We narrowed to 1,744 results for: SUA
-
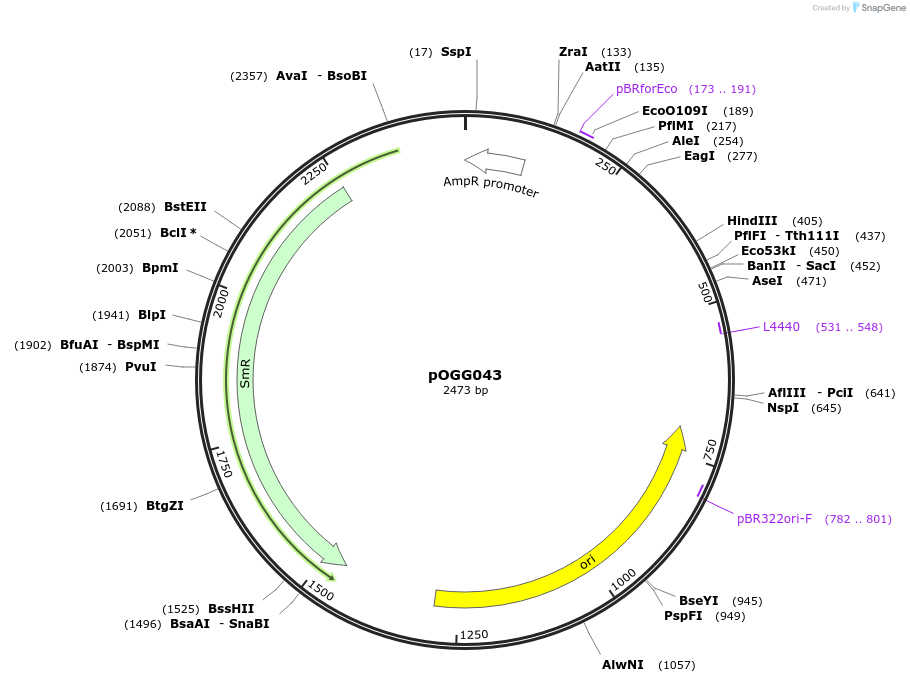
Plasmid#133123PurposeLevel 0 PU module synthesised as a fragment (pL0M-PU-PsNifH). Synthetic consensus promoter to drive nodule-specific expression in biovars and strains of R. leguminosarum.DepositorInsertPsNifH Synthetic consensus nifH promoter
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
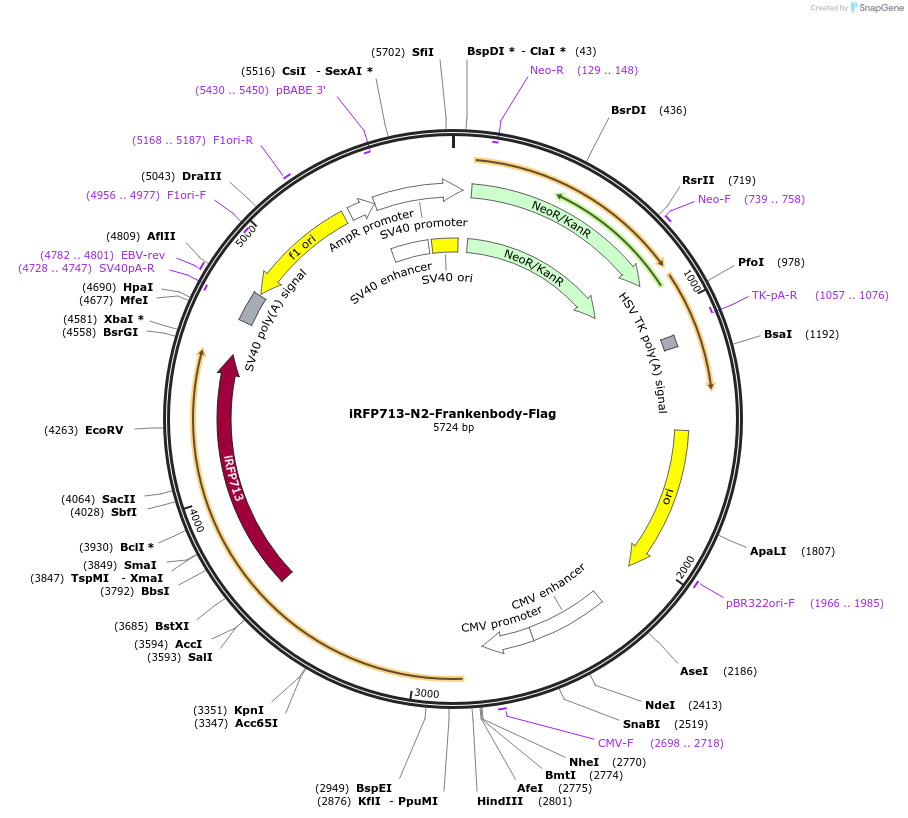
iRFP713-N2-Frankenbody-Flag
Plasmid#175291PurposeVisualize endogeneous loci coupled with ZF probes harboring 10xFLAG tagDepositorInsertanti-FLAG frankenbody fused with iRFP713
TagsiRFP713ExpressionMammalianAvailable SinceDec. 3, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
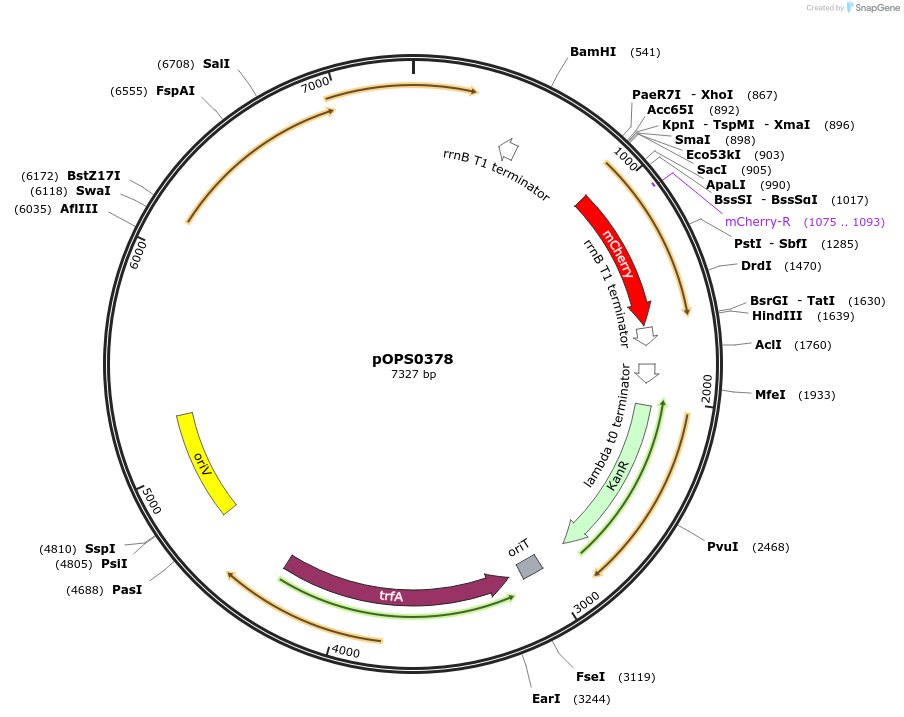
pOPS0378
Plasmid#133229PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with constitutive promoter pNeo (pOGG001), mCherry (EC15071) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
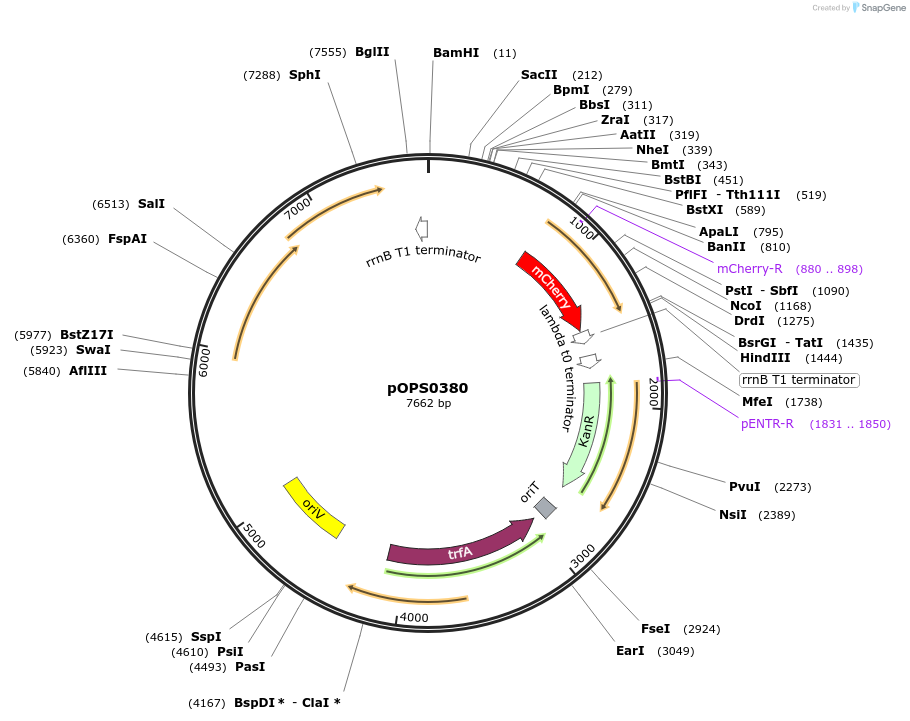
pOPS0380
Plasmid#133230PurposeReporter plasmid for R. leguminosarum suitable for experiments in environments where there is no antibiotic selection present.DepositorInsertconstructed with constitutive promoter Rlv3841pnifH (pOGG082), mCherry (EC15071) and T-pharma (pOGG003)
ExpressionBacterialMutationDomesticated for Golden Gate cloningAvailable SinceApril 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
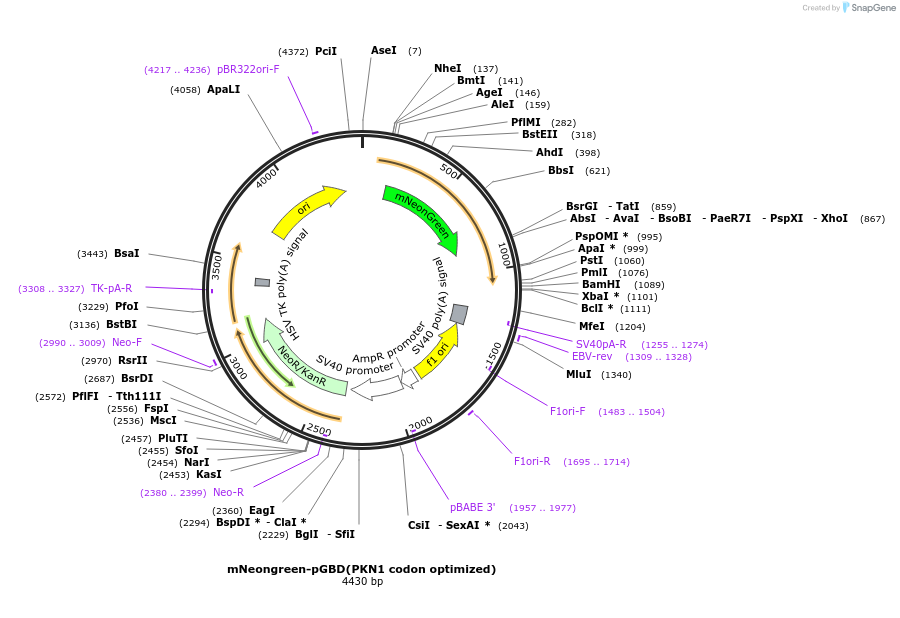
mNeongreen-pGBD(PKN1 codon optimized)
Plasmid#129634PurposeVisualization of active RhoADepositorInsertRhoA binding doamin of PKN1(30 - 100) codon optimized
ExpressionMammalianPromoterCMVdelAvailable SinceFeb. 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
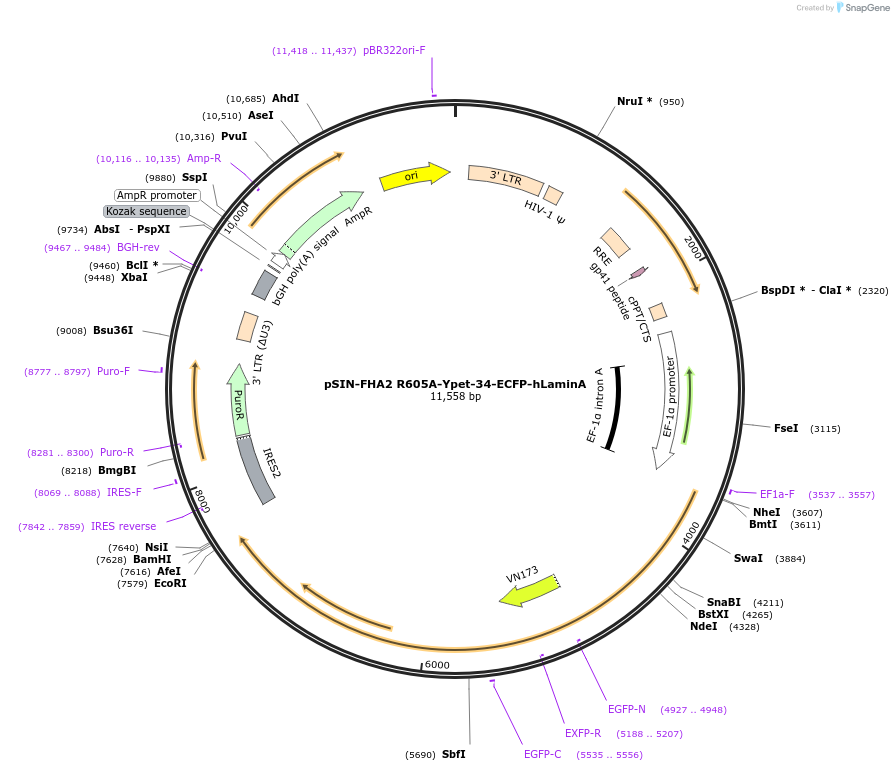
pSIN-FHA2 R605A-Ypet-34-ECFP-hLaminA
Plasmid#235123PurposeFHA2-mutated FRET biosensor R605A; FRET biosensor for live cell visualization of pSer22 Lamin ADepositorInsertFHA2 R605A-Ypet-34-ECFP-hLaminA
UseLentiviralExpressionMammalianMutationFHA2 R605AAvailable SinceAug. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
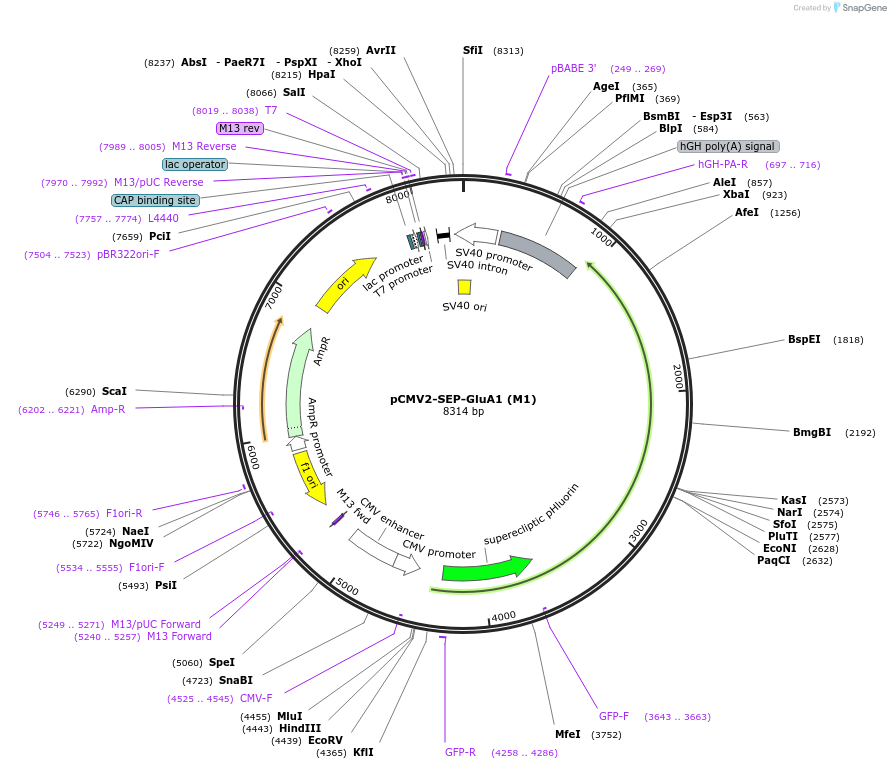
pCMV2-SEP-GluA1 (M1)
Plasmid#64942Purposemammalian expression of rat GluA1DepositorAvailable SinceMay 14, 2015AvailabilityAcademic Institutions and Nonprofits only -
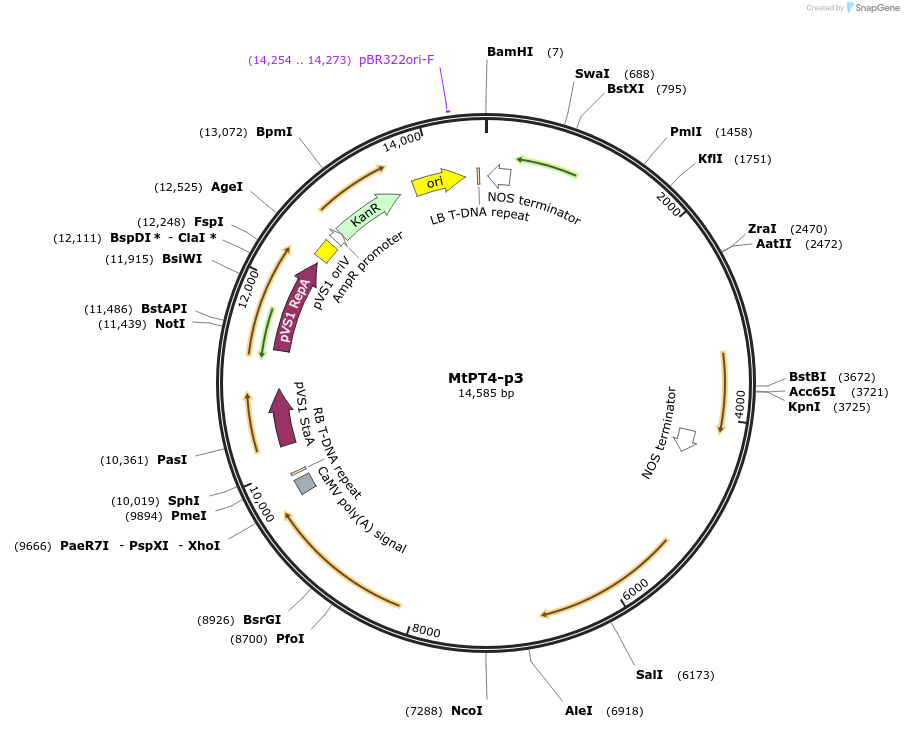
MtPT4-p3
Plasmid#160003PurposeMultigene construct containing the betalain pigment biosynthetic genes under the control of the MtPT4 promoter for visualisation of arbuscular mycorrhizal colonisation in Medicago truncatula roots.DepositorInsertsDsRed (reverse orientation)
DODAα1
CYP76AD1
cDOPA5GT
ExpressionPlantPromoterArabidopsis thaliana Ubiquitin 10 Promoter (AtUb1…Available SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
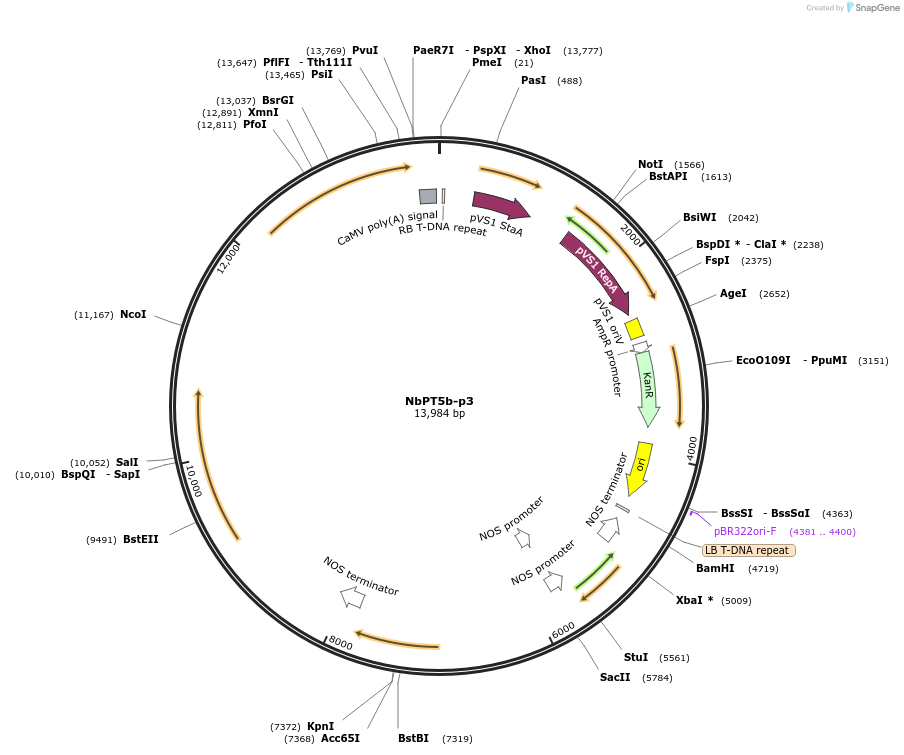
NbPT5b-p3
Plasmid#160001PurposeMultigene construct containing the betalain pigment biosynthetic genes under the control of the NbPT5b promoter for visualisation of arbuscular mycorrhizal colonisation in Nicotiana benthamiana roots.DepositorInsertsBar (reverse orientation)
DODAα1
CYP76AD1
cDOPA5GT
ExpressionPlantPromoterAgrobacterium tumefaciens Nopaline synthase Promo…Available SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
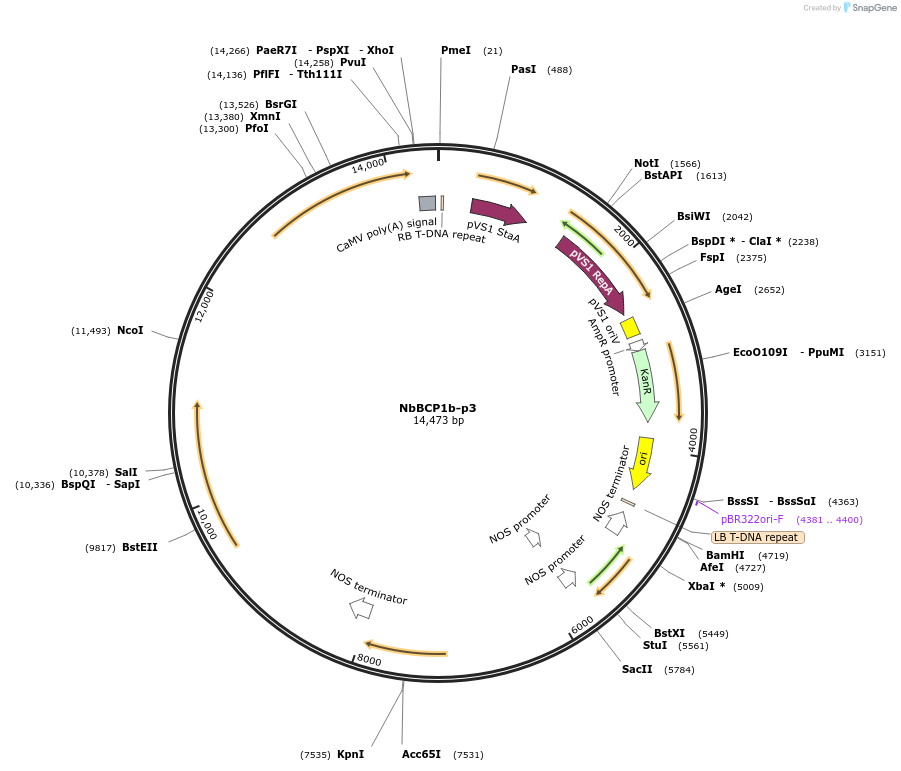
NbBCP1b-p3
Plasmid#160002PurposeMultigene construct containing the betalain pigment biosynthetic genes under the control of the NbBCP1b promoter for visualisation of arbuscular mycorrhizal colonisation in Nicotiana benthamiana rootsDepositorInsertsBar (reverse orientation)
DODAα1
CYP76AD1
cDOPA5GT
ExpressionPlantPromoterAgrobacterium tumefaciens Nopaline synthase Promo…Available SinceJuly 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
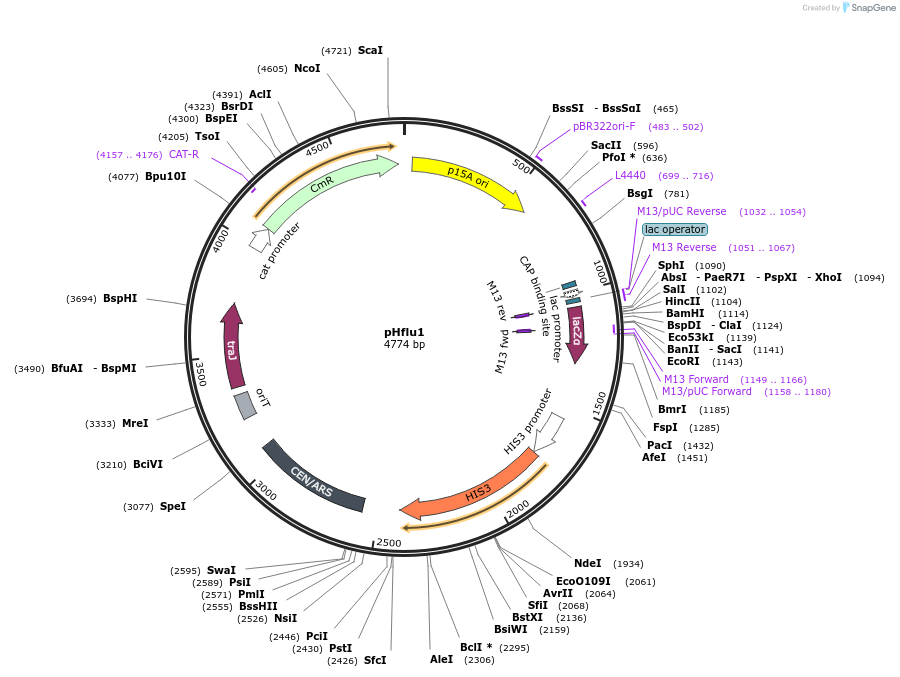
pHflu1
Plasmid#203878PurposepSU20 with addition of CEN6-ARSH4-HIS3 cassette for S. cerevisiae and RK2 origin of conjugal transferDepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailable SinceDec. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
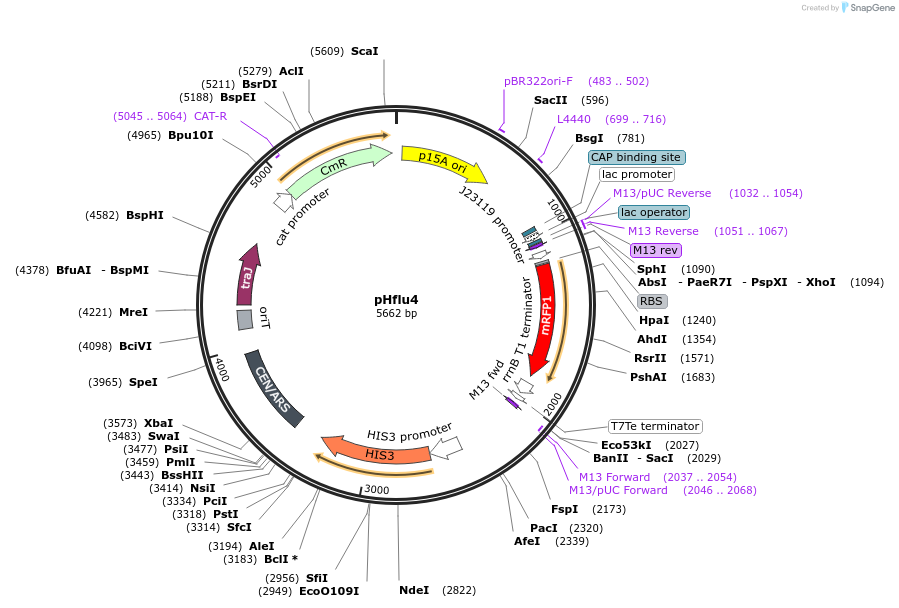
pHflu4
Plasmid#203882PurposepHI1 with insertion of monomeric red fluorescent protein (mRFP)DepositorInsertmonomeric red fluorescent protein
UseSynthetic BiologyExpressionBacterial and YeastAvailable SinceDec. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
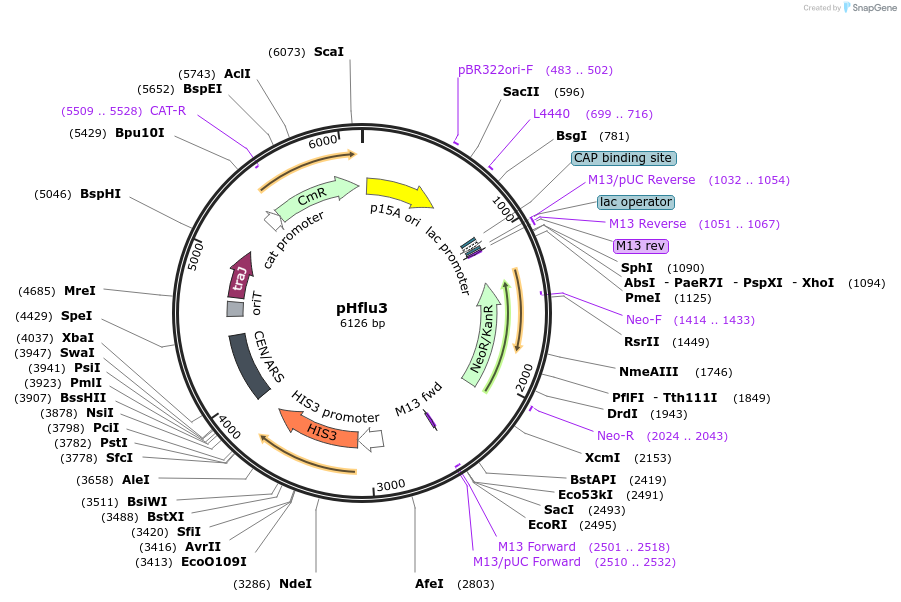
pHflu3
Plasmid#203881PurposepHI1 with insertion of kanamycin-resistance geneDepositorInsertaminoglycoside O-phosphotransferase APH(3')-IIa
UseSynthetic BiologyExpressionBacterial and YeastAvailable SinceDec. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
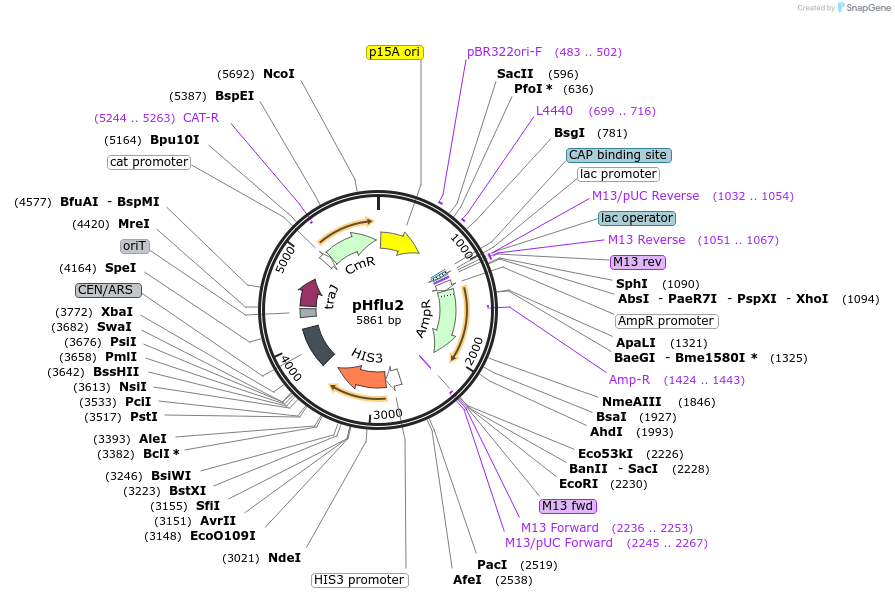
pHflu2
Plasmid#203880PurposepHI1 with insertion of ampicillin-resistance geneDepositorInsertbroad-spectrum class A beta-lactamase TEM-116
UseSynthetic BiologyExpressionBacterial and YeastAvailable SinceDec. 6, 2023AvailabilityAcademic Institutions and Nonprofits only -
pTY1048-S65T-LacI
Plasmid#34853DepositorInsertGFP-LacI
ExpressionWormMutationGFP is mutated: S65T and contains introns, in ord…Available SinceFeb. 21, 2012AvailabilityAcademic Institutions and Nonprofits only -
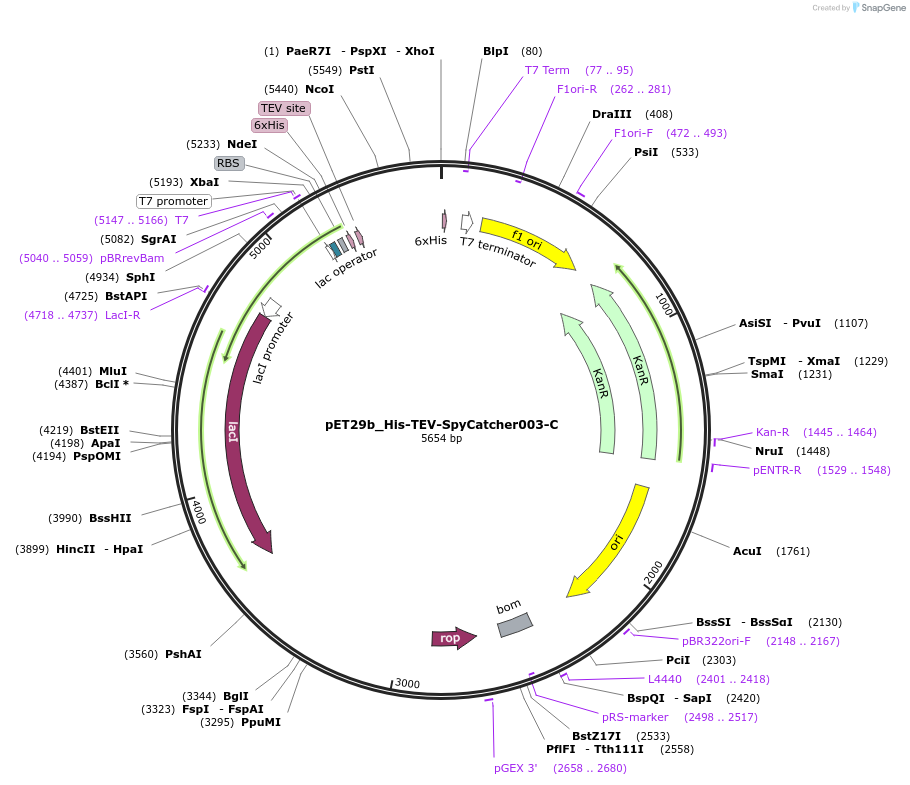
pET29b_His-TEV-SpyCatcher003-C
Plasmid#233339PurposeTo express SpyCatcherDepositorInsertHis-TEV-SpyCatcher003-C
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
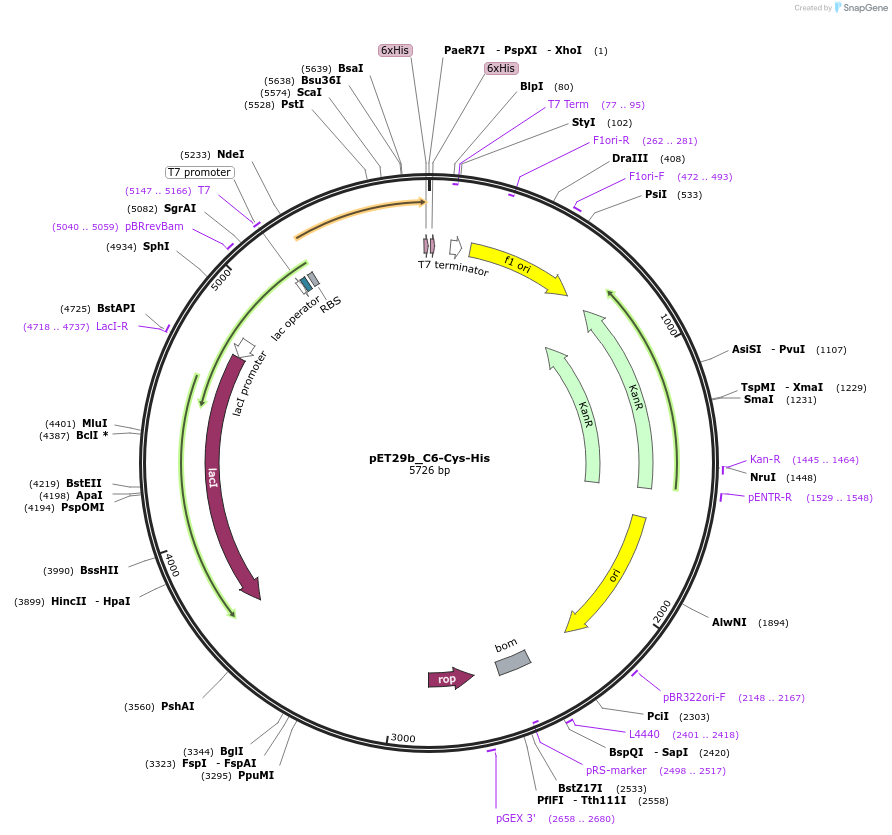
pET29b_C6-Cys-His
Plasmid#233323PurposeTo express nanobody C6 with C terminal cysteineDepositorInsertC6-Cys-His
ExpressionBacterialPromoterT7Available SinceOct. 14, 2025AvailabilityAcademic Institutions and Nonprofits only -
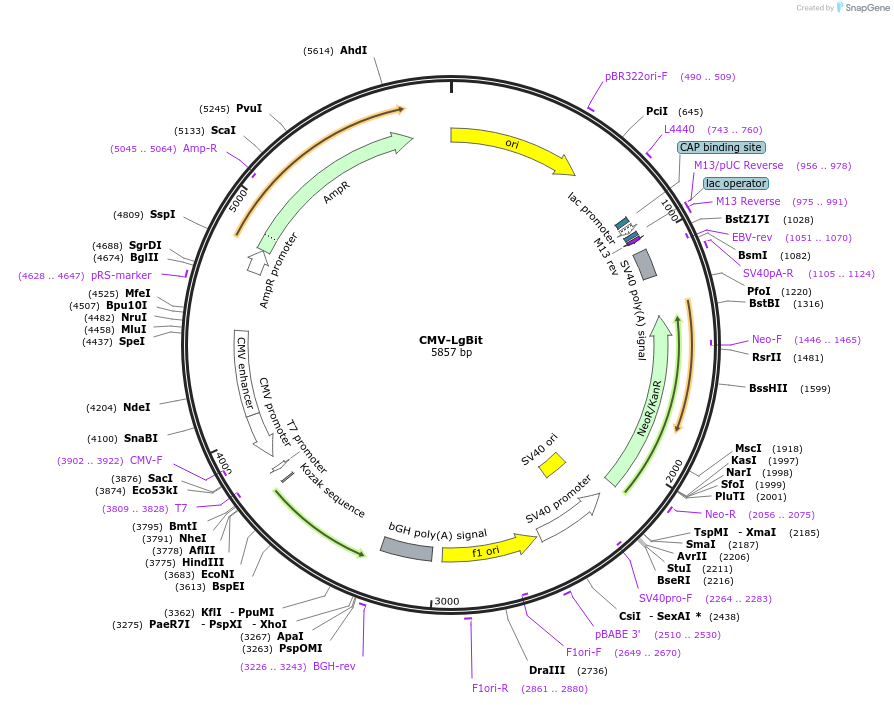
CMV-LgBit
Plasmid#234738PurposeMammalian expression of large NanoBit fragment (LgBit)DepositorInsertLgBit
ExpressionMammalianPromoterCMVAvailable SinceMarch 20, 2025AvailabilityAcademic Institutions and Nonprofits only -
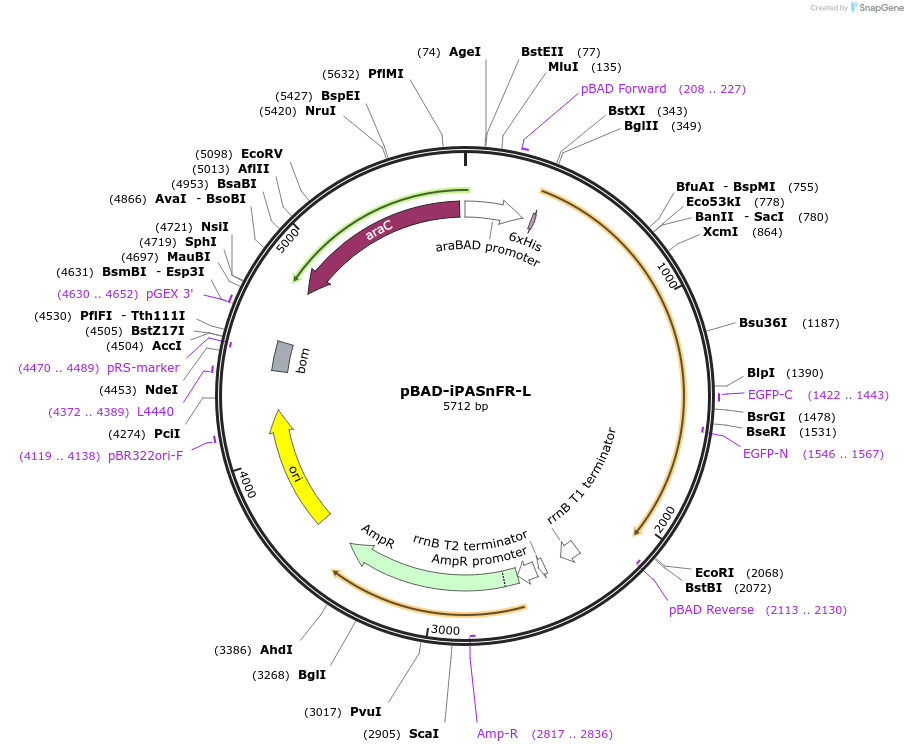
pBAD-iPASnFR-L
Plasmid#225238PurposeBacterial expression of iPASnFR-LDepositorInsertiPASnFR-L
Tags6xHisExpressionBacterialAvailable SinceSept. 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
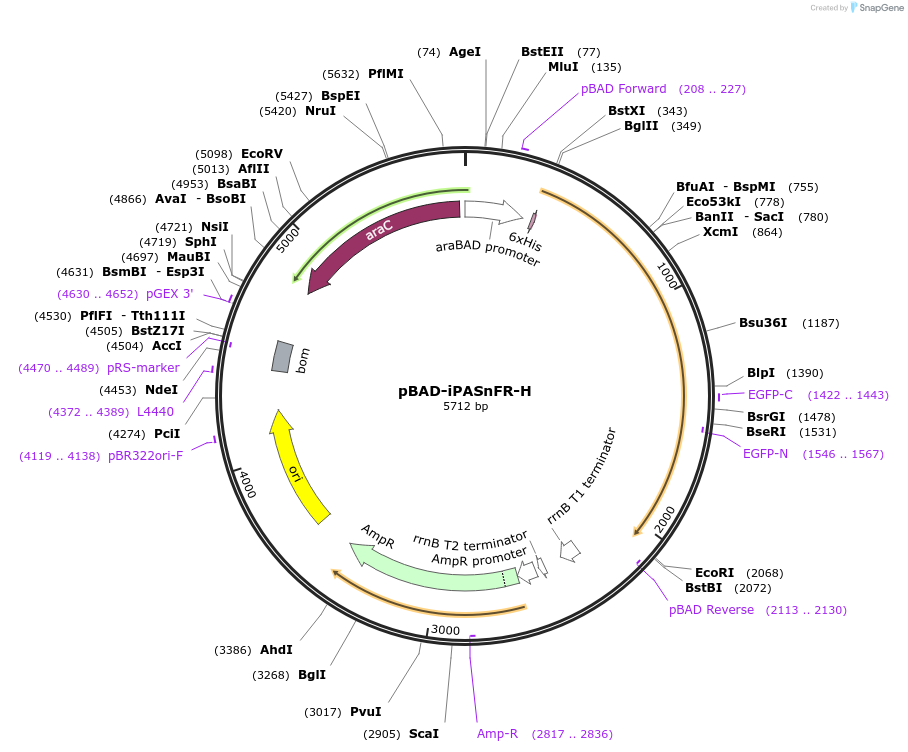
pBAD-iPASnFR-H
Plasmid#225237PurposeBacterial expression of iPASnFR-HDepositorInsertiPASnFR-H
Tags6xHisExpressionBacterialAvailable SinceSept. 16, 2024AvailabilityAcademic Institutions and Nonprofits only