We narrowed to 38,180 results for: CaS
-
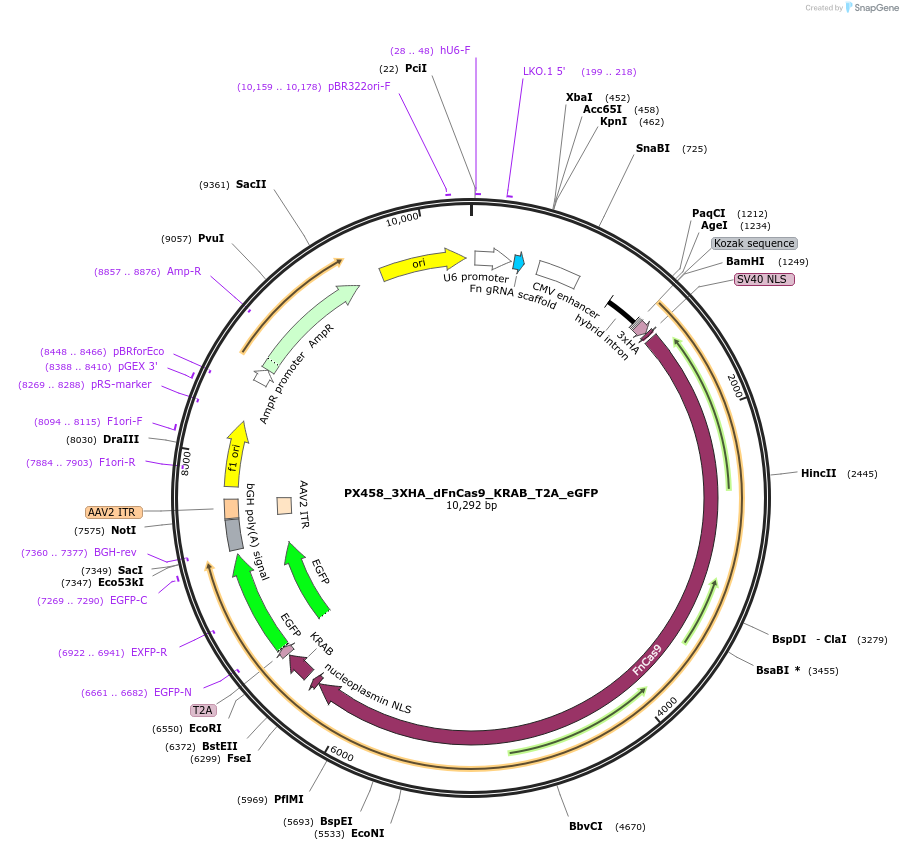
Plasmid#200389Purpose3XHA tagged dead Cas9 from F. novicida with KRAB repressor domain, T2A self-cleaving peptide and enhanced GFP biomarker for mammalian expressionDepositorInsertPX458_3XHA_dFnCas9_KRAB_eGFP
UseCRISPRTags3xHA-NLS and SV40NLS-KRAB-T2A-EGFPExpressionMammalianPromoterCBHAvailable SinceSept. 11, 2023AvailabilityAcademic Institutions and Nonprofits only -
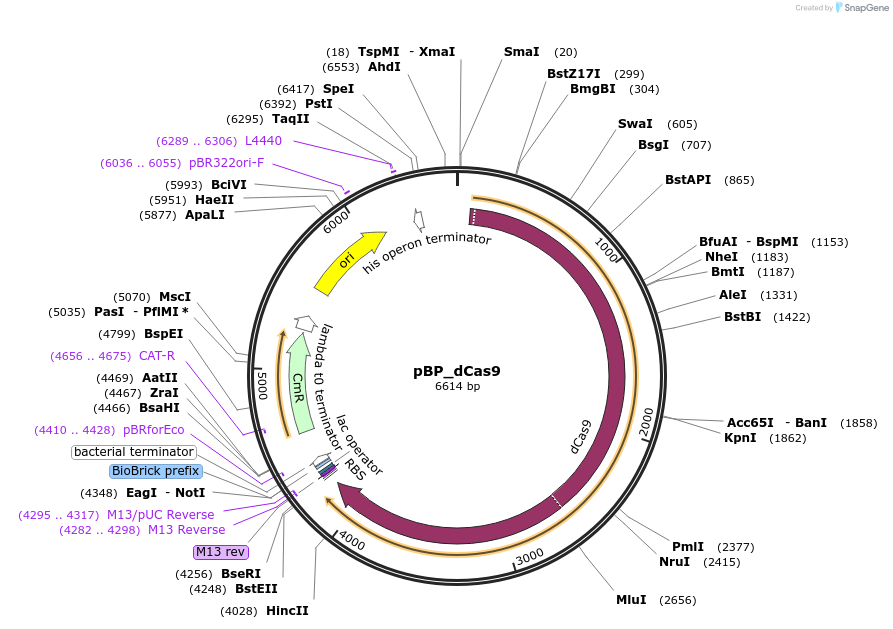
pBP_dCas9
Plasmid#190131PurposeLevel 0 MoClo plasmid containing dCas9DepositorInsertdCas9
UseCRISPRAvailable SinceAug. 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
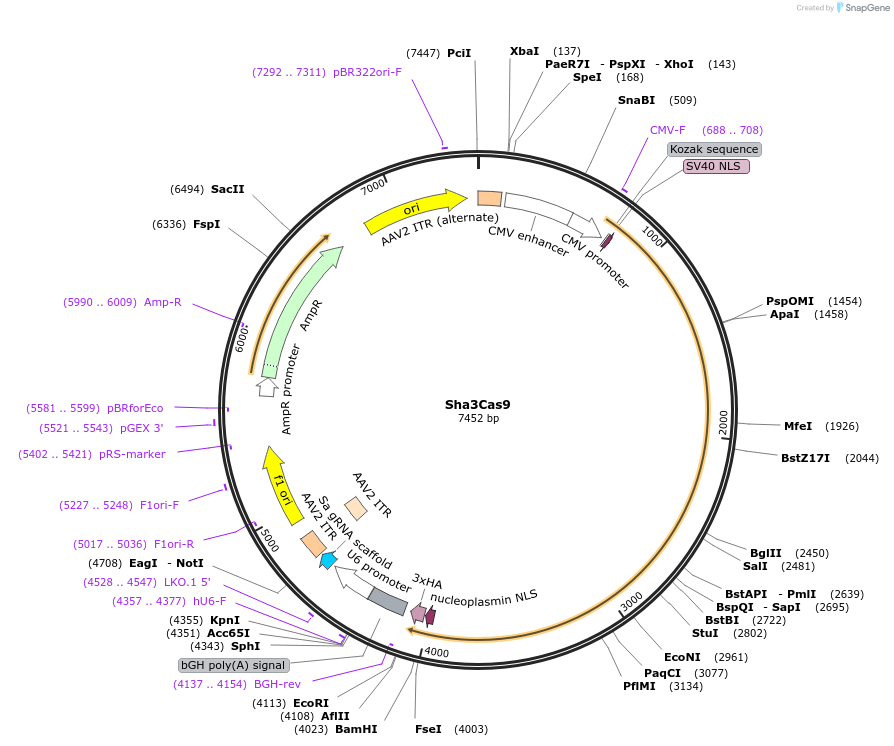
Sha3Cas9
Plasmid#192136PurposeExpresses Sha3Cas9, and cloning backbone for sgRNADepositorInsertSha3Cas9
UseAAVExpressionMammalianAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
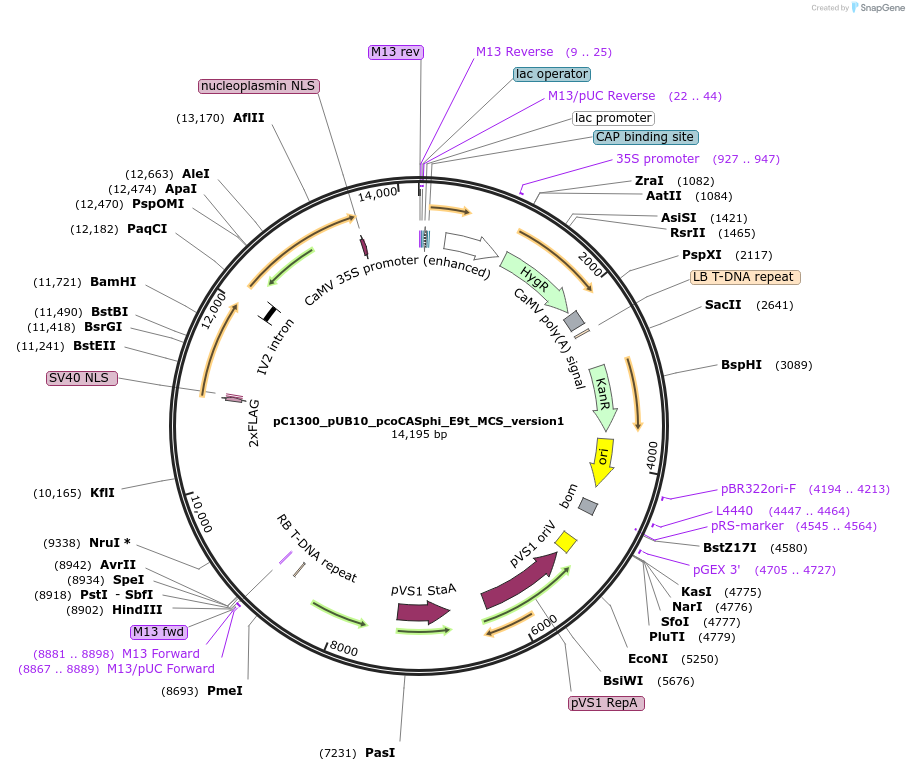
pC1300_pUB10_pcoCASphi_E9t_MCS_version1
Plasmid#197937PurposeT-DNA binary vector to express pcoCasphi driven by UBQ10 gene promoter, with SV40 NLS at the N-terminal and nucleoplasmin NLS at the C-terminal.DepositorInsertpcoCasphi-2
UseCRISPRExpressionPlantPromoterpUBQ10Available SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
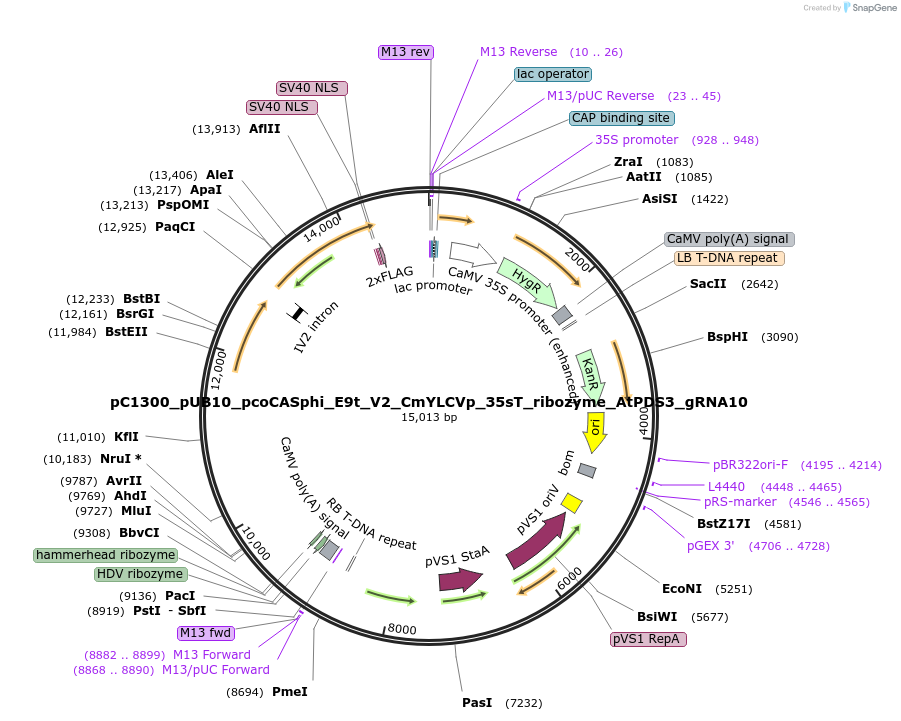
pC1300_pUB10_pcoCASphi_E9t_V2_CmYLCVp_35sT_ribozyme_AtPDS3_gRNA10
Plasmid#197958PurposeT-DNA binary vector to express pcoCasphi driven by UBQ10 gene promoter and a single AtPDS3 gRNA driven by CmYLCVp and flanked by ribozymes.DepositorInsertspcoCasphi-2
AtPDS3 gRNA10
UseCRISPRExpressionPlantPromoterCmYLCVp and pUBQ10Available SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
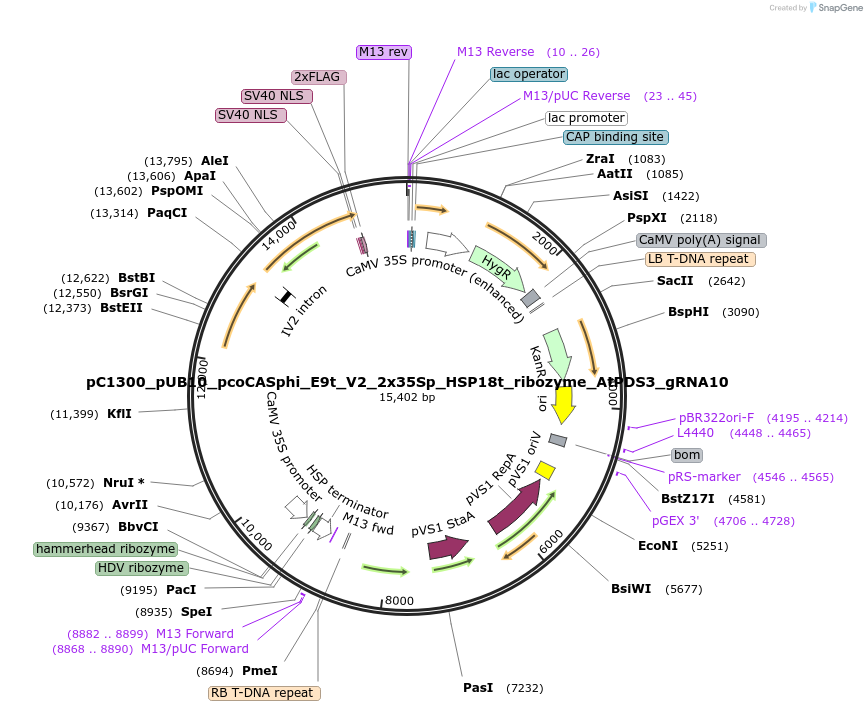
pC1300_pUB10_pcoCASphi_E9t_V2_2x35Sp_HSP18t_ribozyme_AtPDS3_gRNA10
Plasmid#197959PurposeT-DNA binary vector to express pcoCasphi driven by UBQ10 gene promoter and a single AtPDS3 gRNA driven by 2x35Sp and flanked by ribozymes.DepositorInsertspcoCasphi-2
AtPDS3 gRNA10
UseCRISPRExpressionPlantPromoter2x35S promoter and pUBQ10Available SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
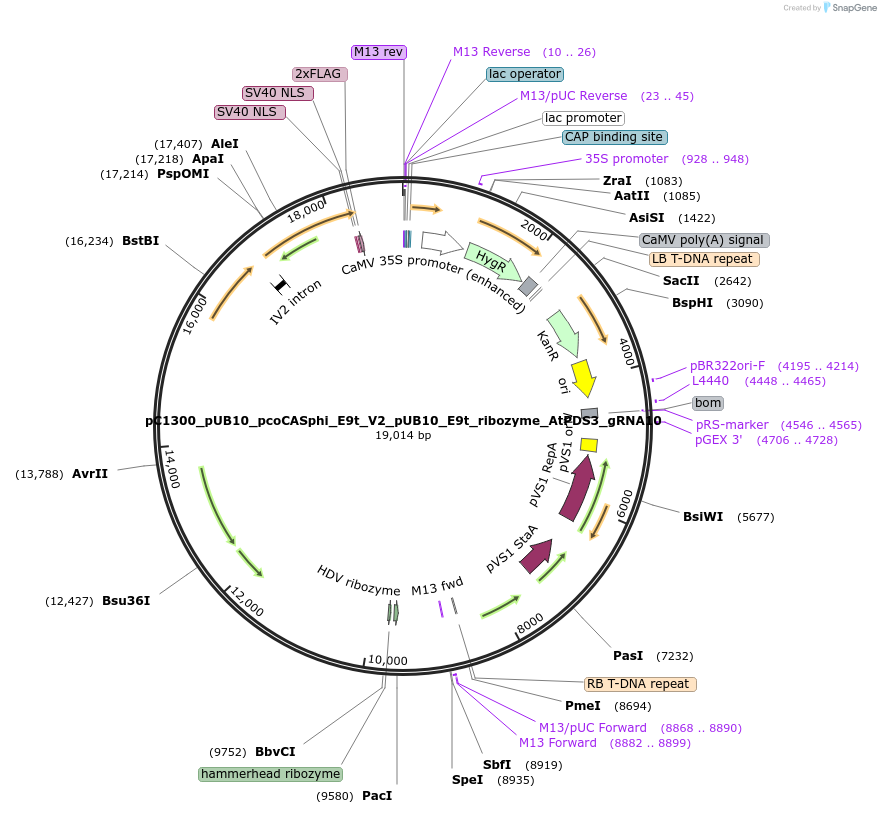
pC1300_pUB10_pcoCASphi_E9t_V2_pUB10_E9t_ribozyme_AtPDS3_gRNA10
Plasmid#197960PurposeT-DNA binary vector to express pcoCasphi driven by UBQ10 gene promoter and a single AtPDS3 gRNA driven by UBQ10 gene promoter and flanked by ribozymes.DepositorInsertspcoCasphi-2
AtPDS3 gRNA10
UseCRISPRExpressionPlantPromoterpUBQ10Available SinceMarch 30, 2023AvailabilityAcademic Institutions and Nonprofits only -
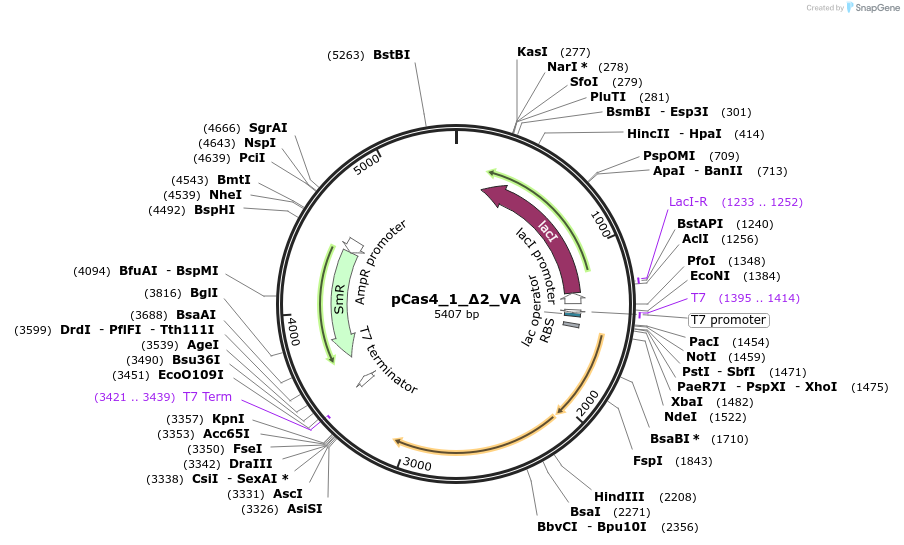
pCas4_1_Δ2_VA
Plasmid#186439PurposeEncodes Cas4, Cas1, leader, repeat and one spacer from F. Novicida (type V-A)DepositorInsertCas4, Cas1 and Cas2
UseCRISPRPromoterT7Available SinceMarch 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
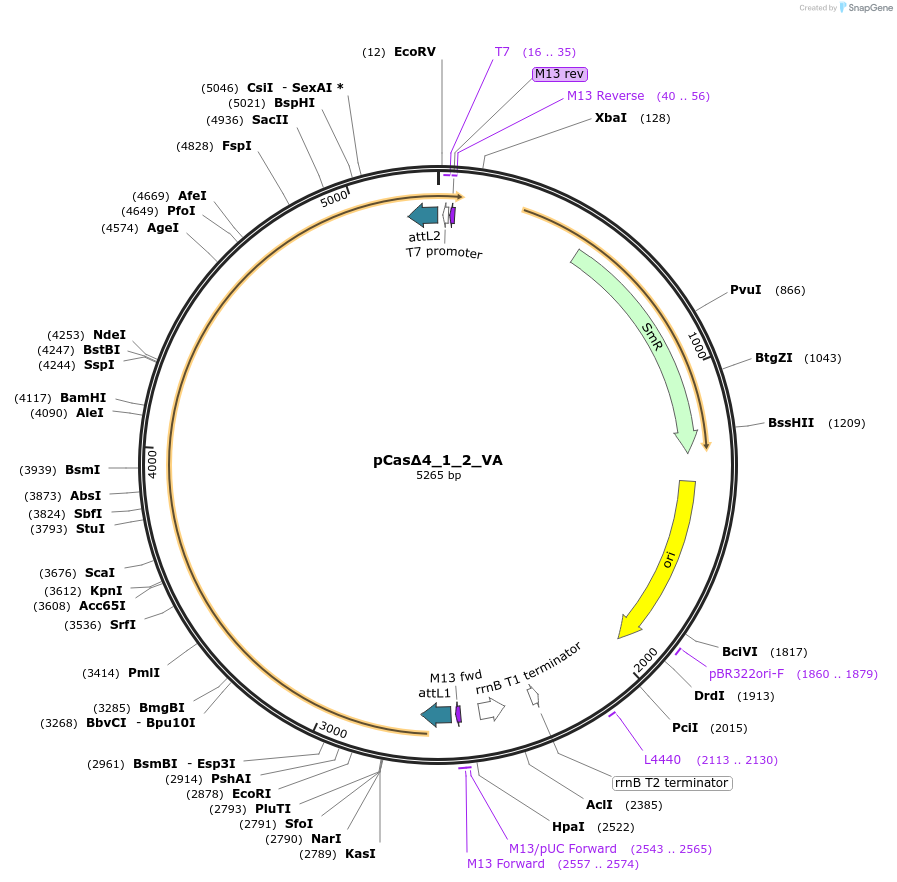
pCasΔ4_1_2_VA
Plasmid#186440PurposeEncodes Cas1, Cas2, leader, repeat and one spacer from F. Novicida (type V-A)DepositorInsertCas1 and Cas2
UseCRISPRPromoterT7Available SinceMarch 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
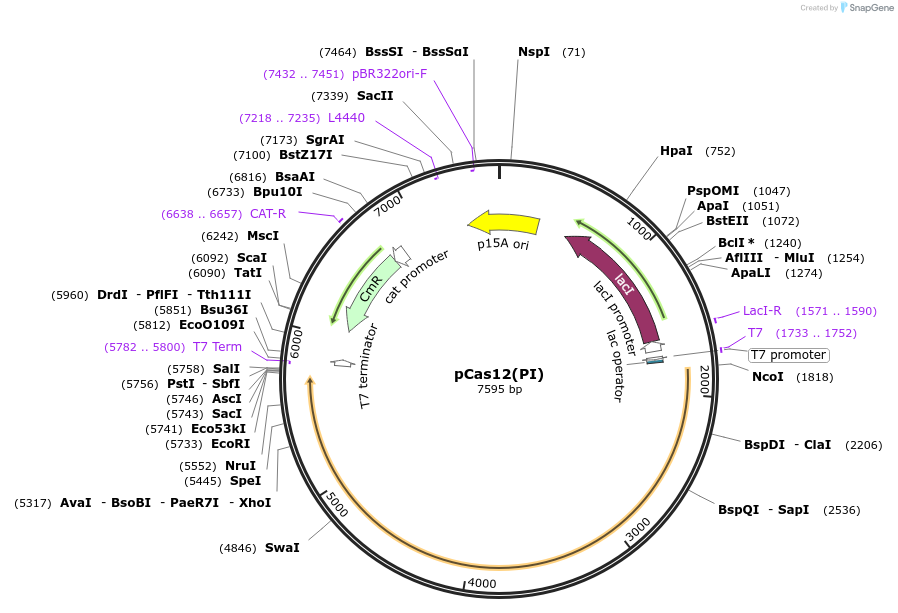
pCas12(PI)
Plasmid#186448PurposeEncodes inactive Cas12a containing mutation in the PI domain (K613A, K617A) from F. novicida (type V-A)DepositorInsertCas12a (PI)
UseCRISPRMutationK613A, K617APromoterT7Available SinceMarch 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
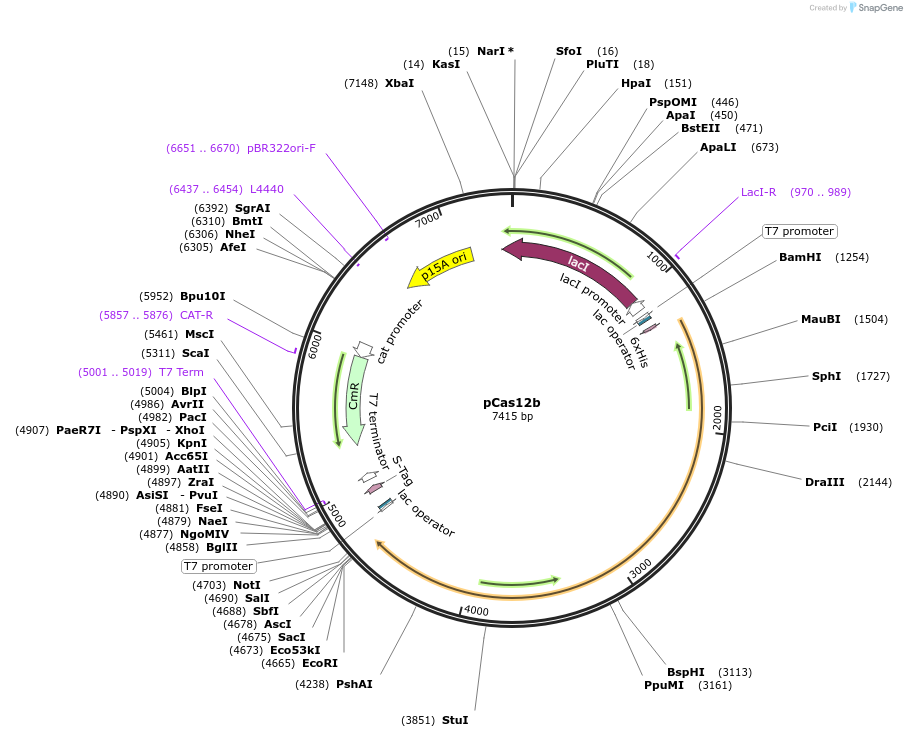
pCas12b
Plasmid#186449PurposeEncodes Cas12b from A. acidoterrestris (type V-B)DepositorInsertCas12b
UseCRISPRPromoterT7Available SinceMarch 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
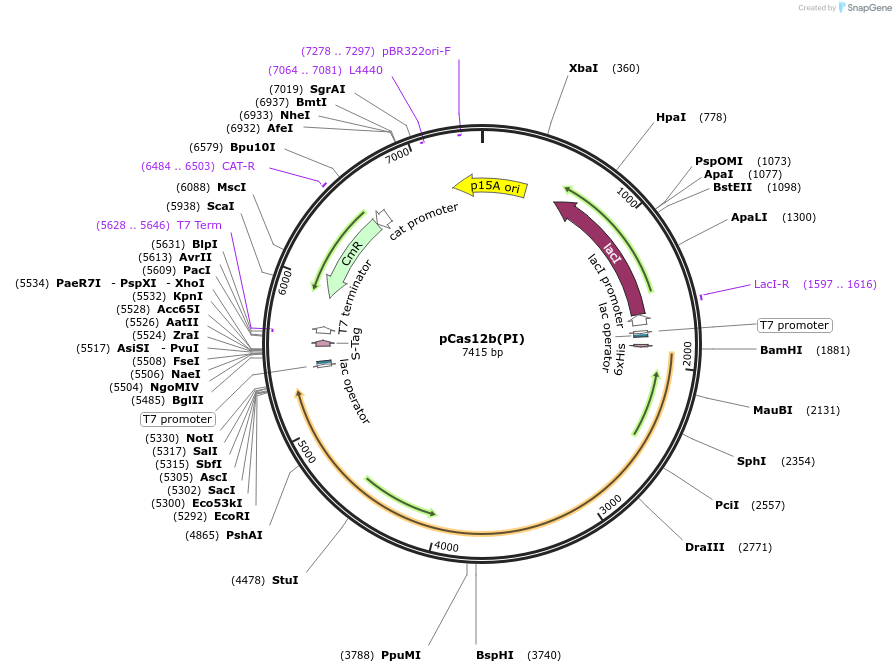
pCas12b(PI)
Plasmid#186451PurposeEncodes inactive Cas12b containing mutation in the PI domain (R122A, G143P) from A. acidoterrestris (type V-B)DepositorInsertCas12b (PI)
UseCRISPRMutationR122A, G143PPromoterT7Available SinceMarch 20, 2023AvailabilityAcademic Institutions and Nonprofits only -
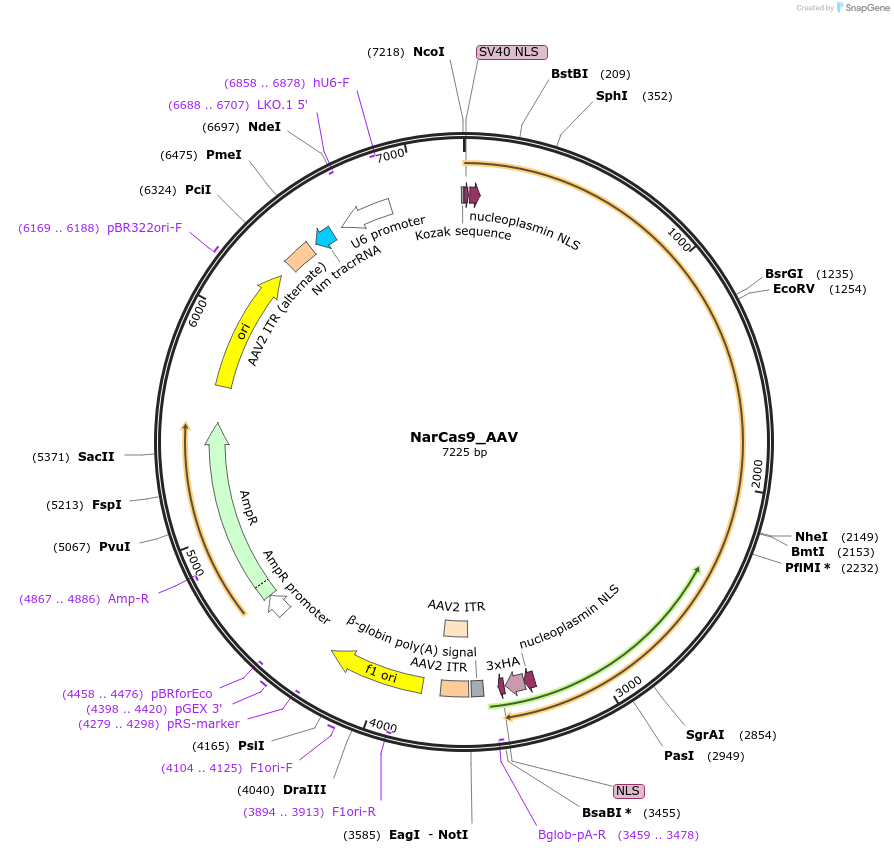
NarCas9_AAV
Plasmid#192145PurposeExpresses NarCas9?and cloning backbone for sgRNADepositorInsertNarCas9
UseAAVExpressionMammalianAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
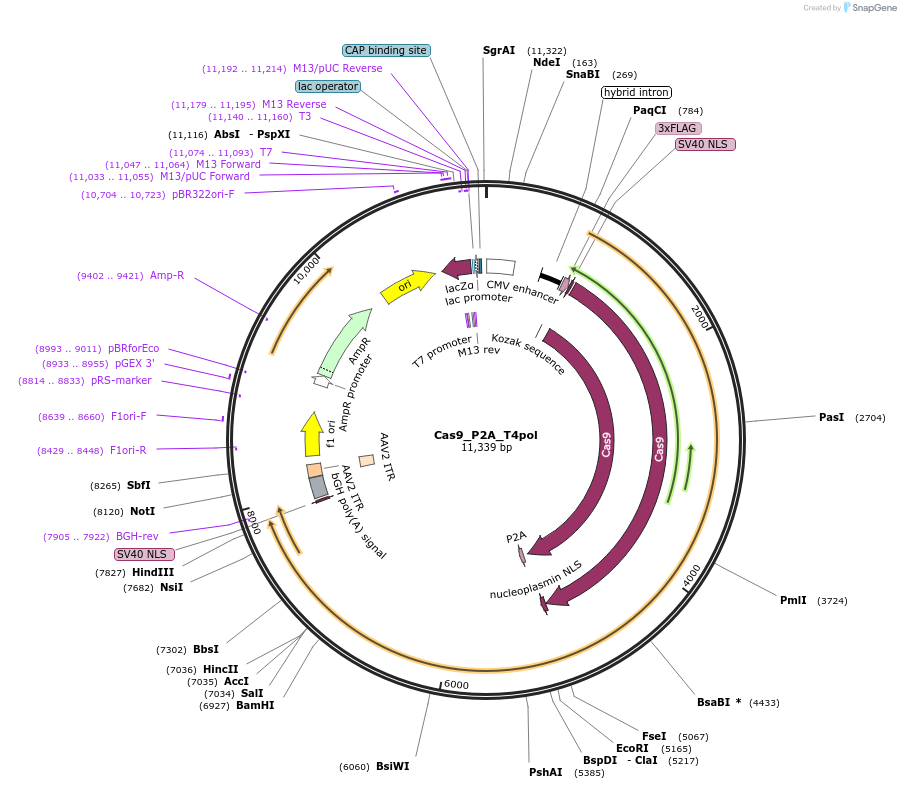
Cas9_P2A_T4pol
Plasmid#190146PurposeExpresses SpCas9 and T4polDepositorInsertSpCas9_P2A_T4pol
UseCRISPRTags3xFLAGExpressionMammalianMutationCodon Optimized for mammalian cellsPromoterCBhAvailable SinceMarch 1, 2023AvailabilityAcademic Institutions and Nonprofits only -
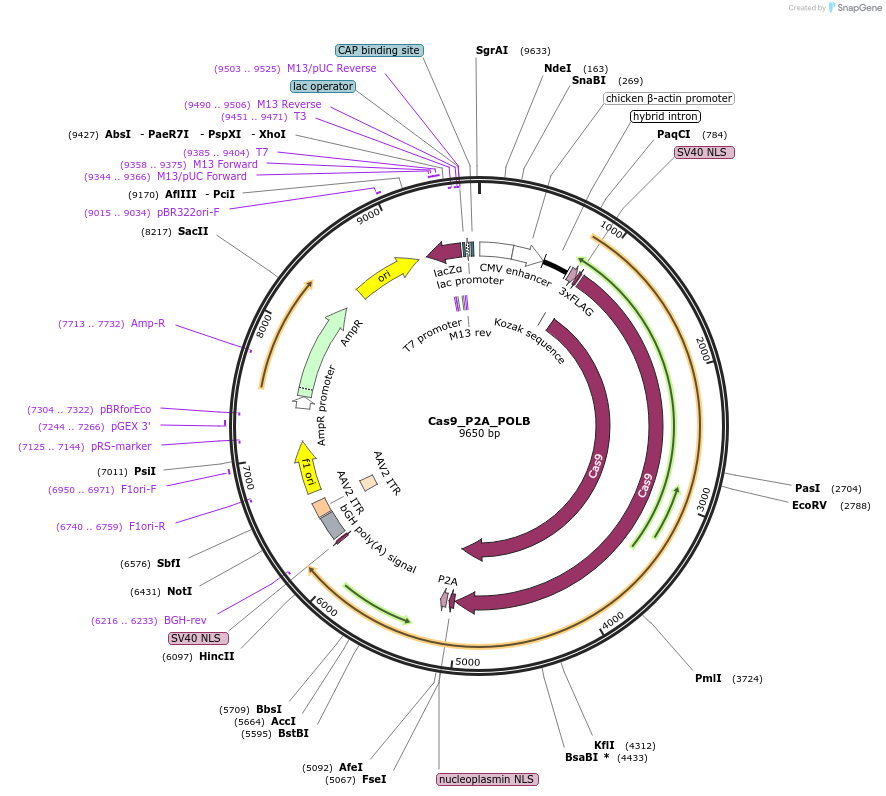
Cas9_P2A_POLB
Plasmid#190145PurposeExpresses SpCas9 and POLBDepositorInsertSpCas9_P2A_POLB
UseCRISPRTags3xFLAGExpressionMammalianMutationCodon Optimized for mammalian cellsPromoterCBhAvailable SinceFeb. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
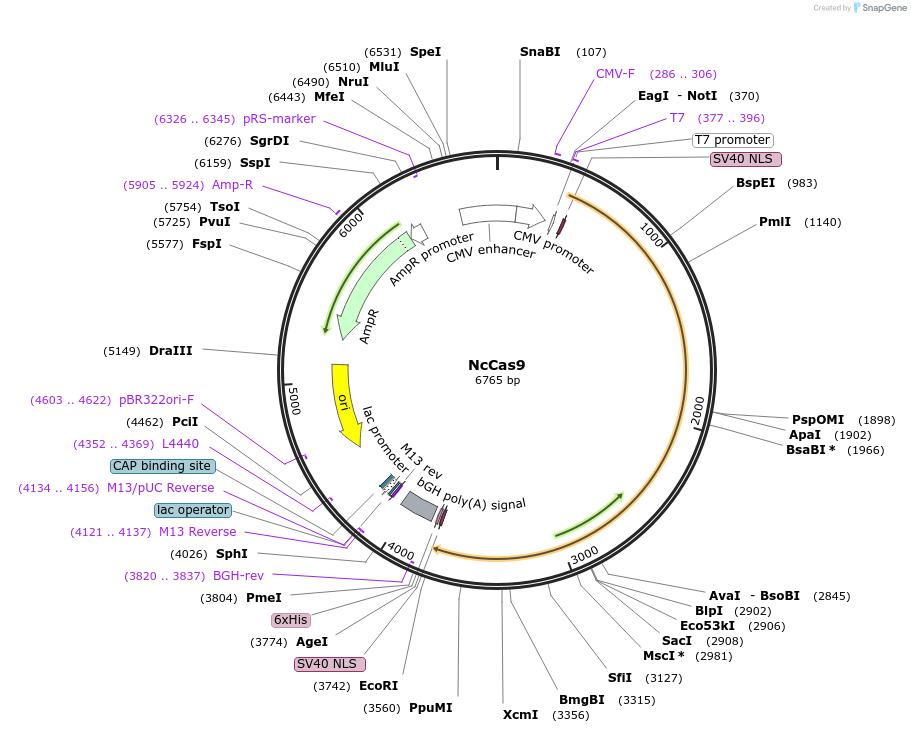
NcCas9
Plasmid#194093PurposeExpresses human codon optimized NcCas9. Genome editing in human cells.DepositorInsertNcCas9
ExpressionMammalianPromoterCMVAvailable SinceJan. 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
eSpCas9-EF
Plasmid#177267PurposeCRISPR vectorDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceDec. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
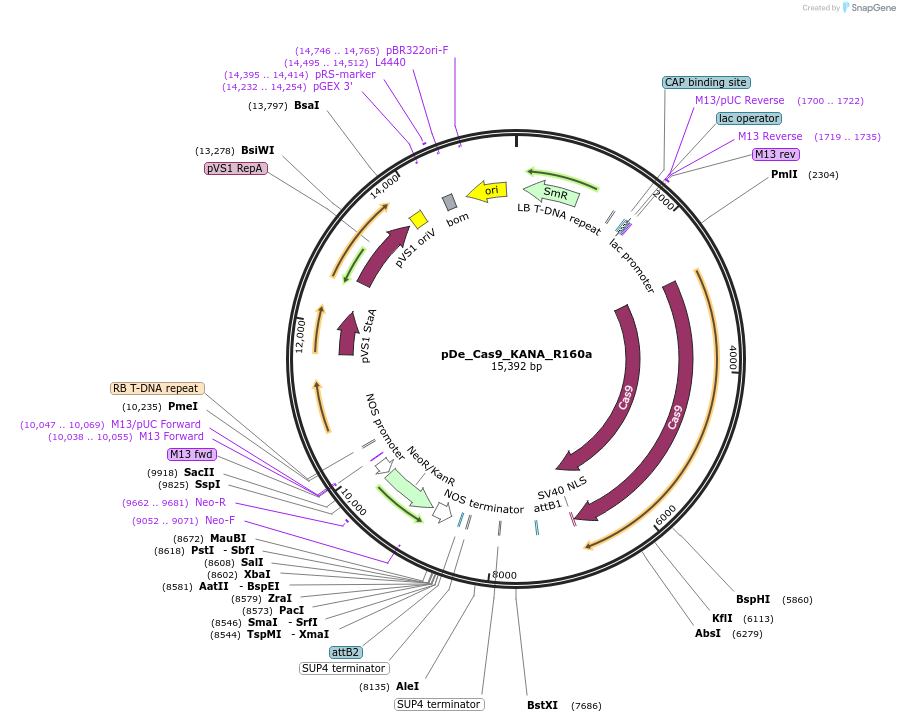
pDe_Cas9_KANA_R160a
Plasmid#173756PurposeBinary expression vector with A.th. codon-optimized Cas9 and sgRNA for miRNA160aDepositorInsertCas9_sgRNA1/R160a_sgRNA2/R160a
UseCRISPRPromoterAtU6 &StU6Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
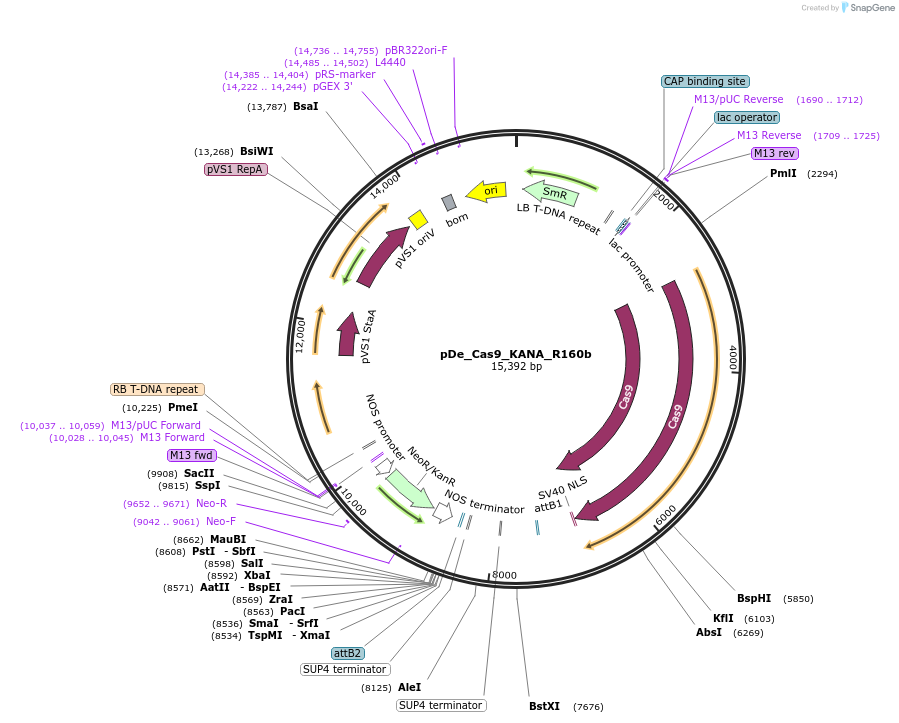
pDe_Cas9_KANA_R160b
Plasmid#173757PurposeBinary expression vector with A.th. codon-optimized Cas9 and sgRNA for miRNA160bDepositorInsertCas9_sgRNA1/R160b_sgRNA2/160b
UseCRISPRPromoterAtU6 &StU6Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only -
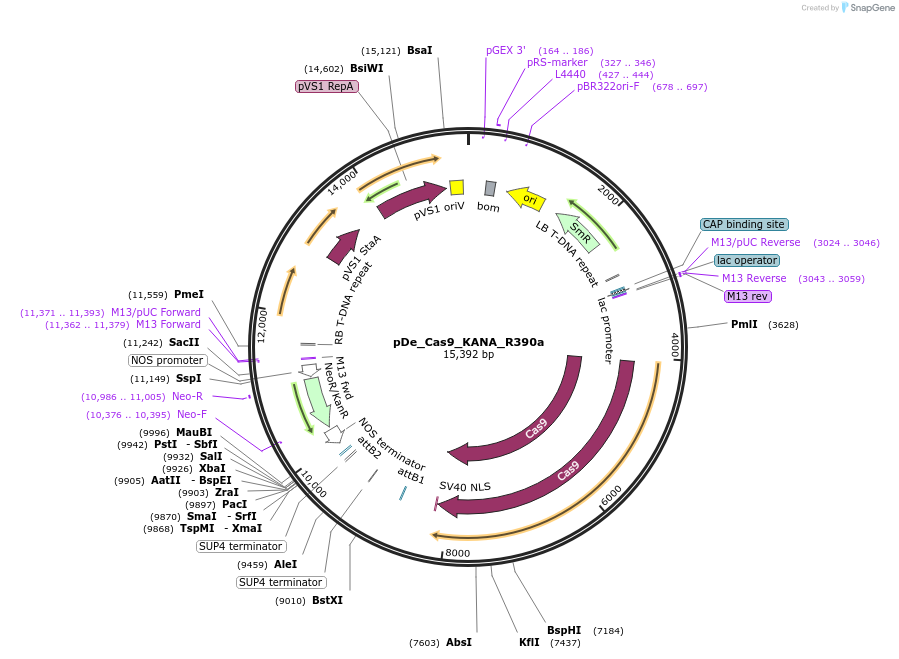
pDe_Cas9_KANA_R390a
Plasmid#173758PurposeBinary expression vector with A.th. codon-optimized Cas9 and sgRNA for miRNA390aDepositorInsertCas9_sgRNA1/390a_sgRNA2/390a
UseCRISPRPromoterAtU6 &StU6Available SinceNov. 3, 2022AvailabilityAcademic Institutions and Nonprofits only