We narrowed to 23,783 results for: c myc
-
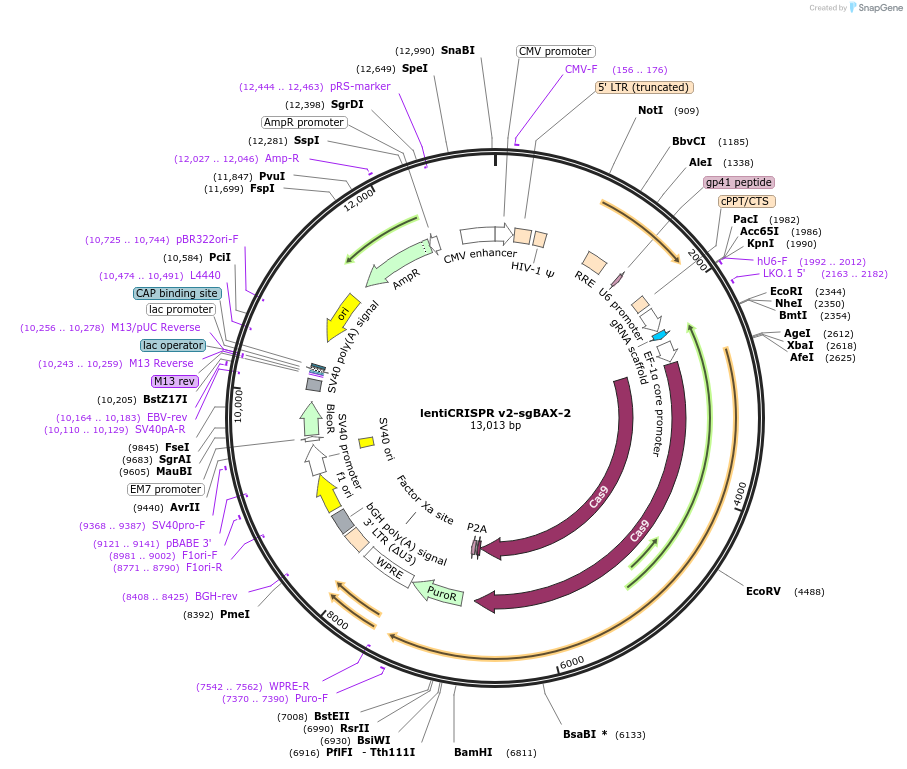
Plasmid#210130Purposeknock out BAX in mammalian cellsDepositorInsertApoptosis regulator BAX (BAX Human)
UseCRISPRAvailable SinceNov. 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
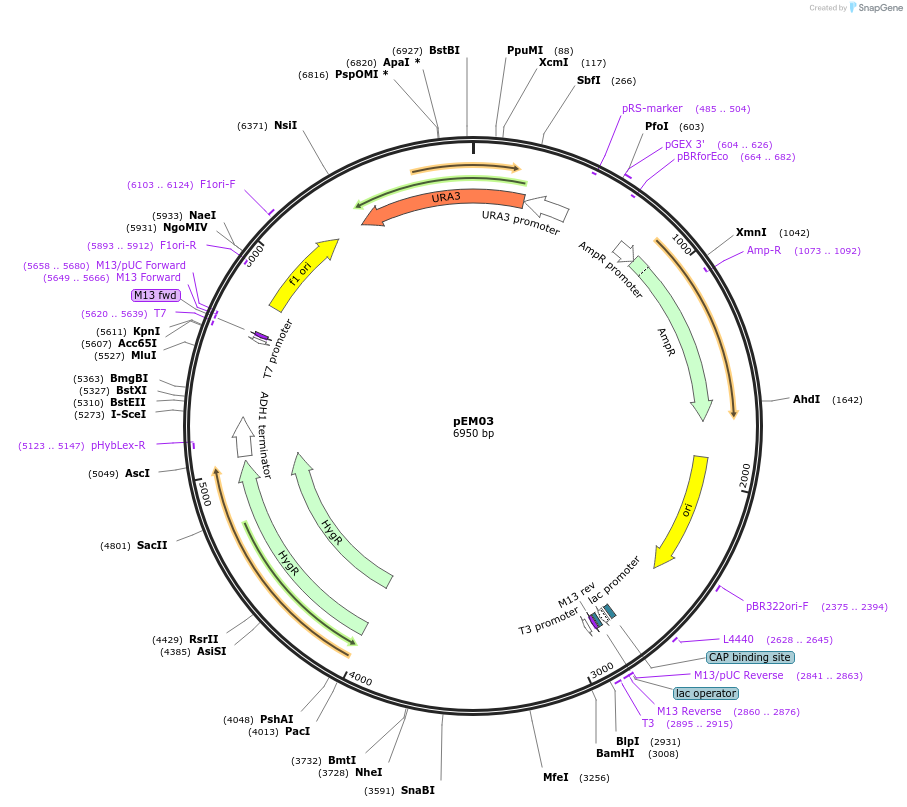
pEM03
Plasmid#135417PurposeExpression of Hygromycin resistance from Spizellomyces Hsp70 promoterDepositorInsertSpunHsp70pr
ExpressionBacterial and YeastPromoterHsp70prAvailable SinceMay 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
Human sgRNA library Gattinara in pRDA_118 backbone
Pooled Library#136986PurposeDesigned for assays with limited cell numbers with 2 guides per gene. Compatible with John Doench’s Brunello human genome-wide library.DepositorExpressionMammalianSpeciesHomo sapiensUseCRISPR and LentiviralAvailable SinceFeb. 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
DNA repair library with 1,573 sgRNAs
Pooled Library#177663PurposeCRISPR-interference (CRISPRi) library of 1,573 sgRNAs targeting 476 genes encoding factors involved in DNA repair and associated processesDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
Mouse sgRNA library Gouda in pRDA_118 backbone
Pooled Library#136987PurposeDesigned for assays with limited cell numbers with 2 guides per gene. Compatible with John Doench’s Brie mouse genome-wide library.DepositorExpressionMammalianSpeciesMus musculusUseCRISPR and LentiviralAvailable SinceFeb. 24, 2020AvailabilityAcademic Institutions and Nonprofits only -
DNA repair library with 336 sgRNAs
Pooled Library#177664PurposeCRISPR-interference (CRISPRi) library of 366 sgRNAs targeting 118 genes encoding factors involved in DNA repair and associated processesDepositorSpeciesHomo sapiensUseCRISPRAvailable SinceNov. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pSMPUW_6.16_ M4_Ef1a-gateway-V5-SV40-Zeo
Plasmid#128452Purposelentiviral destination vector with Ef1a promoter, C-terminal V5 tag, SV40 promoter, and Zeo Resistance geneDepositorInsertEf1a promoter, C-terminal V5 tag, SV40 promoter, and Zeocin Resistance gene
UseLentiviralAvailable SinceJan. 31, 2020AvailabilityAcademic Institutions and Nonprofits only -
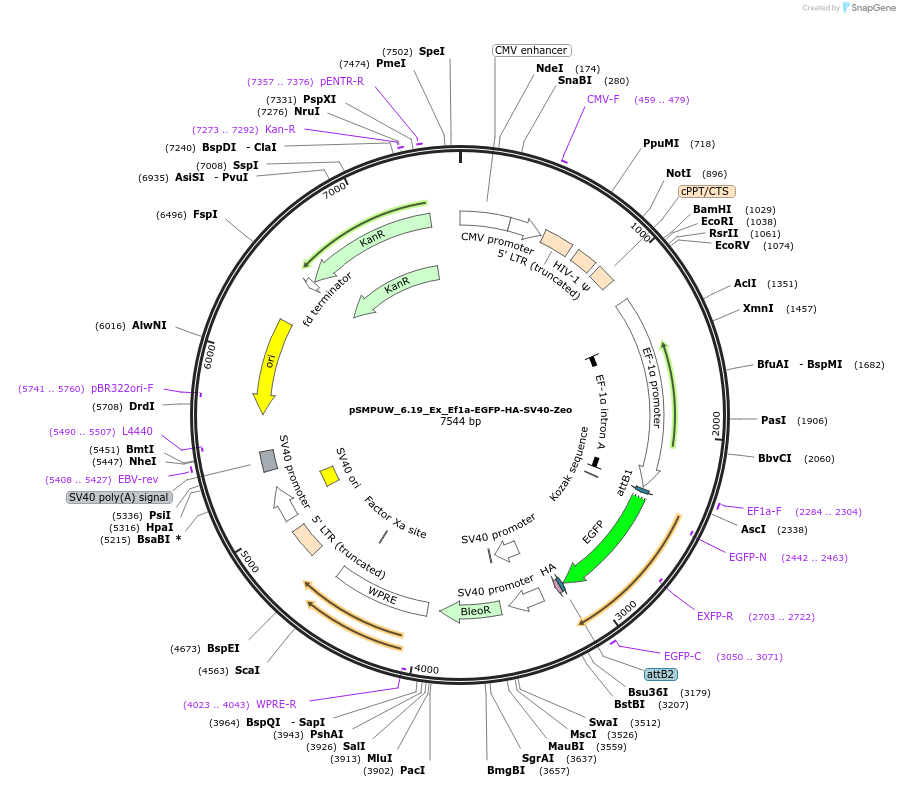
pSMPUW_6.19_Ex_Ef1a-EGFP-HA-SV40-Zeo
Plasmid#128455Purposelentiviral expression vector with Ef1a promoter, EGFP gene, C-terminal HA tag, SV40 promoter, and Zeo Resistance geneDepositorInsertEf1a promoter, EGFP gene, C-terminal HA tag, SV40 promoter, and Zeocin Resistance gene
UseLentiviralAvailable SinceSept. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
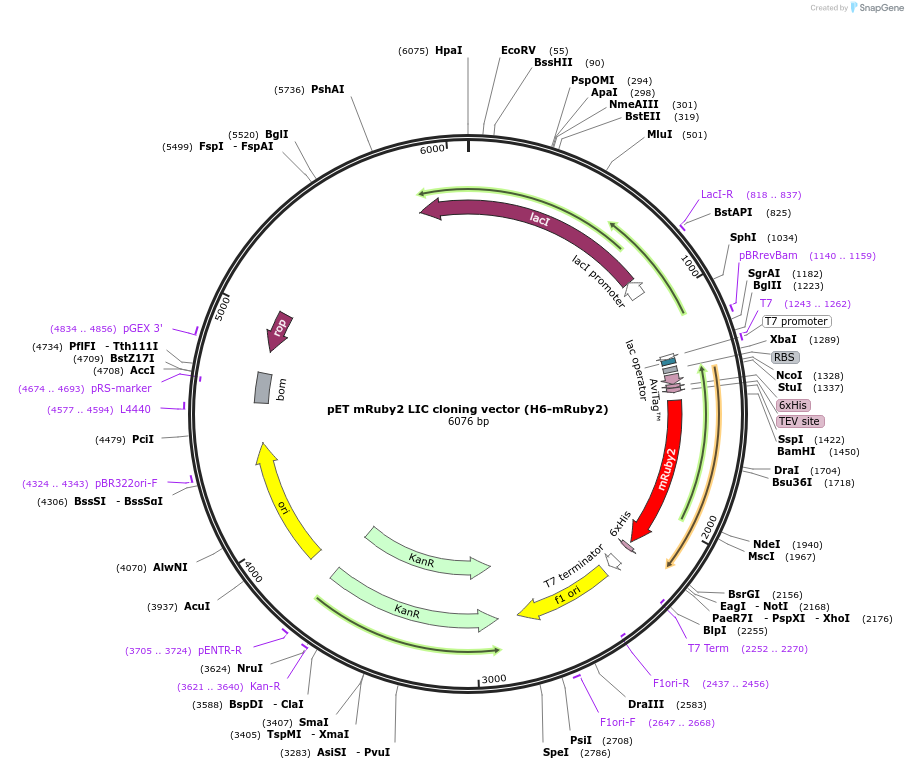
pET mRuby2 LIC cloning vector (H6-mRuby2)
Plasmid#86934PurposepET LIC cloning vector for Biotin-His6-tev-yORF-mRuby2DepositorTypeEmpty backboneTags6xHis, AviTag (biotin), TEV cleavage site, and mR…ExpressionBacterialPromoterT7-lacO (lactose/IPTG inducible)Available SinceMarch 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
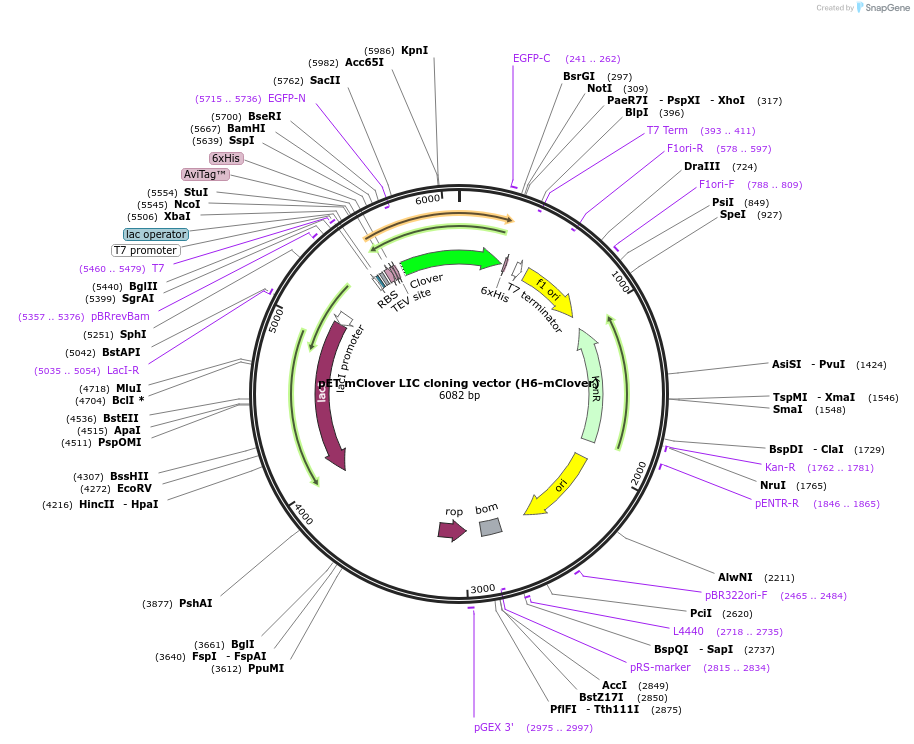
pET mClover LIC cloning vector (H6-mClover)
Plasmid#86931PurposepET LIC cloning vector for Biotin-His6-tev-yORF-mCloverDepositorTypeEmpty backboneTags6xHis, AviTag (biotin), TEV cleavage site, and mC…ExpressionBacterialPromoterT7-lacO (lactose/IPTG inducible)Available SinceMarch 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
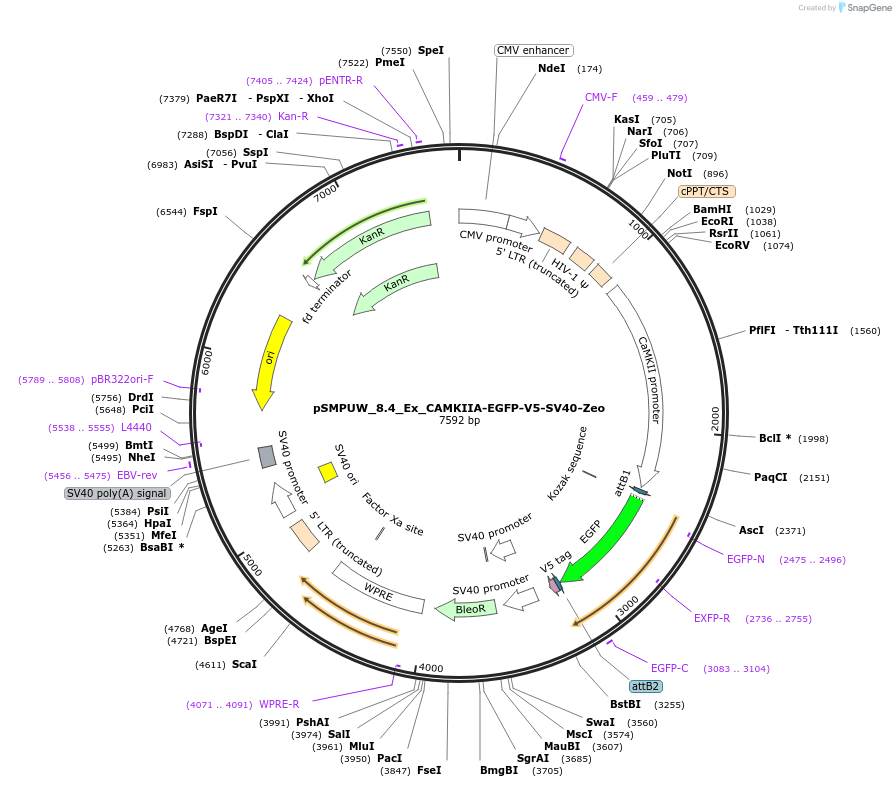
pSMPUW_8.4_Ex_CAMKIIA-EGFP-V5-SV40-Zeo
Plasmid#128464Purposelentiviral expression vector with CAMKIIA promoter, EGFP gene, C-terminal V5 tag, SV40 promoter, and Zeo Resistance geneDepositorInsertCAMKIIA promoter, EGFP gene, C-terminal V5 tag, SV40 promoter, and Zeocin Resistance gene
UseLentiviralAvailable SinceSept. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
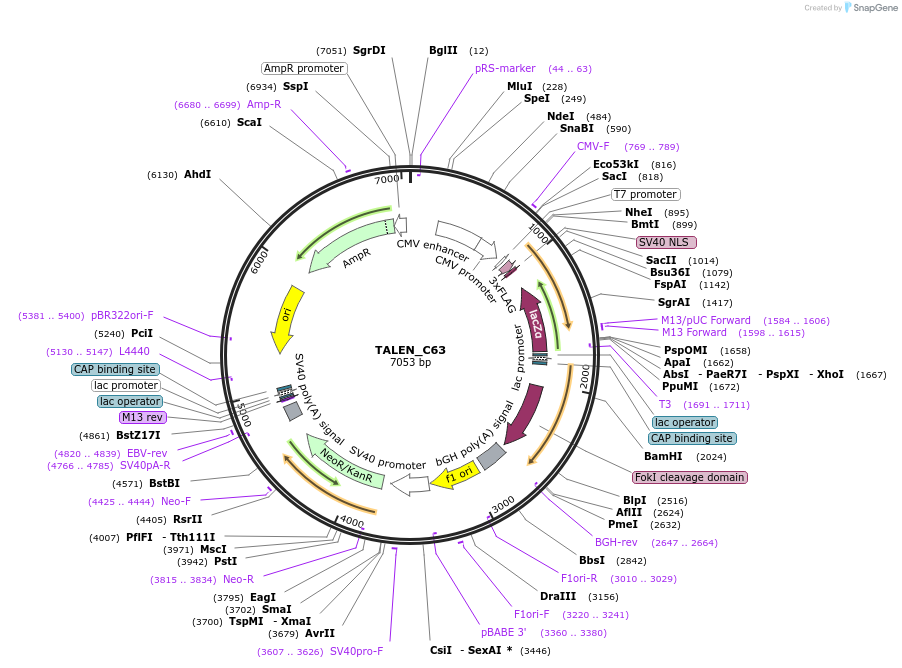
TALEN_C63
Plasmid#40788PurposeGolden Gate Compatible TALEN Construct for site specific modification of genome, +63 C Terminus, Codon Optimized FokIDepositorInsertTranscription Activator Like Effector Nuclease
UseTALENExpressionMammalianMutation+63 C Terminus, Codon Optimized FokI EndonucleasePromoterCMVAvailable SinceDec. 12, 2013AvailabilityIndustry, Academic Institutions, and Nonprofits -
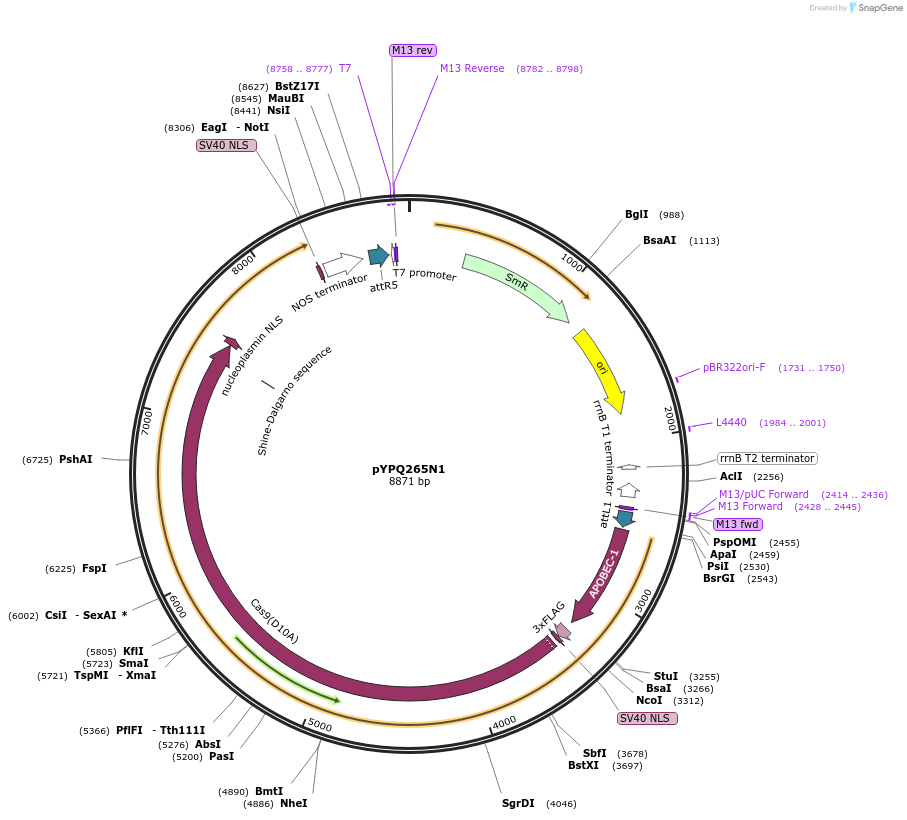
pYPQ265N1
Plasmid#173998PurposeGateway entry clone (attL1 & attR5) rAPOBEC1-zCas9(D10A)-UNG for C-G base editingDepositorInsertrAPOBEC1-zCas9(D10A)-UNG
UseCRISPRTags3X FLAG, NLSExpressionPlantMutationD10AAvailable SinceOct. 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
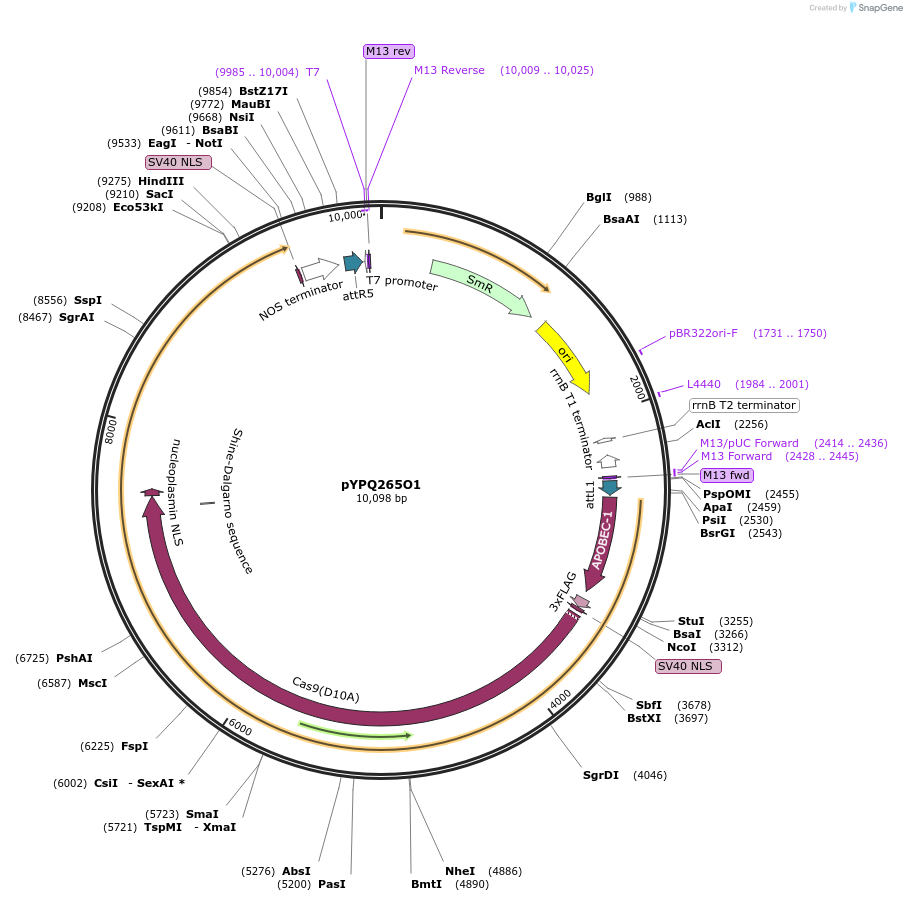
pYPQ265O1
Plasmid#173999PurposeGateway entry clone (attL1 & attR5) rAPOBEC1-zCas9(D10A)-rXRCC1 for C-G base editingDepositorInsertrAPOBEC1-zCas9(D10A)-rXRCC1
UseCRISPRTags3X FLAG, NLSExpressionPlantMutationD10AAvailable SinceOct. 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
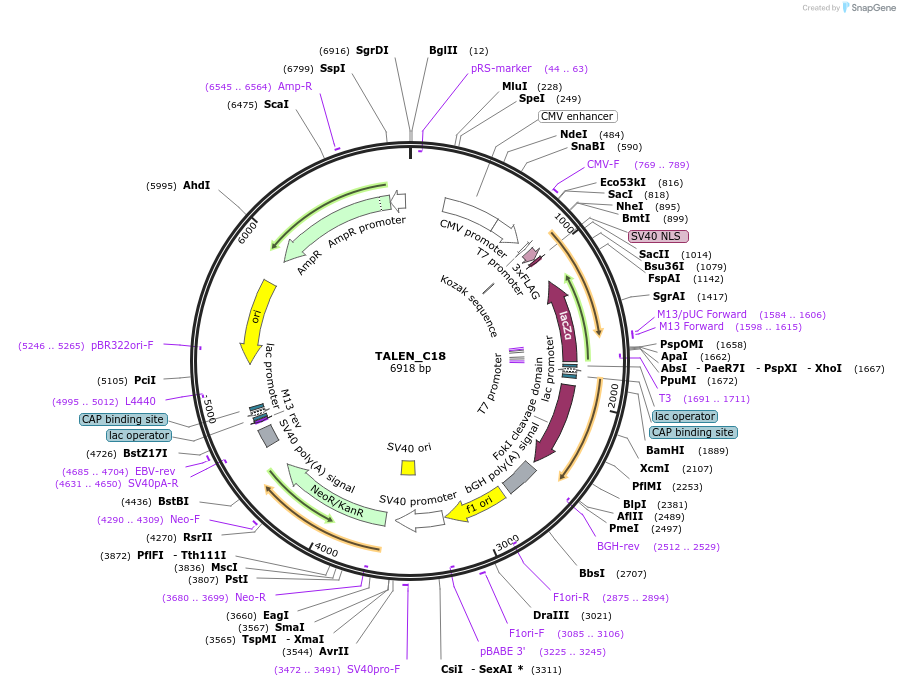
TALEN_C18
Plasmid#47974PurposeGolden Gate Compatible TALEN Construct site specific modification of genome, shortened C Terminus, Codon Optimized FokIDepositorInsertTranscription Activator Like Effector Nuclease
UseTALENExpressionMammalianMutationShortened C-Terminus,Codon Optimized FokI Endonuc…PromoterCMVAvailable SinceDec. 12, 2013AvailabilityIndustry, Academic Institutions, and Nonprofits -
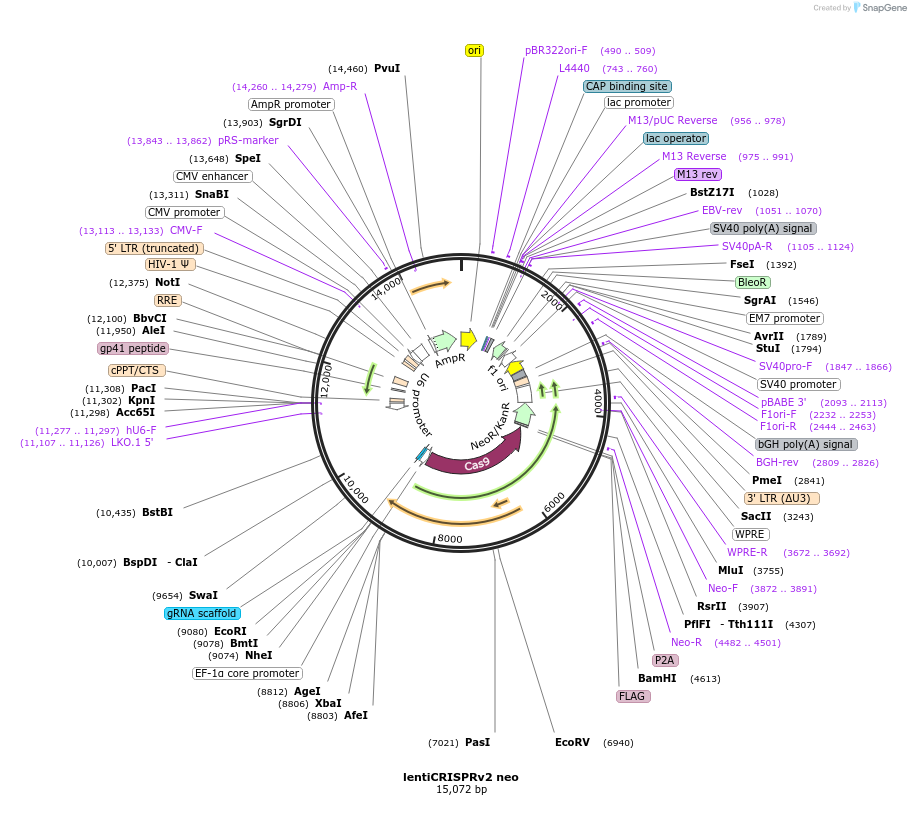
lentiCRISPRv2 neo
Plasmid#98292PurposeVariant of lentiCRISPRv2 that confers G418 resistanceDepositorHas ServiceCloning Grade DNATypeEmpty backboneUseCRISPR and LentiviralPromoterEF-1aAvailable SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
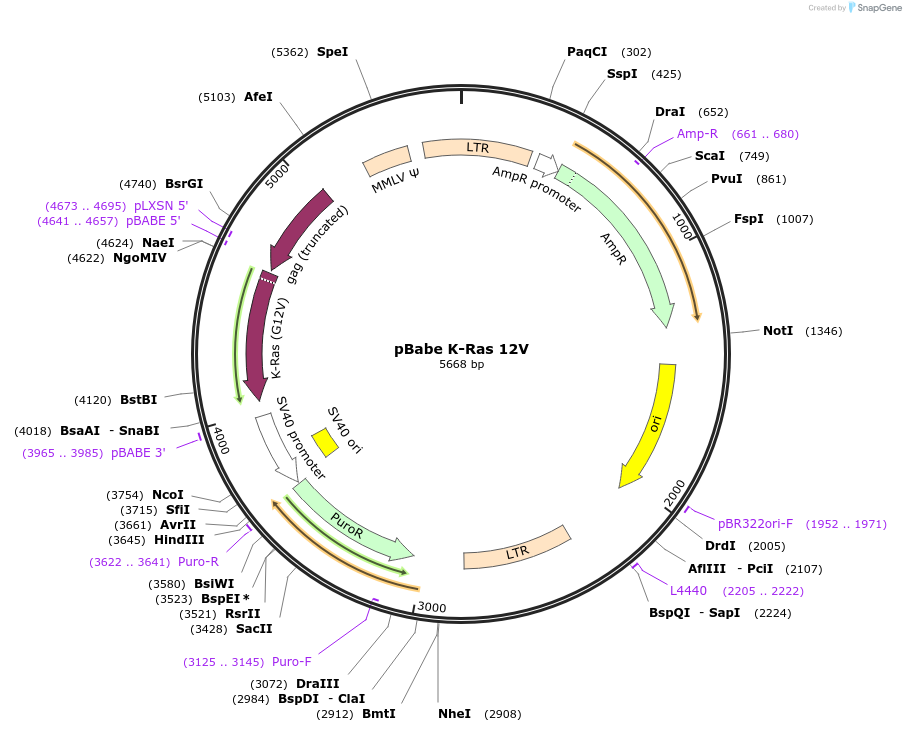
pBabe K-Ras 12V
Plasmid#12544DepositorInsertK-Ras 12V (KRAS Human)
UseRetroviralExpressionMammalianMutation12V constituitively activatedAvailable SinceSept. 8, 2006AvailabilityAcademic Institutions and Nonprofits only -
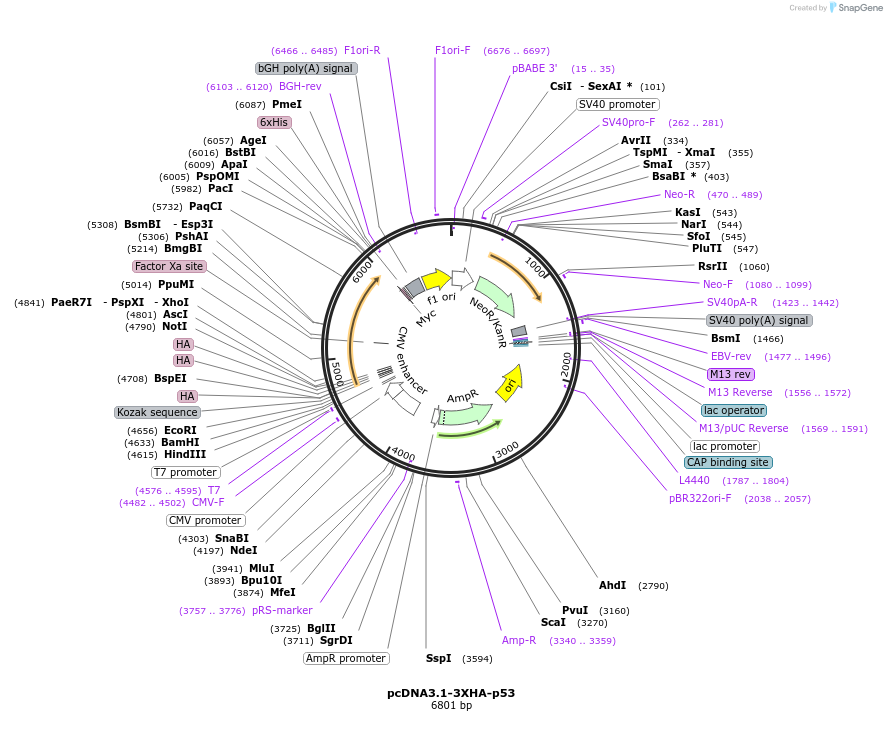
pcDNA3.1-3XHA-p53
Plasmid#196267PurposeMammalian expression of 3xHA tagged wild-type p53DepositorAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
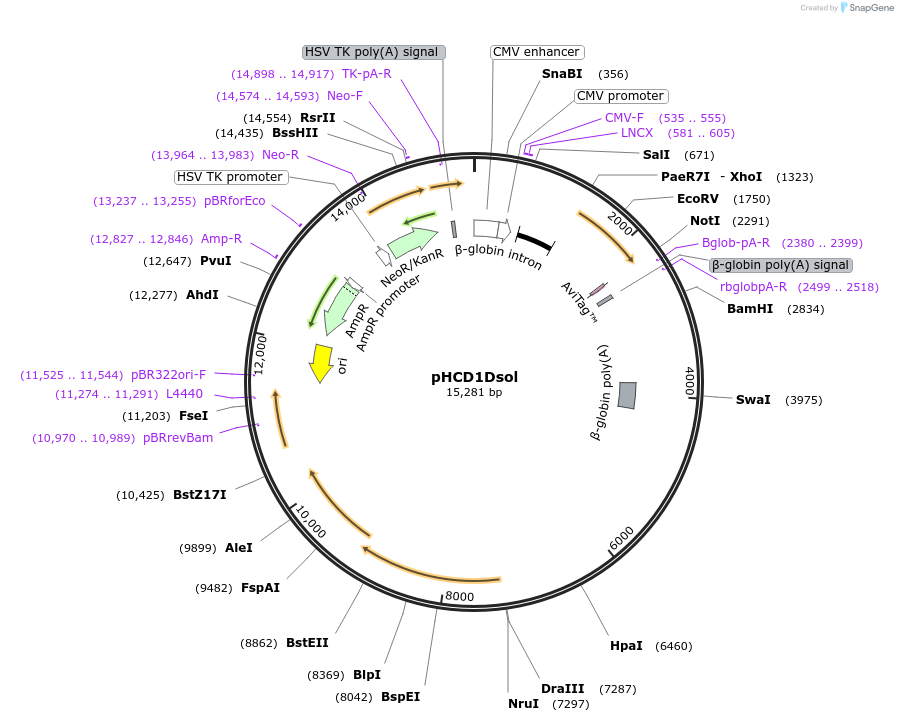
pHCD1Dsol
Plasmid#102955PurposeMammalian expression of human soluble CD1D cDNADepositorInsertHuman soluble CD1D cDNA (CD1D Human)
TagsBirAExpressionMammalianMutationLacks the transmembrane regionPromoterCMVAvailable SinceJune 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
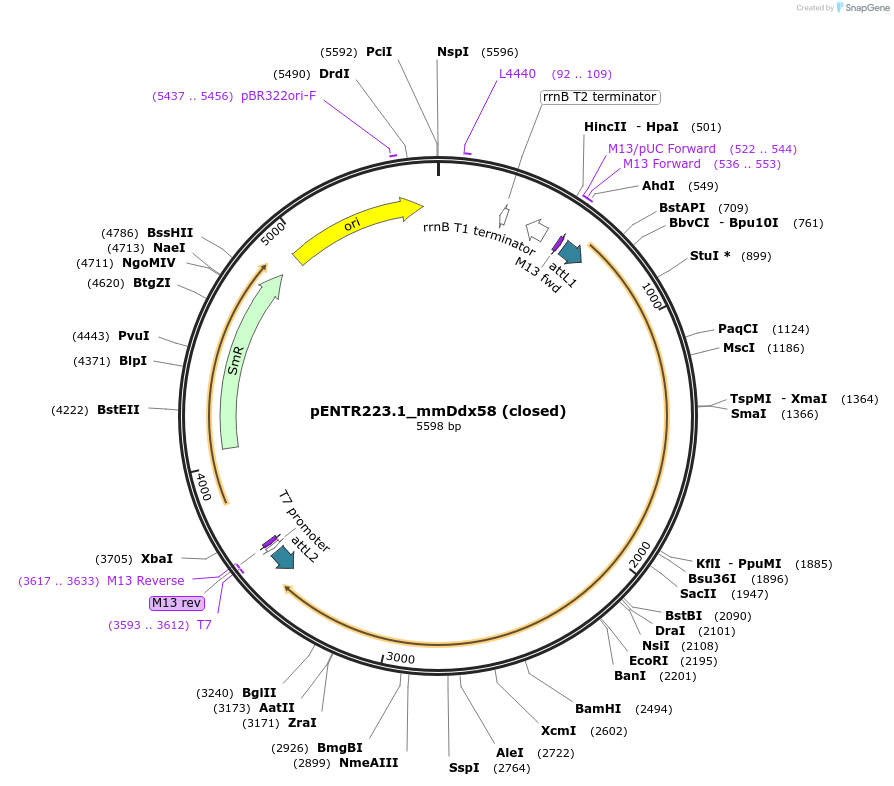
pENTR223.1_mmDdx58 (closed)
Plasmid#160107PurposeExpresses murine Ddx58 in mammalian cellsDepositorAvailable SinceDec. 1, 2020AvailabilityAcademic Institutions and Nonprofits only