8,173 results
-
Plasmid#198651PurposeEasy-MISE toolkit pEM-plasmid containing PDA1 promoter with EF protruding endsDepositorInsertpPDA1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
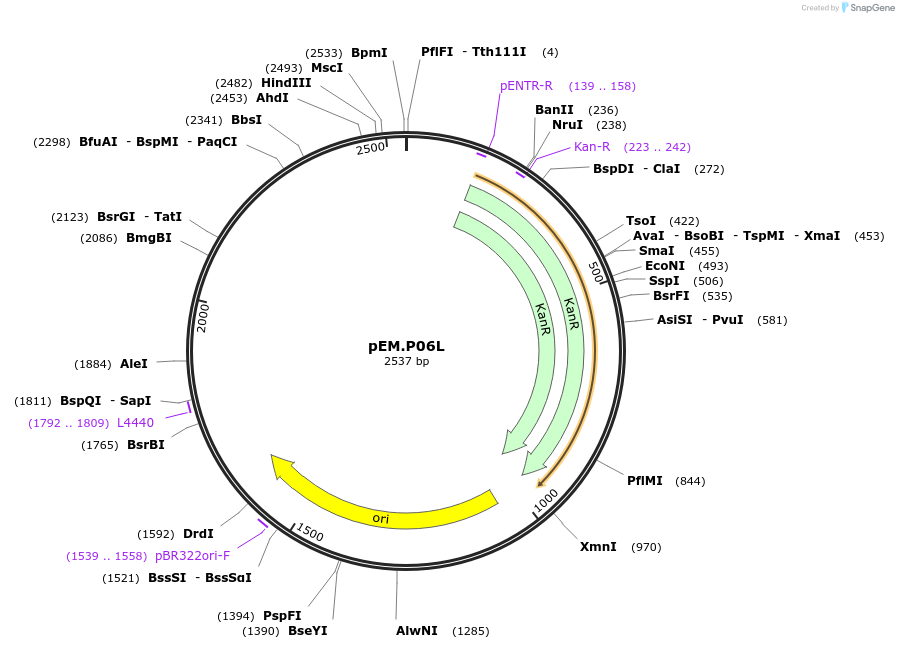
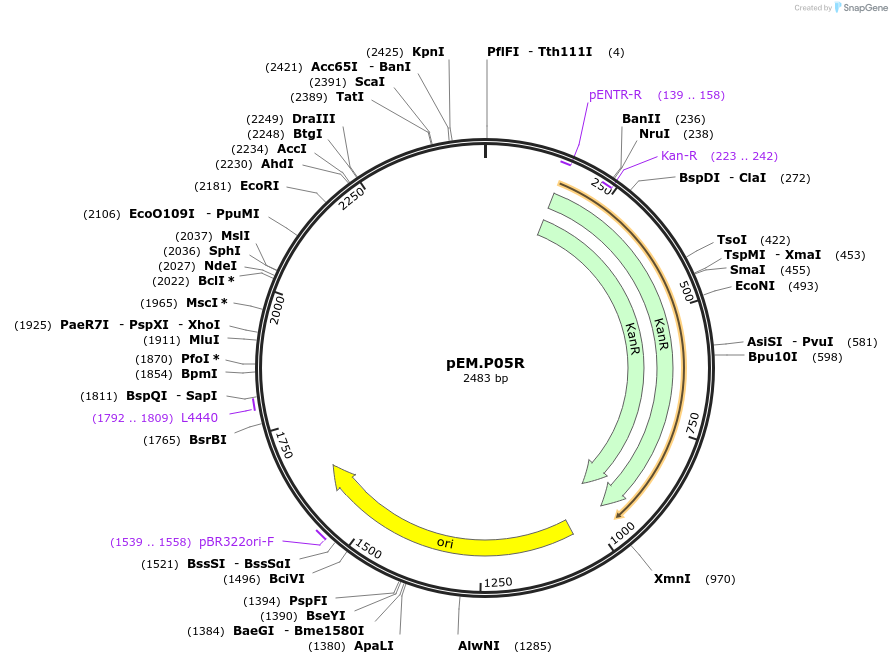
pEM.P05R
Plasmid#198650PurposeEasy-MISE toolkit pEM-plasmid containing CYC1 promoter with FG protruding endsDepositorInsertpCYC1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
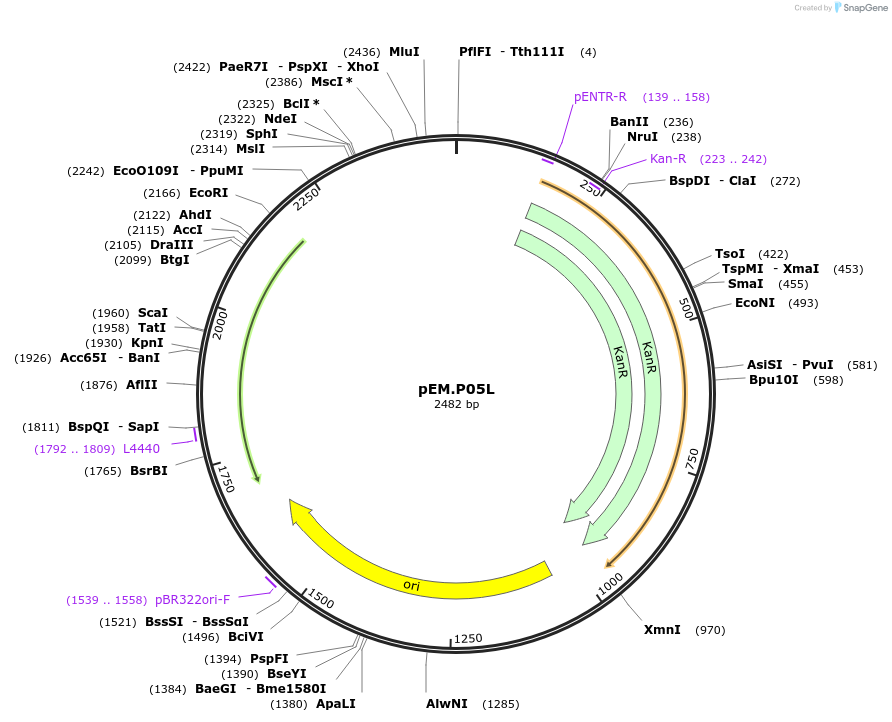
pEM.P05L
Plasmid#198649PurposeEasy-MISE toolkit pEM-plasmid containing CYC1 promoter with EF protruding endsDepositorInsertpCYC1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
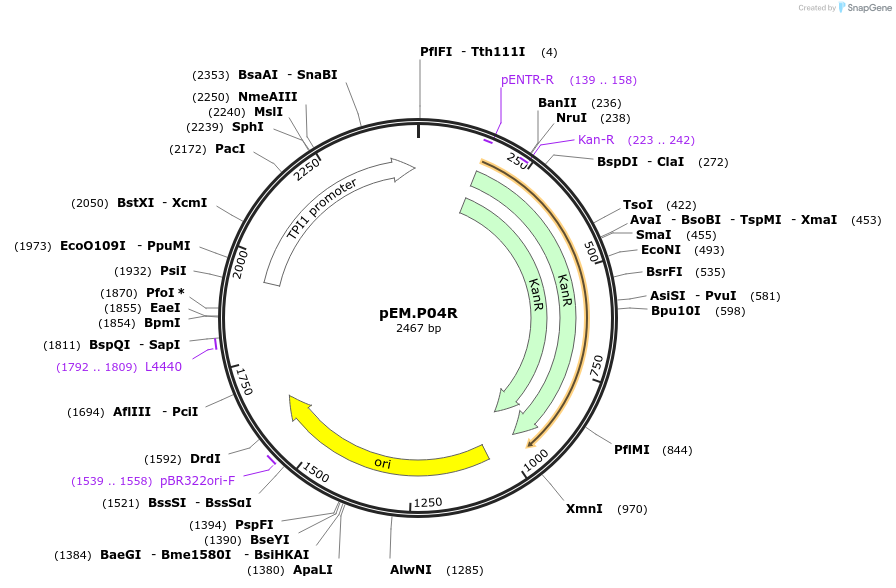
pEM.P04R
Plasmid#198648PurposeEasy-MISE toolkit pEM-plasmid containing TPI1 promoter with FG protruding endsDepositorInsertpTPI1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
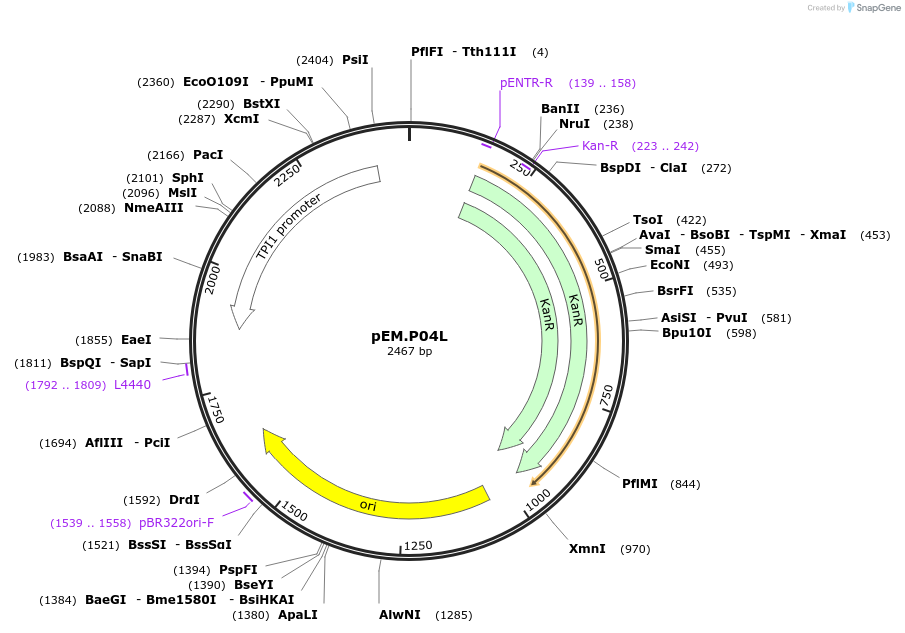
pEM.P04L
Plasmid#198647PurposeEasy-MISE toolkit pEM-plasmid containing TPI1 promoter with EF protruding endsDepositorInsertpTPI1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
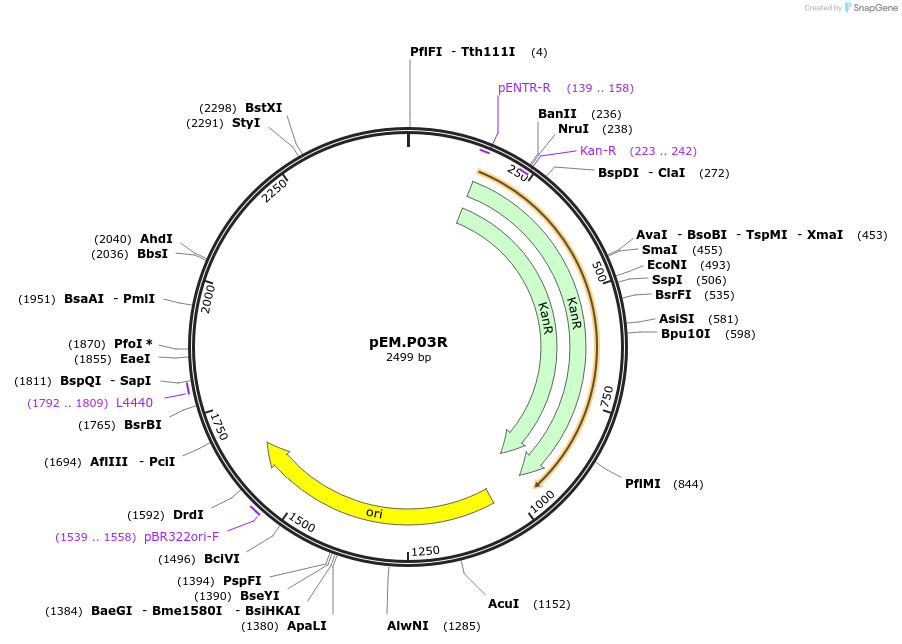
pEM.P03R
Plasmid#198646PurposeEasy-MISE toolkit pEM-plasmid containing PGK1 promoter with FG protruding endsDepositorInsertpPGK1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
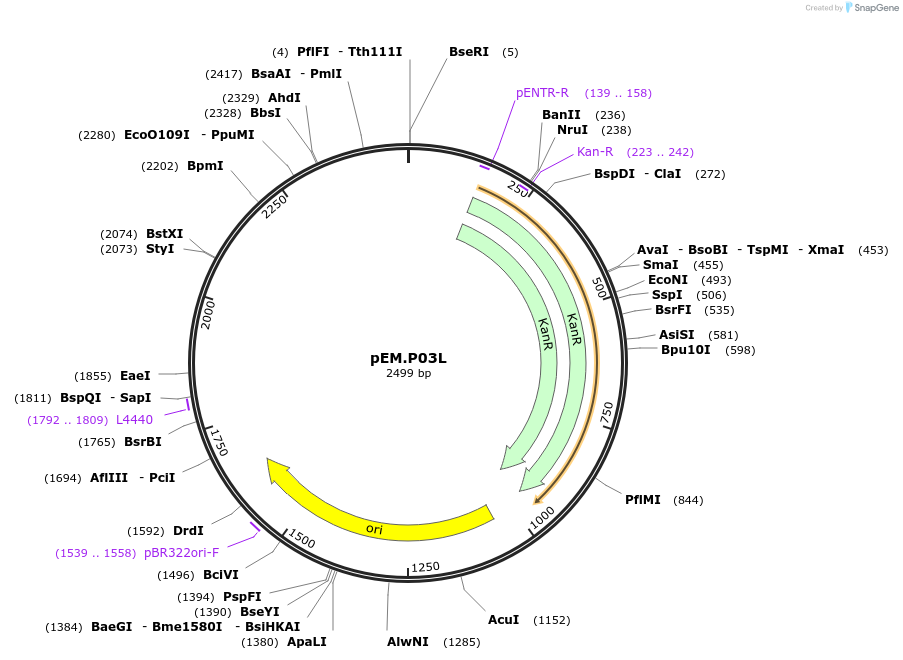
pEM.P03L
Plasmid#198645PurposeEasy-MISE toolkit pEM-plasmid containing PGK1 promoter with EF protruding endsDepositorInsertpPGK1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
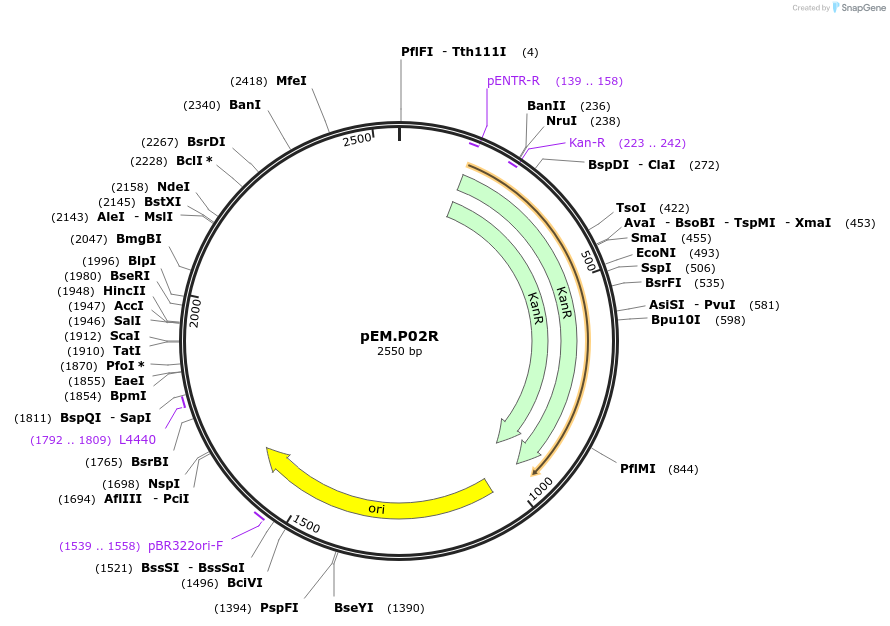
pEM.P02R
Plasmid#198644PurposeEasy-MISE toolkit pEM-plasmid containing ENO2 promoter with FG protruding endsDepositorInsertpENO2
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
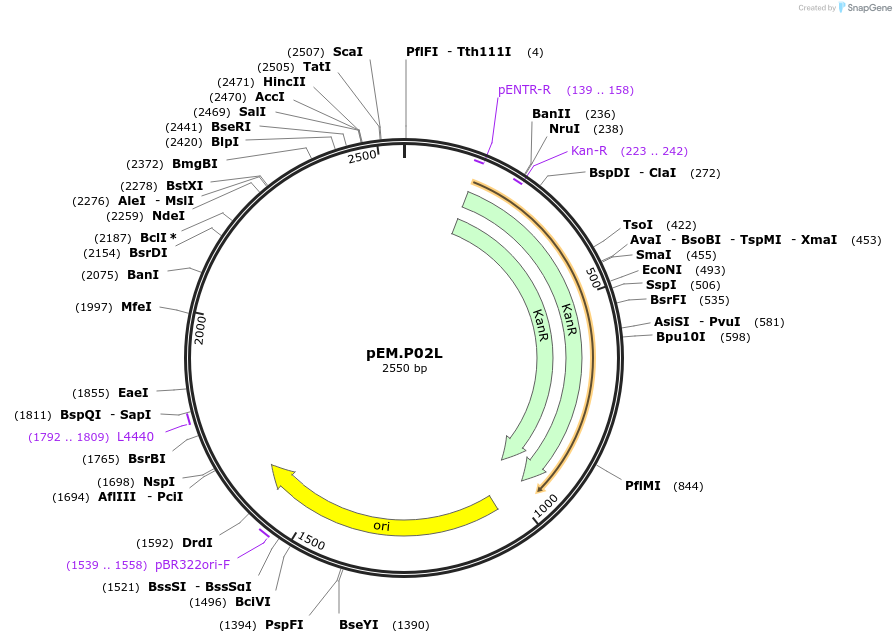
pEM.P02L
Plasmid#198643PurposeEasy-MISE toolkit pEM-plasmid containing ENO2 promoter with EF protruding endsDepositorInsertpENO2
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
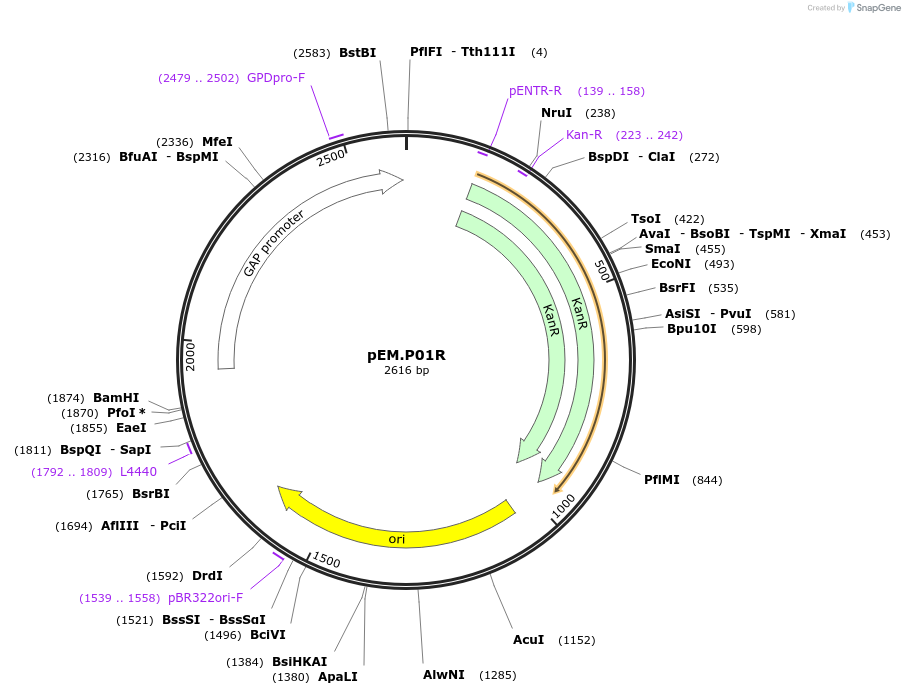
pEM.P01R
Plasmid#198642PurposeEasy-MISE toolkit pEM-plasmid containing TDH3 promoter with FG protruding endsDepositorInsertpTDH3
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
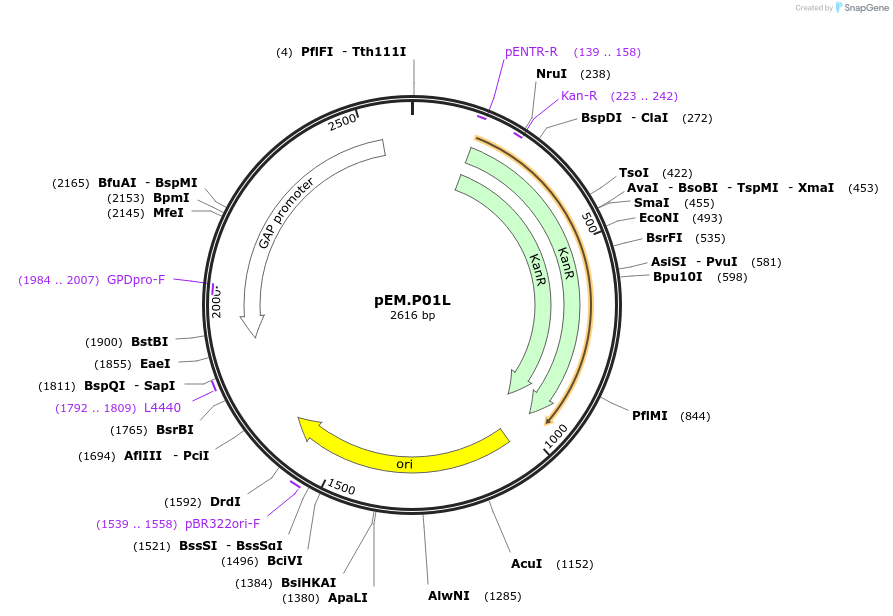
pEM.P01L
Plasmid#198641PurposeEasy-MISE toolkit pEM-plasmid containing TDH3 promoter with EF protruding endsDepositorInsertpTDH3
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
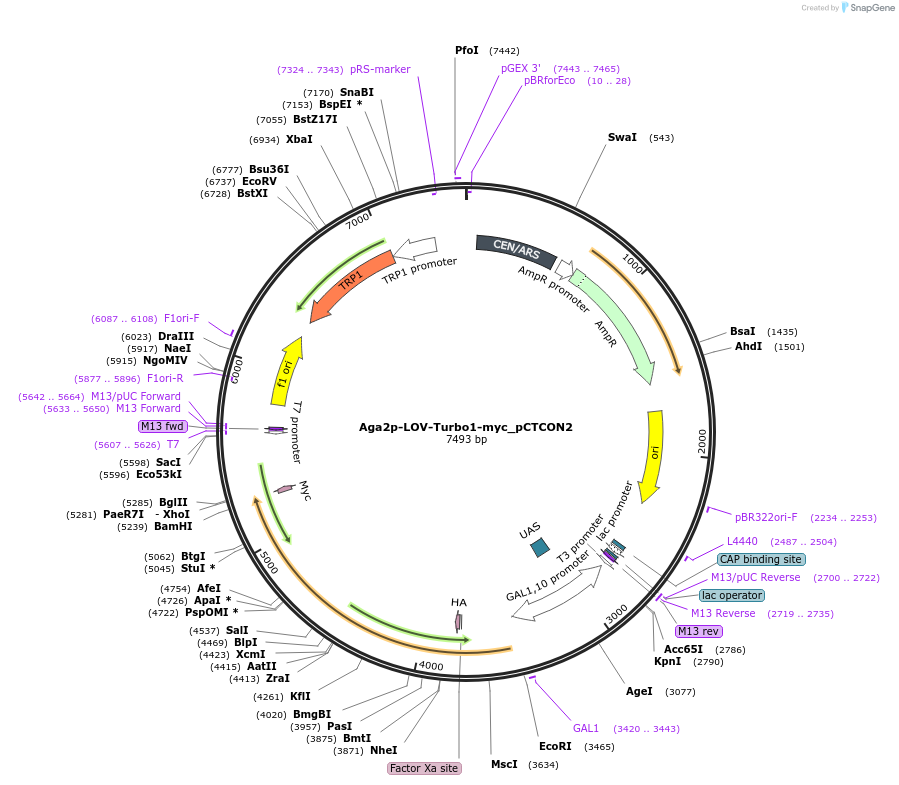
Aga2p-LOV-Turbo1-myc_pCTCON2
Plasmid#199673Purposeexpresses pre-evolved version of LOV-Turbo on the yeast surfaceDepositorInsertLOV-Turbo1
TagsAga2p and MycExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
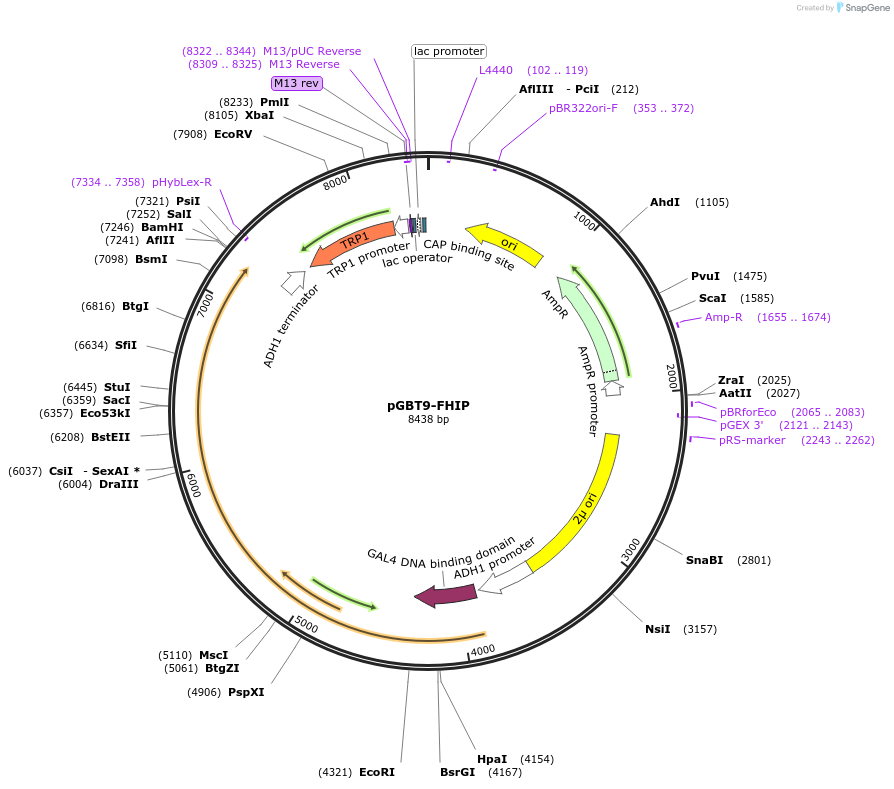
pGBT9-FHIP
Plasmid#198527PurposeExpression of GAL4 DNA-binding domain (BD)-FHIP fusion protein in yeast (yeast two-hybrid assays)DepositorInsertFHIP (FHIP1B Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastMutationsilent mutations in codons 353 and 651 as well as…PromoterADH1Available SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
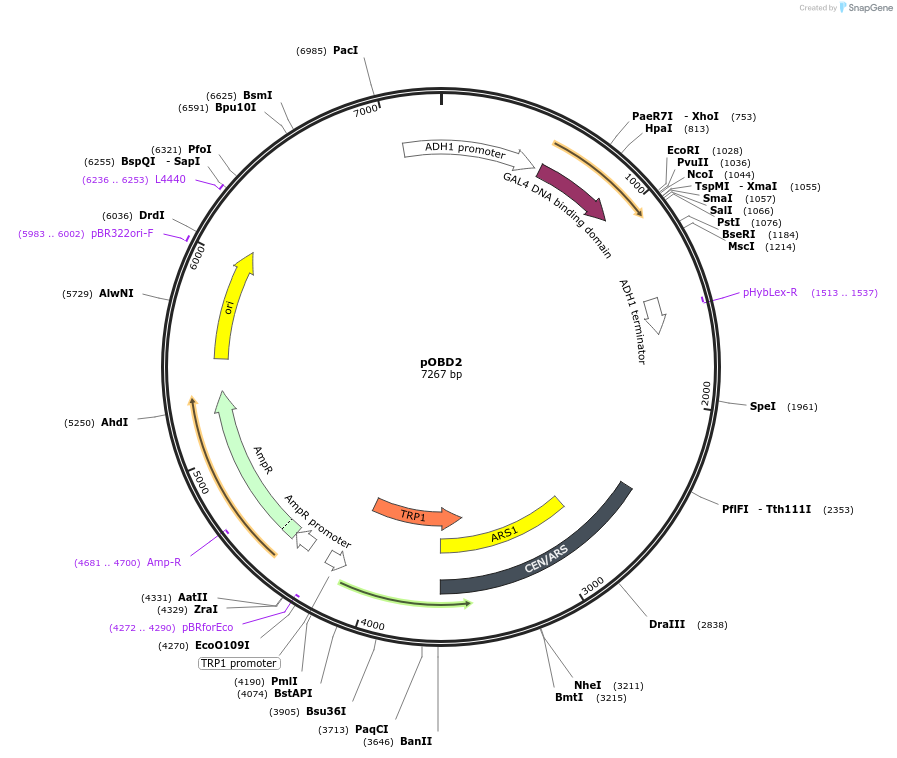
pOBD2
Plasmid#196826PurposeYeast two-hybrid ; Gal4 DNA binding domainDepositorTypeEmpty backboneTagsGal4 DNA binding domainExpressionYeastPromoterADHAvailable SinceMay 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
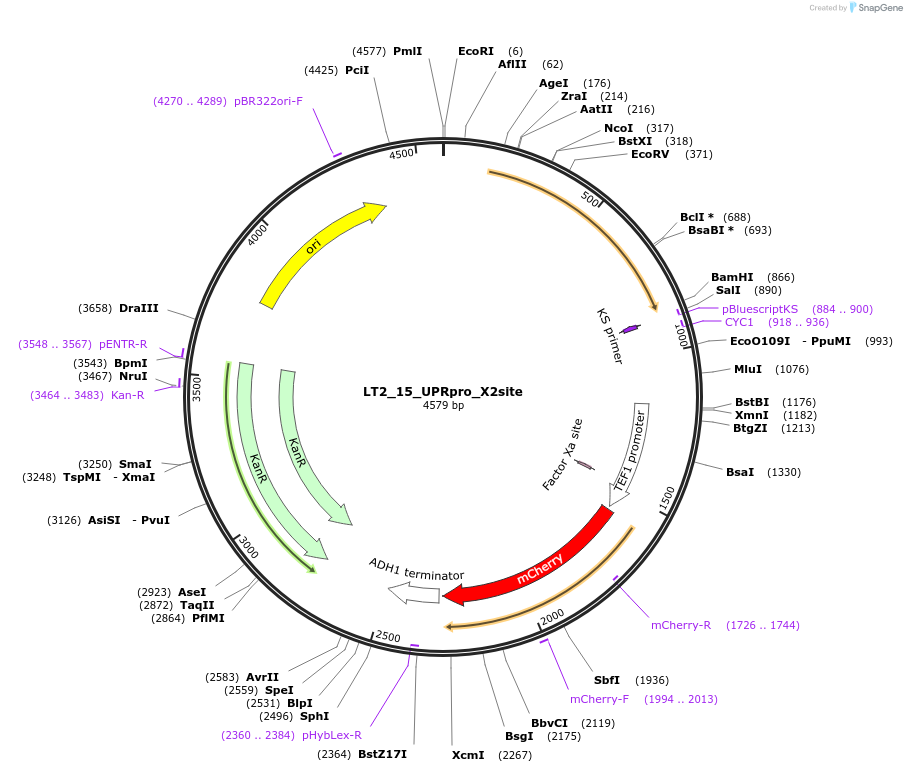
LT2_15_UPRpro_X2site
Plasmid#194427PurposePlasmid for genome integration in X2 site expressing UPRpro - Sensor for Unfolded Protein ResponseDepositorInsertUPRpro
UseUsed as donor dna in genome integration after lin…ExpressionYeastMutationNAAvailable SinceMay 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
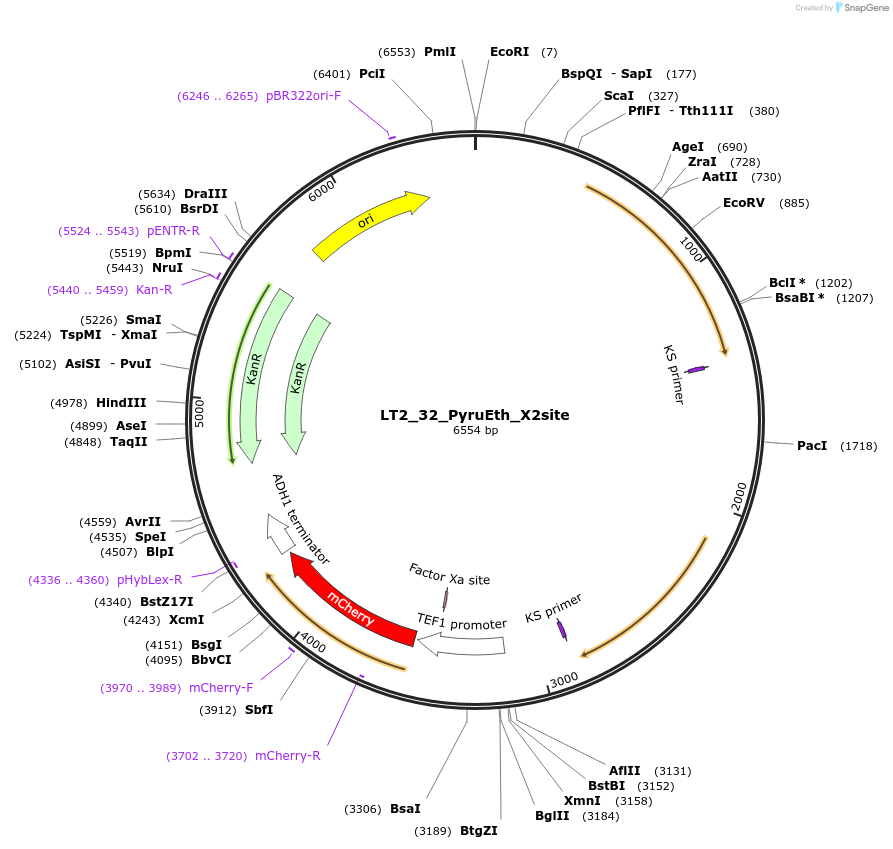
LT2_32_PyruEth_X2site
Plasmid#194428PurposePlasmid for genome integration in X2 site expressing PyruEth (Pyruvate and Ethanol consumption sensors)DepositorInsertPyruEth
UseUsed as donor dna in genome integration after lin…ExpressionYeastMutationNAAvailable SinceMay 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
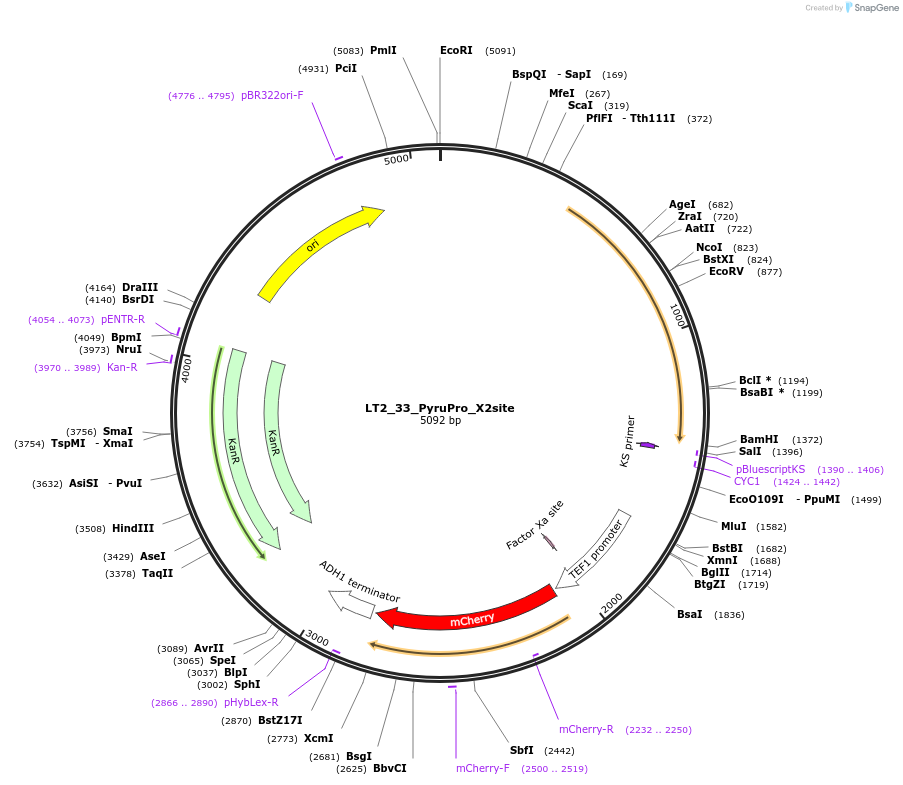
LT2_33_PyruPro_X2site
Plasmid#194429PurposePlasmid for genome integration in X2 site expressing PyruPro (Pyruvate consumption Probe)DepositorInsertPyruPro
UseUsed as donor dna in genome integration after lin…ExpressionYeastMutationNAAvailable SinceMay 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
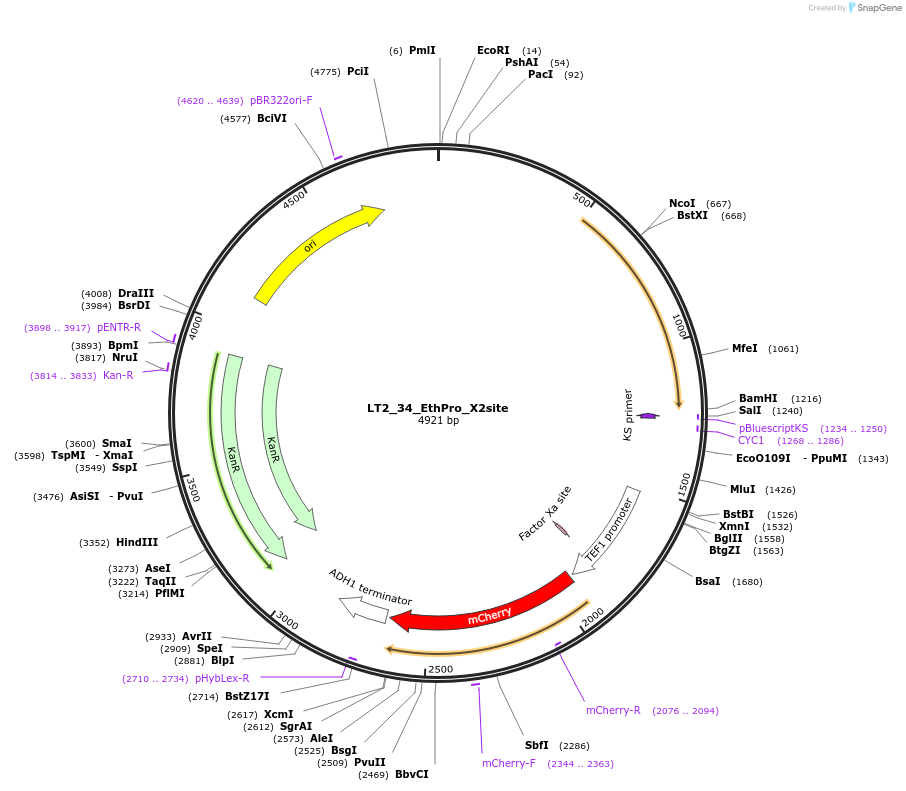
LT2_34_EthPro_X2site
Plasmid#194430PurposePlasmid for genome integration in X2 site expressing ResPro (Ethanol consumption Probe)DepositorInsertEthPro
UseUsed as donor dna in genome integration after lin…ExpressionYeastMutationNAAvailable SinceMay 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
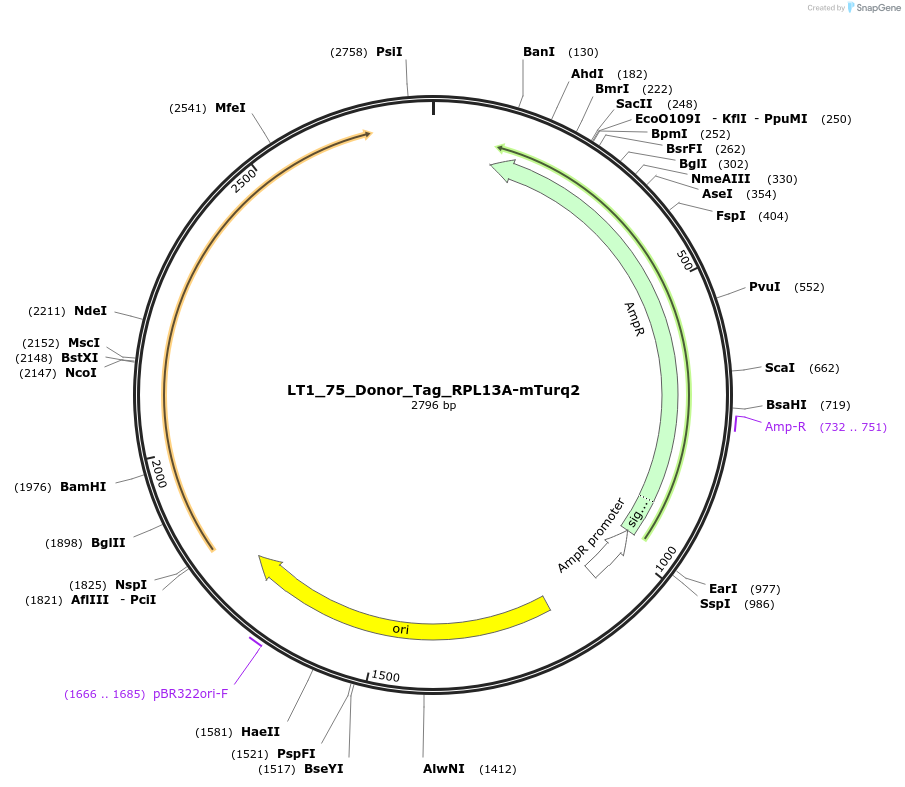
LT1_75_Donor_Tag_RPL13A-mTurq2
Plasmid#194415PurposePlasmid for genome integration of mTurquoise2 CDS to tag native RPL13A for biosensors RibPro and UPRproDepositorInsertmTurquoise2 CDS
UseSynthetic Biology; Used as donor dna in genome in…ExpressionYeastMutationNAAvailable SinceMay 8, 2023AvailabilityAcademic Institutions and Nonprofits only -
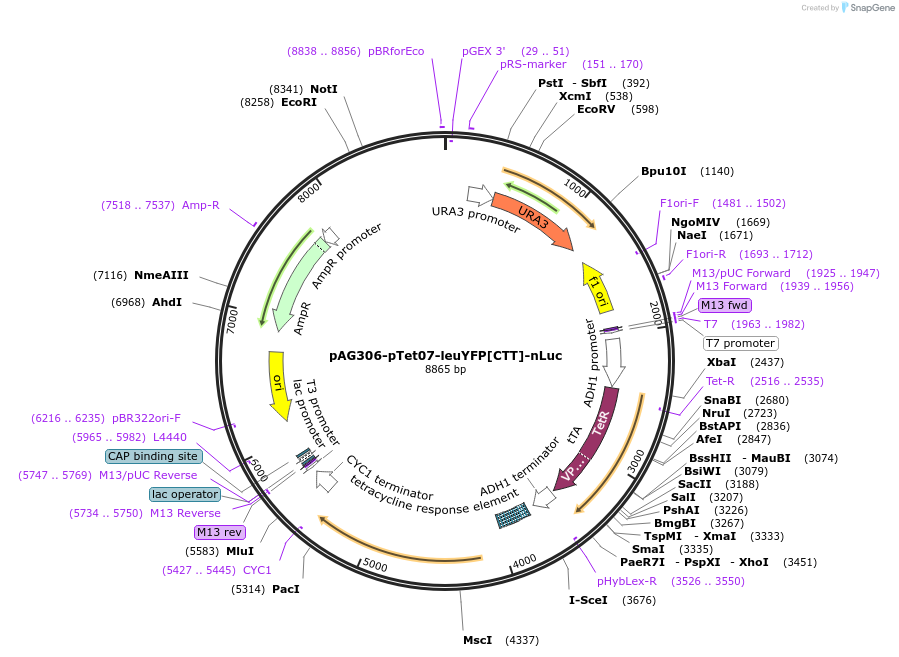
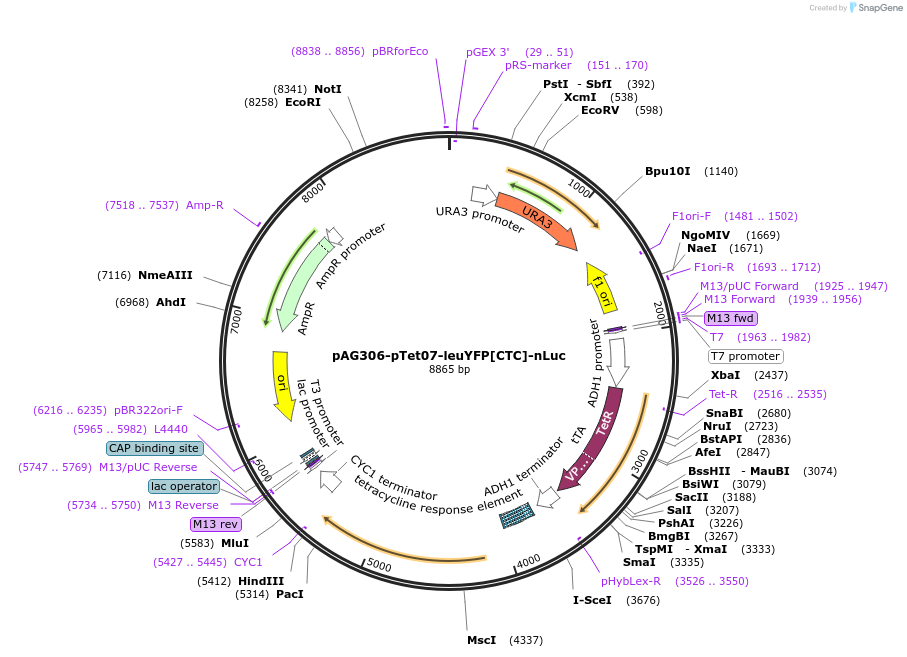
pAG306-pTet07-leuYFP[CTT]-nLuc
Plasmid#199757PurposeElongation reporter construct to quantify elongation of synYFP[CTT]DepositorInsertleuYFP[CTT]-nLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
pAG306-pTet07-leuYFP[CTC]-nLuc
Plasmid#199758PurposeElongation reporter construct to quantify elongation of synYFP[CTC]DepositorInsertleuYFP[CTC]
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
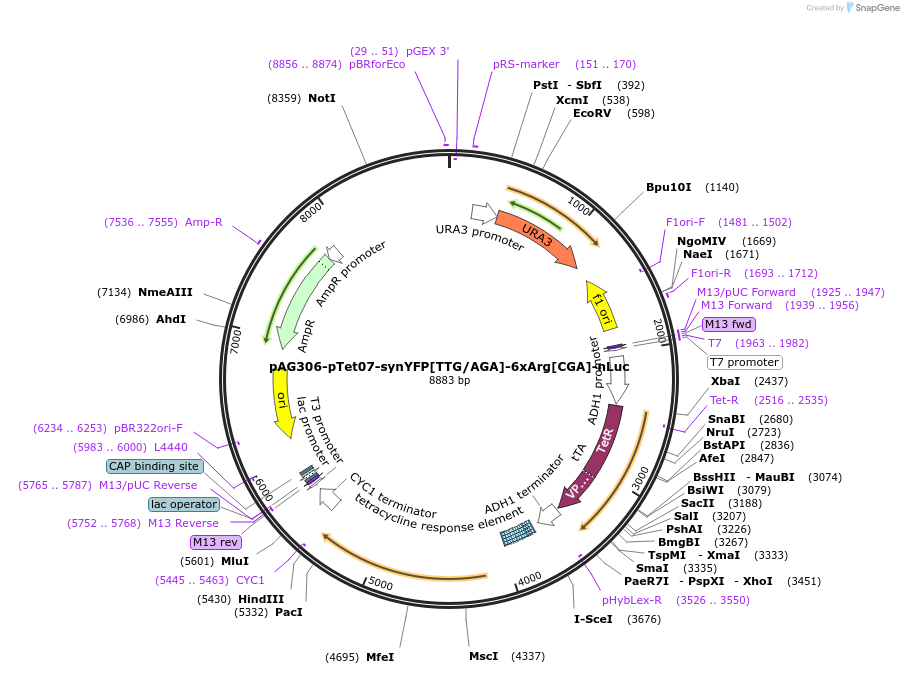
pAG306-pTet07-synYFP[TTG/AGA]-6xArg[CGA]-nLuc
Plasmid#199760PurposeElongation reporter construct to quantify elongation duration of 6_CGA stallingDepositorInsertsynYFP[TTG/AGA]-6xArg[CGA]-nLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
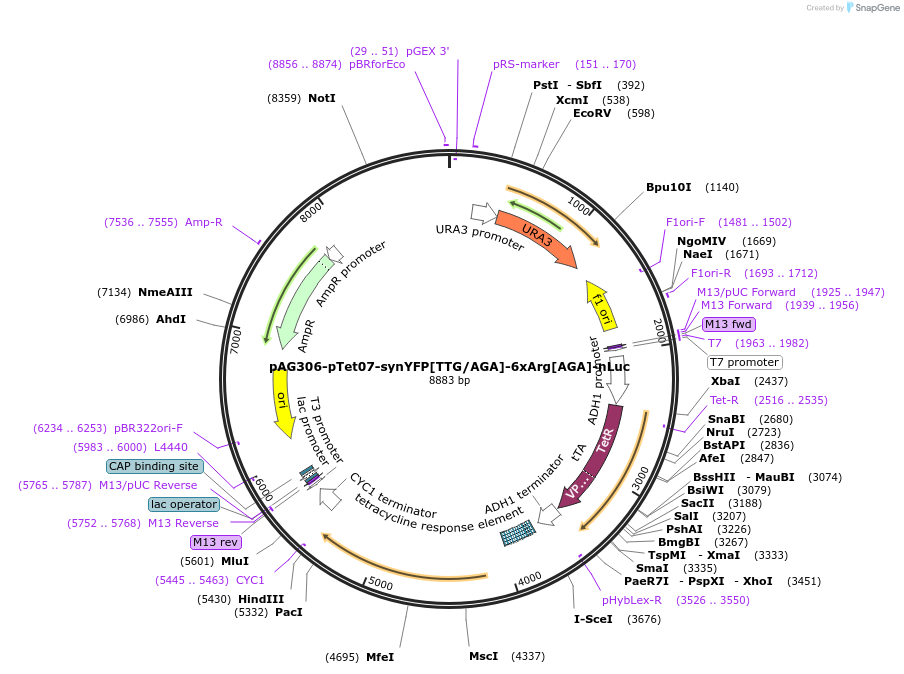
pAG306-pTet07-synYFP[TTG/AGA]-6xArg[AGA]-nLuc
Plasmid#199761PurposeElongation reporter construct to quantify elongation duration of 6_AGA stallingDepositorInsertsynYFP[TTG/AGA]-6xArg[AGA]-cLuc
ExpressionYeastPromoterTetO7Available SinceMay 5, 2023AvailabilityAcademic Institutions and Nonprofits only -
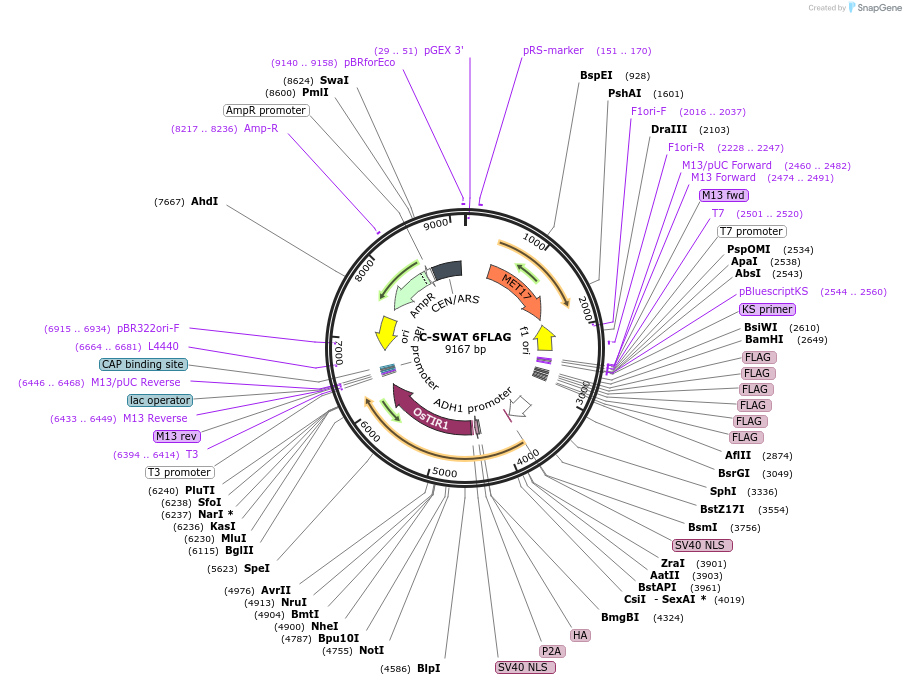
C-SWAT 6FLAG
Plasmid#182509PurposeC-SWAT compatible vector for C-terminal tagging with 6 FLAGDepositorInsert6 FLAG
ExpressionYeastAvailable SinceApril 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
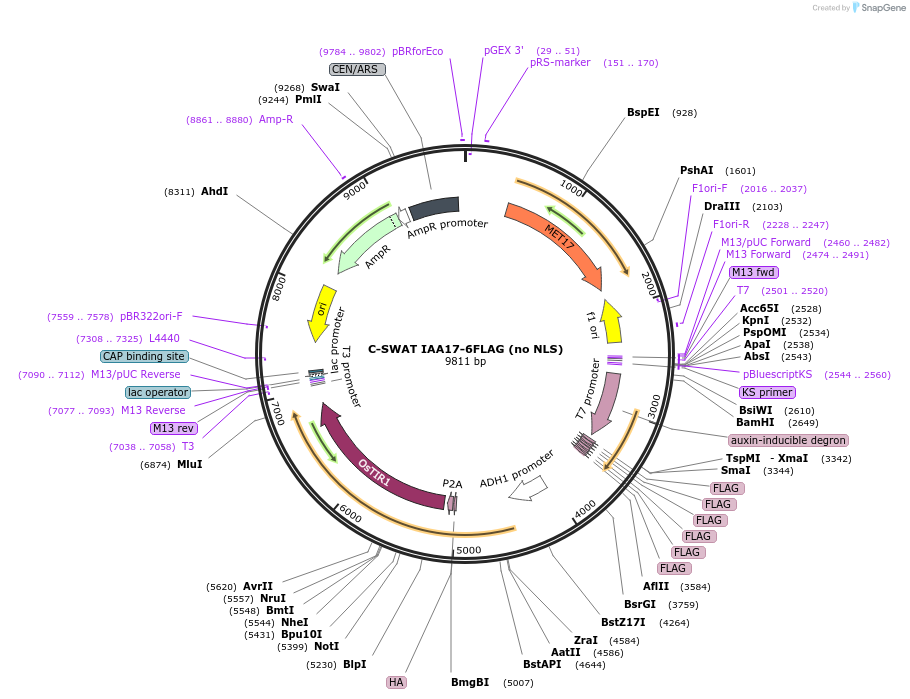
C-SWAT IAA17-6FLAG (no NLS)
Plasmid#193322PurposeC-SWAT compatible vector for C-terminal tagging with IAA17 (Auxin-Inducible Degron) and 6FLAG. ARF16(PB1)-P2A-osTIR1 has SIR4 homology arms for genome integration and does not have NLS tags..DepositorInsertIAA17
Tags6FLAGExpressionYeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
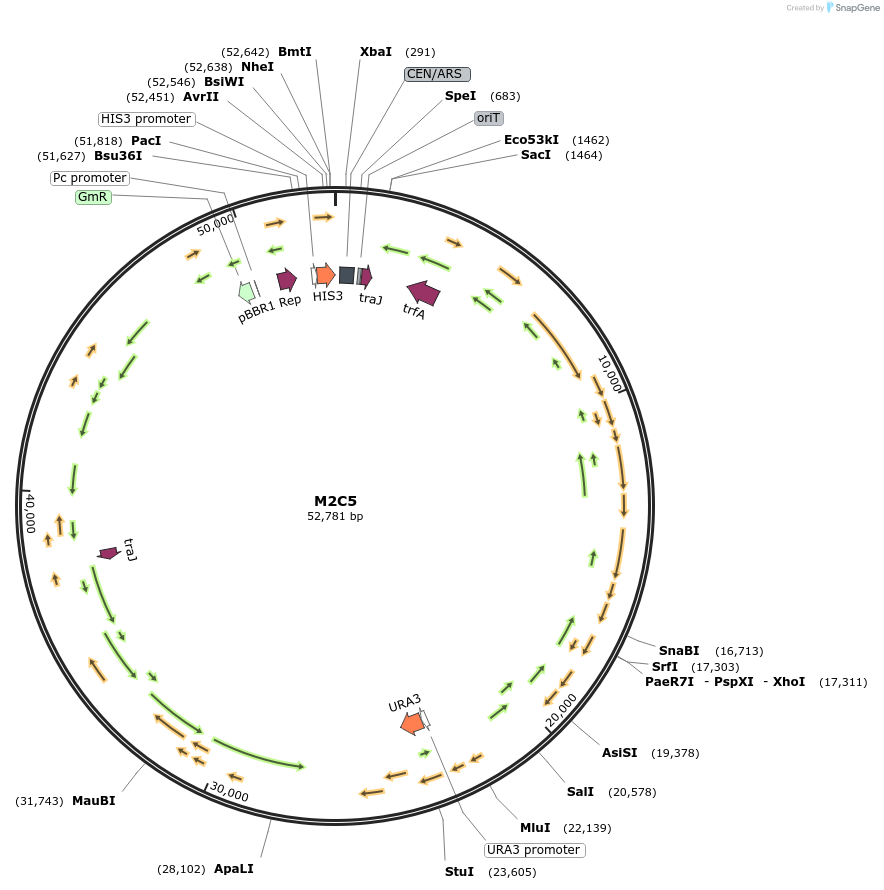
M2C5
Plasmid#188606PurposeMinimal Conjugative Plasmid 2DepositorTypeEmpty backboneUseSynthetic Biology; Diatom expressionExpressionBacterial and YeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
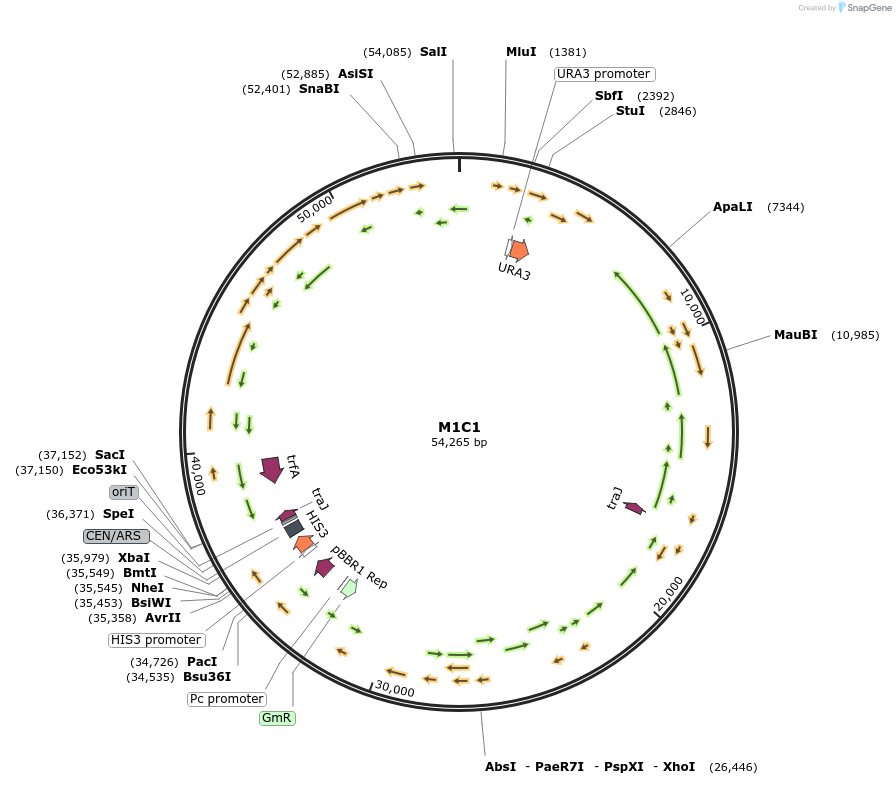
M1C1
Plasmid#188609PurposeMinimal Conjugative Plasmid 1DepositorTypeEmpty backboneUseSynthetic Biology; Diatom expressionExpressionBacterial and YeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
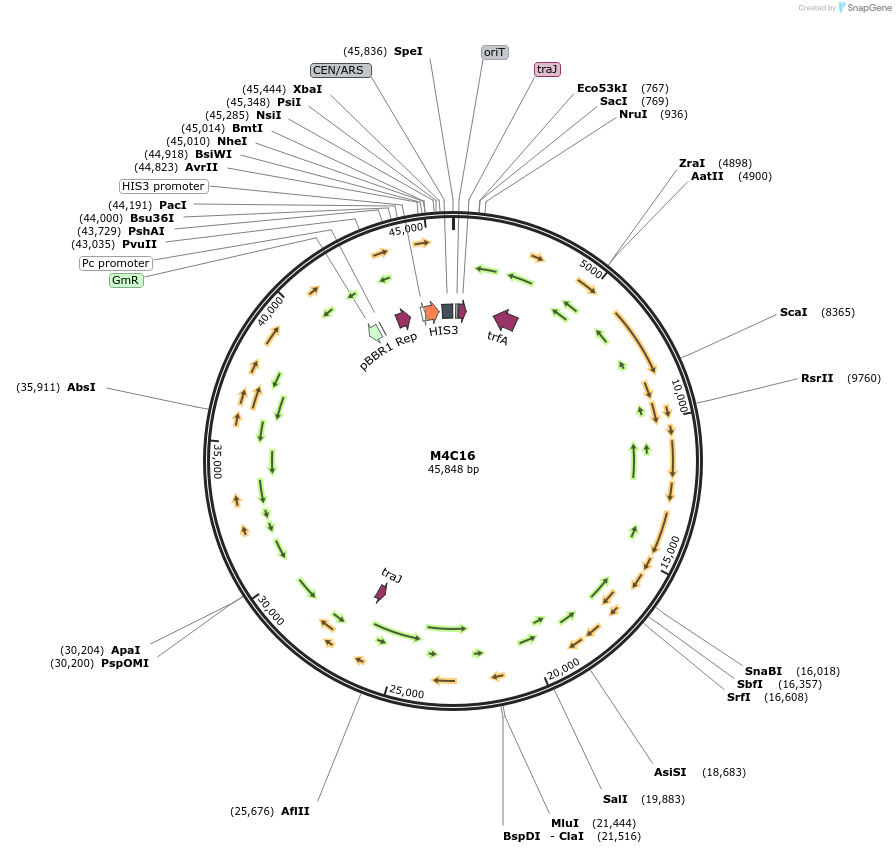
M4C16
Plasmid#188610PurposeMinimal Conjugative Plasmid 4DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
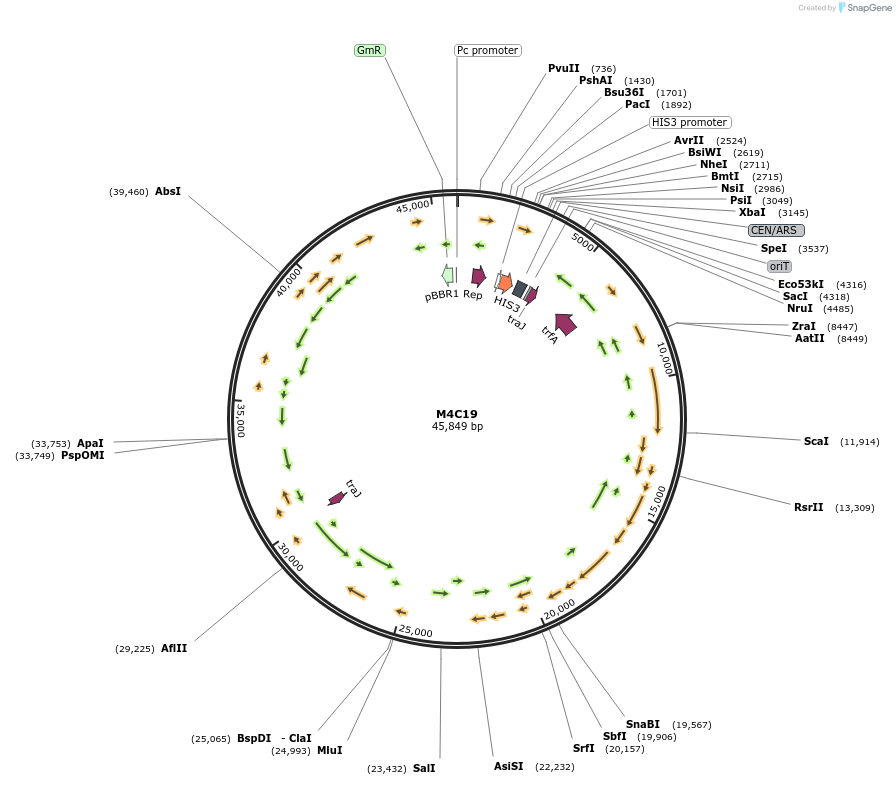
M4C19
Plasmid#188611PurposeMinimal Conjugative Plasmid 4DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailable SinceApril 24, 2023AvailabilityAcademic Institutions and Nonprofits only -
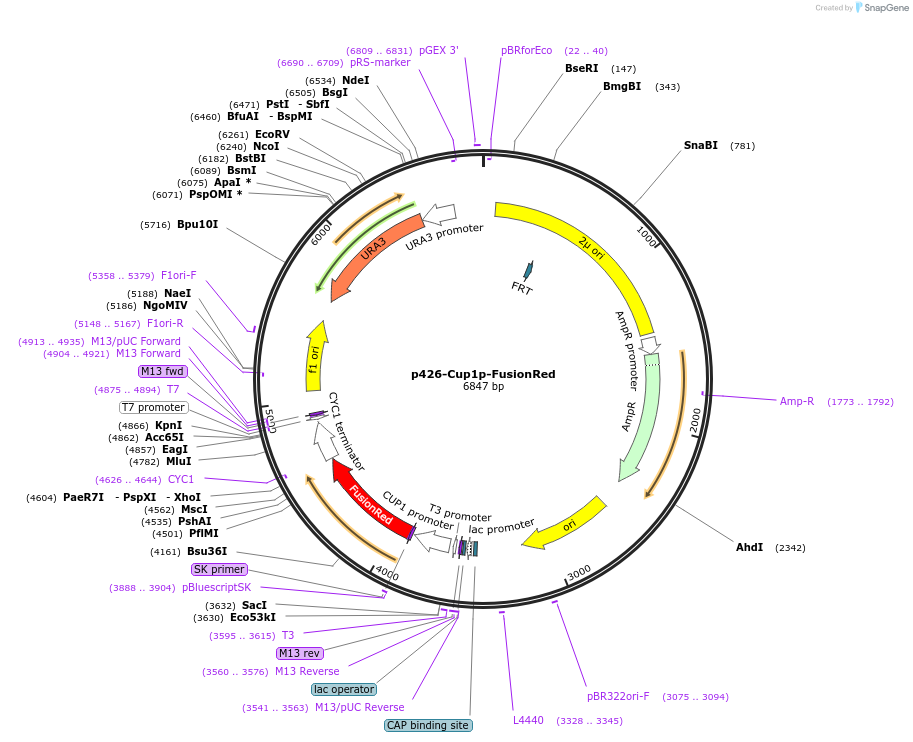
p426-Cup1p-FusionRed
Plasmid#188392PurposeCu2+-dependent expression of red fluorescent protein in yeast cellsDepositorInsertFusionRed
ExpressionYeastPromoterCUP1 promoterAvailable SinceApril 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
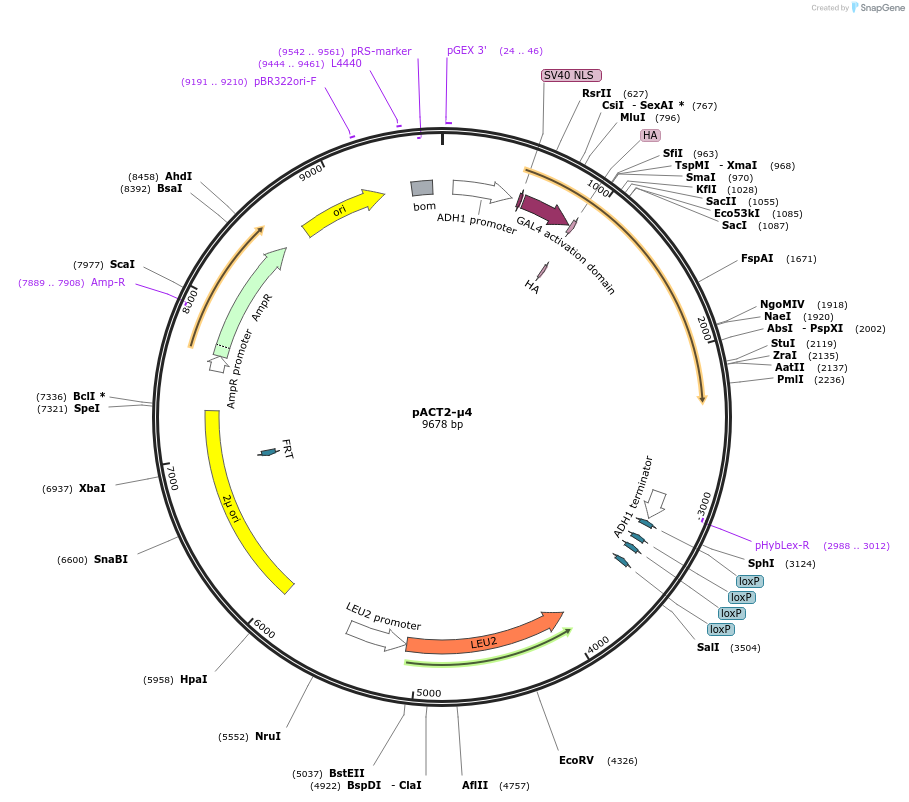
pACT2-μ4
Plasmid#198176PurposeExpression of GAL4 transcriptional activation domain (AD)-AP-4 μ4 fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-4 μ4
TagsGAL4 transcriptional activation domain (AD) fragm…ExpressionYeastPromoterADH1Available SinceApril 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
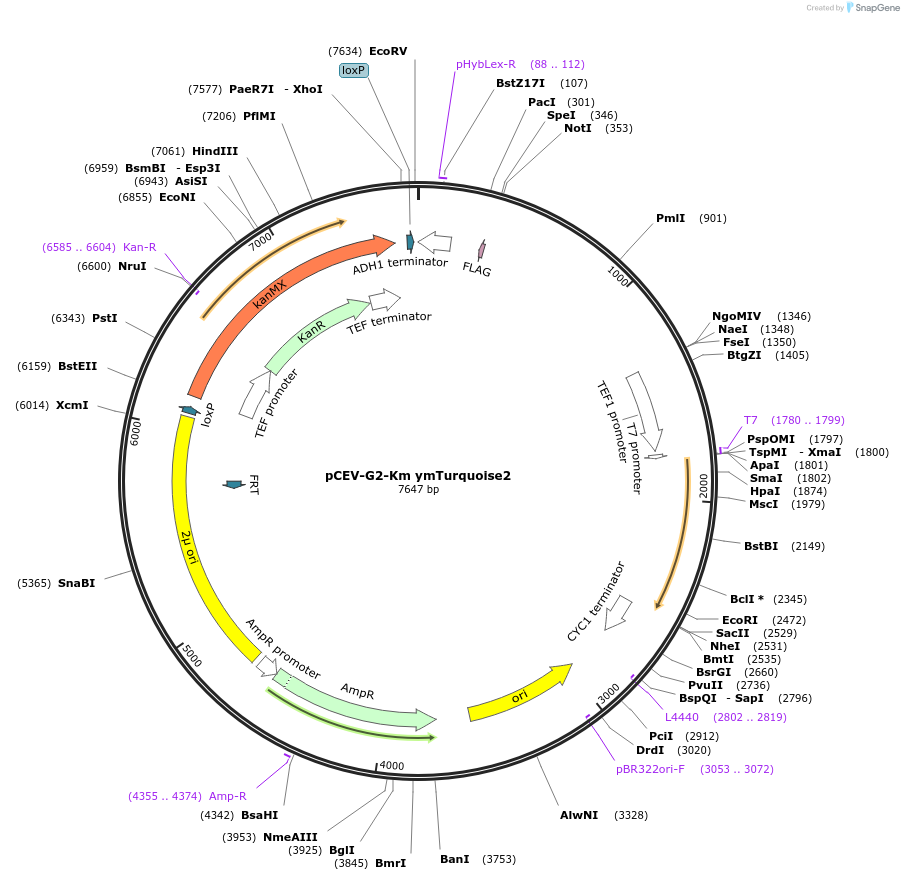
pCEV-G2-Km ymTurquoise2
Plasmid#193958PurposeTEF1 promoter controlled expression plasmid with G418 resistance for expressing ymTurquoise2 in budding yeastDepositorInsertymTurquoise2
ExpressionYeastPromoterTEF1Available SinceApril 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
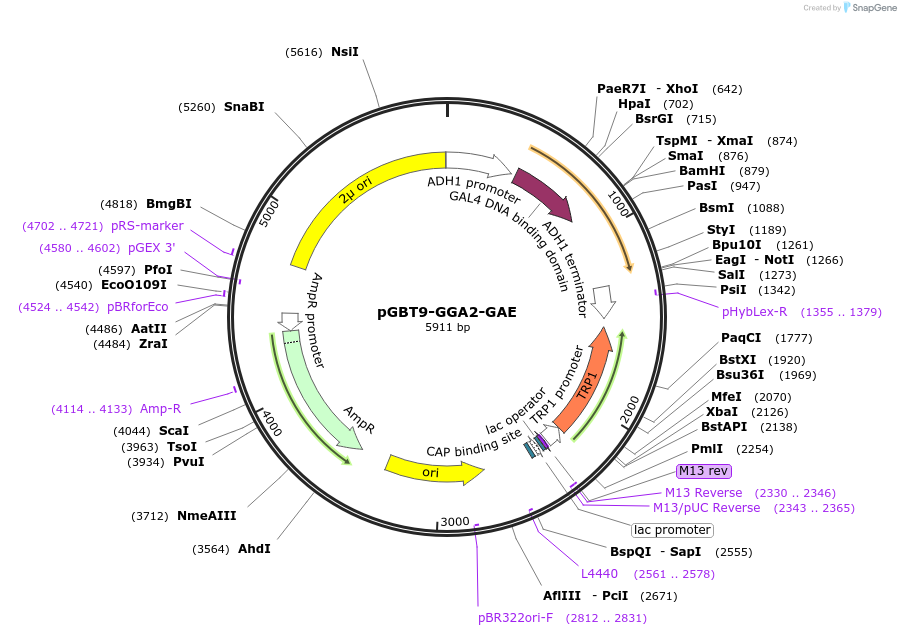
pGBT9-GGA2-GAE
Plasmid#199175PurposeExpression of GAL4 DNA-binding domain (BD)-GGA2-GAE domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertGGA2-GAE domain (GGA2 Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastMutationContains Lys 545 Gln substitutionPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
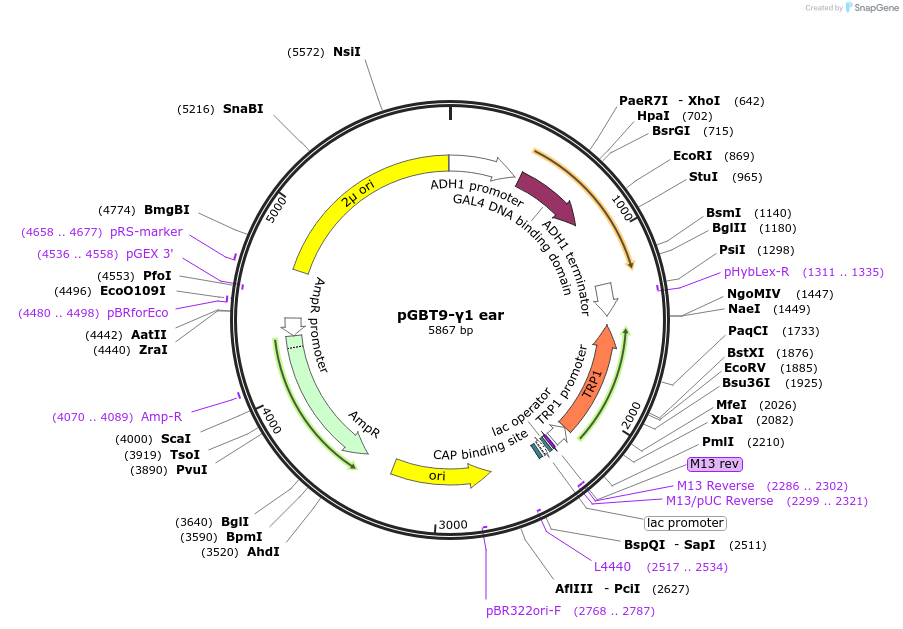
pGBT9-γ1 ear
Plasmid#199177PurposeExpression of GAL4 DNA-binding domain (BD)-AP-1 γ1 ear domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-1 γ1 ear domain (Ap1g1 Mouse)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
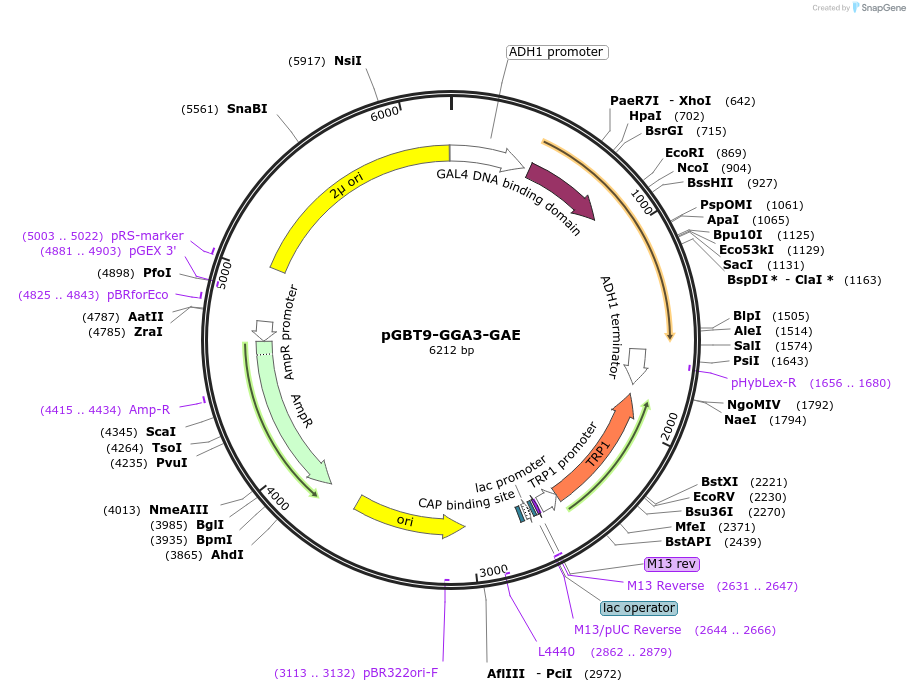
pGBT9-GGA3-GAE
Plasmid#199176PurposeExpression of GAL4 DNA-binding domain (BD)-GGA3-GAE domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertGGA3-GAE domain (GGA3 Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
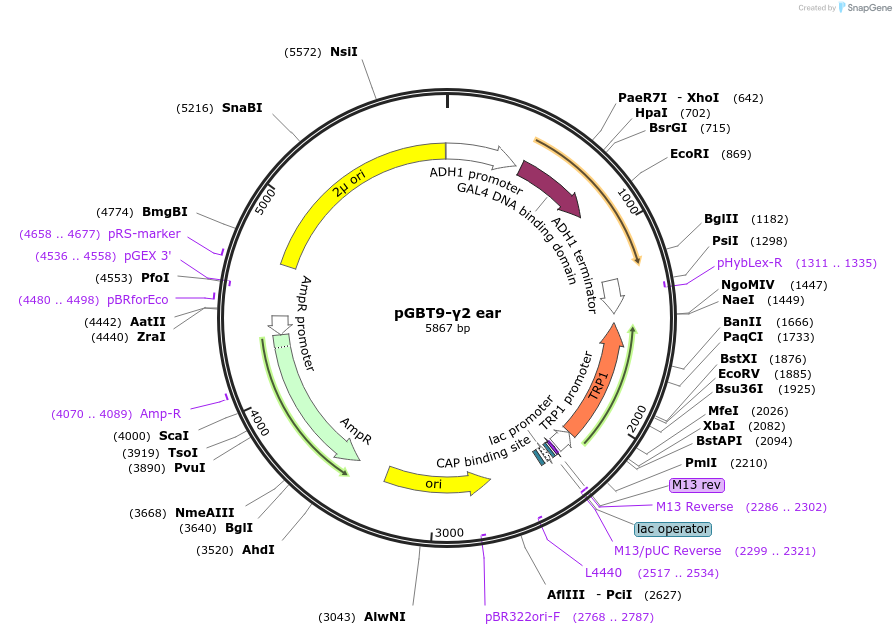
pGBT9-γ2 ear
Plasmid#199178PurposeExpression of GAL4 DNA-binding domain (BD)-AP-1 γ2 ear domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertAP-1 γ2 ear domain (AP1G2 Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
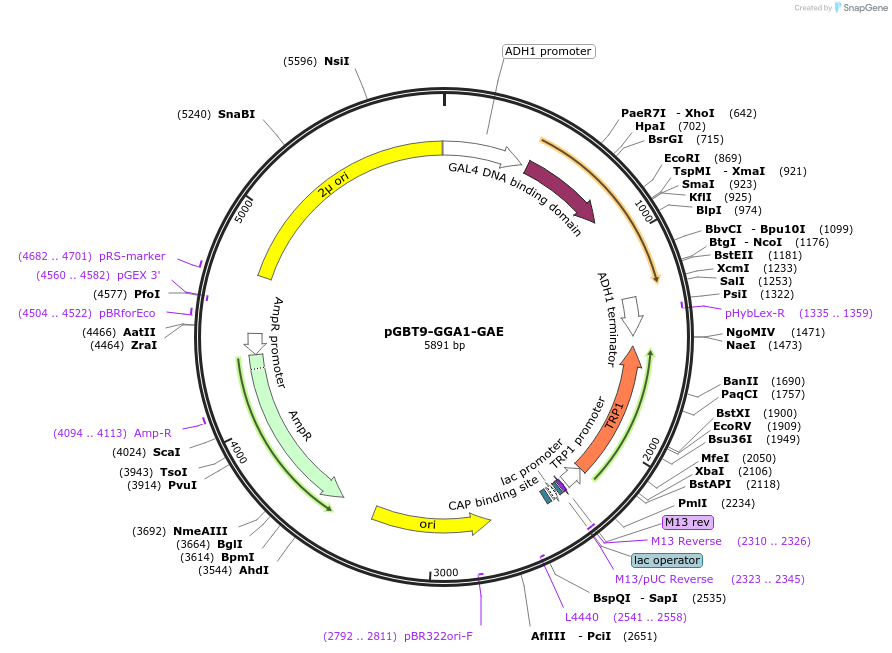
pGBT9-GGA1-GAE
Plasmid#199174PurposeExpression of GAL4 DNA-binding domain (BD)-GGA1-GAE domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertGGA1-GAE domain (GGA1 Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 10, 2023AvailabilityAcademic Institutions and Nonprofits only -
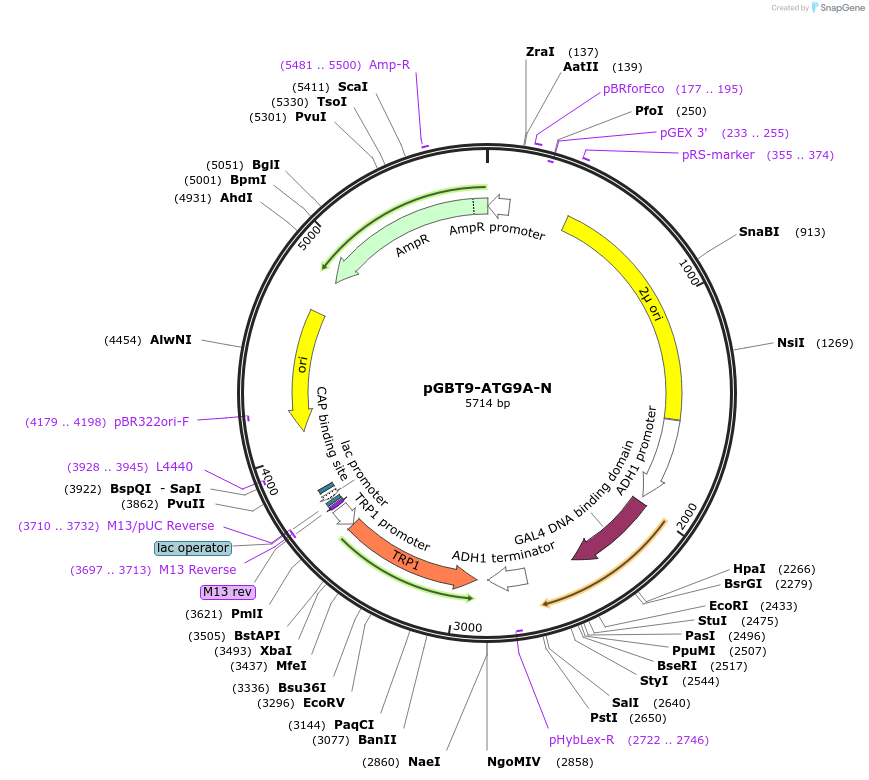
pGBT9-ATG9A-N
Plasmid#198530PurposeExpression of GAL4 DNA-binding domain (BD)-ATG9A-N terminal cytosolic domain fusion protein in yeast (yeast two-hybrid assays)DepositorInsertATG9A N-terminal cytosolic tail (residues 1-66) (ATG9A Human)
TagsGAL4-DNA binding domain fragmentExpressionYeastPromoterADH1Available SinceApril 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
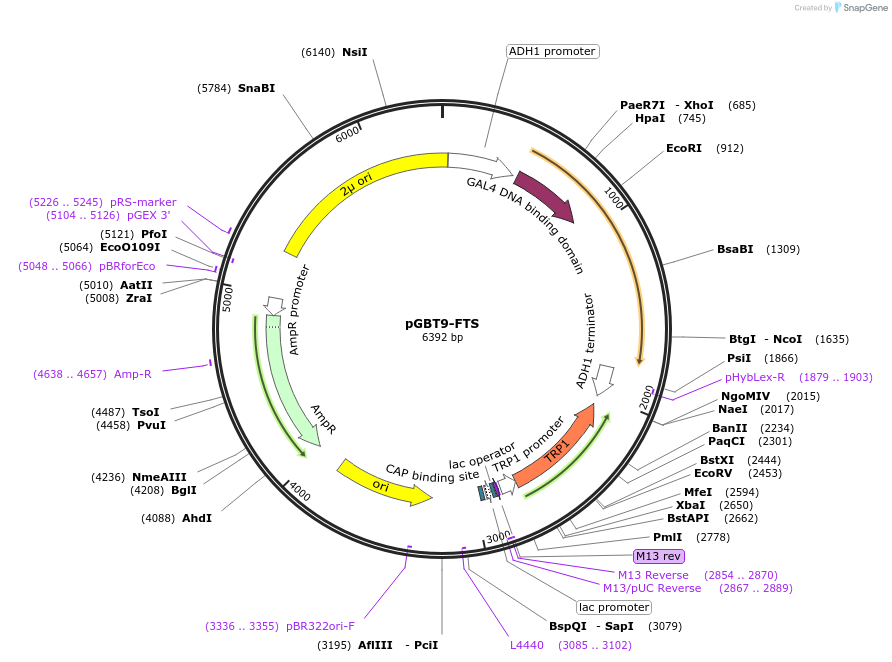
pGBT9-FTS
Plasmid#198526PurposeExpression of GAL4 DNA-binding domain (BD)-FTS fusion protein in yeast (yeast two-hybrid assays)DepositorAvailable SinceApril 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
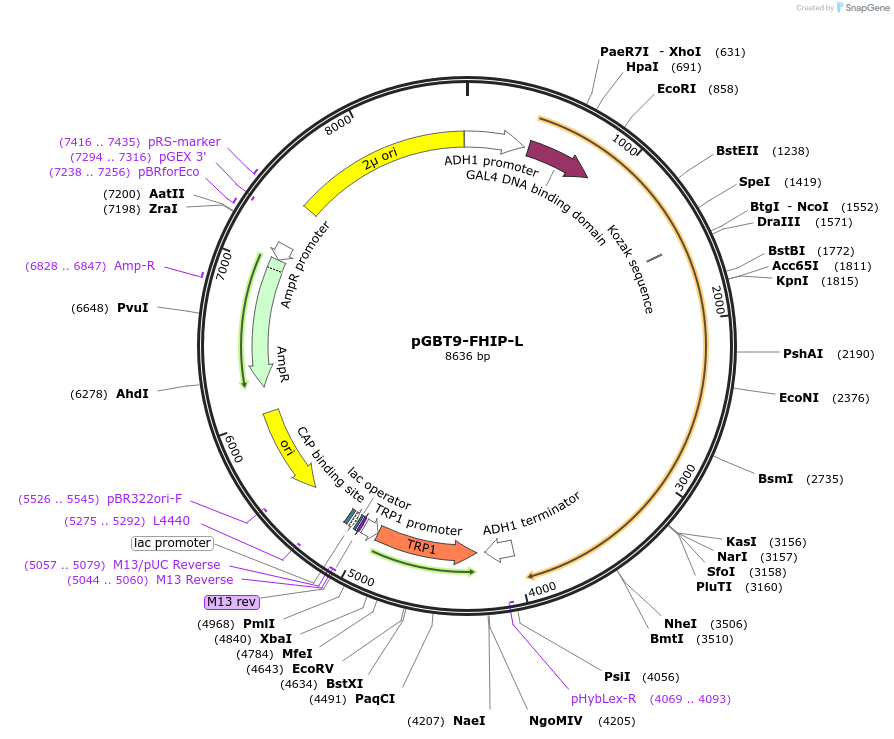
pGBT9-FHIP-L
Plasmid#198528PurposeExpression of GAL4 DNA-binding domain (BD)-FHIP-L fusion protein in yeast (yeast two-hybrid assays)DepositorAvailable SinceApril 3, 2023AvailabilityAcademic Institutions and Nonprofits only