We narrowed to 11,312 results for: ENA
-
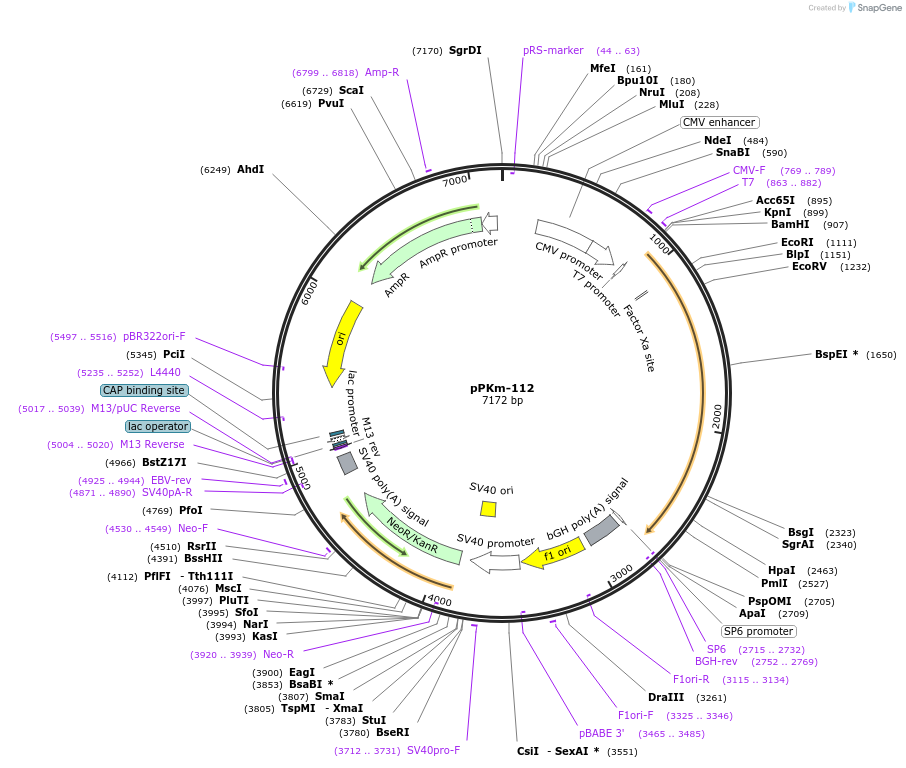
Plasmid#90494PurposepcDNA3 - pCMV - MTAD - PIF3, expresses Phytochrome interacting factor 3 (PIF3) and minimal transactivation domain (MTAD), under CMV promoterDepositorInsertMTAD-PIF3
ExpressionMammalianAvailable SinceDec. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
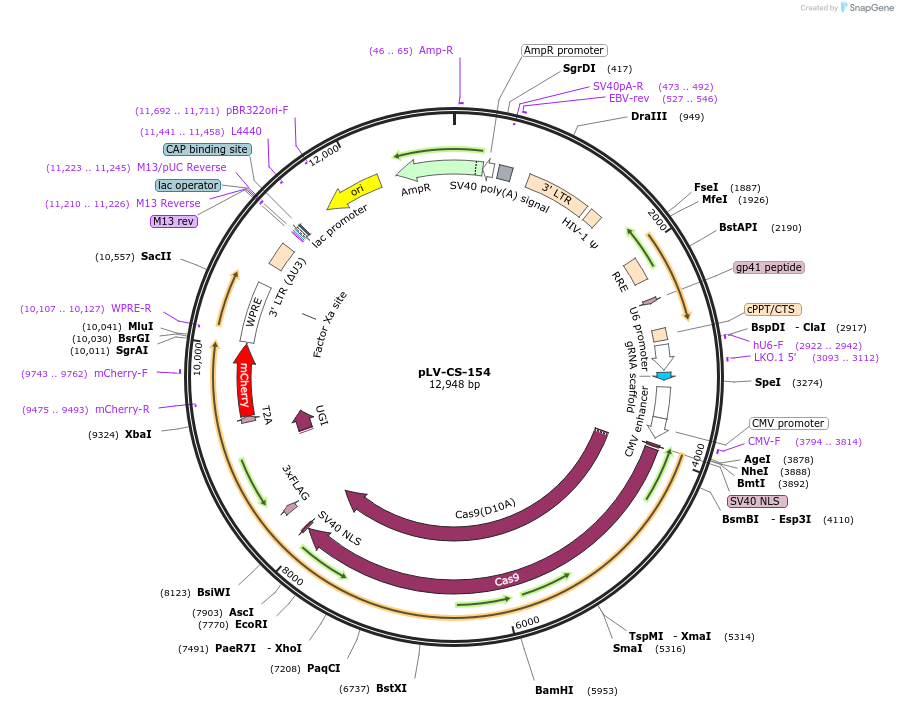
pLV-CS-154
Plasmid#166230Purposelenti-U6-gRNA::CMVp-nCas9-PmCDA1-UGI-2A-mCherryDepositorInsertsU6-gRNA EGFP targetting
nCas9-PmCDA1-UGI-2A-mCherry
UseCRISPR and LentiviralPromoterCMV promoter and Human U6Available SinceDec. 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
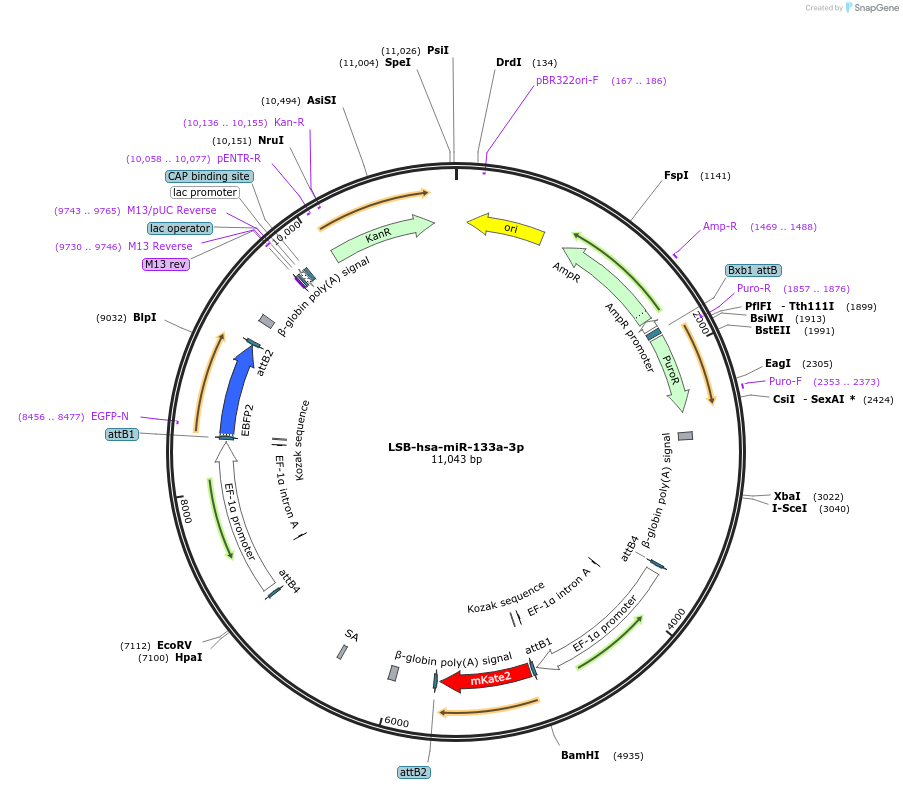
LSB-hsa-miR-133a-3p
Plasmid#103221PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-133a-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-133a-3p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
pChlr-8SE
Plasmid#52172PurposeEncodes module 8 with amino acids 12S-16E for recognition of nt 1GDepositorInsertPumilio homology domain amino acids 284-319 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in module 8Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SYR
Plasmid#52158PurposeEncodes module 4 with amino acids 12S-13Y-16R for recognition of nt 5CDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S, 13H to 13Y, and 16Q to 16R in…Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5SR
Plasmid#52162PurposeEncodes module 5 with amino acids 12S-16R for recognition of nt 4CDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16R in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6SE
Plasmid#52164PurposeEncodes module 6 with amino acids 12S-16E for recognition of nt 3GDepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
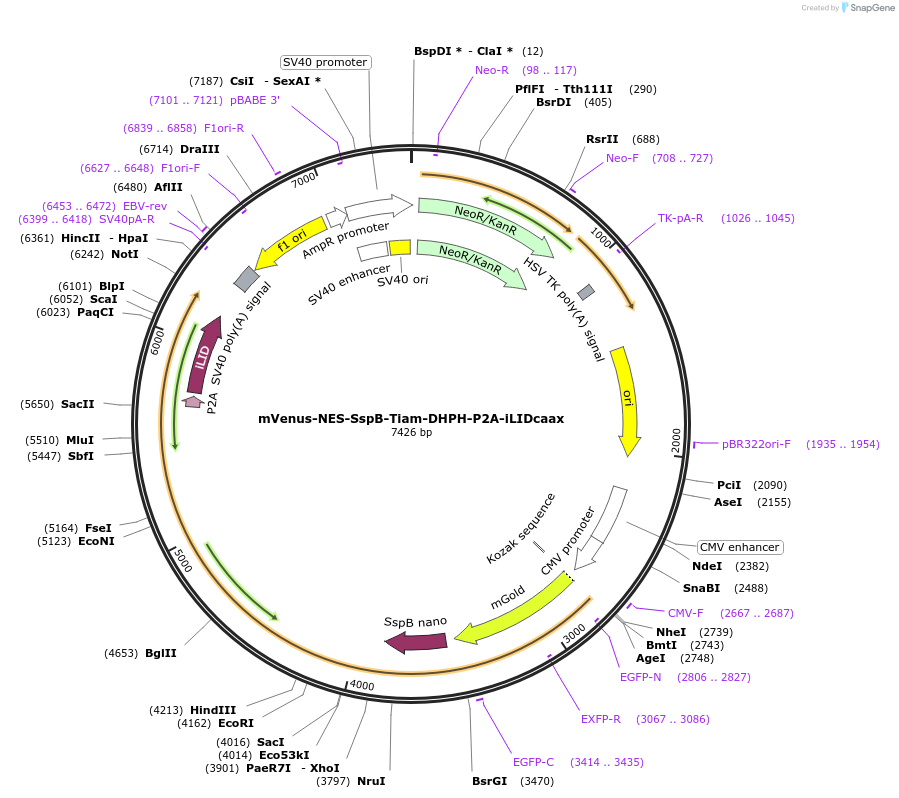
mVenus-NES-SspB-Tiam-DHPH-P2A-iLIDcaax
Plasmid#173865PurposeMammalian expression of mVenus-NES-SspB-Tiam-DHPH-P2A-iLIDcaax (opto-Rac1)DepositorInsertmVenus-NES-SspB-Tiam-DHPH-P2A-iLIDcaax
TagsC-terminal CAAX-box and mVenusExpressionMammalianPromoterCMVAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
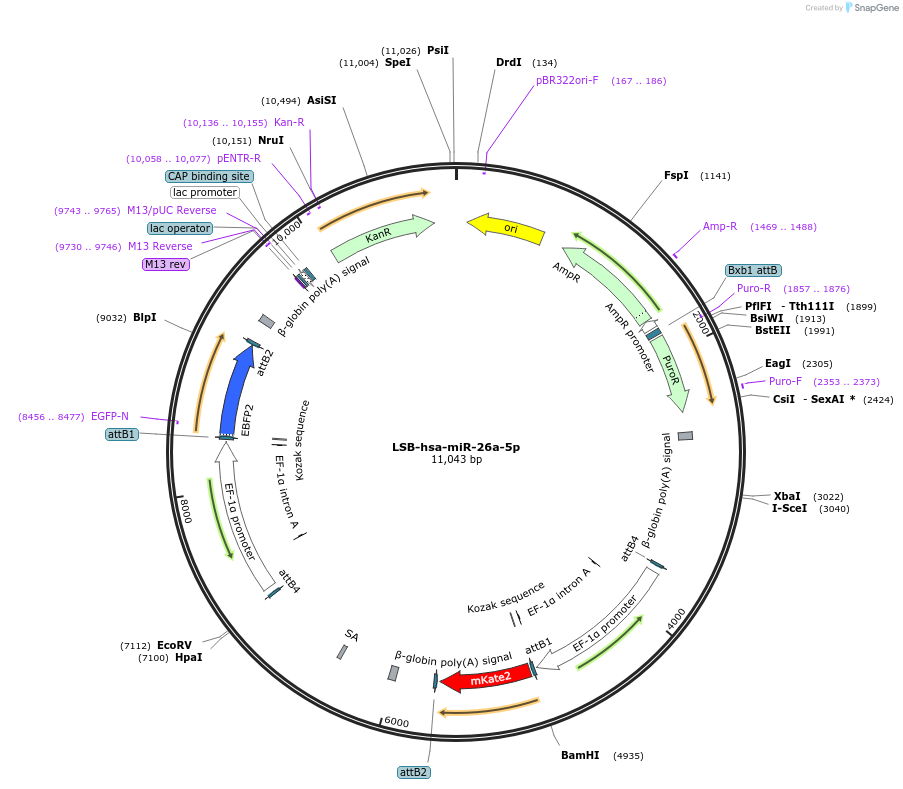
LSB-hsa-miR-26a-5p
Plasmid#103379PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-26a-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-26a-5p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceDec. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
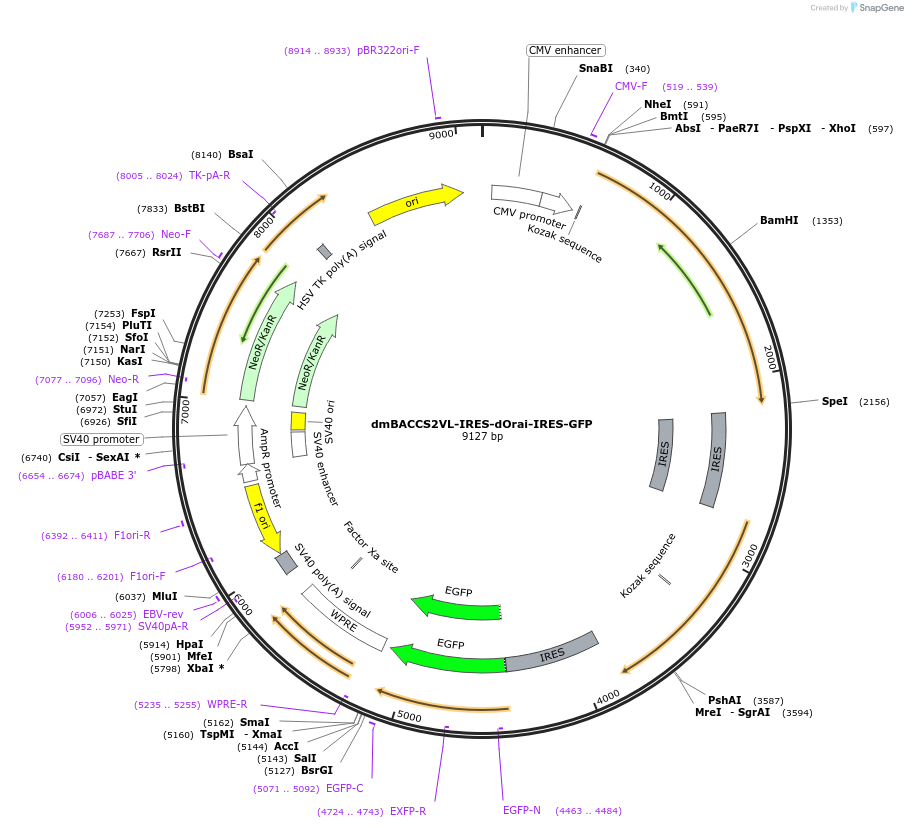
dmBACCS2VL-IRES-dOrai-IRES-GFP
Plasmid#72897PurposeMammalian expression of dmBACCS2VL, a modified Drosophila melanogaster blue light-activated Ca2+ channel switch, along with dOrai and GFPDepositorInsertdmBACCS2VL-IRES-dOrai-IRES-GFP
TagsIRES-GFPExpressionMammalianPromoterCMVAvailable SinceJan. 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
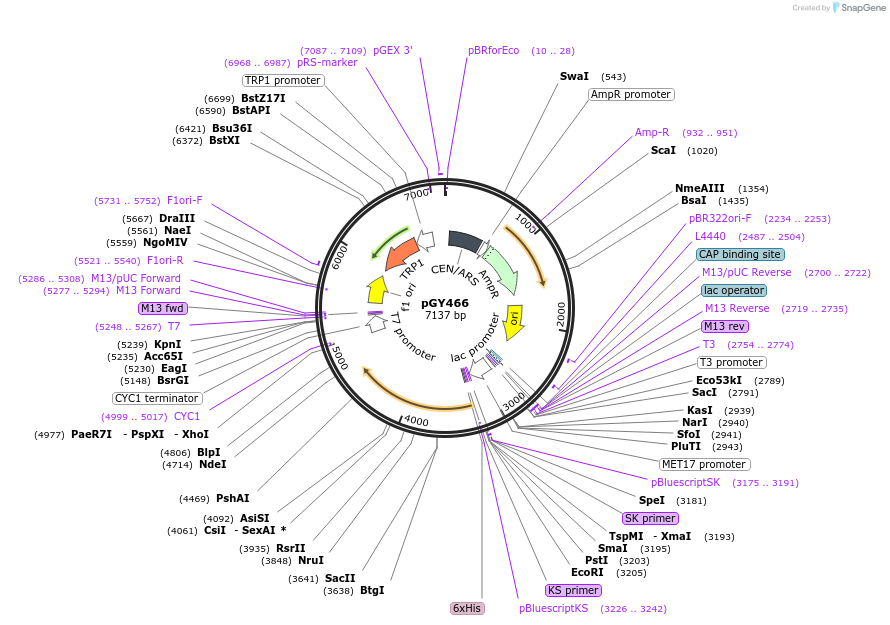
pGY466
Plasmid#166661PurposeFor light-inducible Cre recombinase (LiCre) expression under the Met17 promoter in yeastDepositorInsertLiCre
UseCre/LoxExpressionYeastMutationCre: E340A, D341APromoterMET17Available SinceApril 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
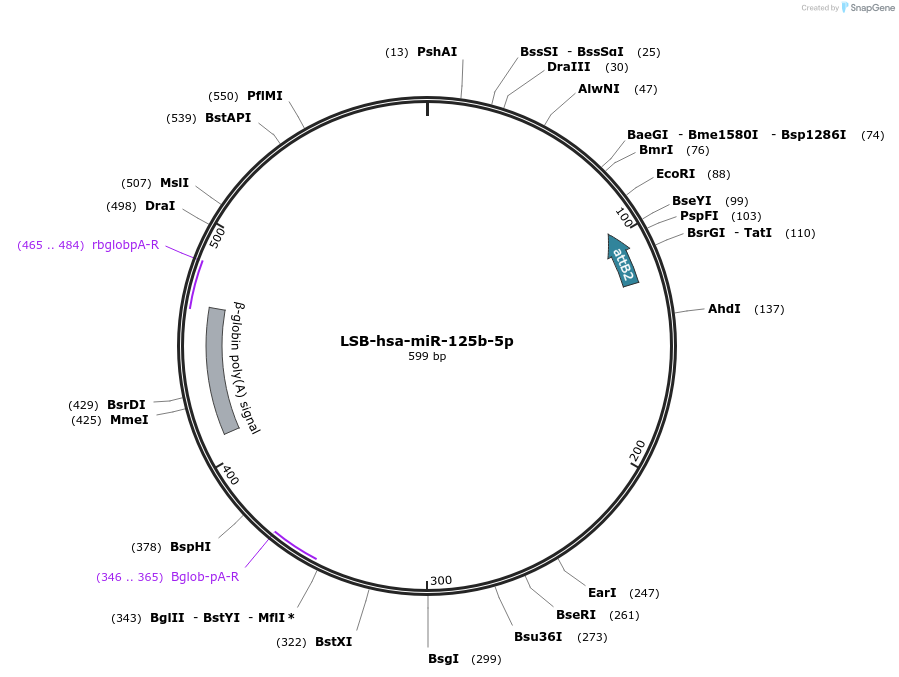
LSB-hsa-miR-125b-5p
Plasmid#103191PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-125b-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-125b-5p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
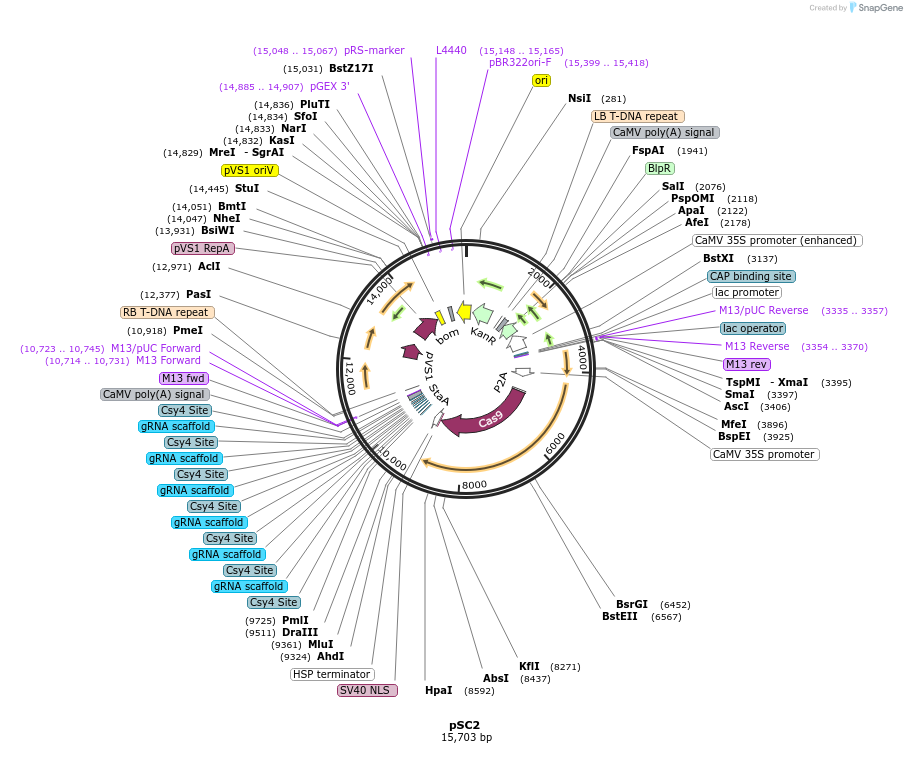
pSC2
Plasmid#91180PurposeT-DNA vector for combinatorial deletion of NCR genes in medicago truncatula (Csy4 array of 6 gRNAs under the control of CmYLCV promoter)DepositorInsertgRNAs targeting NCR genes
UseCRISPRExpressionPlantAvailable SinceSept. 29, 2017AvailabilityAcademic Institutions and Nonprofits only -
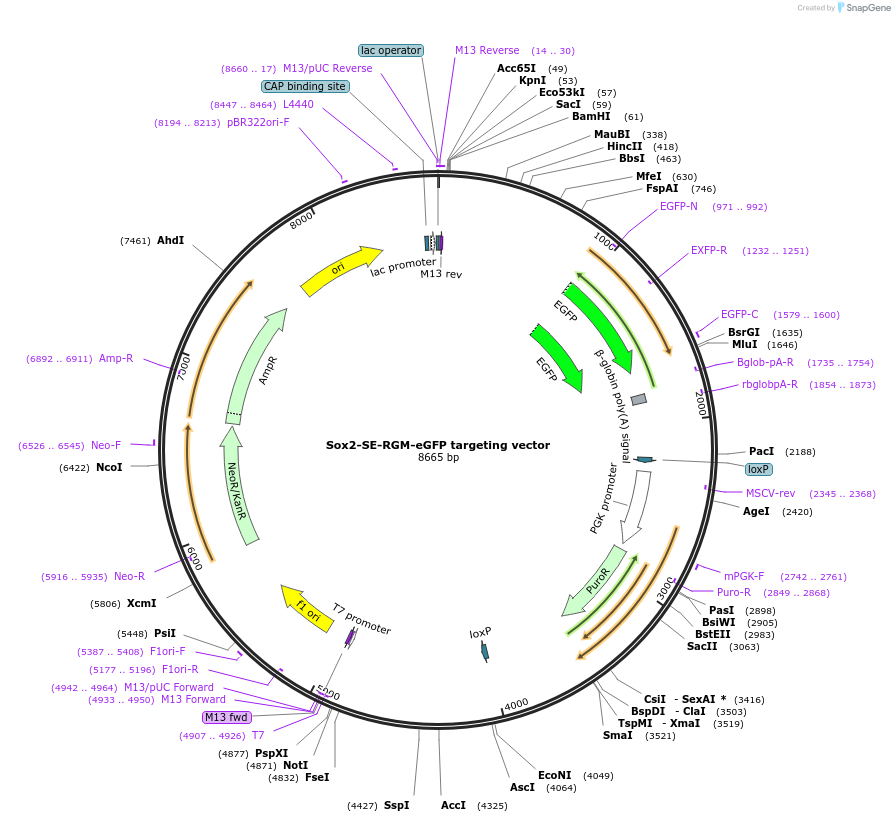
Sox2-SE-RGM-eGFP targeting vector
Plasmid#133045PurposeTargeting vector of RGM-eGFP with homologous arms around Sox2 super-enhancer DMRDepositorInsertSox2-SE-5arm-RGM-Sox2-SE-3arm
UseMouse TargetingAvailable SinceNov. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
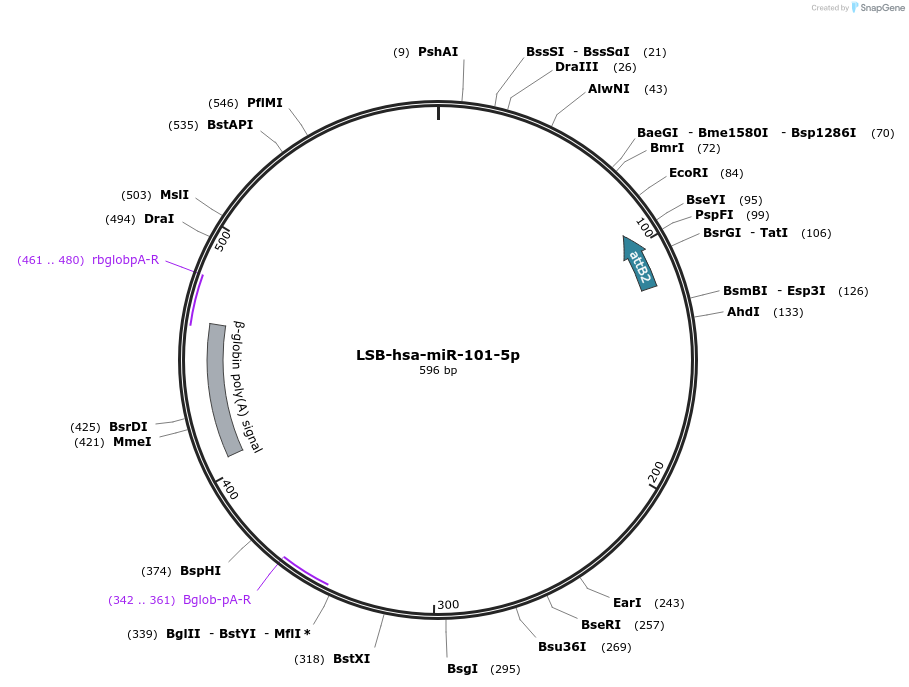
LSB-hsa-miR-101-5p
Plasmid#103166PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-101-5p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-101-5p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
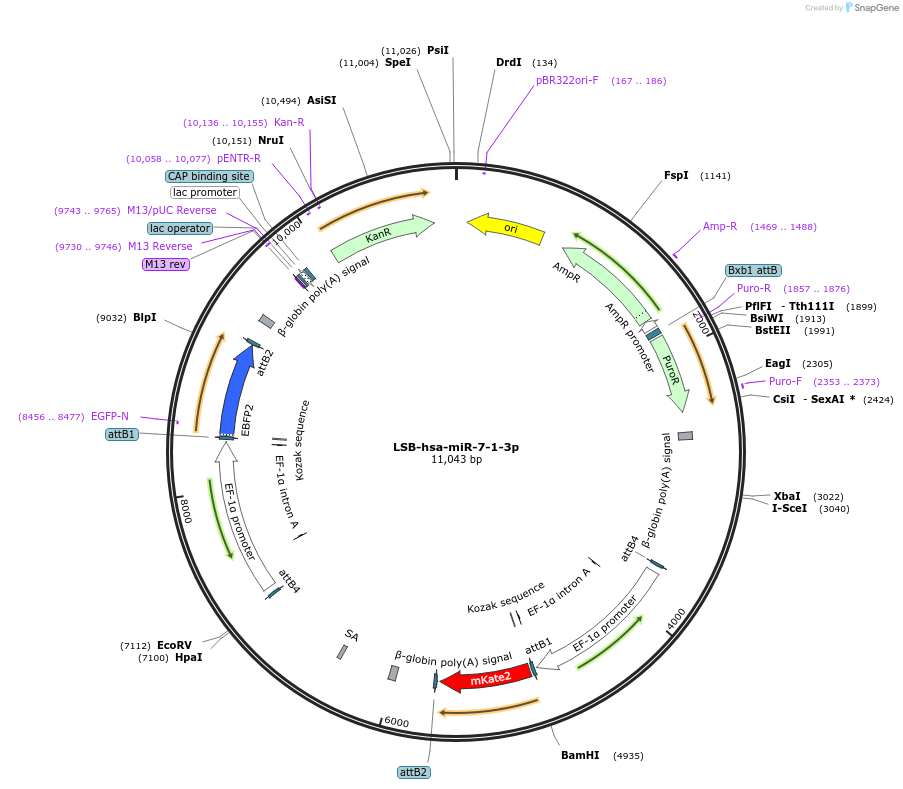
LSB-hsa-miR-7-1-3p
Plasmid#103722PurposeUsed to sense miRNA activity using fluorescence based tools. hsa-miR-7-1-3p target in 3' UTR of of mKate. miRNA activity can be measured by the repression of mKate2 relative to EBFP2 fluorescent proteins . Plasmid is inherently unstable; please see Depositor CommentsDepositorInserthsa-miR-7-1-3p target
UseSynthetic BiologyExpressionMammalianPromoterEF-1aAvailable SinceNov. 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SE
Plasmid#52152PurposeEncodes module 4 with amino acids 12S-16E for recognition of nt 5GDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 4Available SinceMay 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-8SR
Plasmid#52174PurposeEncodes module 8 with amino acids 12S-16R for recognition of nt 1CDepositorInsertPumilio homology domain amino acids 284-319 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 8Available SinceMay 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7CQ
Plasmid#52167PurposeEncodes module 7 with amino acids 12C-16Q for recognition of nt 2ADepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12C and 16E to 16Q in Module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7NQ
Plasmid#52169PurposeEncodes module 7 with amino acids 12N-16Q for recognition of nt 2UDepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12N and 16E to 16Q in module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only