We narrowed to 20,907 results for: OMP;
-
Plasmid#8755DepositorInsertBcl-XL (Exon 1) (Bcl2l1 Mouse)
ExpressionBacterialAvailable SinceJuly 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
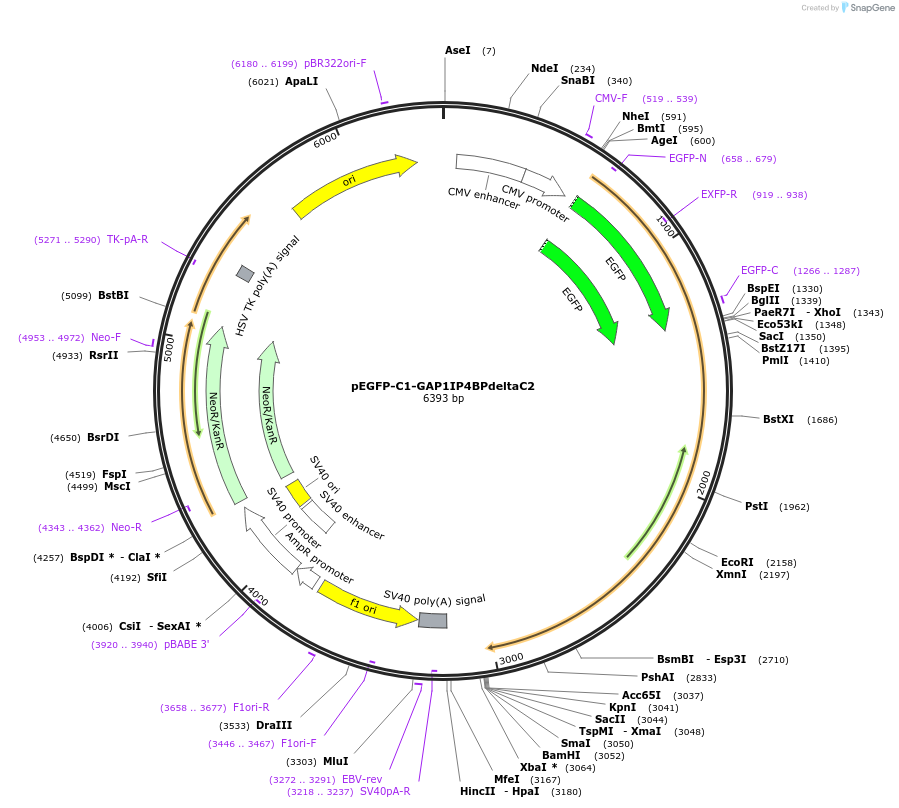
pEGFP-C1-GAP1IP4BPdeltaC2
Plasmid#20199DepositorInsertGAP1IP4BP-detlaC2 (RASA3 Human)
TagsGFPExpressionMammalianMutationDeletion of C2 domainsAvailable SinceFeb. 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
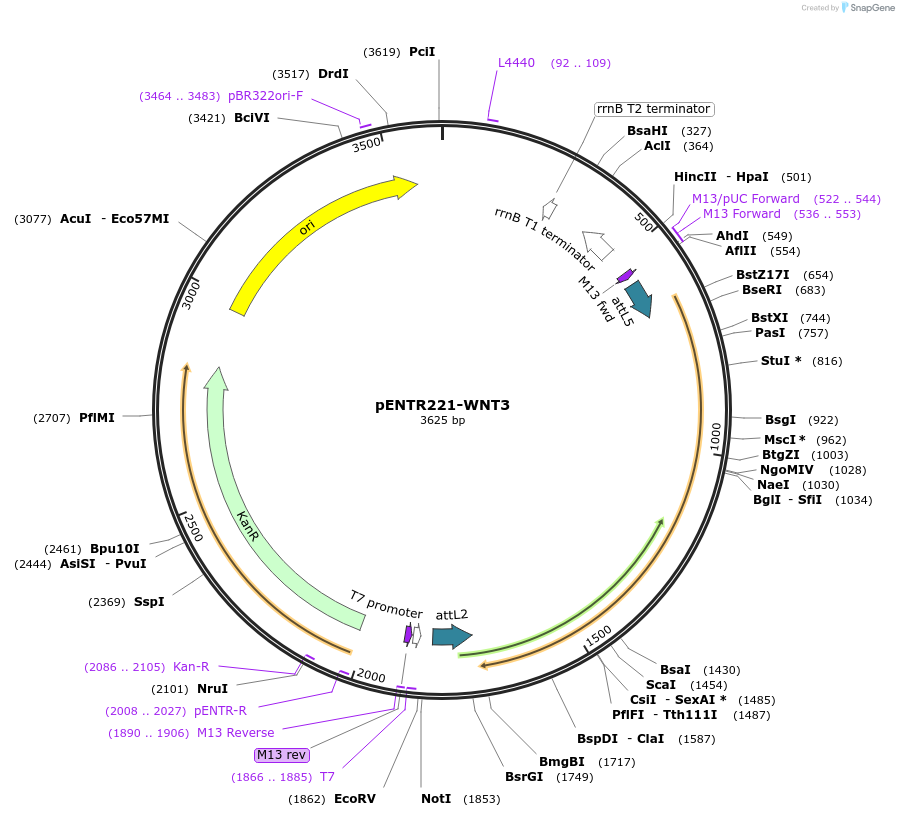
pENTR221-WNT3
Plasmid#79500PurposeMultiSite Gateway entry clones, second fragment (attL5, attL2) Human WNT3 CDSDepositorAvailable SinceMay 10, 2017AvailabilityAcademic Institutions and Nonprofits only -
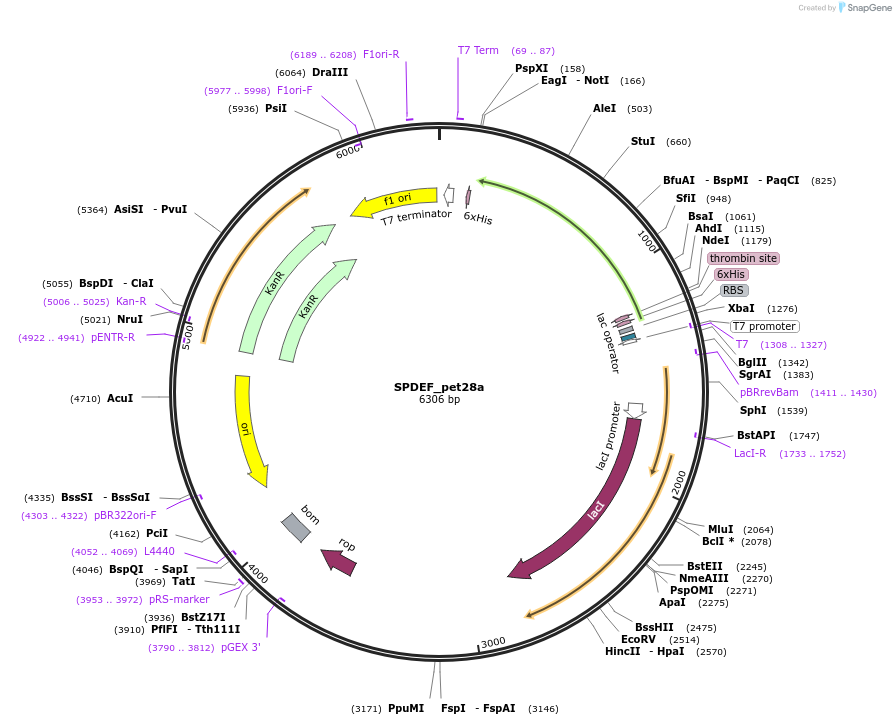
SPDEF_pet28a
Plasmid#131643PurposeBacterial expression of ETS gene SPDEFDepositorAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
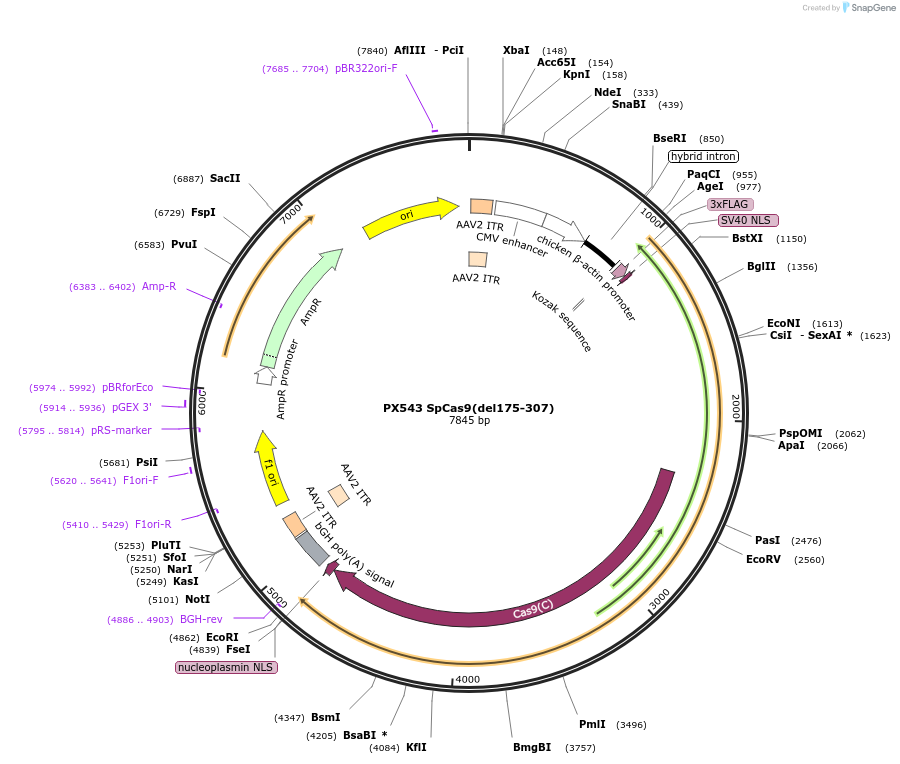
PX543 SpCas9(del175-307)
Plasmid#58888PurposeTruncated form of SpCas9 without REC2 domainDepositorInserthSpCas9
UseCRISPRTags3XFLAGExpressionMammalianMutationdel amino acids 175-307Available SinceSept. 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
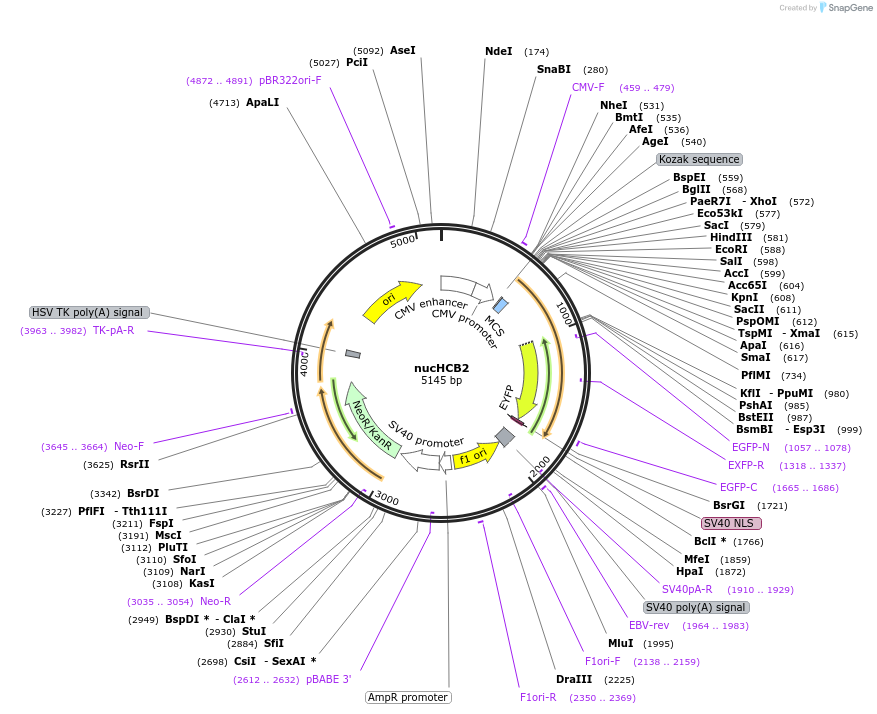
nucHCB2
Plasmid#98581PurposeNuclear Huntingtin Chromobody 2: "iVHH4" intrabody recognizing amino acids 49–148 of huntingtin, fused to eYFP and nuclear localization signal.DepositorInsertHCB2
TagsNLS and YFPExpressionMammalianPromoterCMVAvailable SinceJuly 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
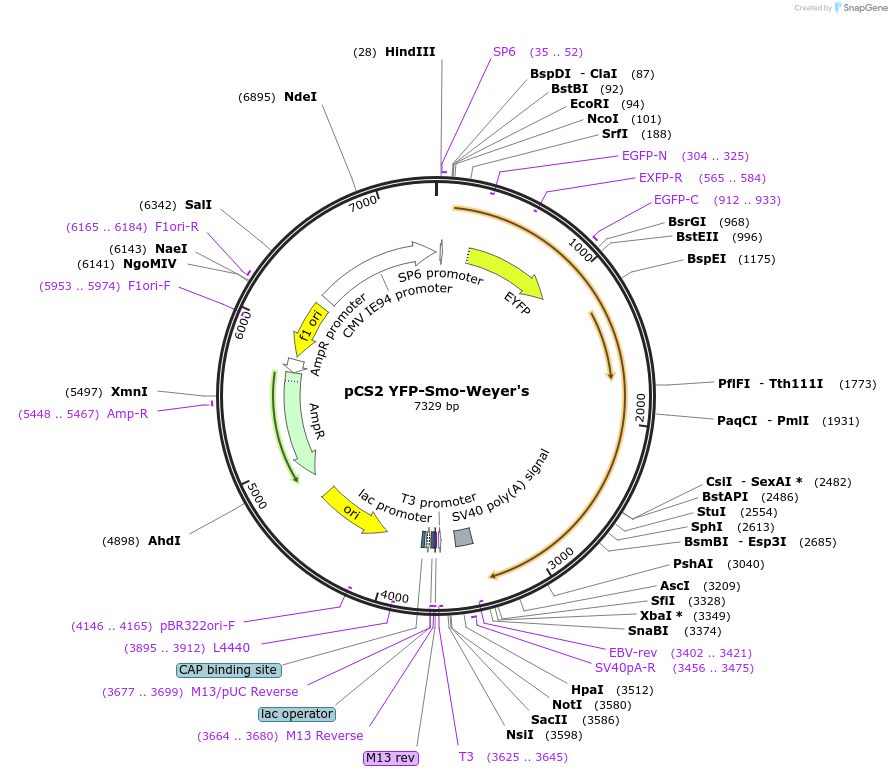
pCS2 YFP-Smo-Weyer's
Plasmid#41014DepositorInsertsmoothened homolog (Smo Mouse)
TagsEVC2 (aa 1177-1220) and YFPExpressionMammalianMutationYFP inserted after Smo signal sequence. Transfer…Available SinceNov. 16, 2012AvailabilityAcademic Institutions and Nonprofits only -
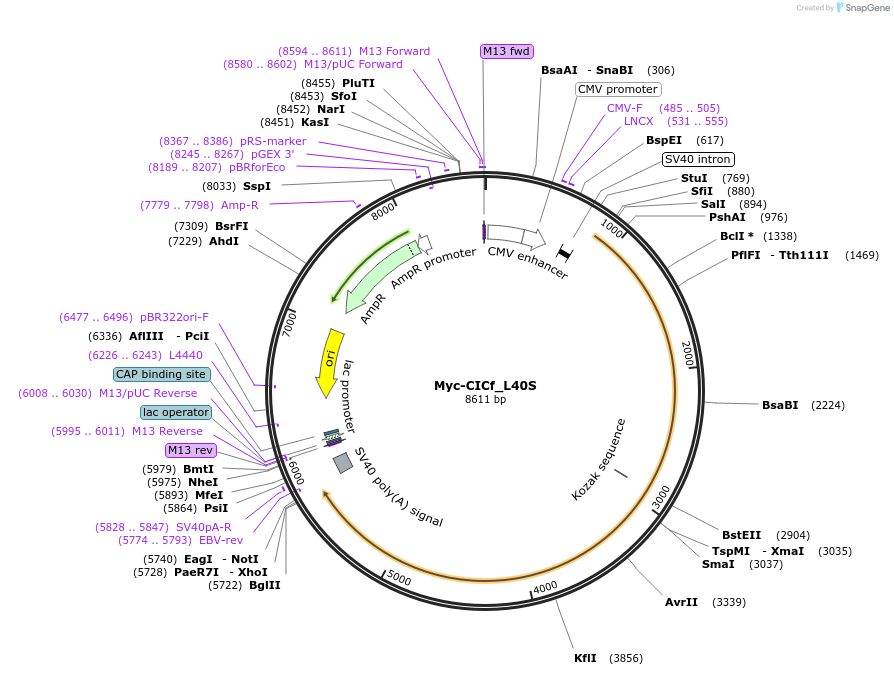
Myc-CICf_L40S
Plasmid#48187PurposeExpresses L40S mutated CicDepositorAvailable SinceOct. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
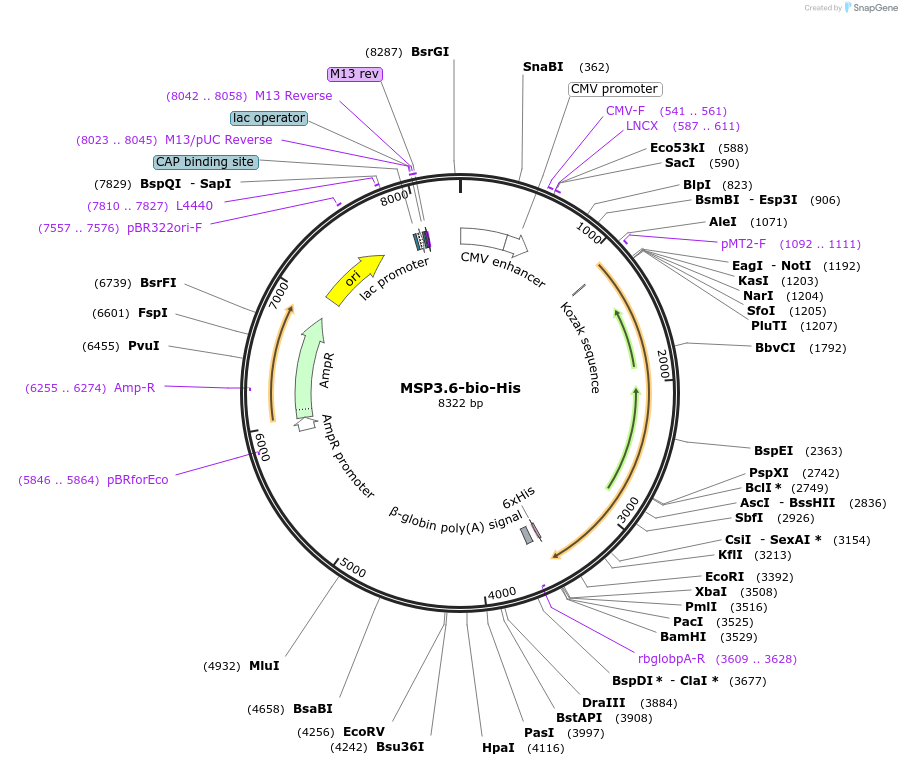
MSP3.6-bio-His
Plasmid#50804PurposeExpresses enzymatically monobiotinylated full length MSP3.6DepositorInsertMSP3.6
TagsratCD4d3+4,biotinylation sequence, 6xHisExpressionMammalianMutationall potential N-linked glycosylation sites (N-X-S…PromoterCMVAvailable SinceFeb. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
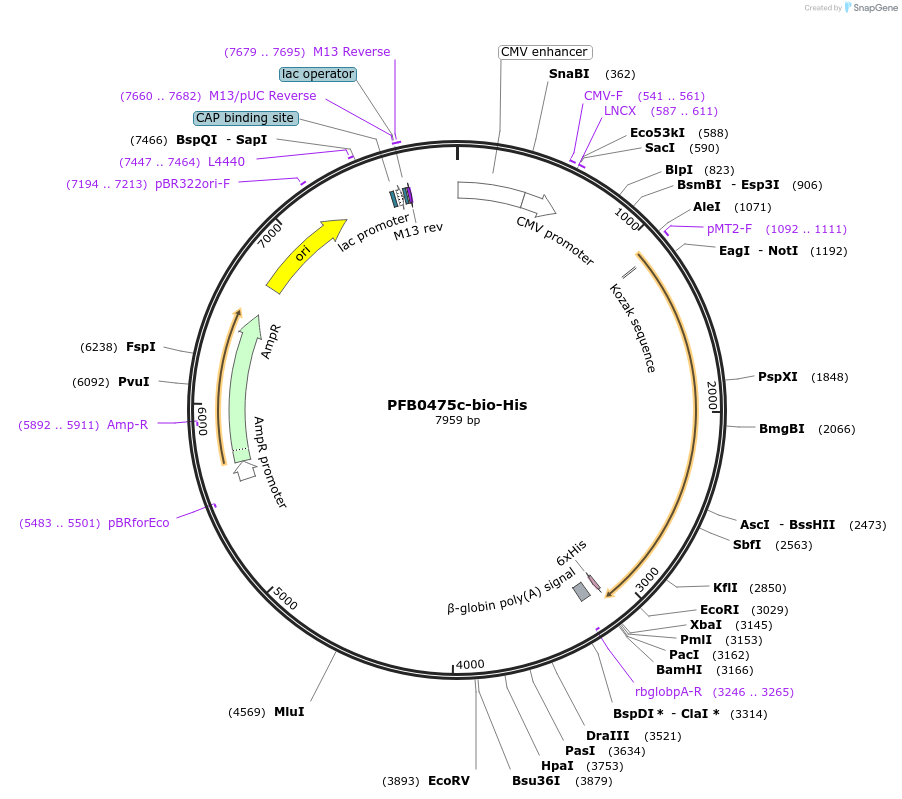
PFB0475c-bio-His
Plasmid#50822PurposeExpresses enzymatically monobiotinylated full length PFB0475cDepositorInsertPFB0475c
TagsratCD4d3+4,biotinylation sequence, 6xHisExpressionMammalianMutationall potential N-linked glycosylation sites (N-X-S…PromoterCMVAvailable SinceFeb. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
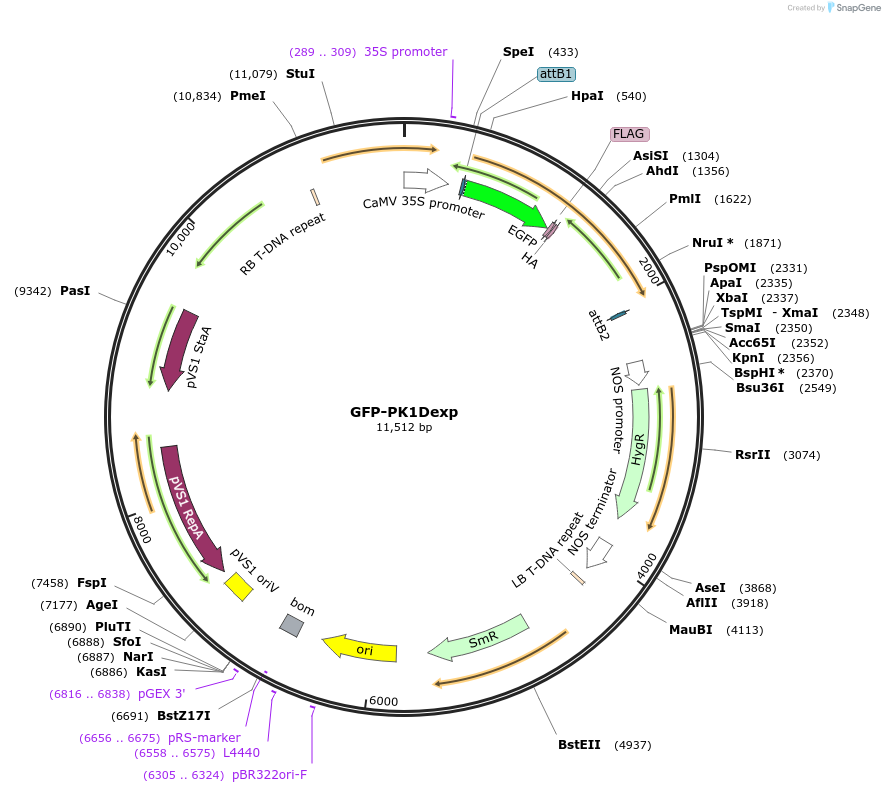
GFP-PK1Dexp
Plasmid#130834PurposeExpression of plant LINC fluorescent fusion protein for cell biologyDepositorInsertMLKG1
TagseGFP-FLAG-HAExpressionPlantPromoterCaMV 35SAvailable SinceJune 19, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
psiCHECK-2-Snail1-3'UTR
Plasmid#61781PurposeLuciferase reporter containing the 3'UTR of mouse Snail1DepositorInsertmouse Snail1 3'UTR
UseLuciferaseAvailable SinceMay 14, 2015AvailabilityAcademic Institutions and Nonprofits only -
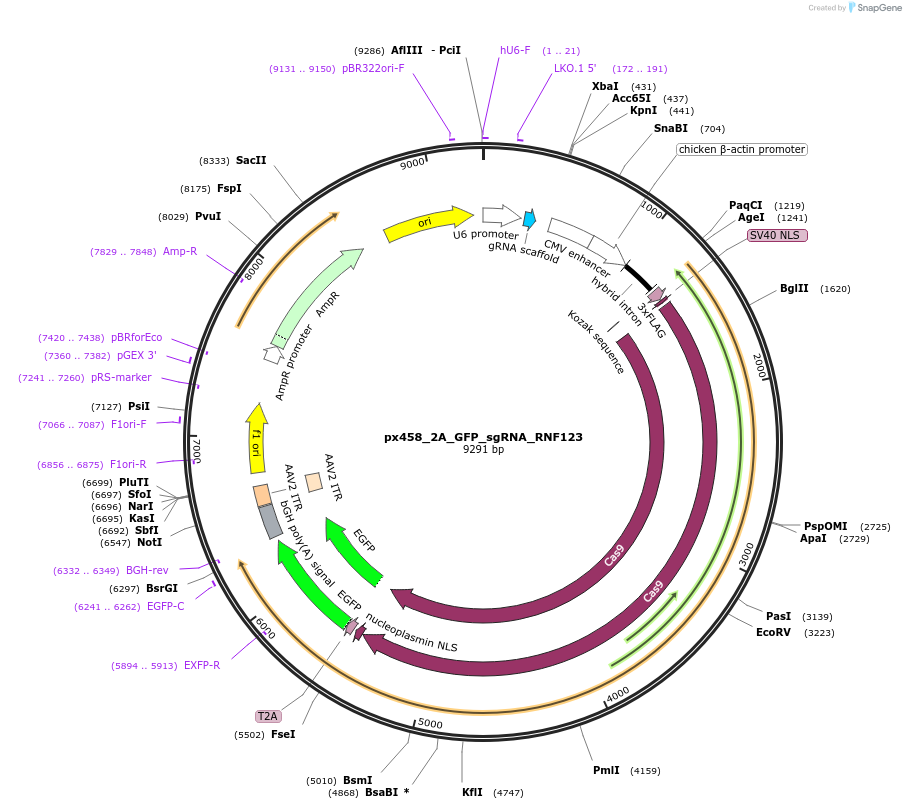
px458_2A_GFP_sgRNA_RNF123
Plasmid#127122DepositorInsertgRNA RNF123 (RNF123 Human)
UseCRISPRAvailable SinceJuly 25, 2019AvailabilityAcademic Institutions and Nonprofits only -
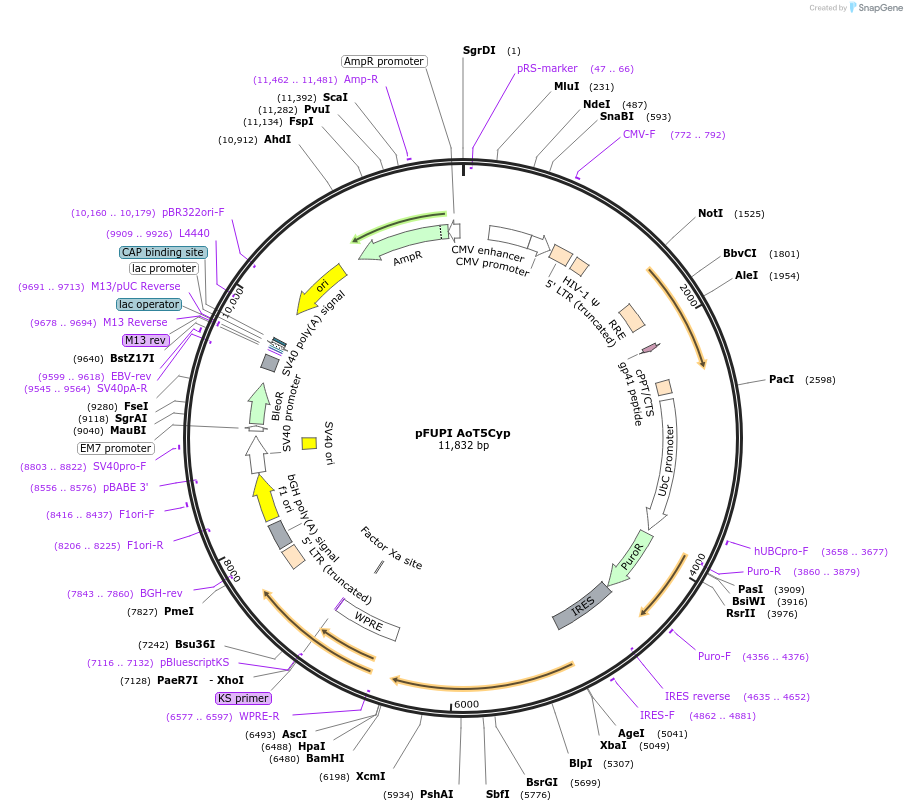
pFUPI AoT5Cyp
Plasmid#80280PurposeExpresses Aotus TRIM5 CypA fusionDepositorInsertTRIM5 CypA fusion
UseLentiviralExpressionMammalianAvailable SinceAug. 3, 2016AvailabilityAcademic Institutions and Nonprofits only -
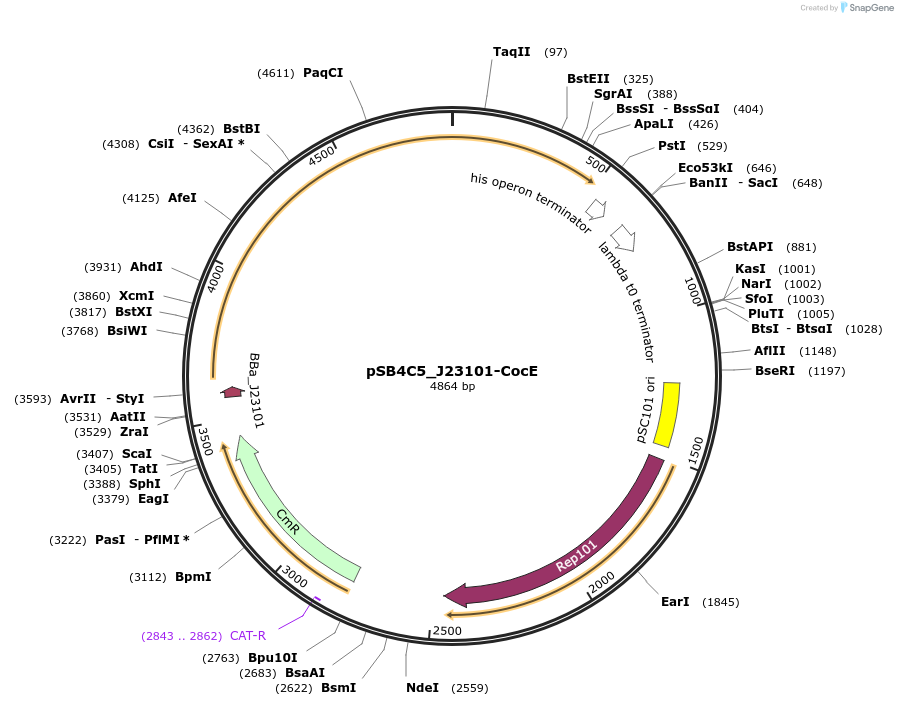
pSB4C5_J23101-CocE
Plasmid#128129PurposeExpressing CocE enzyme (cocaine to benzoate) gene under control of the constitutive promoter J23101, and RBS B0032DepositorInsertCocE
UseSynthetic BiologyExpressionBacterialPromoterJ23101Available SinceSept. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
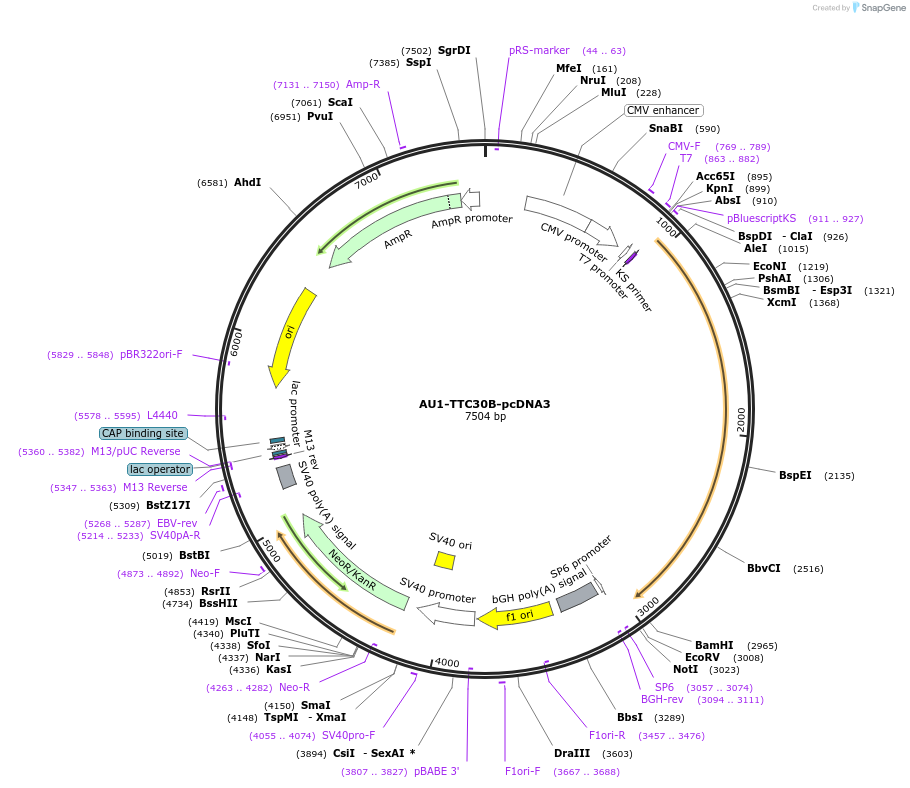
AU1-TTC30B-pcDNA3
Plasmid#47325PurposeExpresses AU1 epitope-tagged TTC30B in mammalian cellsDepositorAvailable SinceSept. 3, 2013AvailabilityAcademic Institutions and Nonprofits only -
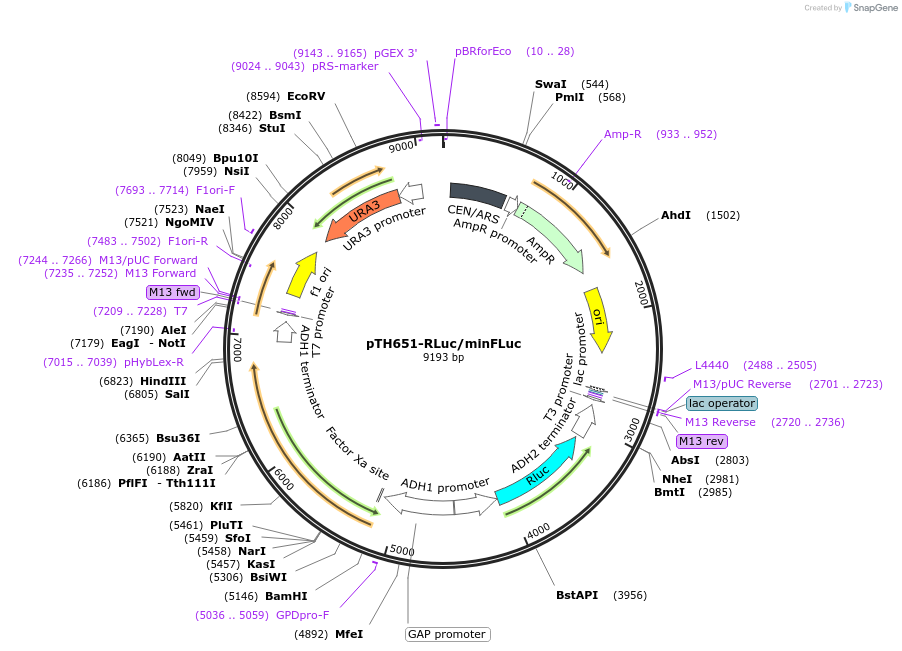
pTH651-RLuc/minFLuc
Plasmid#29697DepositorInsertFirefly Luciferase
ExpressionYeastMutationAll codons of the original FLuc gene were exchang…Available SinceAug. 25, 2011AvailabilityAcademic Institutions and Nonprofits only -
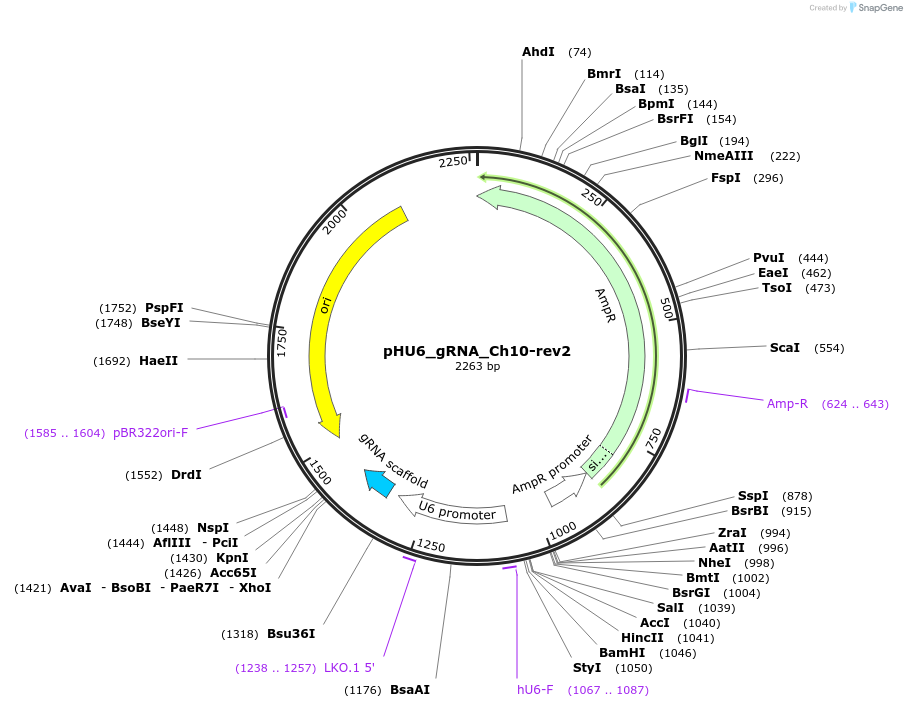
pHU6_gRNA_Ch10-rev2
Plasmid#81207Purposefor targeting Ch10 psuedo gix site in PCDH15 gene (five base pairs from the cas9 site and 5' of gix site)DepositorInserthU6 expression of gRNA
ExpressionMammalianPromoterU6Available SinceNov. 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
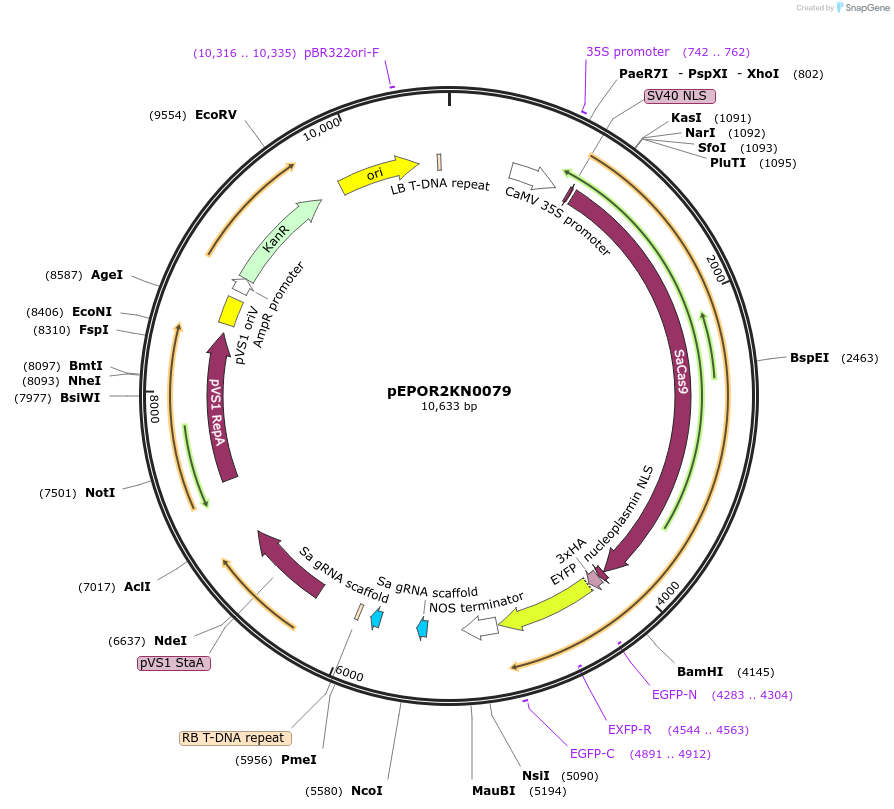
pEPOR2KN0079
Plasmid#117646PurposeLevel2 MoClo construct, containing level1 plant expression cassettes for SaCas9(pEPOR1CB0015) and sgRNA_ENTH/ANT {AT1G14910}(pEPOR1CB0100)and sgRNA_ENTH/ANT {AT2G01600}(pEPOR1CB0111)DepositorInsert[35S:SaCas9(pEPOR1CB0015) ] +[AtU6-26:sgRNA]
ExpressionPlantAvailable SinceNov. 16, 2018AvailabilityAcademic Institutions and Nonprofits only -
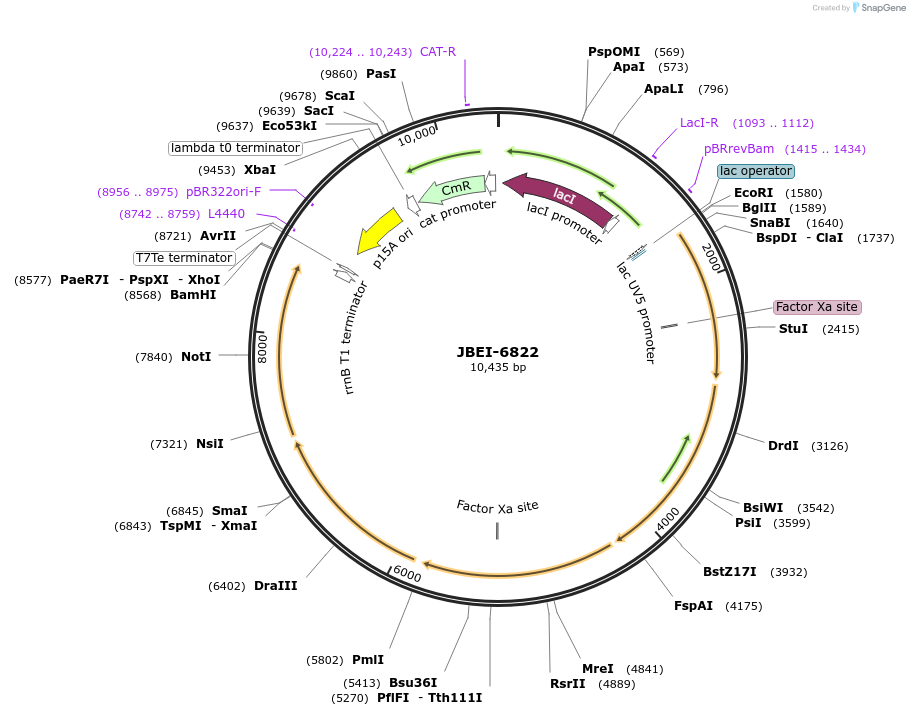
JBEI-6822
Plasmid#84985PurposeFor expression of pBbA5c-MevTaco-MKco-PMKco(aka 2A) under lacUV5 promoter for isopentenol productionDepositorInsertMevTaco-MKco-PMKco
ExpressionBacterialPromoterLacUV5Available SinceJan. 25, 2017AvailabilityAcademic Institutions and Nonprofits only