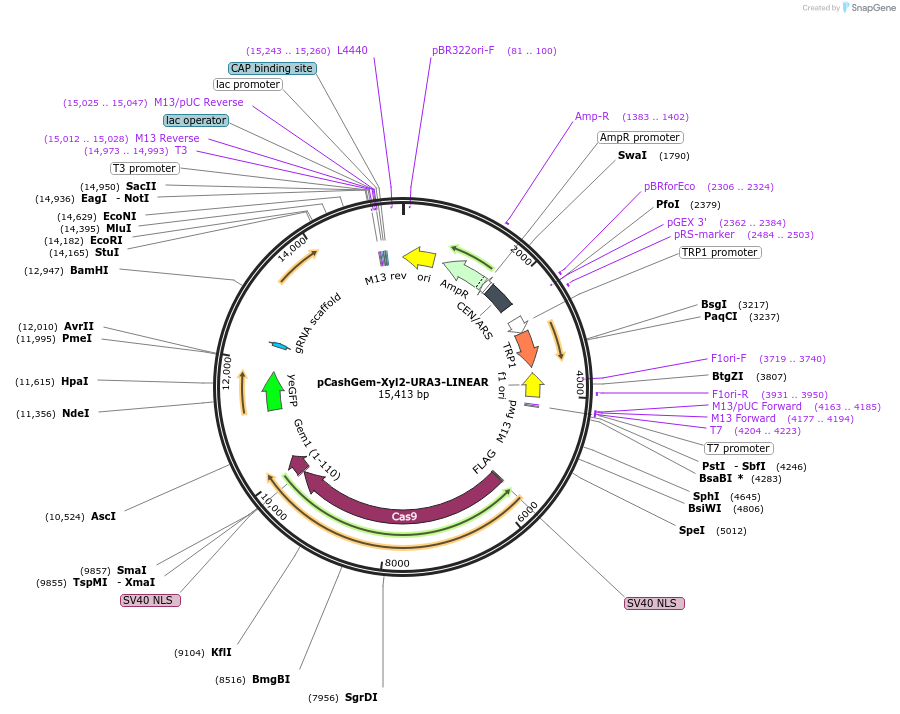
pCashGem-Xyl2-URA3-LINEAR
Plasmid#174840PurposepRS414 backbone containing LINEAR fragment targeting XYL2 in S. stipitisDepositorInsertCodon optimized Cas9:hGem
UseCRISPR and Synthetic BiologyTagshGeminin tagExpressionBacterial and YeastPromoterENO1pAvailable SinceJan. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
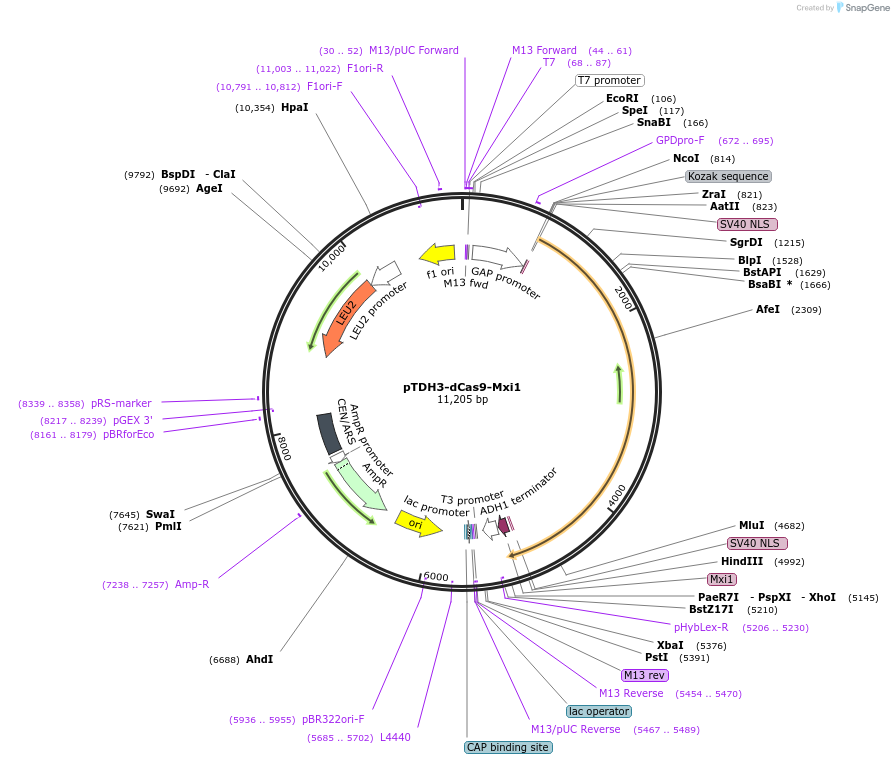
pTDH3-dCas9-Mxi1
Plasmid#46921PurposeYeast CEN/ARS vector (Leu2) that contains dCas9 fused to NLS and Mxi1 domain controlled by TDH3 promoterDepositorInsertdCas9-Mxi1
UseCRISPRExpressionYeastPromoterTDH3Available SinceOct. 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
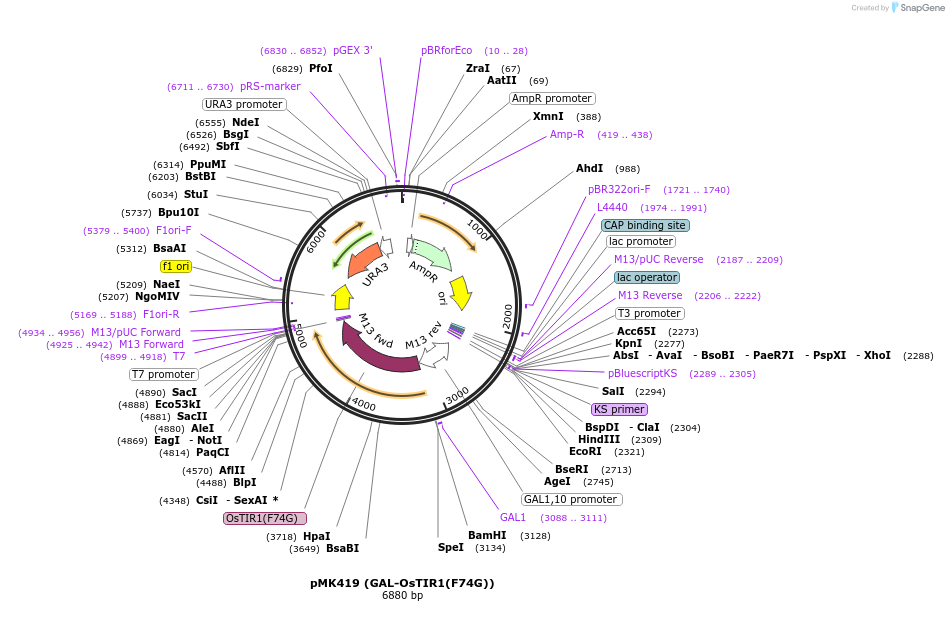
pMK419 (GAL-OsTIR1(F74G))
Plasmid#140656PurposeGAL-OsTIR1(F74G)DepositorInsertGAL-OsTIR1(F74G)
ExpressionYeastAvailable SinceNov. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
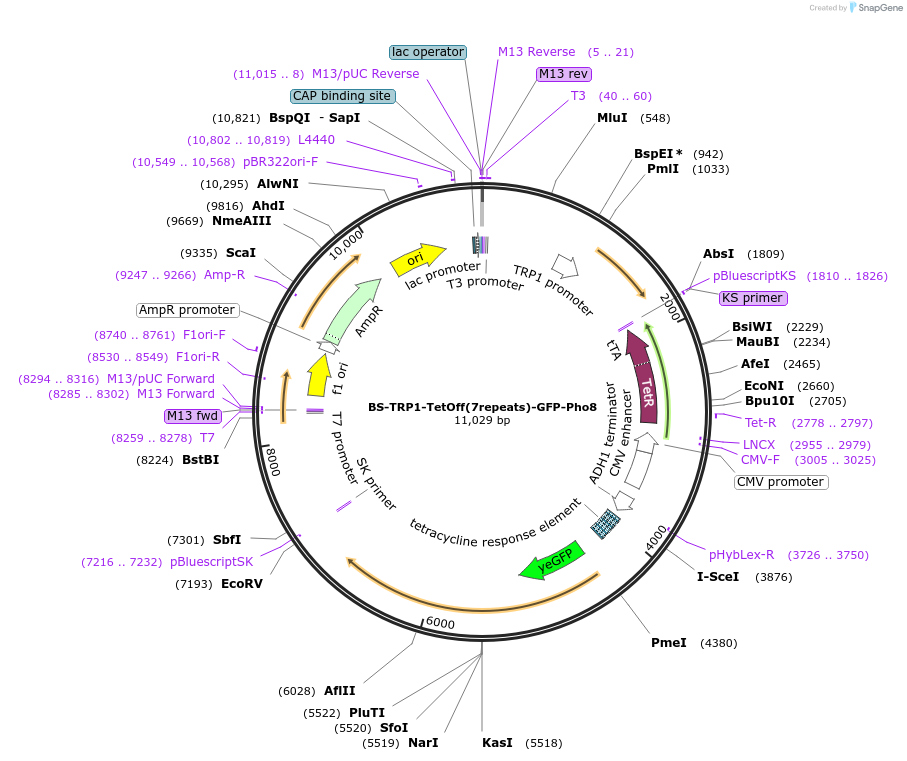
BS-TRP1-TetOff(7repeats)-GFP-Pho8
Plasmid#207011PurposeTet-Off controlled GFP-Pho8 with TRP1 selection. Tetracycline-controlled transactivator (tTA) present on same plasmid.DepositorInsertPho8
TagsGFPExpressionYeastAvailable SinceJune 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
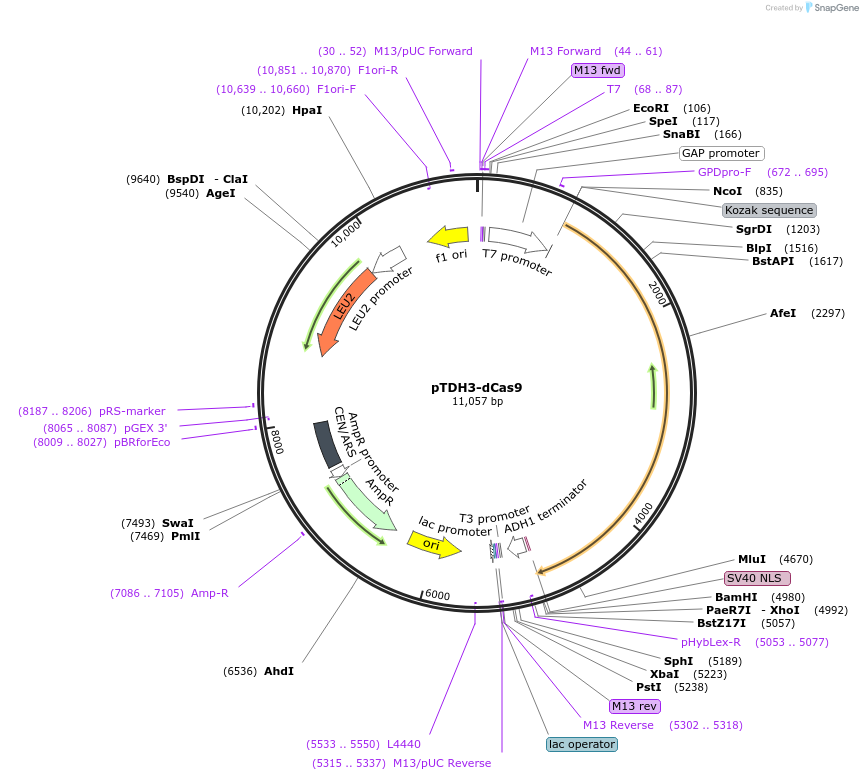
pTDH3-dCas9
Plasmid#46920PurposeYeast CEN/ARS vector (Leu2) that contains dCas9 fused to NLS controlled by TDH3 promoterDepositorInsertdCas9
UseCRISPRExpressionYeastPromoterTDH3Available SinceOct. 1, 2013AvailabilityAcademic Institutions and Nonprofits only -
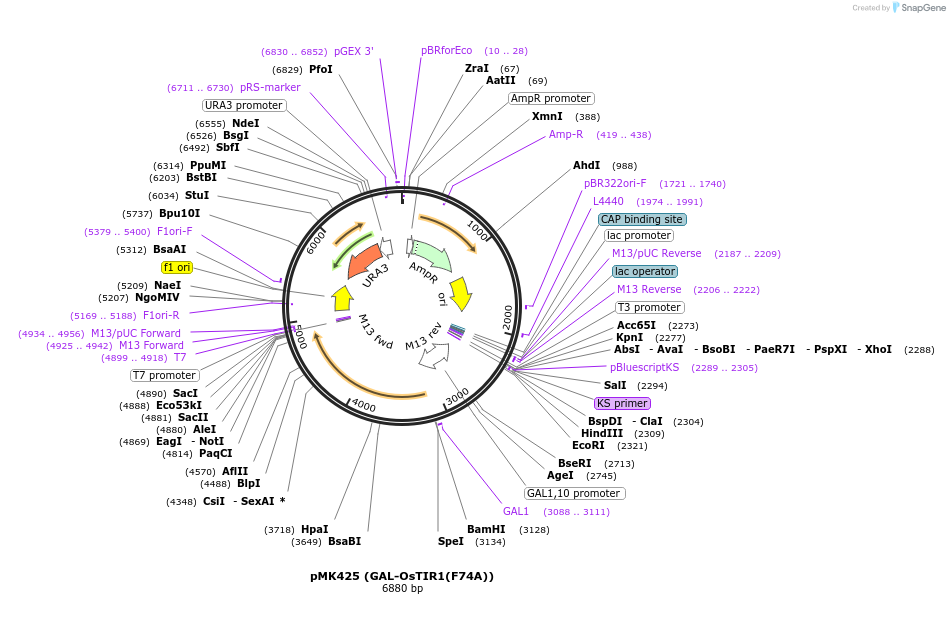
pMK425 (GAL-OsTIR1(F74A))
Plasmid#140657PurposeGAL-OsTIR1(F74A)DepositorInsertGAL-OsTIR1(F74A)
ExpressionYeastAvailable SinceNov. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
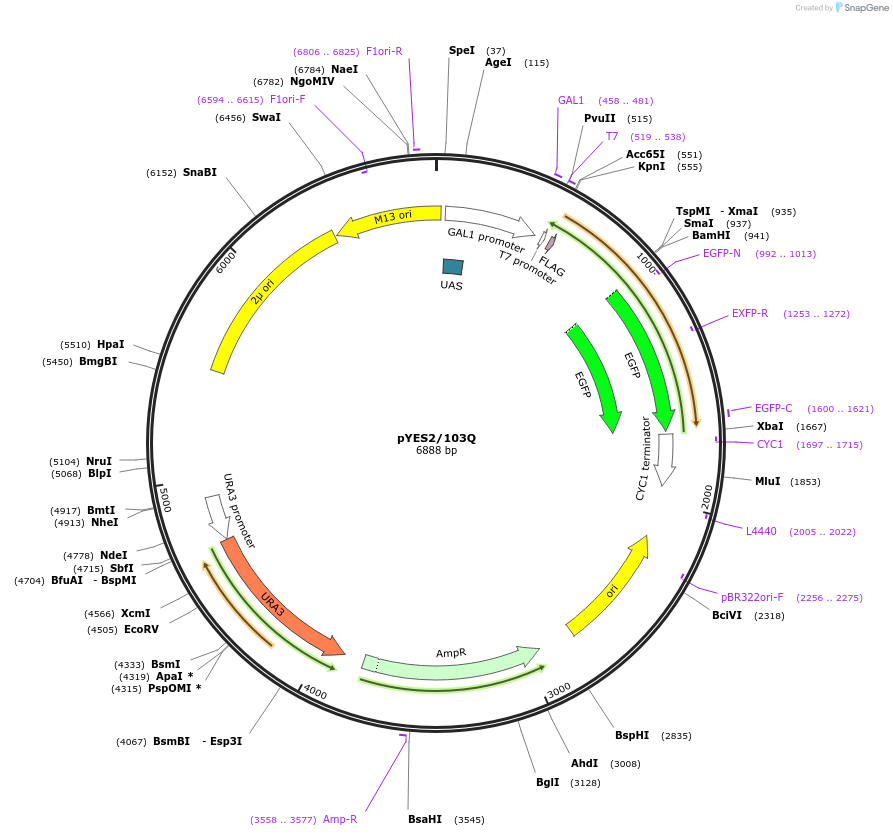
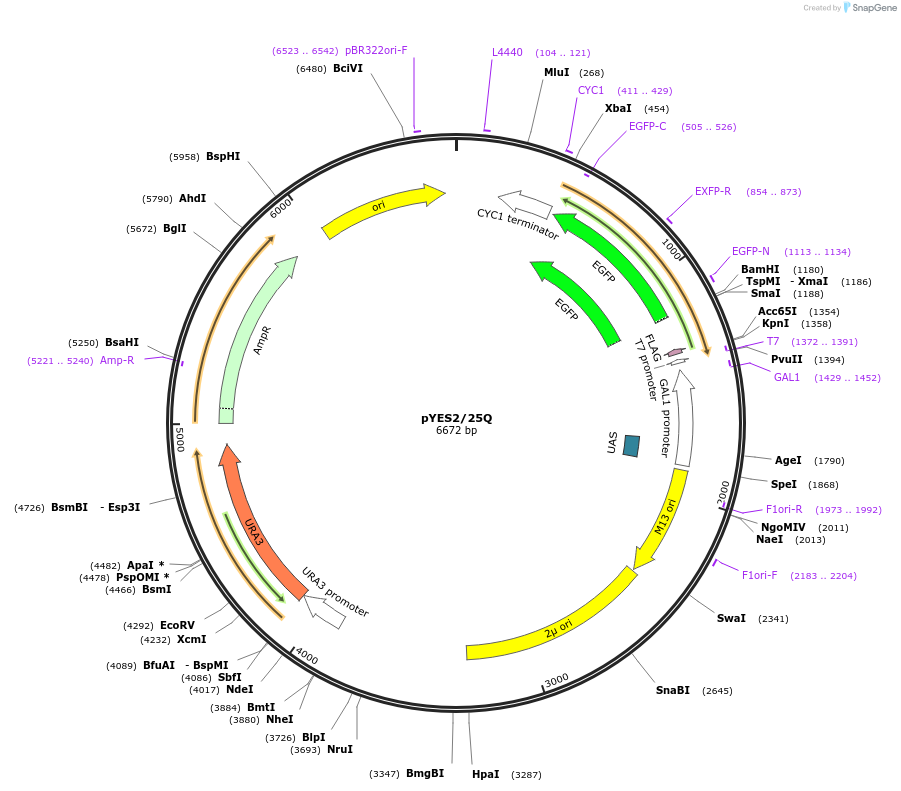
pYES2/103Q
Plasmid#1385DepositorInserthuntingtin (103Q) (HTT Human)
TagsEGFP and FLAGExpressionYeastMutationExon1 sequences containing the first 17 amino aci…Available SinceFeb. 12, 2009AvailabilityAcademic Institutions and Nonprofits only -
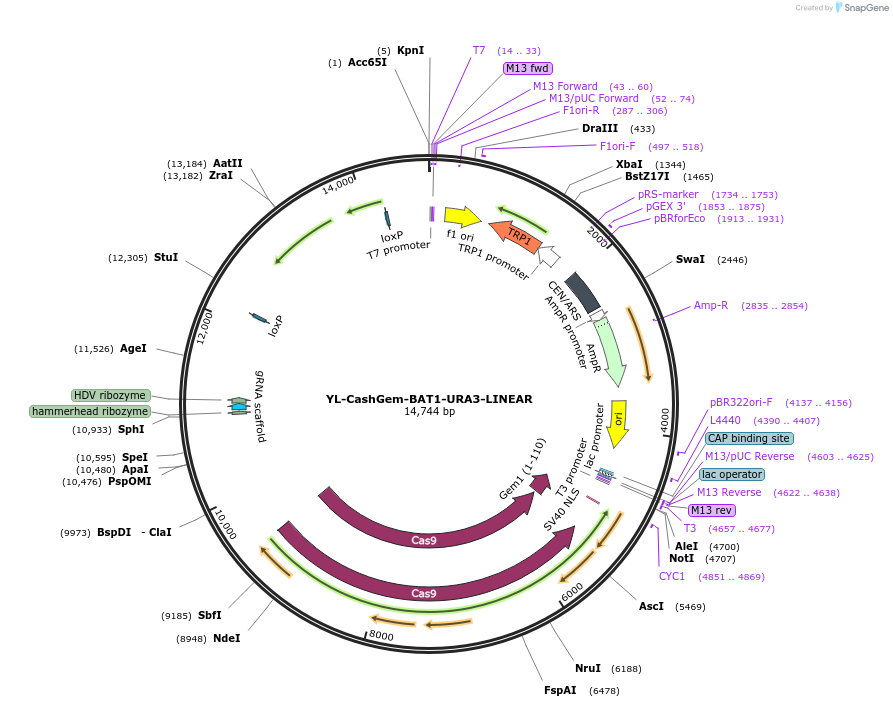
YL-CashGem-BAT1-URA3-LINEAR
Plasmid#174837PurposepRS414 backbone containing LINEAR fragment targeting BAT1 in Y. lipolyticaDepositorInsertCodon optimized Cas9:hGem
UseCRISPR and Synthetic BiologyTagshGeminin tagExpressionBacterial and YeastPromoterGPDpAvailable SinceJan. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
-
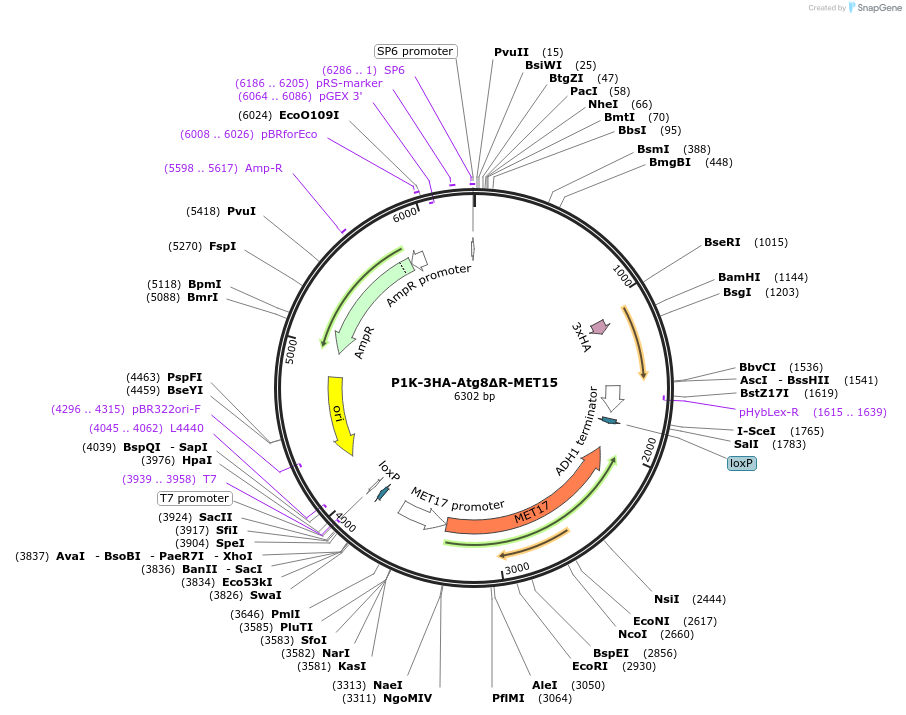
P1K-3HA-Atg8∆R-MET15
Plasmid#207034PurposeExpression of 3HA-Atg8∆R. This variant bypasses the need of Atg4 for initial C-terminal processing.DepositorInsertATG8
Tags3xHAExpressionYeastMutation∆RPromoterpATG8Available SinceFeb. 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
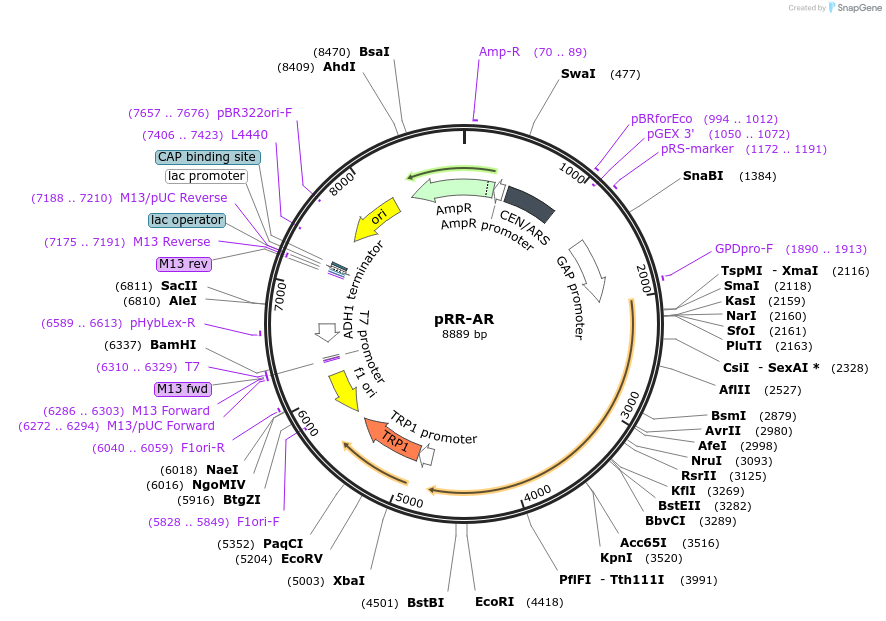
pRR-AR
Plasmid#73045PurposeConstitutively expresses the androgen receptor, which is required for the activation of the promoter in pLAREG (Addgene plasmid #73045)DepositorInsertAndrogen receptor (AR Human)
ExpressionYeastMutationplease see depositor comments belowPromoterglyceraldehyde phosphate dehydrogenase (GPD)Available SinceFeb. 2, 2016AvailabilityAcademic Institutions and Nonprofits only -
pFA6a-kanMX6-P41nmt1-GFP
Plasmid#39290DepositorInsertnmt1 promoter (nmt1 Fission Yeast)
UseYeast genomic targetingTagsA. gossypii translation elongation factor 1a gene…Mutation-1163 to +6 of nmt1 locus with a 4bp deletion (TA…Available SinceAug. 20, 2012AvailabilityAcademic Institutions and Nonprofits only -
pRR-ERbeta-5Z
Plasmid#23062DepositorInsertERbeta (ESR2 Human)
ExpressionYeastAvailable SinceJan. 27, 2010AvailabilityAcademic Institutions and Nonprofits only -
pFA6a-kanMX6-P81nmt1-GFP
Plasmid#39291DepositorInsertnmt1 promoter (nmt1 Fission Yeast)
UseYeast genomic targetingTagsA. gossypii translation elongation factor 1a gene…Mutation-1163 to +6 of nmt1 locus with a 7bp deletion (AT…Available SinceAug. 20, 2012AvailabilityAcademic Institutions and Nonprofits only -
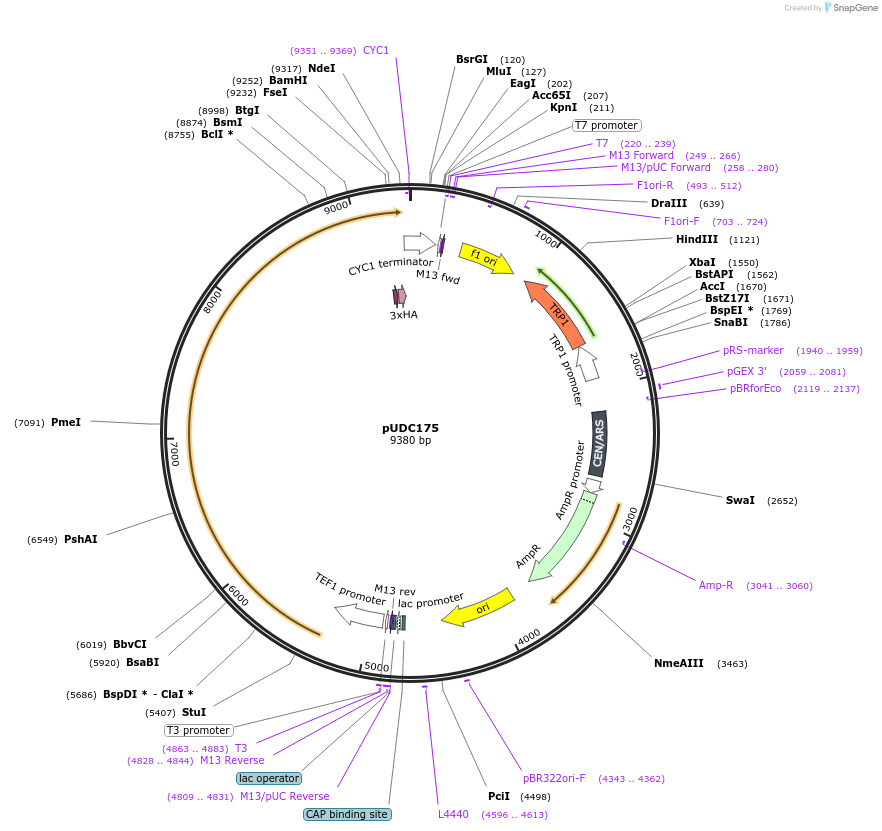
pUDC175
Plasmid#103019PurposeS. cerevisae centromic plasmid harboring Fncpf1 under control of TEF1 promoterDepositorInsertFncpf1
Tags3HA and NLSExpressionYeastMutationhuman codon optimizedPromoterTEF1Available SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only