We narrowed to 38,511 results for: NAM
-
Plasmid#219748PurposeDVK_AF CIDAR vector encoding mutant of Neonothopanus nambi hispidin-3-hydroxylase nnH3H_v2 under control of CMV promoter, for mammalian expressionDepositorInsertpCMV - nnH3H_v2 - tSV40
ExpressionMammalianAvailable SinceJuly 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
pX037
Plasmid#167156PurposeMoClo-compatible Level2 vector for autonomous bioluminescence in plants encoding kanamycin resistance cassette, nnHispS, nnH3H, nnLuz and nnCPH. Does not contain NpgA.DepositorInsertcaffeic acid cycle genes
ExpressionPlantPromoterpNOS, p35sAvailable SinceJune 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCDF-Mm2
Plasmid#197100PurposeTo express the variant of Methanosarcina mazei PylRS specific for pyrrolysine derivatives (Boc-Lys, Az-ZLys, AzAmZLys) and its cognate Pyl tRNA and allow UAG codon to be translated into these derivatiDepositorInsertPylRS variant, Pyl tRNA, kanamycin resistance gene
ExpressionBacterialMutationLys61, Glu131, Ala306, Phe384, Glu444 (PylRS)Available SinceJune 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
GFP-Drosha270-1374S300/302A
Plasmid#62533PurposeN-terminal aa1-269 of Drosha is deleted and serines 300 &302 are mutated to alanineDepositorInserttruncated human Drosha mutant (DROSHA Human)
TagsGFPExpressionMammalianMutationdeleted amino acids 1-269 and changed serines 300…Available SinceMarch 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
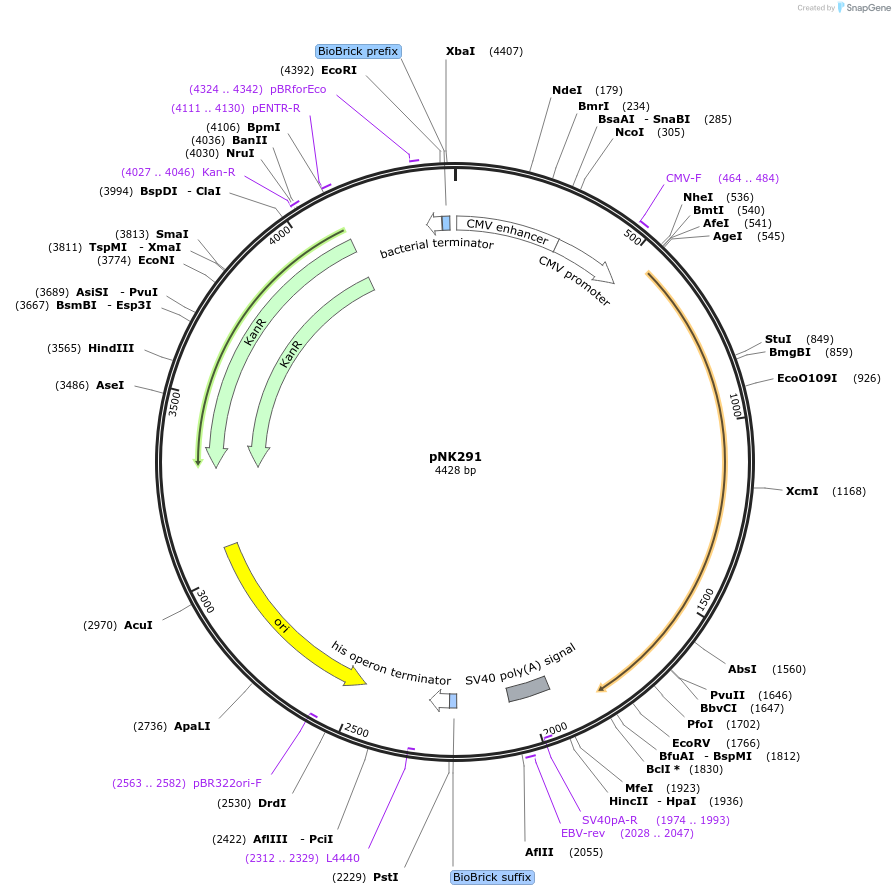
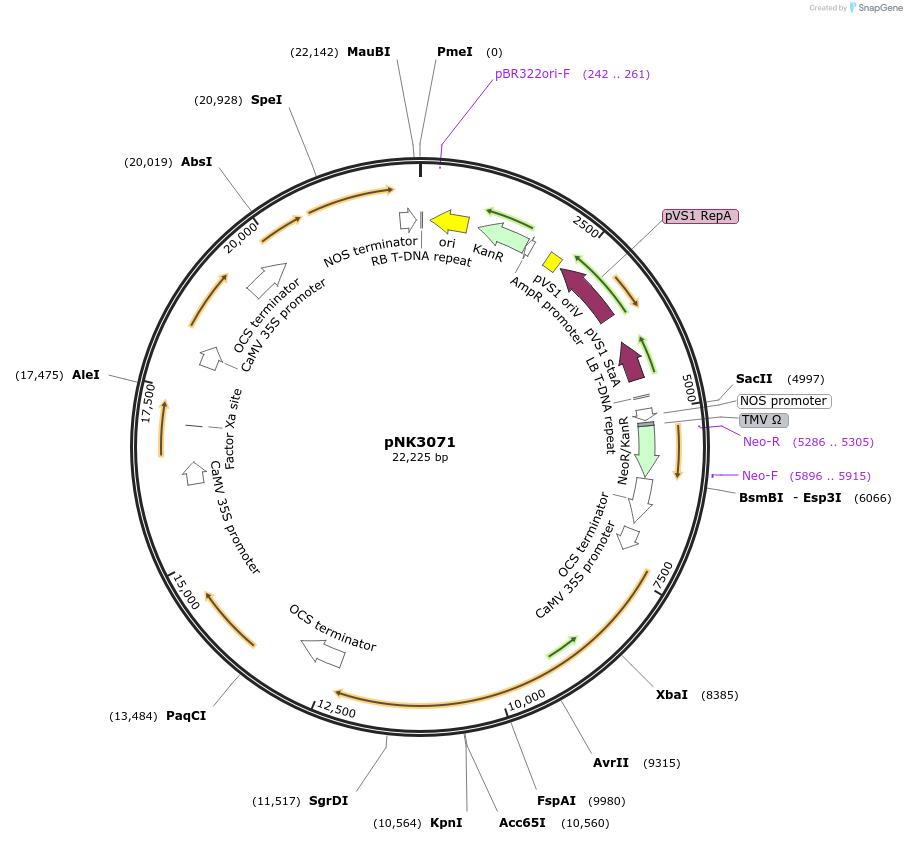
pNK3071
Plasmid#219755PurposeMoClo-compatible Level P vector for improved autonomous bioluminescence in plants encoding kanamycin resistance cassette, mcitHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH.DepositorInsertmcitHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH
ExpressionPlantAvailable SinceOct. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
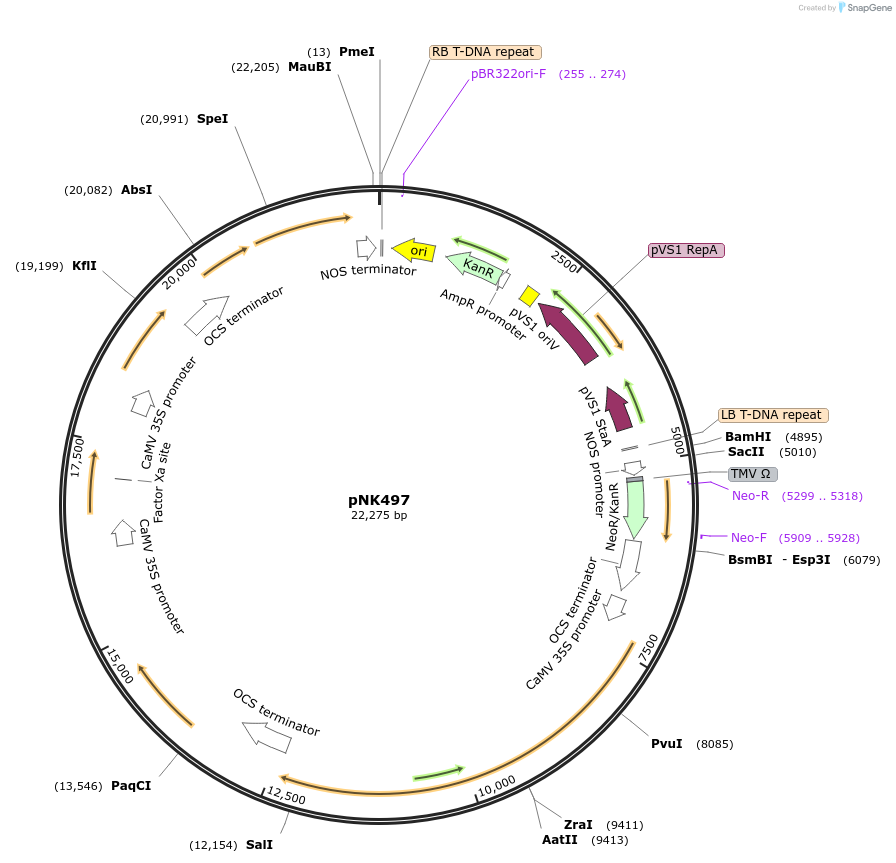
pNK497
Plasmid#219754PurposeMoClo-compatible Level P vector for improved autonomous bioluminescence in plants encoding kanamycin resistance cassette, nnHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH.DepositorInsertnnHispS, NpgA, nnH3H_v2, nnLuz_v4 and nnCPH
ExpressionPlantAvailable SinceJan. 21, 2025AvailabilityAcademic Institutions and Nonprofits only -
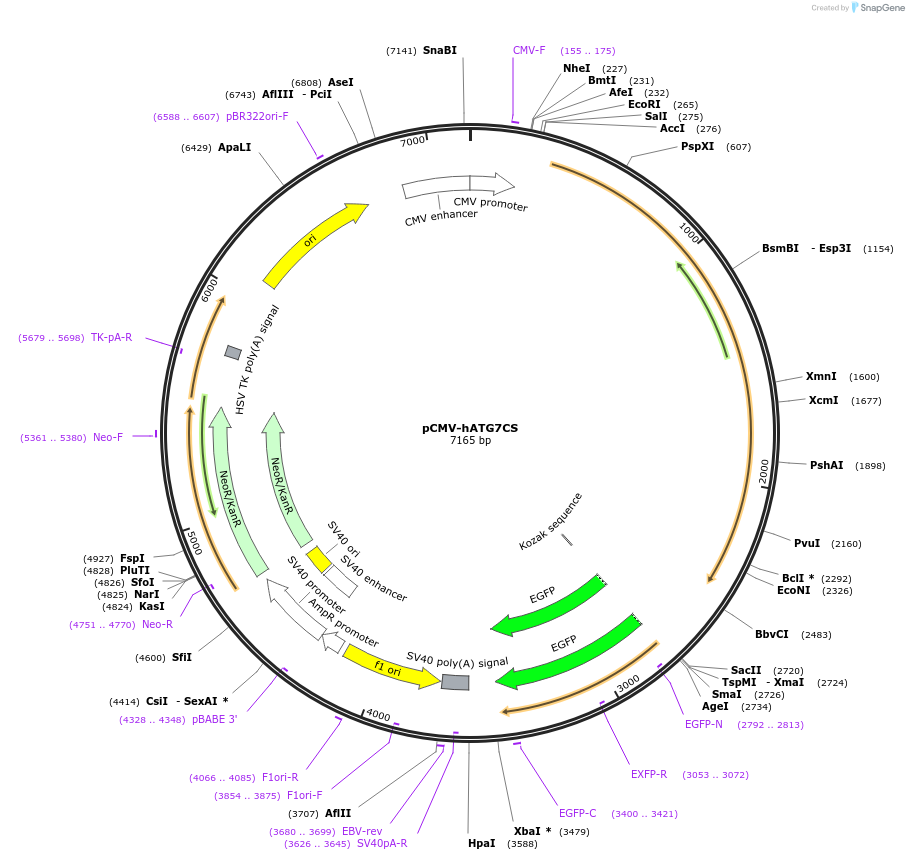
pCMV-hATG7CS
Plasmid#87868PurposeExpress a active-site mutant human Atg7/Apg7 C572S in mammalian cellsDepositorAvailable SinceAug. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
GFP-DroshaS300E/S302D
Plasmid#62531PurposeSerine 300 of Drosha is mutated to glutamic acid and serine 302 is mutated to aspartic acidDepositorInserthuman Drosha with serine 300 changed to glutamic acid and serine 302 changed to aspartic acid (DROSHA Human)
TagsGFPExpressionMammalianMutationchanged serine 300 to glutamic acid and serine 3…Available SinceMarch 20, 2015AvailabilityAcademic Institutions and Nonprofits only -
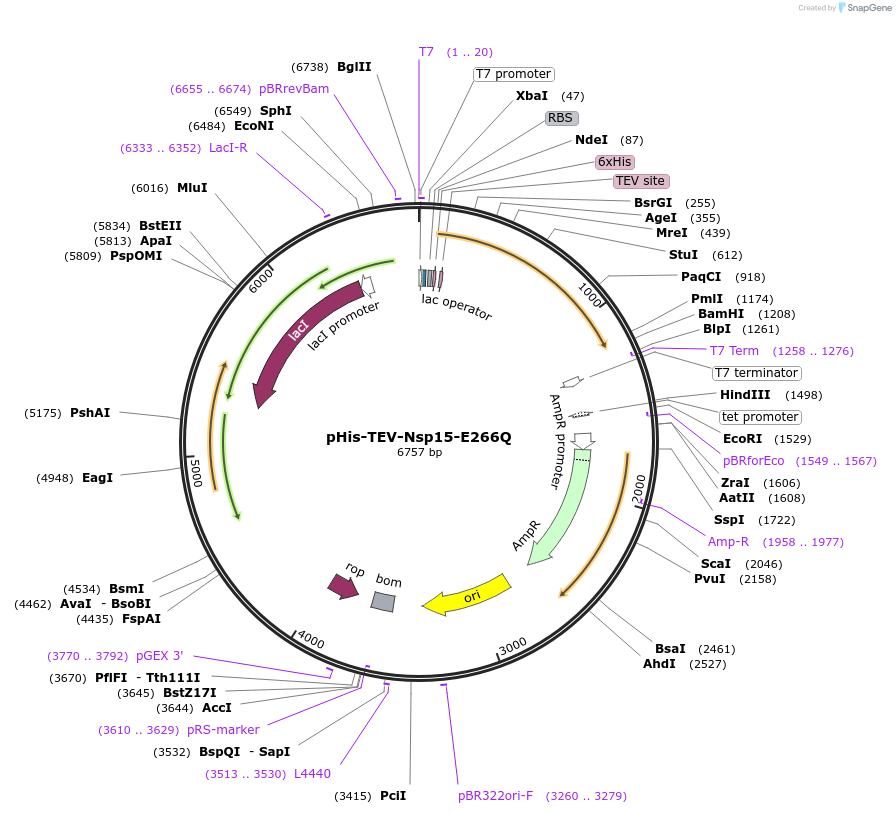
pHis-TEV-Nsp15-E266Q
Plasmid#218803PurposeE. coli plasmid (T7lac promoter) for full-length, SARS CoV-2 Nsp15-E266Q (uridine-specific nidoviral endoribonuclease, expression-optimized gene) with N-terminal TEV protease cleavable His6-tagDepositorInsertNsp15-E266Q (N Severe acute respiratory syndrome coronavirus 2)
TagsTEV protease cleavable His6-tagExpressionBacterialMutationE266Q (G to C)Available SinceMay 27, 2025AvailabilityAcademic Institutions and Nonprofits only -
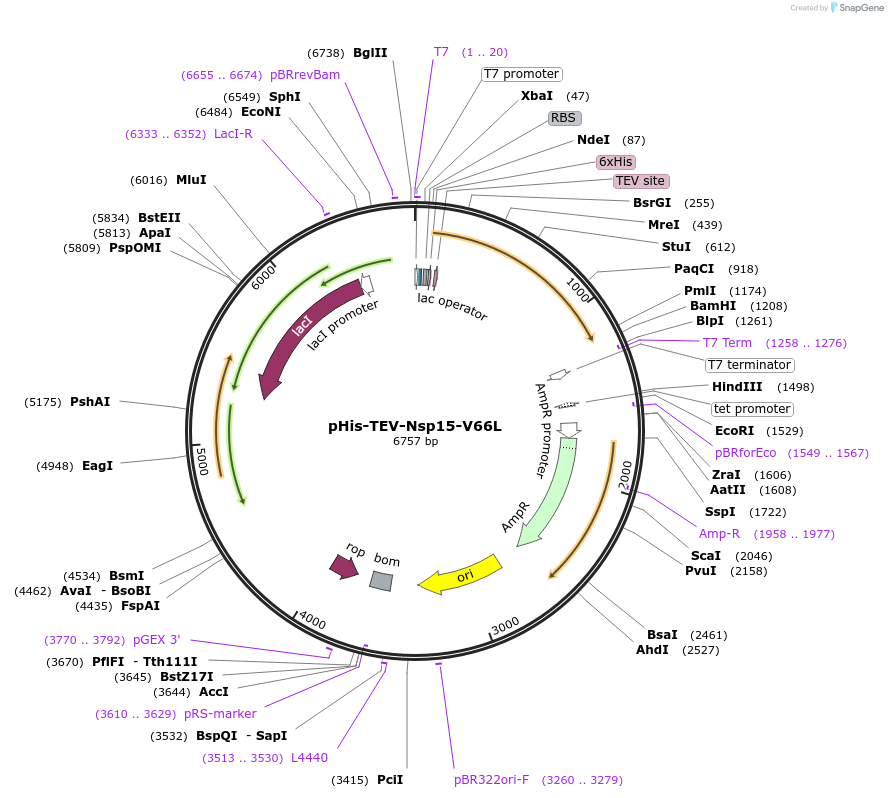
pHis-TEV-Nsp15-V66L
Plasmid#219524PurposeE. coli plasmid (T7lac promoter) for full-length, SARS CoV-2 Nsp15-V66L (uridine-specific nidoviral endoribonuclease, expression-optimized gene) with N-terminal TEV protease cleavable His6-tagDepositorInsertNsp15-V66L (N Severe acute respiratory syndrome coronavirus 2)
TagsTEV protease cleavable His6-tagExpressionBacterialMutationV66L (G to C)Available SinceMay 27, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
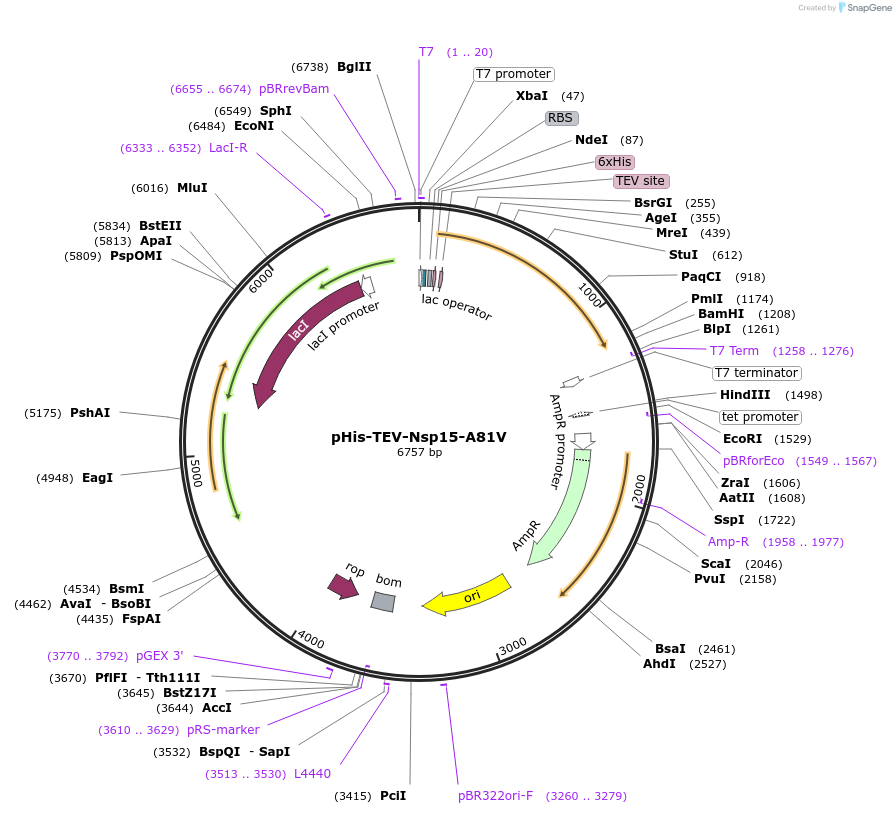
pHis-TEV-Nsp15-A81V
Plasmid#219525PurposeE. coli plasmid (T7lac promoter) for full-length, SARS CoV-2 Nsp15-A81V (uridine-specific nidoviral endoribonuclease, expression-optimized gene) with N-terminal TEV protease cleavable His6-tagDepositorInsertNsp15-A81V (N Severe acute respiratory syndrome coronavirus 2)
TagsTEV protease cleavable His6-tagExpressionBacterialMutationA81V (C to T)Available SinceMay 27, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
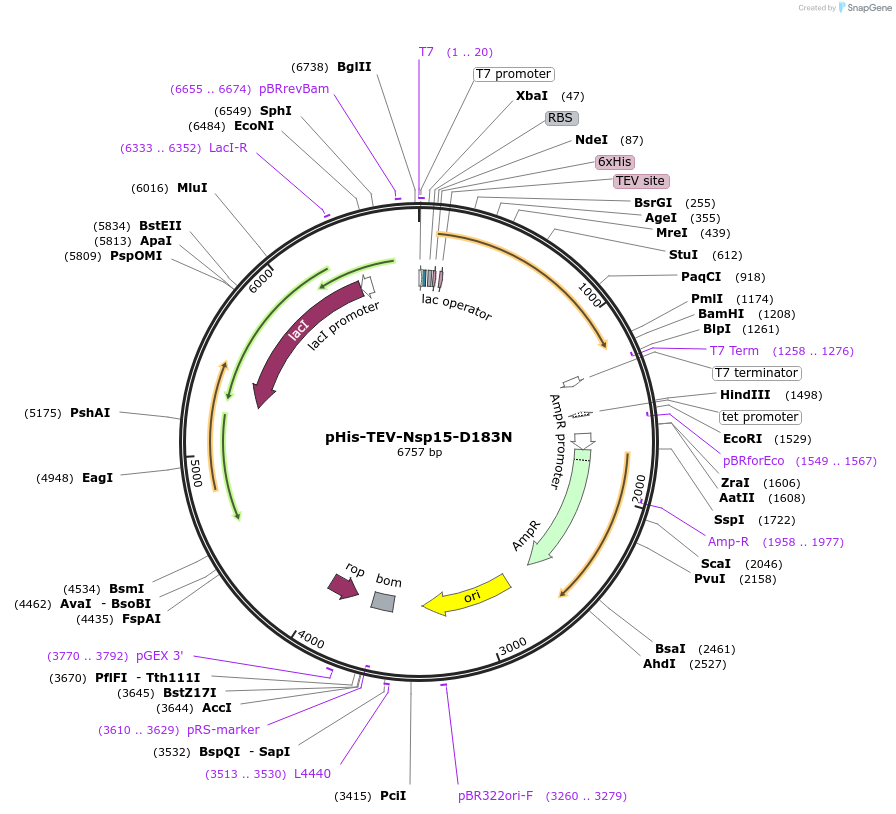
pHis-TEV-Nsp15-D183N
Plasmid#219526PurposeE. coli plasmid (T7lac promoter) for full-length, SARS CoV-2 Nsp15-D183N (uridine-specific nidoviral endoribonuclease, expression-optimized gene) with N-terminal TEV protease cleavable His6-tagDepositorInsertNsp15-D183N (N Severe acute respiratory syndrome coronavirus 2)
TagsTEV protease cleavable His6-tagExpressionBacterialMutationD183N (G to A)Available SinceMay 27, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
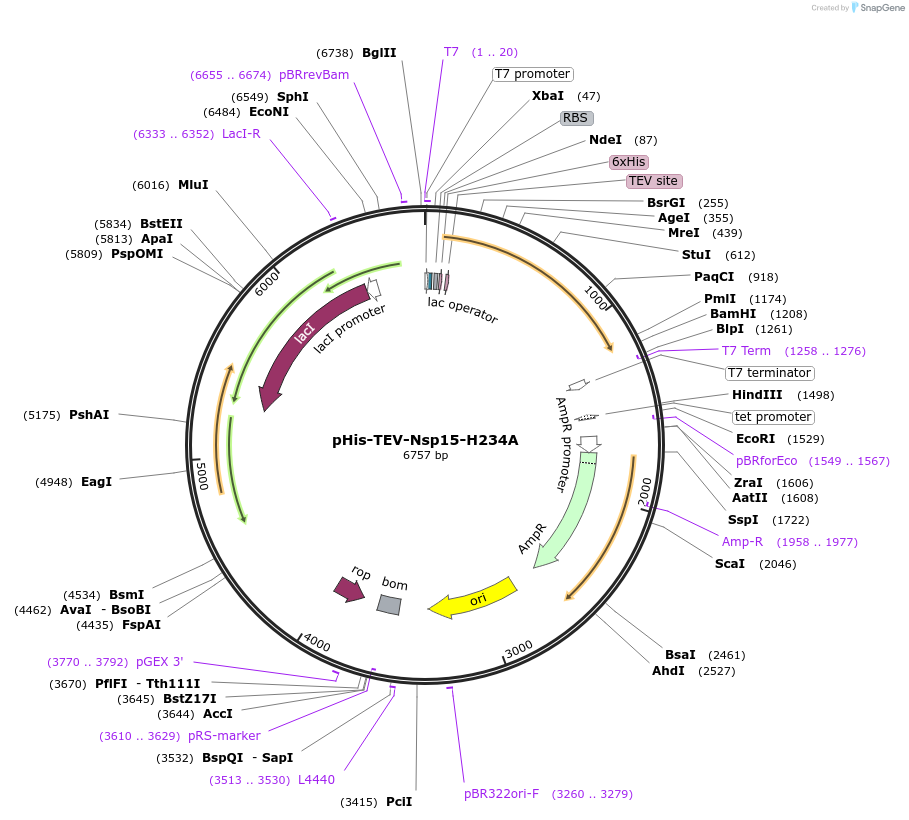
pHis-TEV-Nsp15-H234A
Plasmid#219529PurposeE. coli plasmid (T7lac promoter) for full-length, SARS CoV-2 Nsp15-H234A (uridine-specific nidoviral endoribonuclease, expression-optimized gene) with N-terminal TEV protease cleavable His6-tagDepositorInsertNsp15-H234A (N Severe acute respiratory syndrome coronavirus 2)
TagsTEV protease cleavable His6-tagExpressionBacterialMutationH234A (CAC to GCG)Available SinceMay 27, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
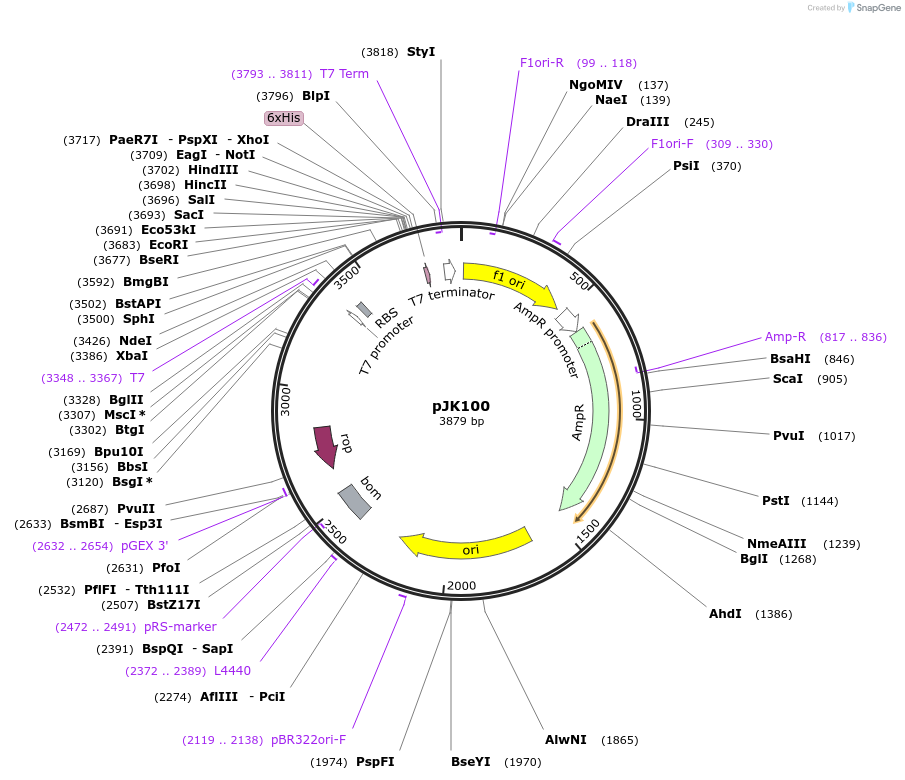
pJK100
Plasmid#73264PurposeProduces Bacillus subtilis phosphoribosylformylglycinamidine synthase small subunit (BsPurS)DepositorInsertphosphoribosylformylglycinamidine synthase subunit PurS
ExpressionBacterialPromoterT7Available SinceFeb. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
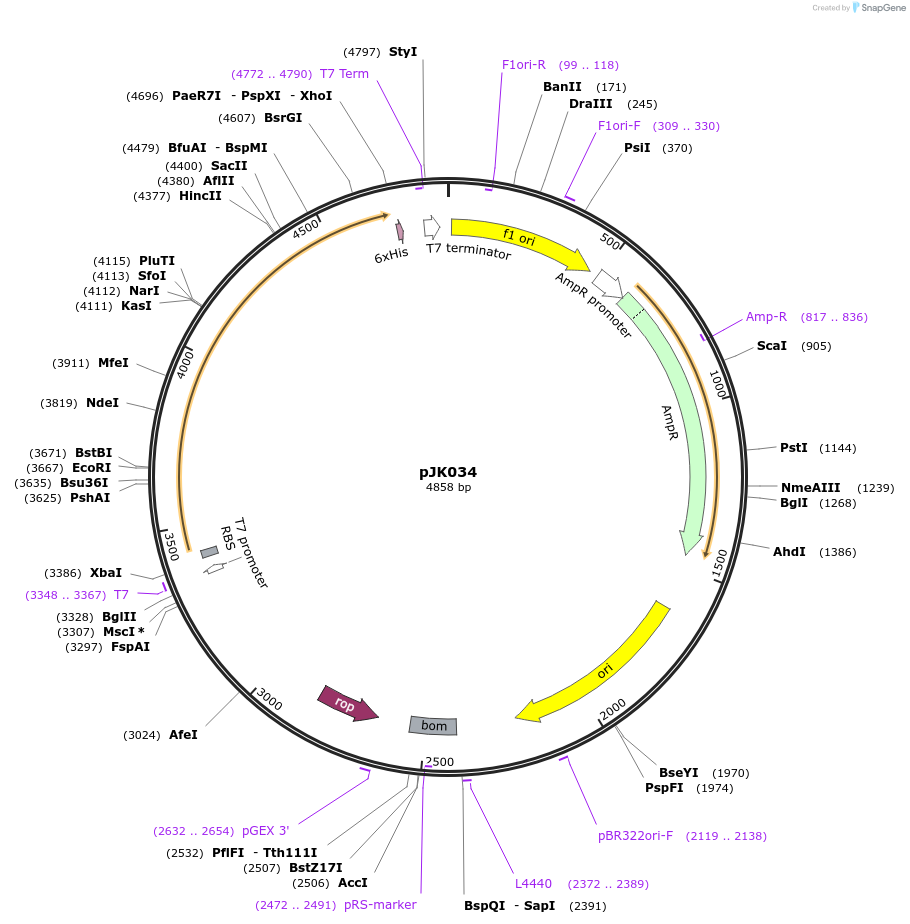
pJK034
Plasmid#72383PurposeProduces Bacillus subtilis glycinamide ribonucleotide synthetase (PurD)DepositorInsertphosphoribosylglycinamide synthetase
ExpressionBacterialPromoterT7Available SinceApril 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
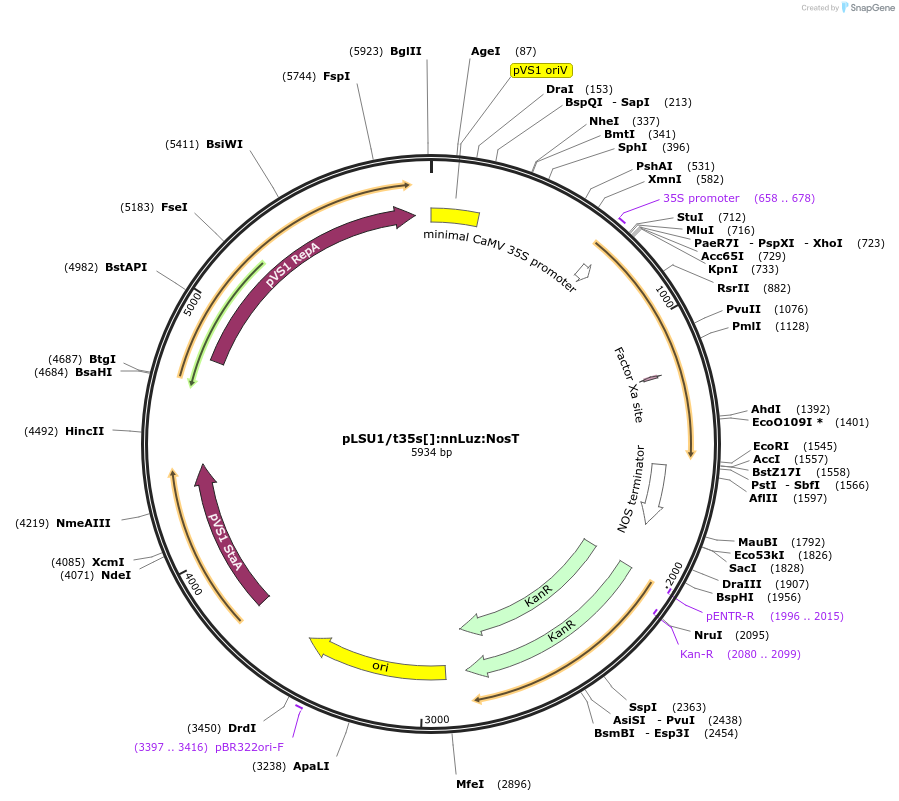
pLSU1/t35s[]:nnLuz:NosT
Plasmid#212183PurposeThis binary vector expresses mushroom luciferase (nnLuz) with the truncated 35sCAMV promoterDepositorInsertnnLuz
ExpressionPlantMutationcodon modified for Physcomitrella patens expressi…Promotertruncated 35sCaMVAvailable SinceAug. 30, 2024AvailabilityAcademic Institutions and Nonprofits only -
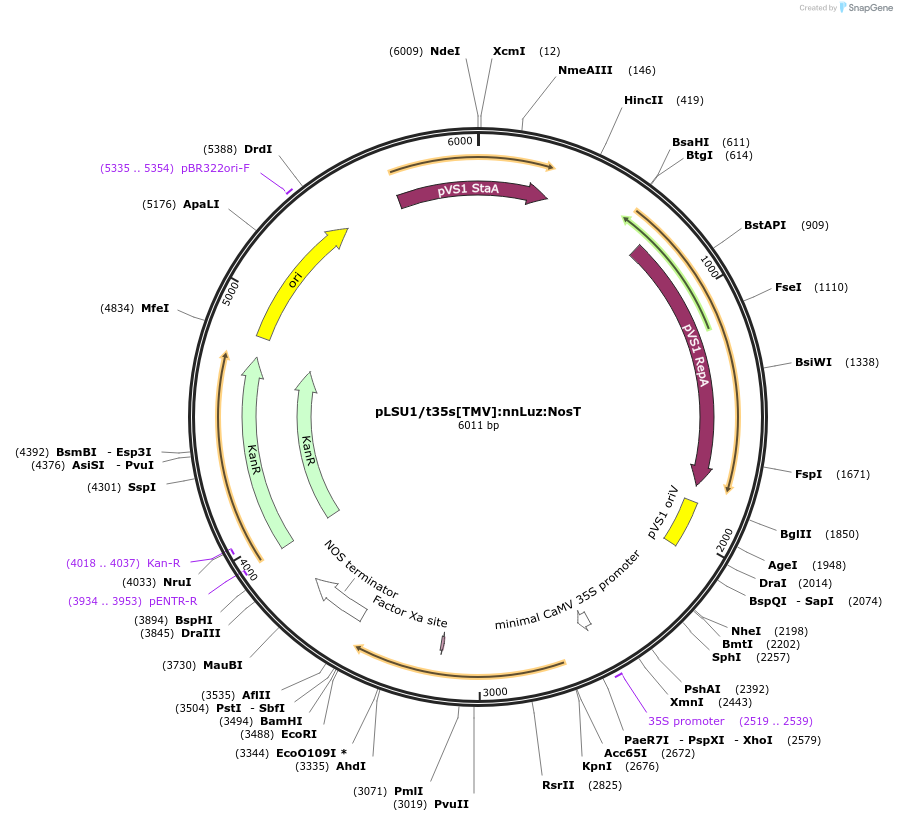
pLSU1/t35s[TMV]:nnLuz:NosT
Plasmid#212188PurposeThis binary vector expresses mushroom luciferase (nnLuz) with the truncated 35sCAMV promoter and Tomato mosaic virus UTRDepositorInsertnnLuz
ExpressionPlantPromotertruncated 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
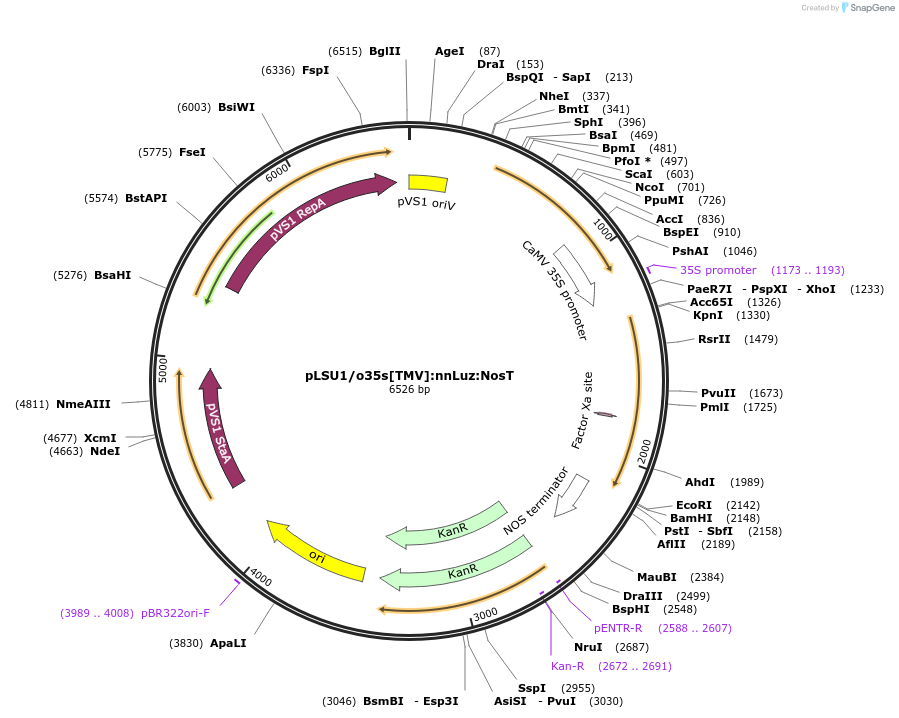
pLSU1/o35s[TMV]:nnLuz:NosT
Plasmid#212189PurposeThis binary vector expresses mushroom luciferase (nnLuz) with the original 35sCAMV promoter and Tomato mosaic virus UTRDepositorInsertnnLuz
ExpressionPlantPromoteroriginal 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
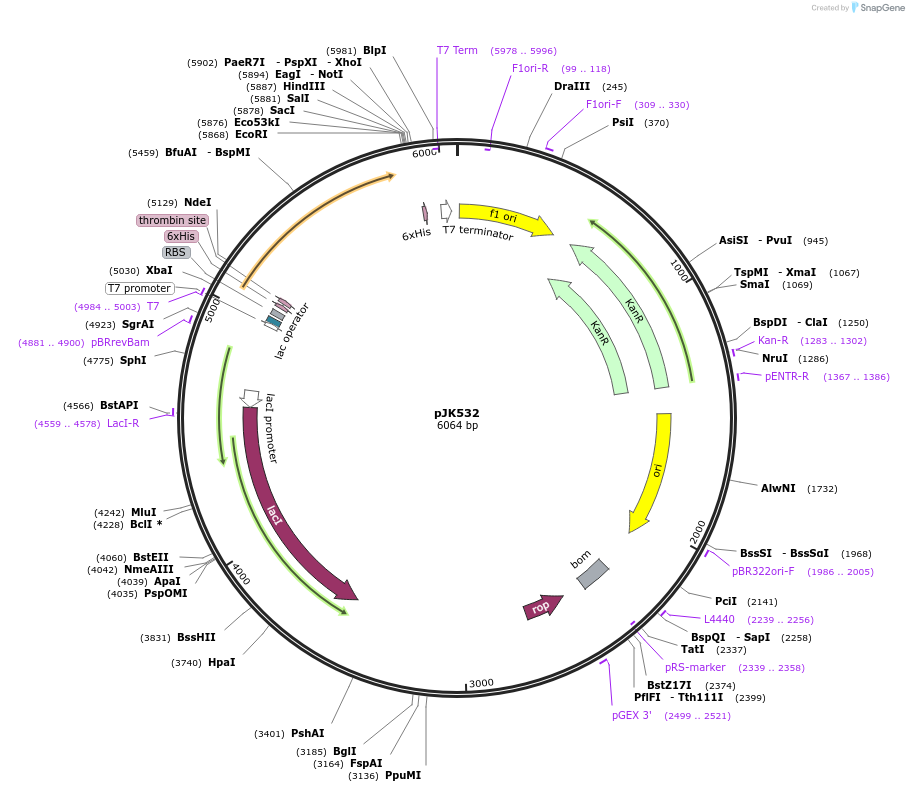
pJK532
Plasmid#72513PurposeProduces Acetobacter aceti 1023 formylglycinamidine ribonucleotide synthetase glutaminase subunit with N-terminal His6 tag (H6AaPurQ)DepositorInsertformylglycinamidine ribonucleotide synthetase, glutaminase subunit
TagsHis6ExpressionBacterialPromoterT7Available SinceMarch 17, 2016AvailabilityAcademic Institutions and Nonprofits only -
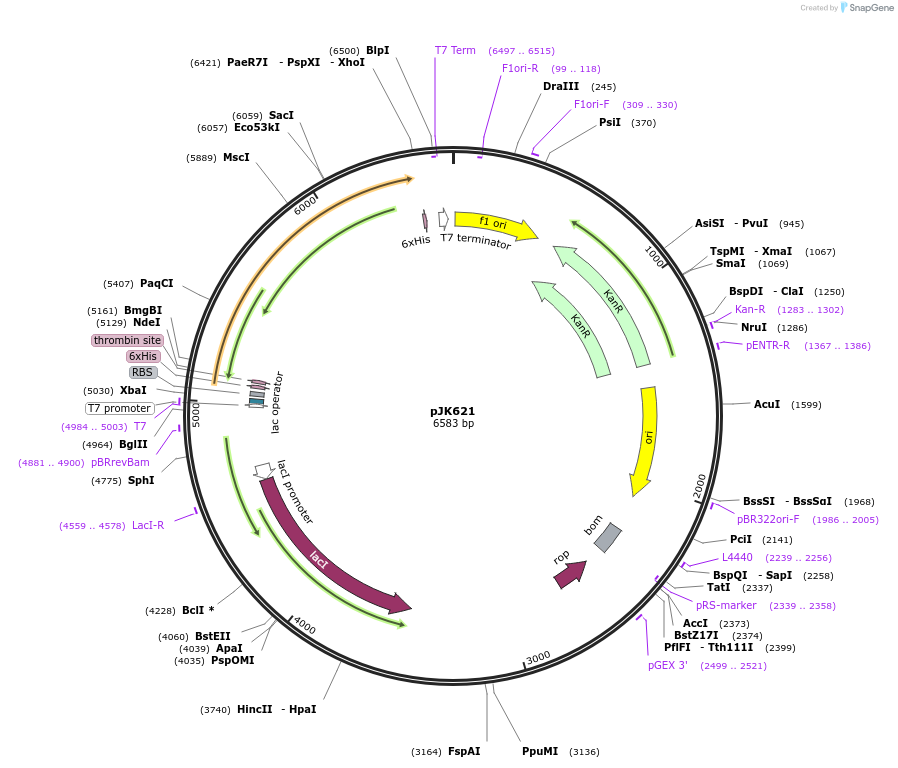
pJK621
Plasmid#72388PurposeProduces Escherichia coli glycinamide ribonucleotide synthetase (EcPurDH6)DepositorInsertphosphoribosylamine-glycine ligase
TagsHis6ExpressionBacterialMutationcontains 3 silent mutations in codons 9-11 to fac…PromoterT7Available SinceFeb. 24, 2016AvailabilityAcademic Institutions and Nonprofits only