We narrowed to 19,943 results for: INO
-
Plasmid#67757PurposeExpresses N-terminal kinase domain of Chk1 with C-term GFP fluorescent tagDepositorInsertzebrafish Chk1
TagsGFPMutationonly encodes N-term kinase domain (amino acids 1-…Available SinceNov. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
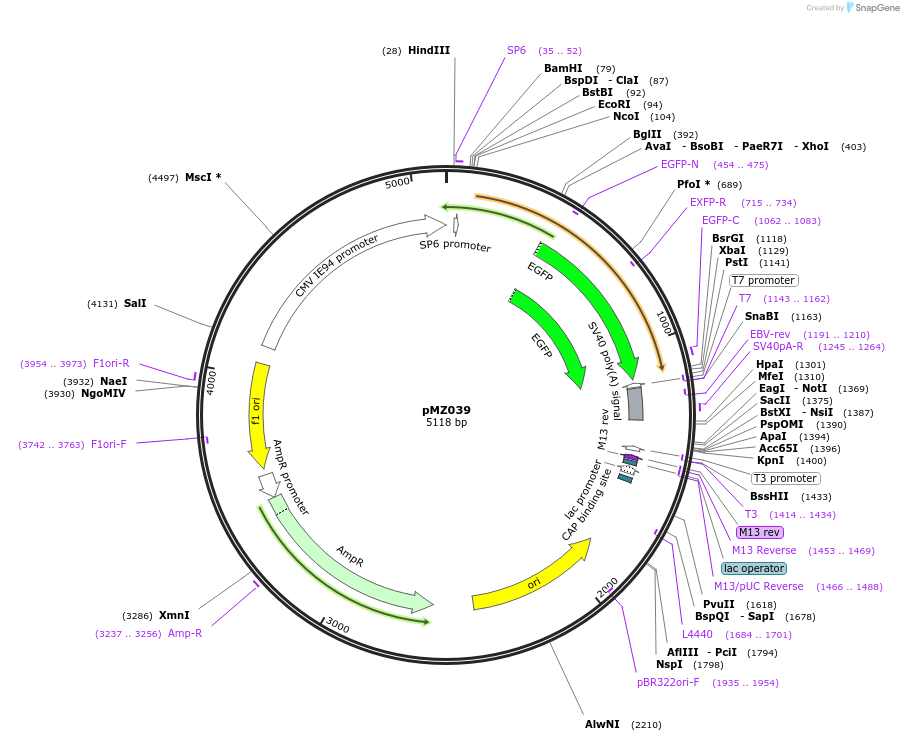
pICH44233
Plasmid#50288DepositorInsert5'UTR CMV2 (Cucumber Mosaic Virus)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
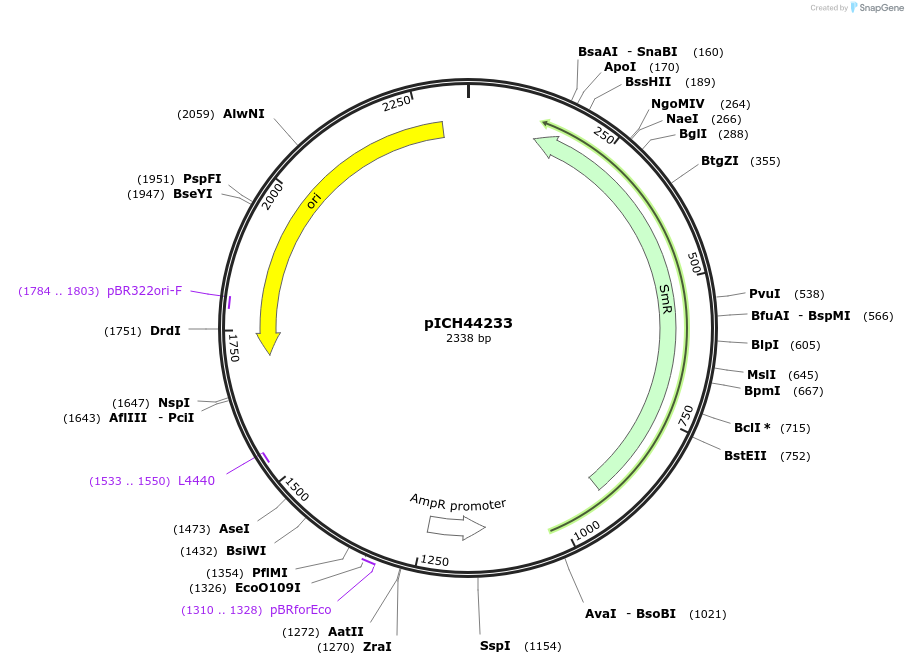
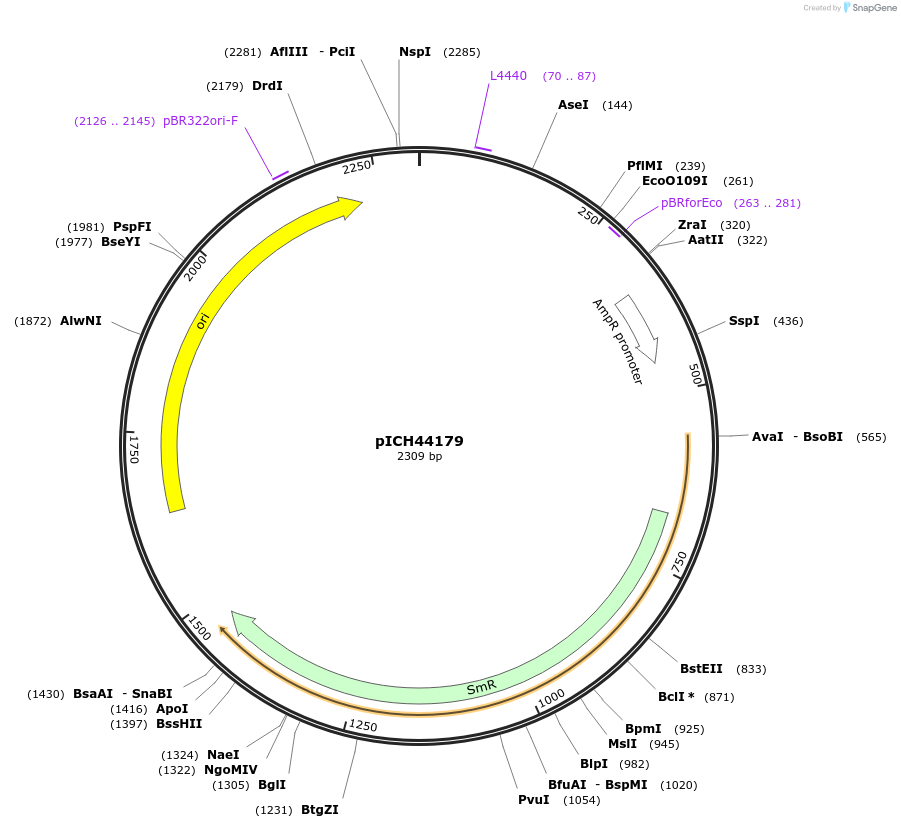
pICH44179
Plasmid#50290DepositorInsert5' UTR, RbcS2B (AT5g38420, A. thaliana)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
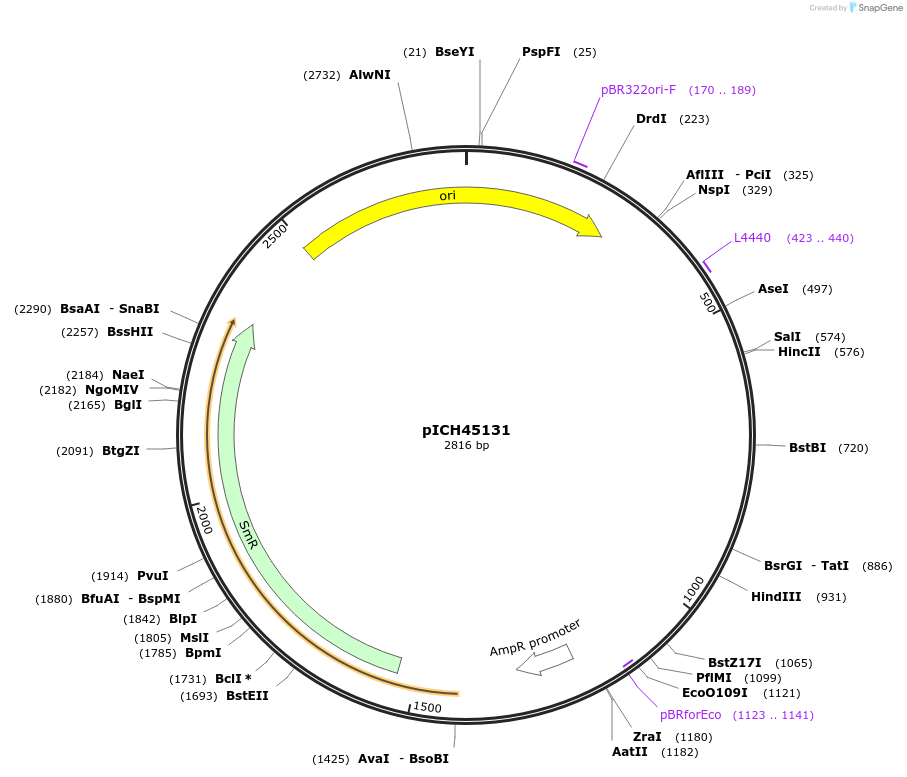
pICH45131
Plasmid#50262DepositorInsertpromoter, LHB1B2 (AT2g34420, A. thaliana)
UsePlant expressionMutationBsaI/BbsI sites removed by point-mutationAvailable SinceFeb. 18, 2014AvailabilityAcademic Institutions and Nonprofits only -
JMJD2C
Plasmid#38974PurposeBacterial expression for structure determination; may not be full ORFDepositorAvailable SinceMarch 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
Venus Cam KII alpha 1-315aa
Plasmid#29431DepositorInsertVenus CAM KII Alpha
TagsVenusExpressionMammalianMutationExpresses CamKIIa amino acids 1-315Available SinceJuly 5, 2011AvailabilityAcademic Institutions and Nonprofits only -
GIMAP4
Plasmid#25337PurposeBacterial expression for structure determination; may not be full ORFDepositorInsertGIMAP4:G20-M240 (GIMAP4 Human)
TagsHisExpressionBacterialMutationContains amino acids G20-M240Available SinceAug. 17, 2010AvailabilityIndustry, Academic Institutions, and Nonprofits -
p6112 pSG5-HPV31-d3-12-E8^E2C(HA)
Plasmid#24743DepositorInsertHPV31 d3-12 E8^E2C (HA)
TagsHA tag in hinge regionExpressionMammalianMutationAmino acids 3-12 missing.Available SinceJune 30, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F15
Plasmid#23064DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 deleted amino acids 4 - 19Available SinceMarch 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
pCMVmyc-ORF73SOCS
Plasmid#21546DepositorInsertORF73 (ORF73 MuHV-4)
TagsMycExpressionMammalianMutationamino acids at positions 199, 202, 203 and 206 we…Available SinceAug. 12, 2009AvailabilityAcademic Institutions and Nonprofits only -
BS.YxiN.Twin1.a1-368.E152Q
Plasmid#17517DepositorInsertB. subtilis DEAD-box helicase YxiN first two domains
Tagsintein-chitin binding domainExpressionBacterialMutationdeleted amino acid residues 369-479; E152Q mutat…Available SinceMarch 21, 2008AvailabilityAcademic Institutions and Nonprofits only -
pGEX Rad C297
Plasmid#11230DepositorAvailable SinceMarch 1, 2006AvailabilityAcademic Institutions and Nonprofits only -
pGEX Rad N88
Plasmid#11235DepositorAvailable SinceMarch 1, 2006AvailabilityAcademic Institutions and Nonprofits only -
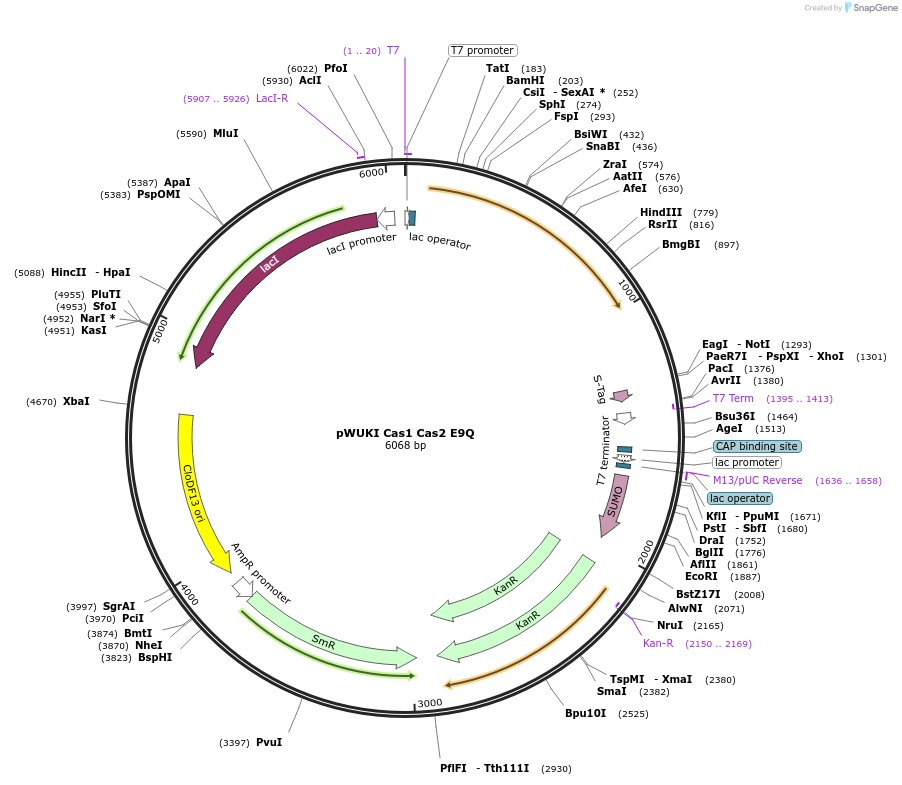
pWUKI Cas1 Cas2 E9Q
Plasmid#80091PurposeEncodes Cas1 and Cas2 E9QDepositorExpressionBacterialMutationE9QPromoterT7AvailabilityAcademic Institutions and Nonprofits only -
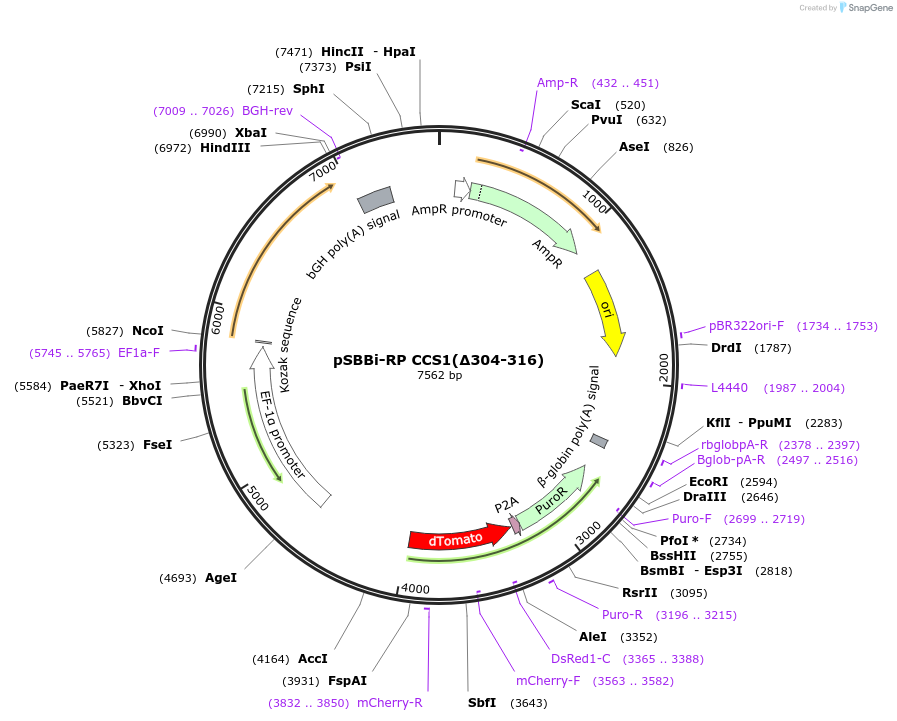
pSBBi-RP CCS1(Δ304-316)
Plasmid#139163PurposeSB-transposon with constitutive bi-directional promoter. One side contains coffee caffeine synthase 1 (CCS1) CCS-CTS deletion mutant; the other side contains RFP and puromycin resistance genes.DepositorInsertcaffeine synthase 1 [Coffea arabica] CCS-CTS region deletion
UseTransposonExpressionMammalianMutationdeleted amino acids 304-319PromoterEF1αAvailabilityAcademic Institutions and Nonprofits only -
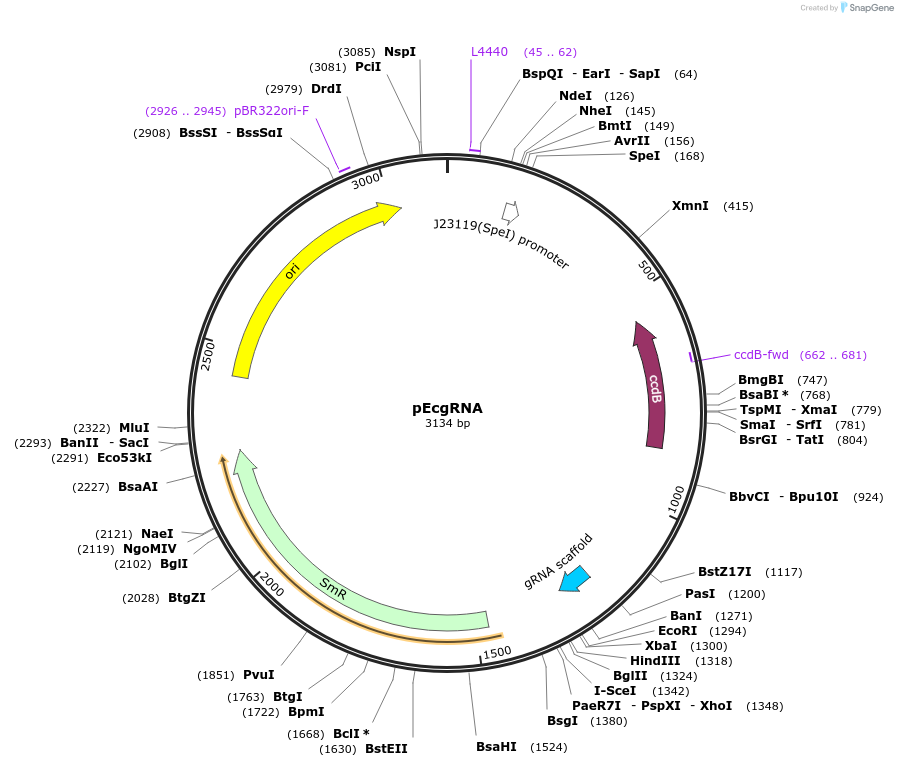
pEcgRNA
Plasmid#166581PurposeCRISPR-Cas9-assisted genome editing in Escherichia coliDepositorInsertccdB
ExpressionBacterialAvailable SinceApril 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
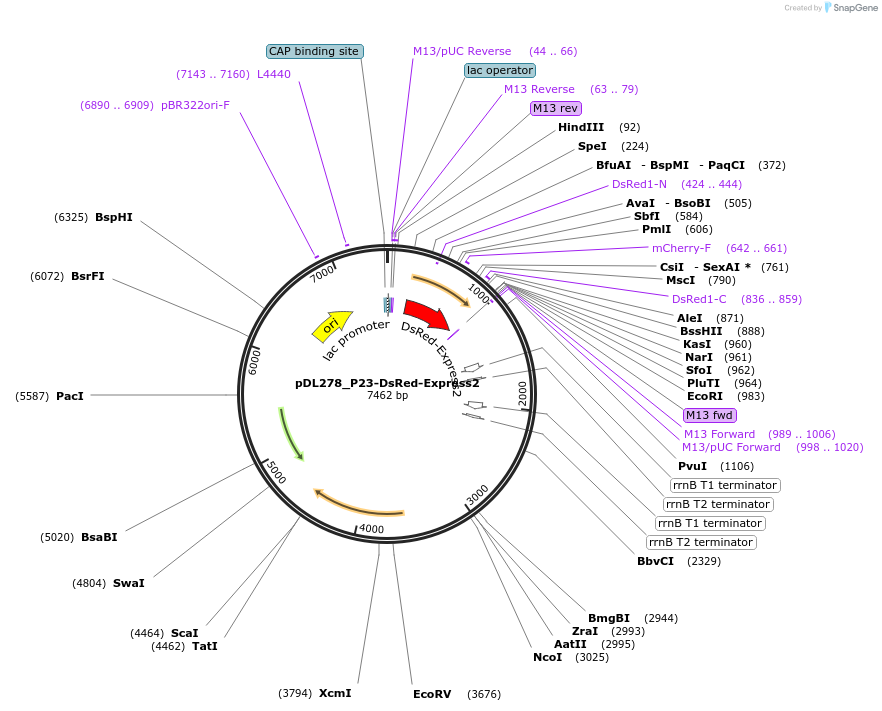
pDL278_P23-DsRed-Express2
Plasmid#121505PurposeE. coli-streptococcal shuttle vector that expresses rfpDepositorInsertDsRed-Express2
UseE. coli streptococcal shuttle vectorPromoterP23Available SinceJuly 29, 2019AvailabilityAcademic Institutions and Nonprofits only -
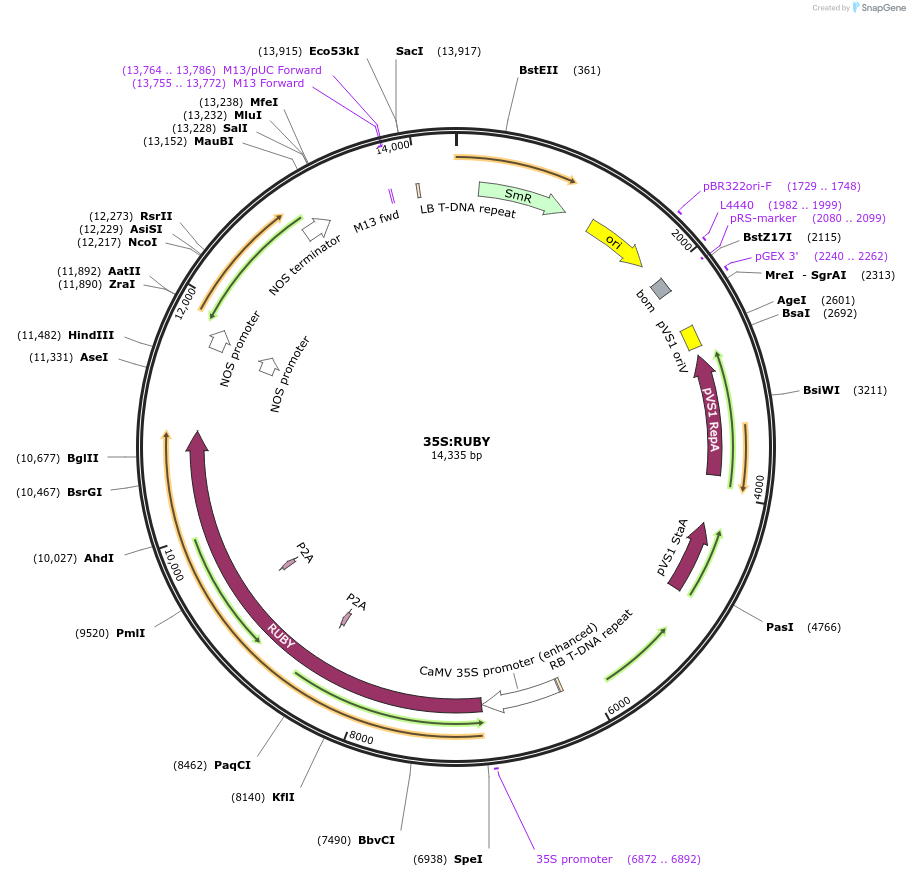
35S:RUBY
Plasmid#160908PurposeRUBY under the control of CaMV 35S promoter/marker for transformationDepositorInsertRUBY and CaMV 35S promoter
ExpressionPlantPromoterCaMV 35SAvailable SinceNov. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
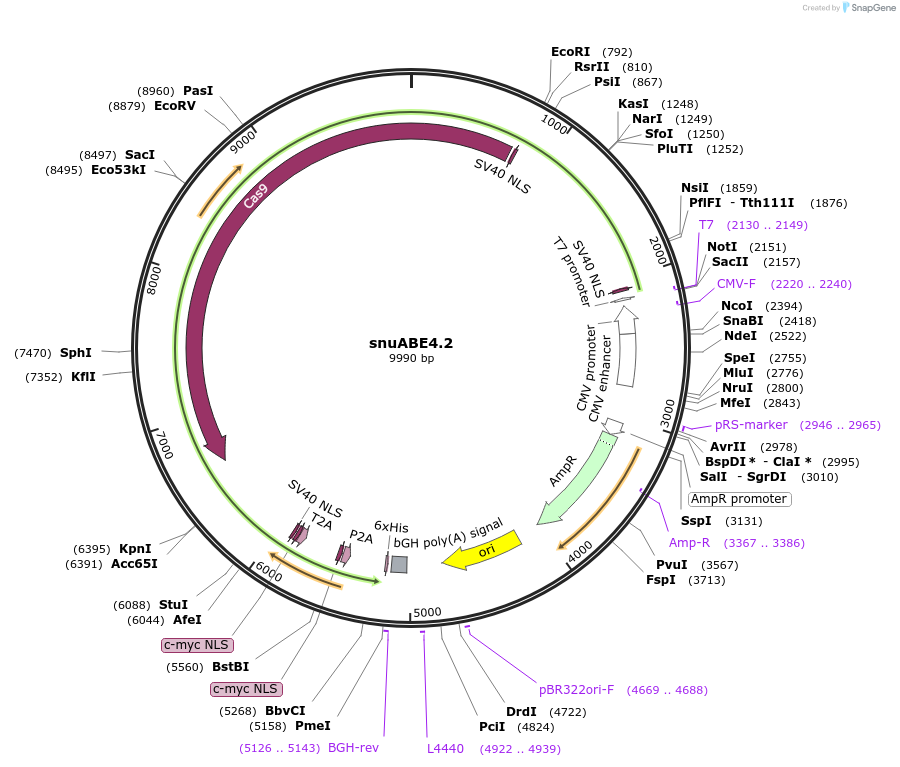
snuABE4.2
Plasmid#246014PurposesnuABE4.1 with MLH1-SB fused to the C-terminus via P2A.DepositorInsertPediculus humanus ADAR deaminase domain (E438Q, I536S, E637F mutant)
TagsMLH1-SBExpressionMammalianMutationE438Q, I536S, E637F, with 10 amino acids truncati…PromoterCMVAvailable SinceNov. 7, 2025AvailabilityAcademic Institutions and Nonprofits only