We narrowed to 45,509 results for: cha;
-
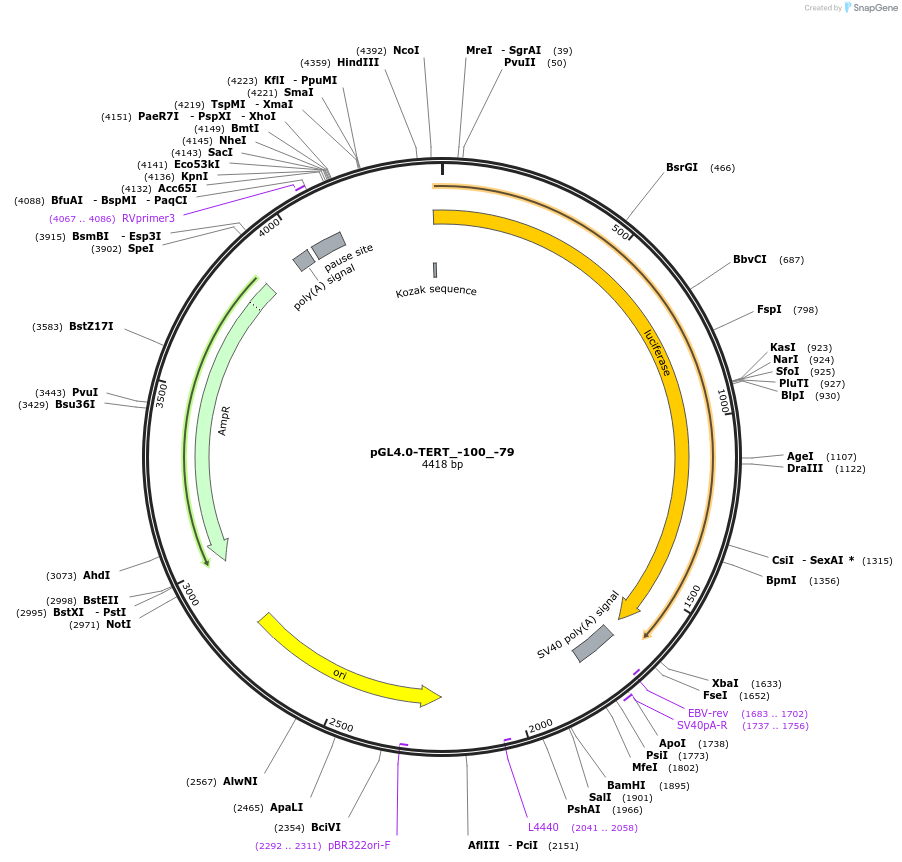
Plasmid#193785Purposeluciferase reporter for duplicated TERT promoterDepositorAvailable SinceAug. 8, 2023AvailabilityAcademic Institutions and Nonprofits only
-
iCAP-BI-65-KanR
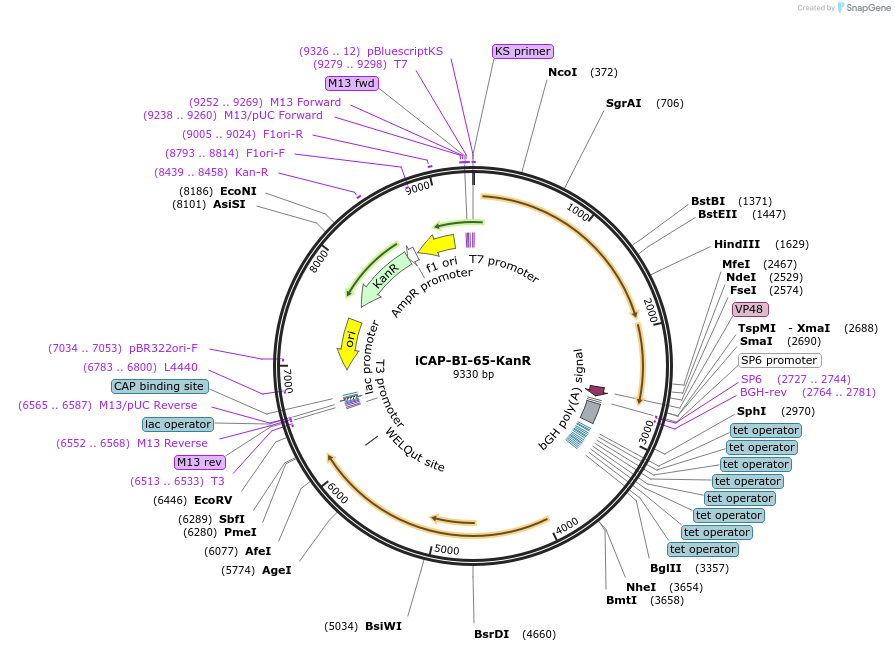
Plasmid#203536PurposeRepCap for AAV productionDepositorInsertAAV-BI65 Cap
UseAAVExpressionMammalianAvailable SinceJuly 28, 2023AvailabilityAcademic Institutions and Nonprofits only -
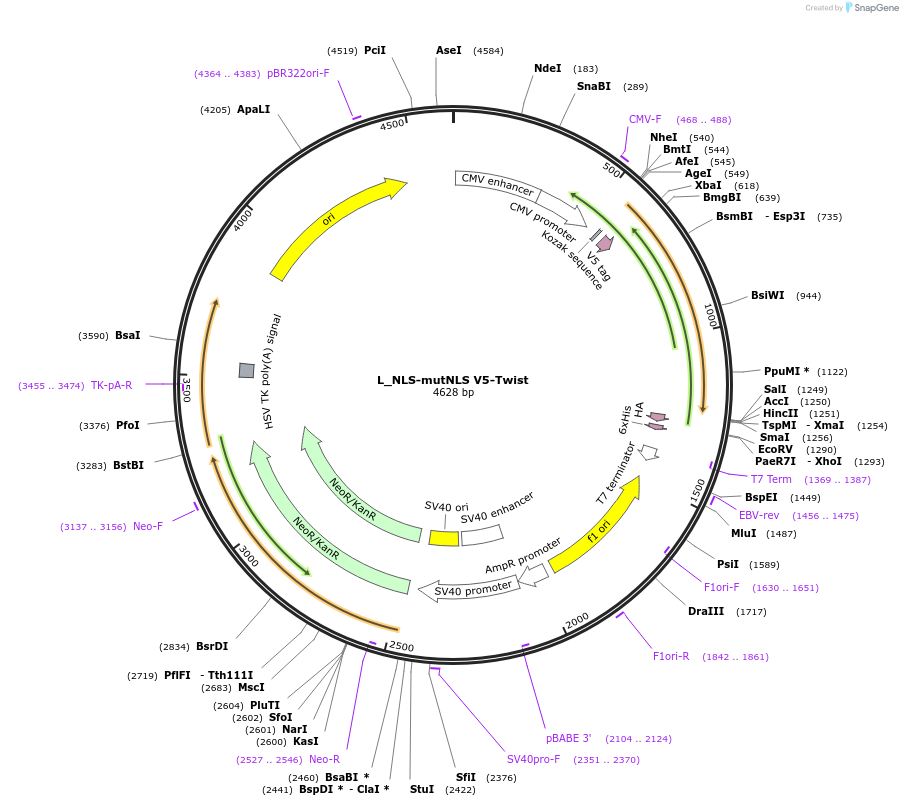
L_NLS-mutNLS V5-Twist
Plasmid#201381PurposeExpresses Twist mutant for nucleocytoplasmic transport studiesDepositorAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
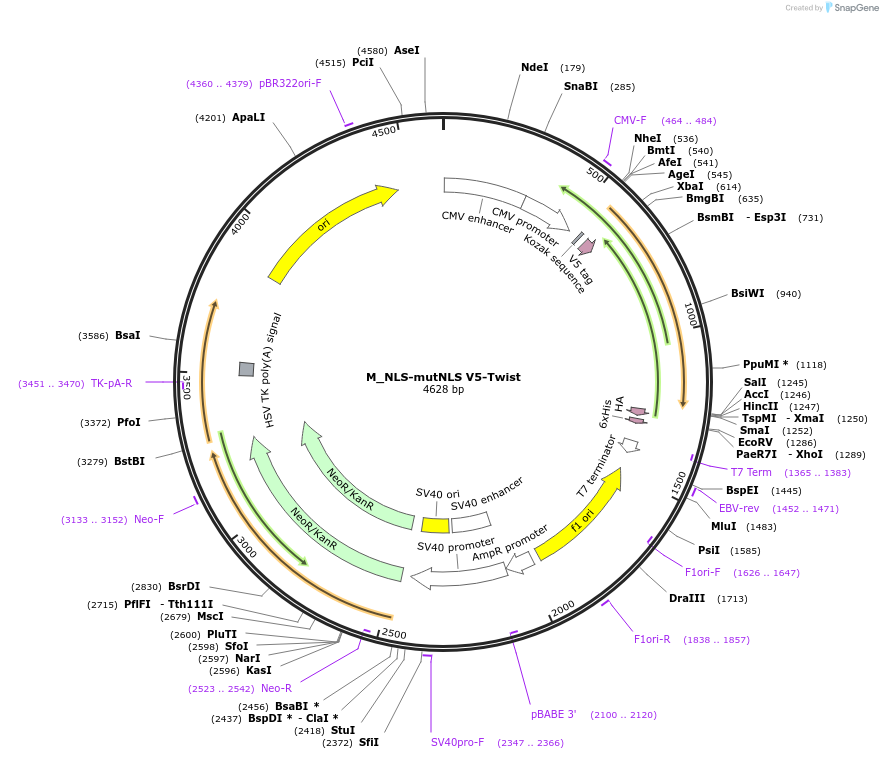
M_NLS-mutNLS V5-Twist
Plasmid#201379PurposeExpresses Twist mutant for nucleocytoplasmic transport studiesDepositorAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
mutNLS V5-Twist
Plasmid#201380PurposeExpresses Twist mutant for nucleocytoplasmic transport studiesDepositorAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
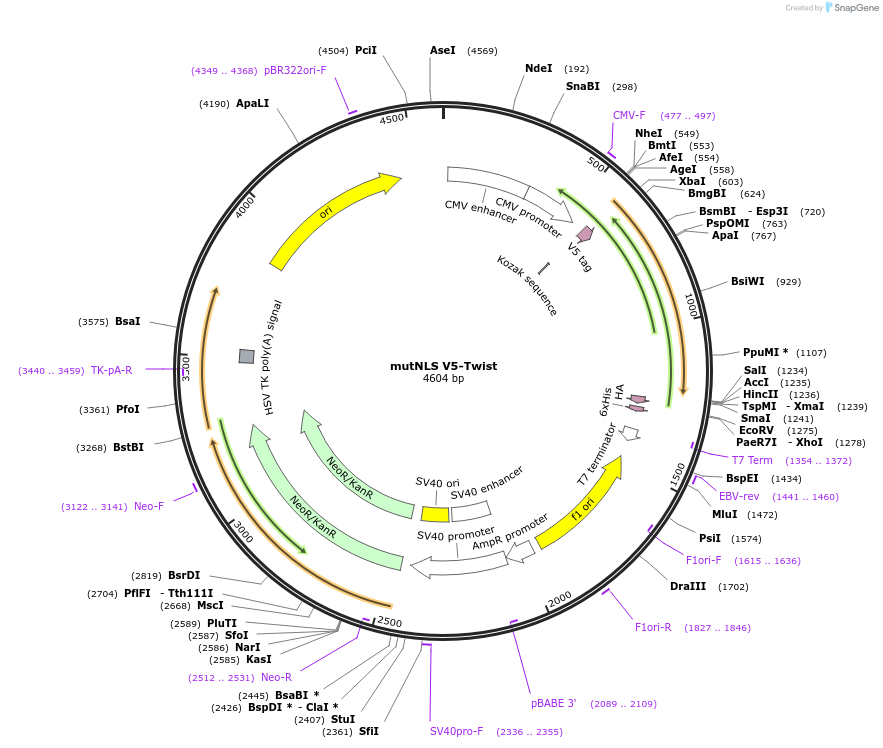
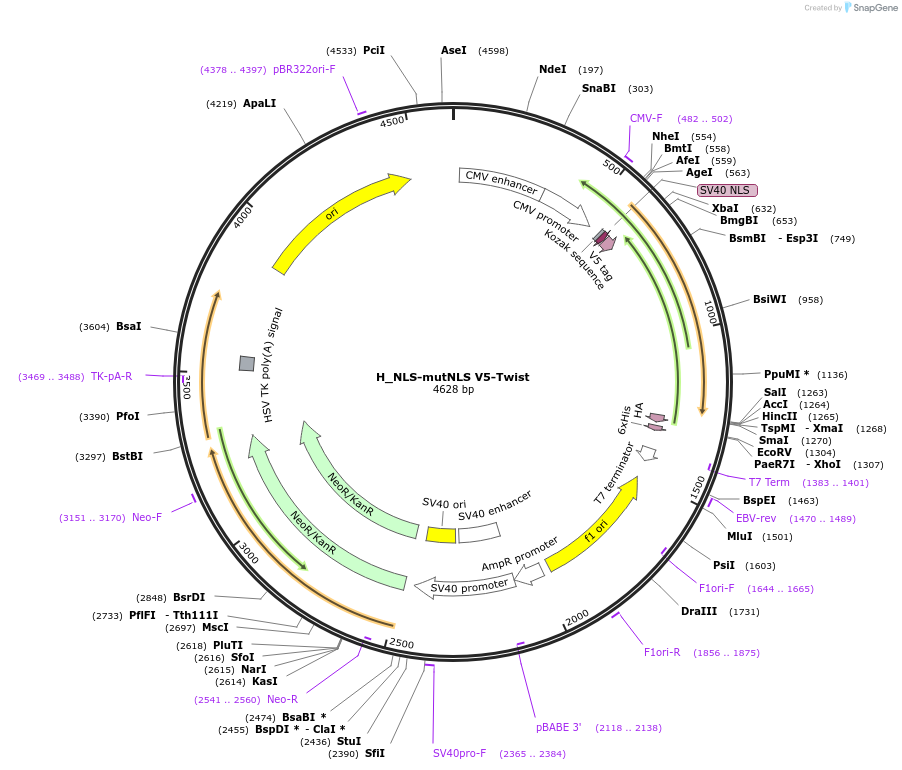
H_NLS-mutNLS V5-Twist
Plasmid#201378PurposeExpresses Twist mutant for nucleocytoplasmic transport studiesDepositorAvailable SinceJuly 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
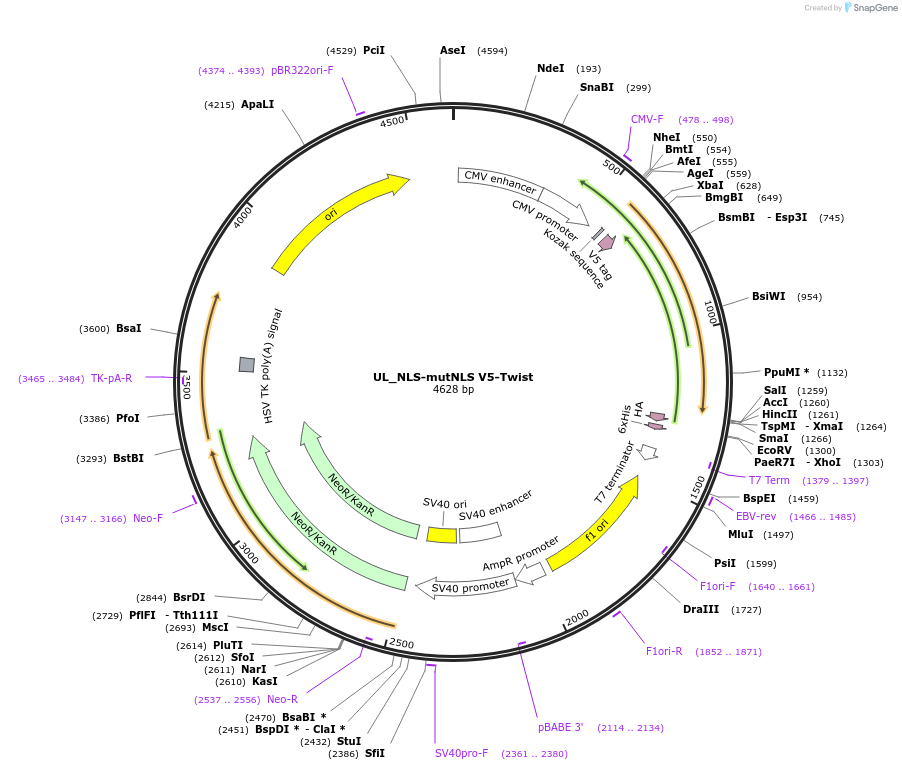
UL_NLS-mutNLS V5-Twist
Plasmid#201382PurposeExpresses Twist mutant for nucleocytoplasmic transport studiesDepositorAvailable SinceJuly 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
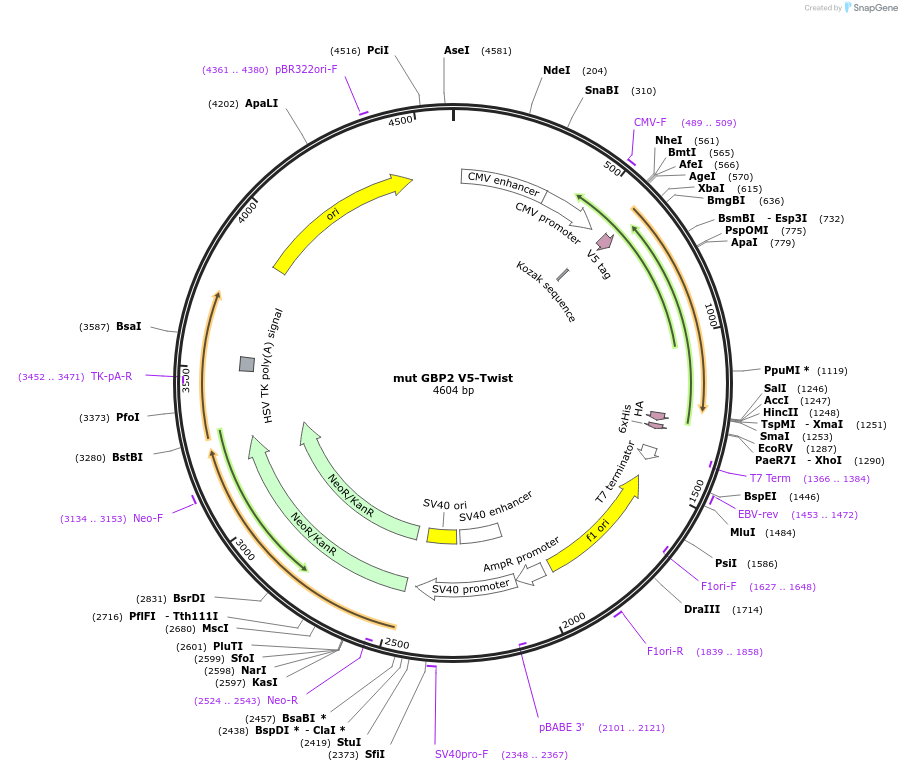
mut GBP2 V5-Twist
Plasmid#201377PurposeExpresses Twist mutant for nucleocytoplasmic transport studiesDepositorAvailable SinceJuly 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
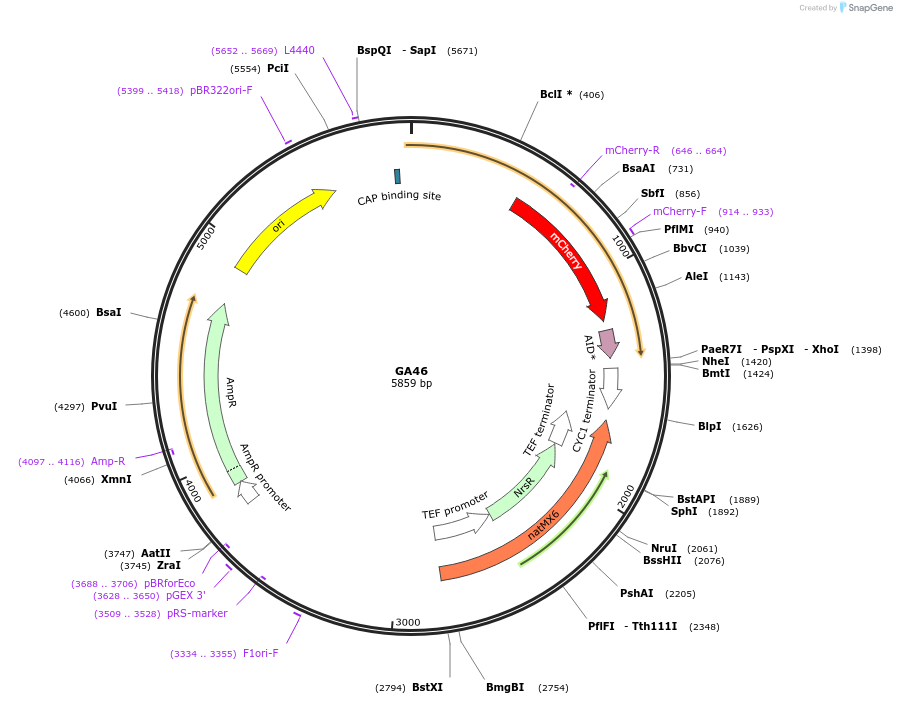
GA46
Plasmid#196614PurposePlasmid used for the C-terminal tagging of Atp3 with mCherry and the auxin-inducible degron in yeastDepositorInsertATP3::mCherry-AID(71-114)-NatMX (ATP3 Budding Yeast)
TagsAID(71-114) and mCherryExpressionYeastAvailable SinceMarch 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
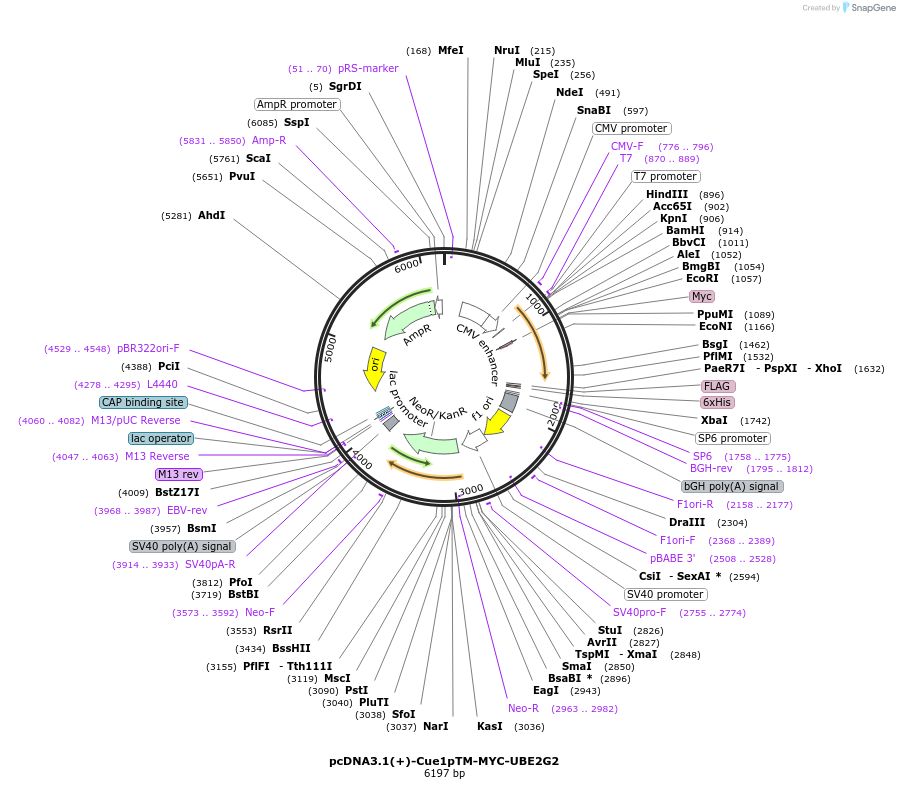
pcDNA3.1(+)-Cue1pTM-MYC-UBE2G2
Plasmid#185346PurposeMembrane anchoring of UBE2G2. Encodes transmembrane domain of S. Cerevisiae Cue1p fused to MYC-UBE2G2DepositorInsertCue1p/Ube2G2 fusion (internal MYC tag)
ExpressionMammalianAvailable SinceFeb. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
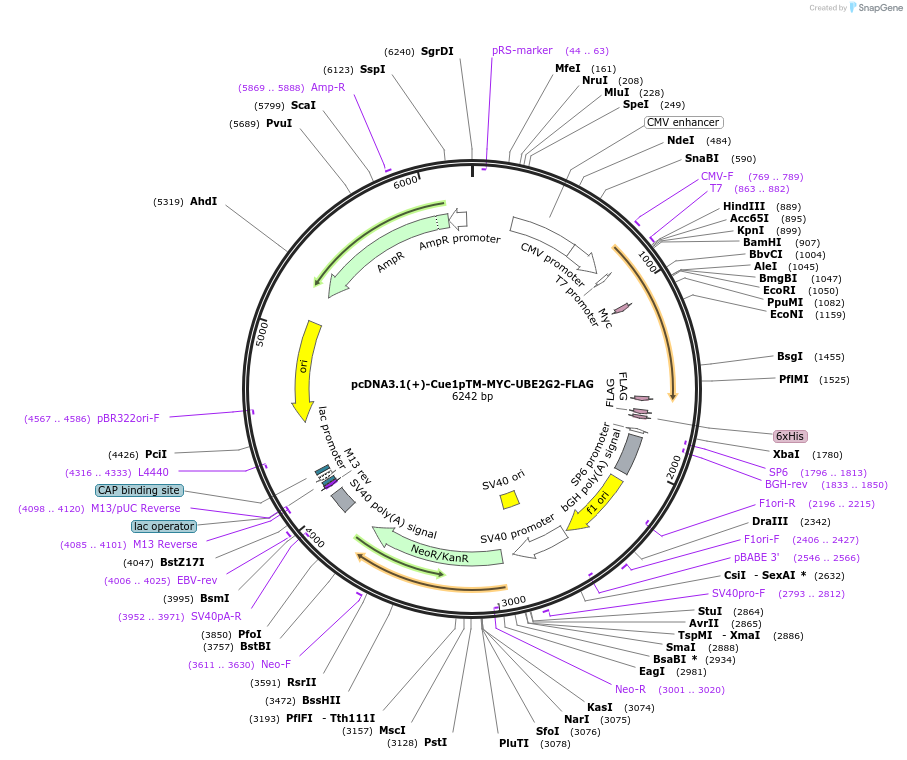
pcDNA3.1(+)-Cue1pTM-MYC-UBE2G2-FLAG
Plasmid#185347PurposeMembrane anchoring of UBE2G2 and co-IP. Encodes transmembrane domain of S. Cerevisiae Cue1p fused to MYC-UBE2G2 with C-terminal FLAG tagDepositorInsertCue1p/Ube2G2 fusion (internal MYC tag)
TagsFLAGExpressionMammalianAvailable SinceFeb. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
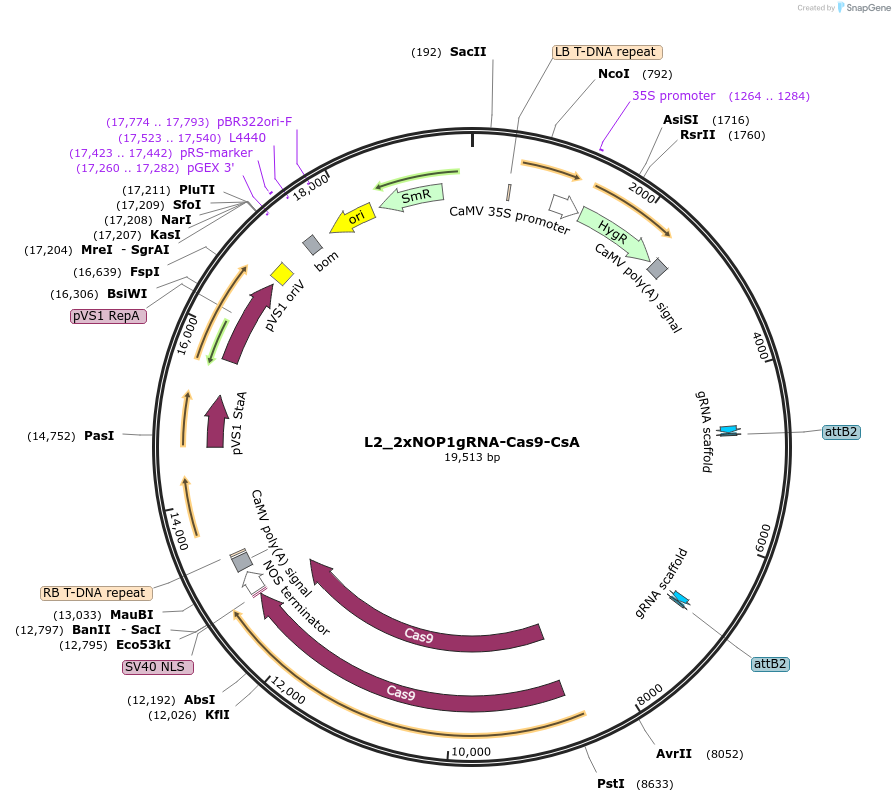
L2_2xNOP1gRNA-Cas9-CsA
Plasmid#136140PurposePlasmid with 2 different NOP1-gRNA. Targets 500 bp apart in the genome. To tranform Mp as CRISPR control. Contains Cas9 and HygRDepositorInsertp5-35S:HygR p5-MpU6:NOP1gRNA2 p5-MpU6:NOP1gRNA1 p5-MpEF1a:Cas9-NLS
UseSynthetic BiologyExpressionPlantMutationBsaI/ SapI domesticatedAvailable SinceNov. 30, 2021AvailabilityAcademic Institutions and Nonprofits only -
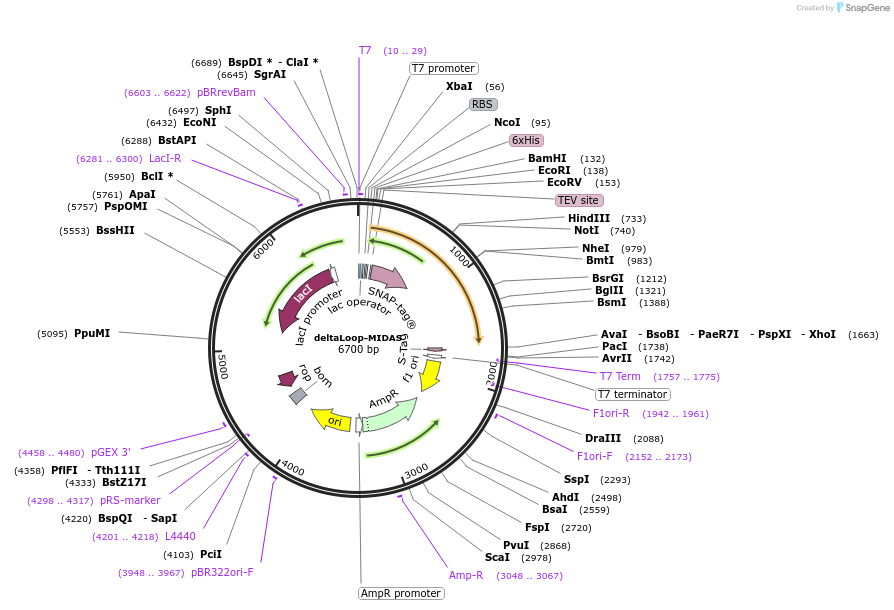
deltaLoop-MIDAS
Plasmid#171605PurposeExpresses the SNAP-tagged MIDAS domain of S. pombe Mdn1 (a.a. 4381-4717) without the loop region (a.a. 4458-4496) in bacteria.DepositorInsertMIDAS domain of S. pombe Mdn1 (a.a. 4381-4717)
TagsHis6 tag and SNAP tagExpressionBacterialMutationReplaced amino acids 4458-4496 with ASGSGS linkerAvailable SinceJuly 19, 2021AvailabilityAcademic Institutions and Nonprofits only -
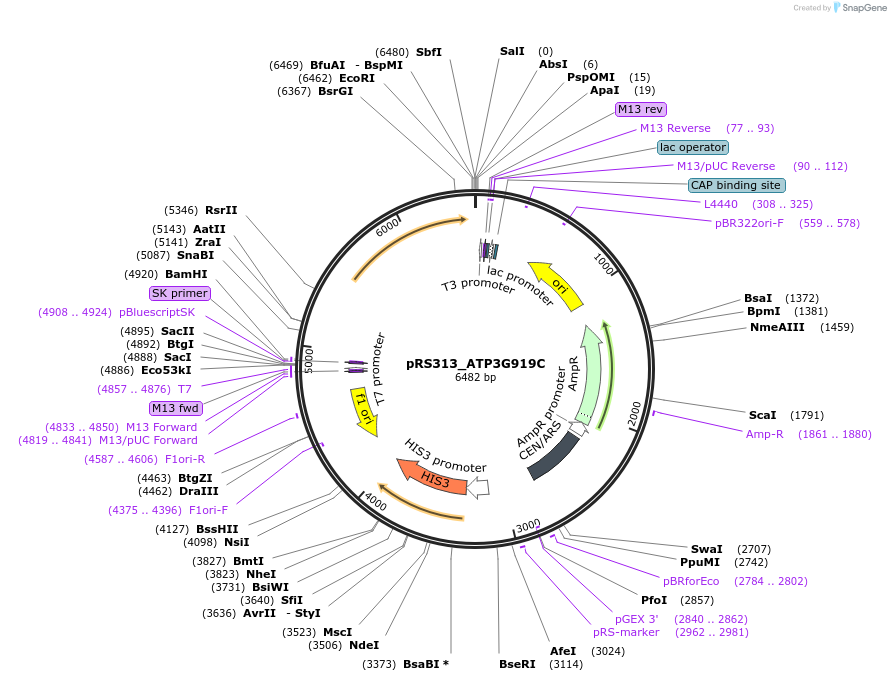
pRS313_ATP3G919C
Plasmid#120255PurposeExpression of ATP3-7 in yeastDepositorInsertATP3 (ATP3 Budding Yeast)
ExpressionYeastMutationG919 changed to C; Alanine 307 changed to ProlinePromoterATP3Available SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
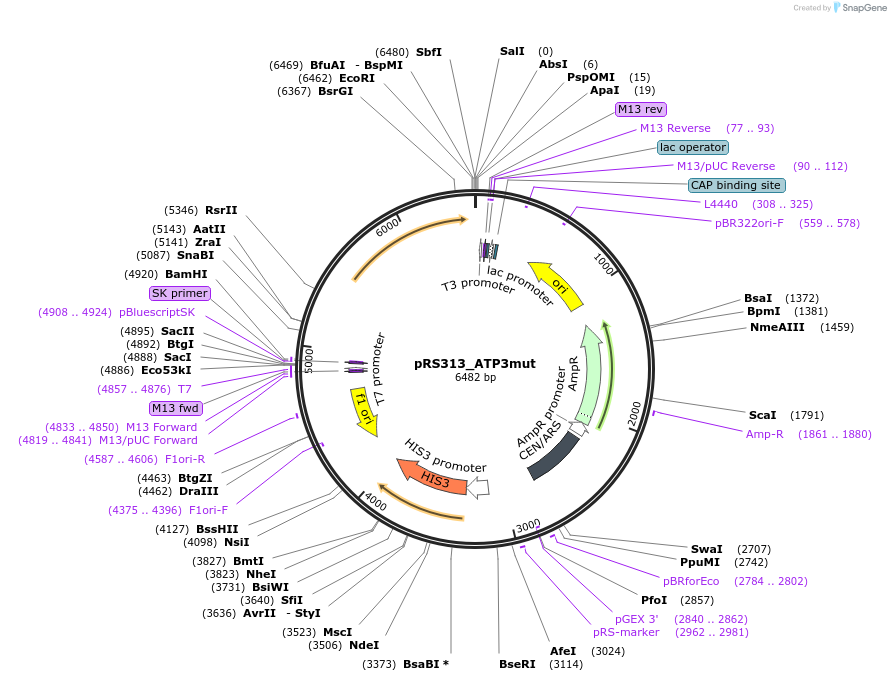
pRS313_ATP3mut
Plasmid#120256PurposeExpression of ATP3-67 in yeastDepositorInsertATP3 (ATP3 Budding Yeast)
ExpressionYeastMutationIsoleucine 304 changed to Asparagine; Alanine 307…PromoterATP3Available SinceApril 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
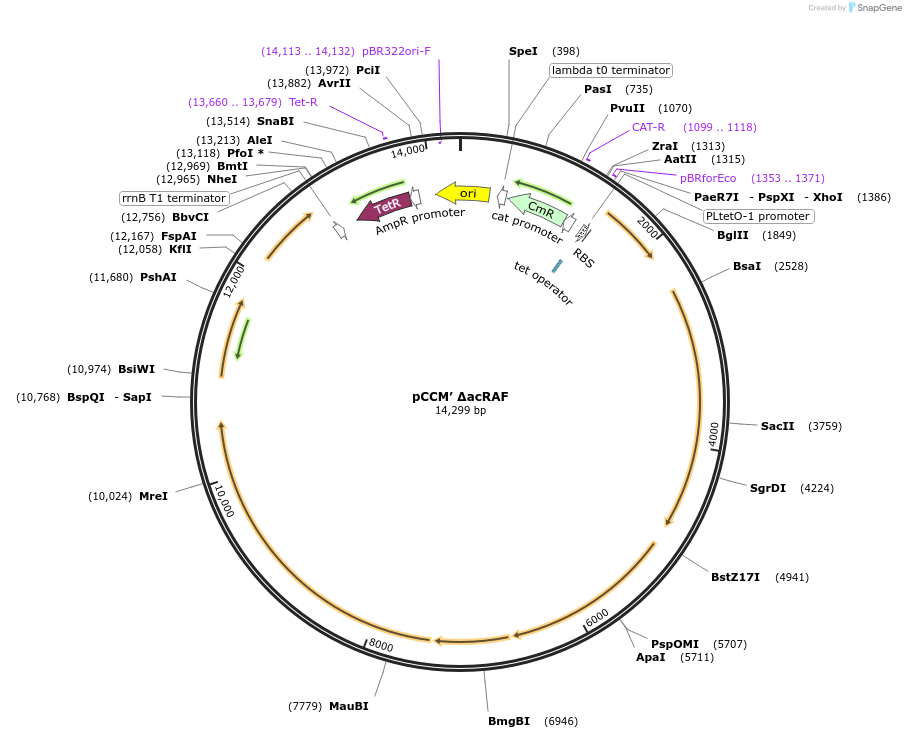
pCCM’ ΔacRAF
Plasmid#162710PurposeSecond CCM operon lacking acRAF; Deletion of putative rubisco chaperone, acRAFDepositorInsertSecond CCM operon cloned from H. neapolitanus
ExpressionBacterialMutationpCCM' with a deletion of putative rubisco ch…PromoterPLtet0-1 promoterAvailable SinceFeb. 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
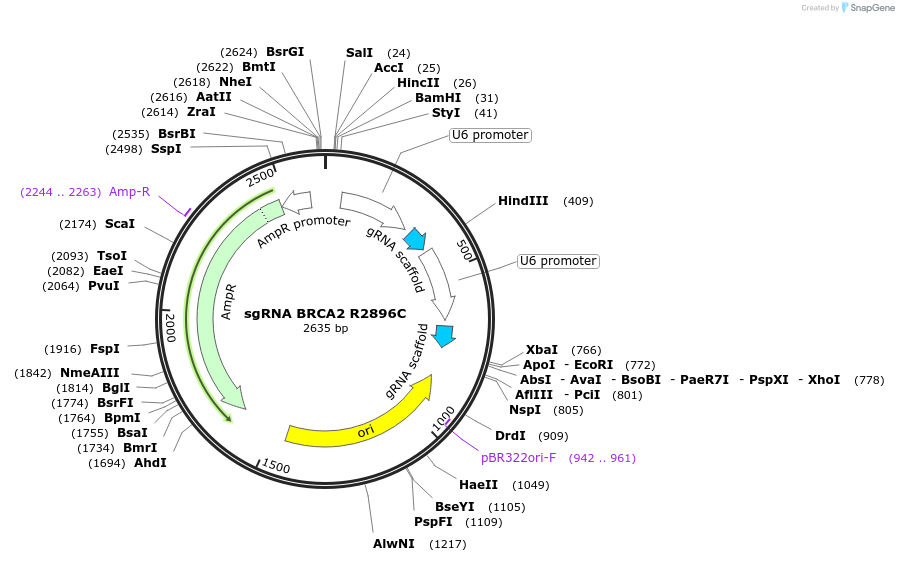
sgRNA BRCA2 R2896C
Plasmid#139511PurposePlasmid expressing a sgRNA to introduce BRCA2 R2896C using base editingDepositorInsertsgRNA to insert BRCA2 E2896C using base editing
ExpressionMammalianPromoterU6Available SinceMay 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
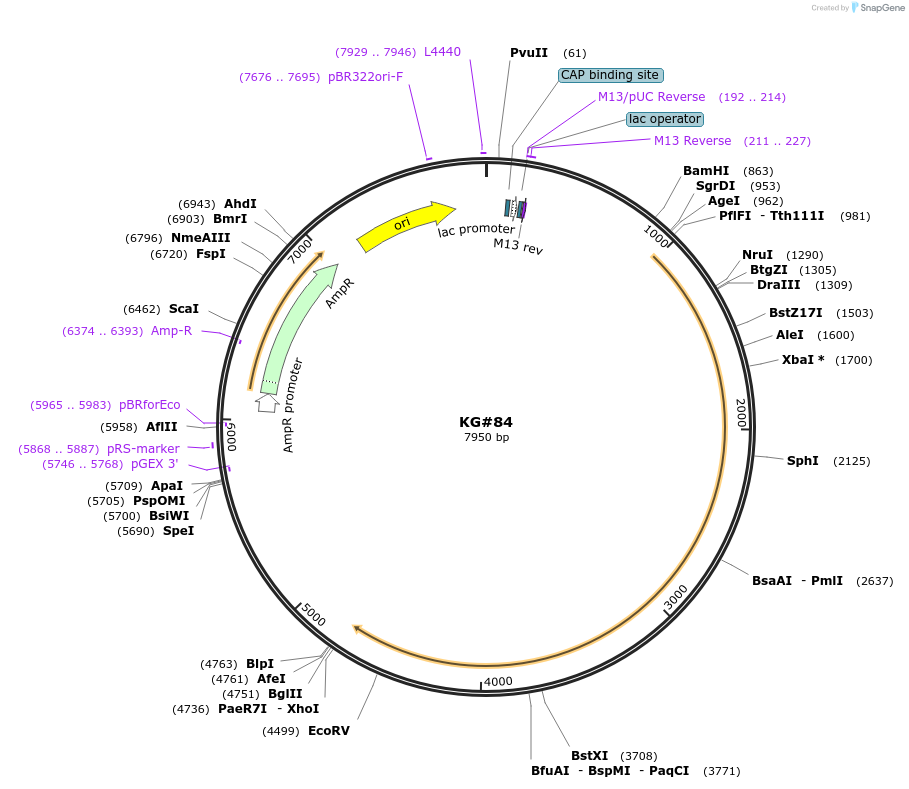
KG#84
Plasmid#110878PurposeExpresses the C. elegans acy--1 P260S gain-of-function cDNA in ventral cord cholinergic motor neuronsDepositorInsertsunc-17beta promoter
acy-1 P260S gain-of-function cDNA
unc-54 3' control region with 1 artificial intron just upstream and 1 artificial intron in control region
ExpressionBacterialMutationChanged Proline to Serine at amino acid 260Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
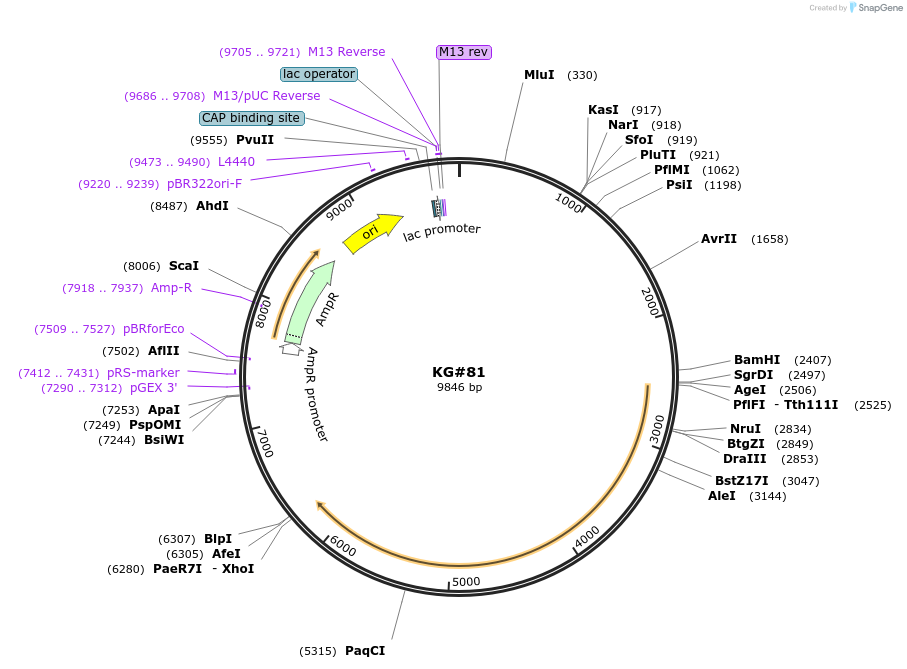
KG#81
Plasmid#110876PurposeExpresses the C. elegans acy-1 P260S gain-of-function cDNA in body wall muscleDepositorInsertsmyo-3 promoter
acy-1 P260S gain-of-function cDNA
unc-54 3' control region with 1 artificial intron just upstream and 1 artificial intron in control region
ExpressionBacterialMutationChanged Proline to Serine at amino acid 260Available SinceJune 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
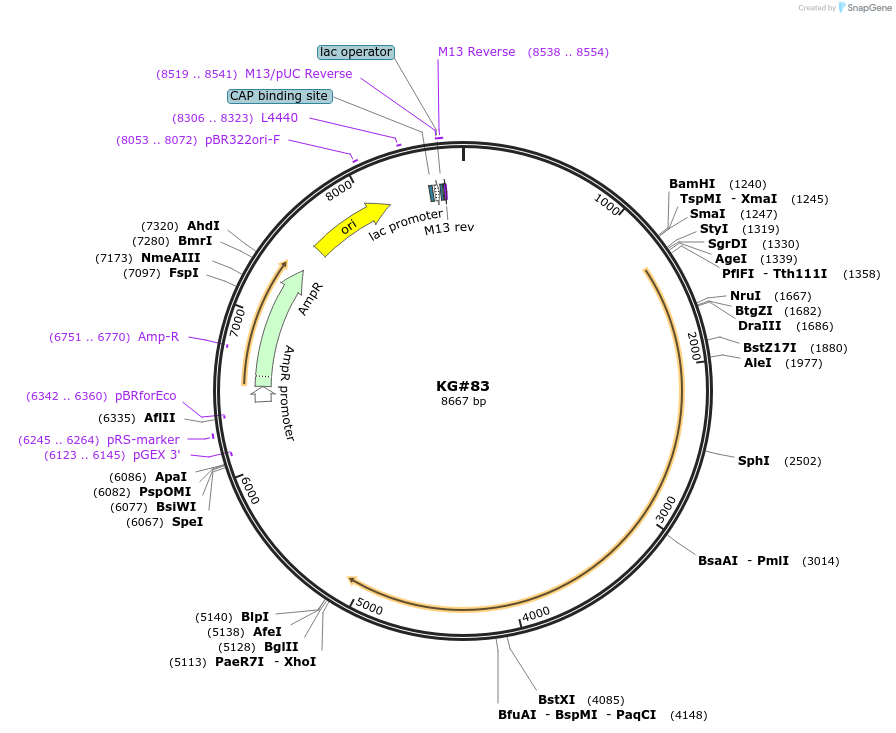
KG#83
Plasmid#110877PurposeExpresses the C. elegans acy--1 P260S gain-of-function cDNA pan-neuronallyDepositorInsertsrab-3 promoter
acy-1 P260S gain-of-function cDNA
unc-54 3' control region with 1 artificial intron just upstream and 1 artificial intron in control region
ExpressionBacterialMutationChanged Proline to Serine at amino acid 260Available SinceJune 11, 2018AvailabilityAcademic Institutions and Nonprofits only