We narrowed to 12,964 results for: BASE
-
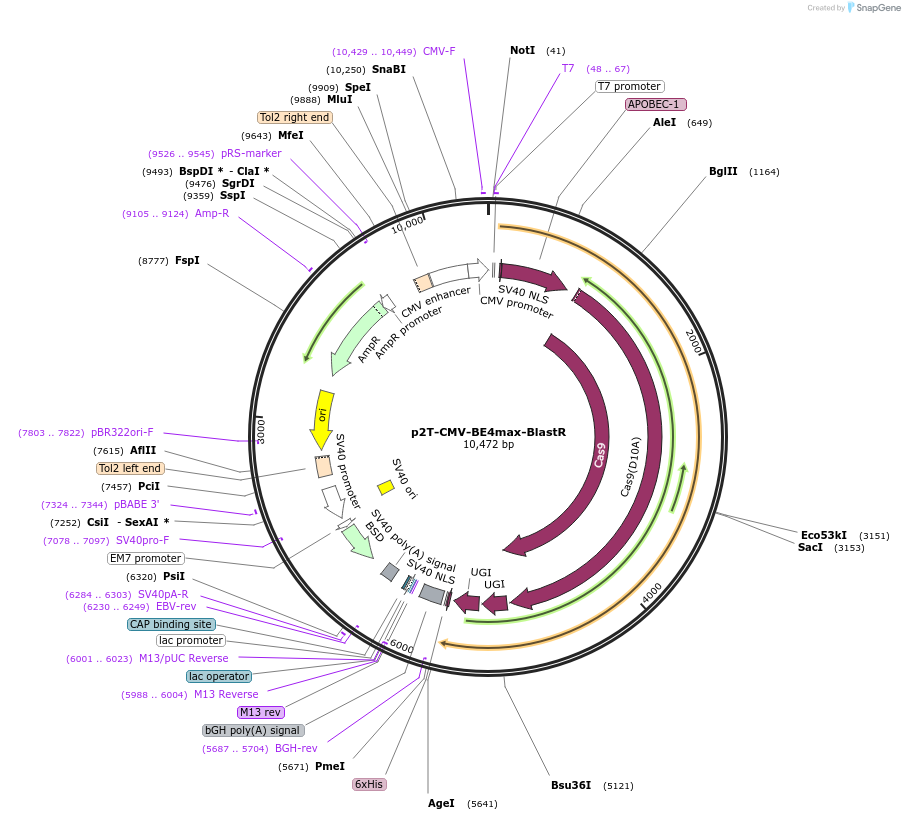
Plasmid#152991PurposeC-to-T base editorDepositorInsertBE4-nSpCas9-UGI-UGI
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceJuly 1, 2020AvailabilityAcademic Institutions and Nonprofits only -
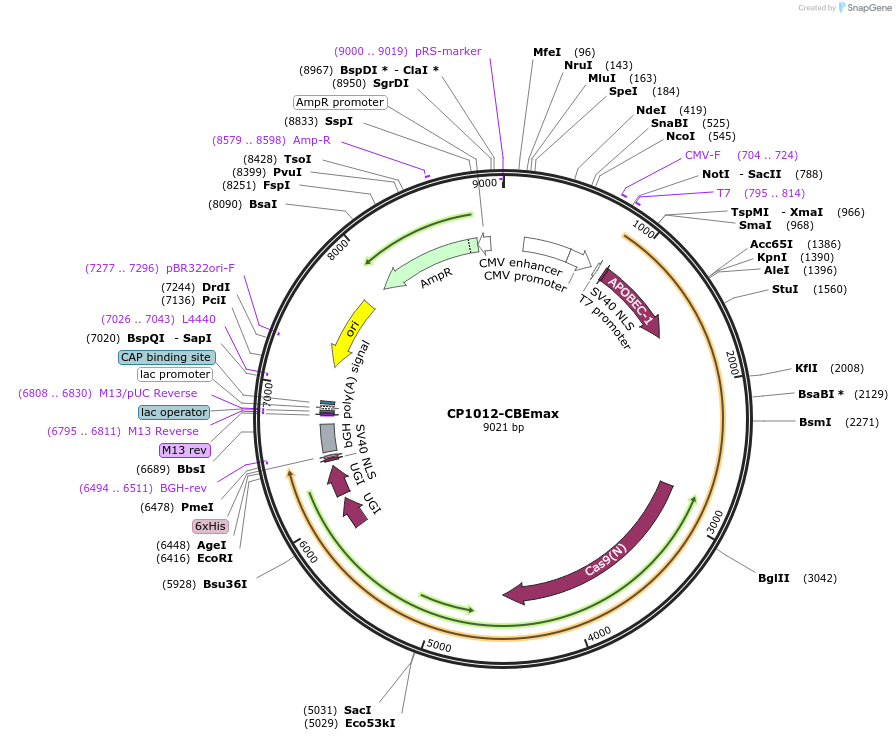
CP1012-CBEmax
Plasmid#119801PurposeC-to-T base editorDepositorInsertrApobec1-nSpCas9 (CP1012) -UGI-UGI
ExpressionMammalianAvailable SinceMay 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
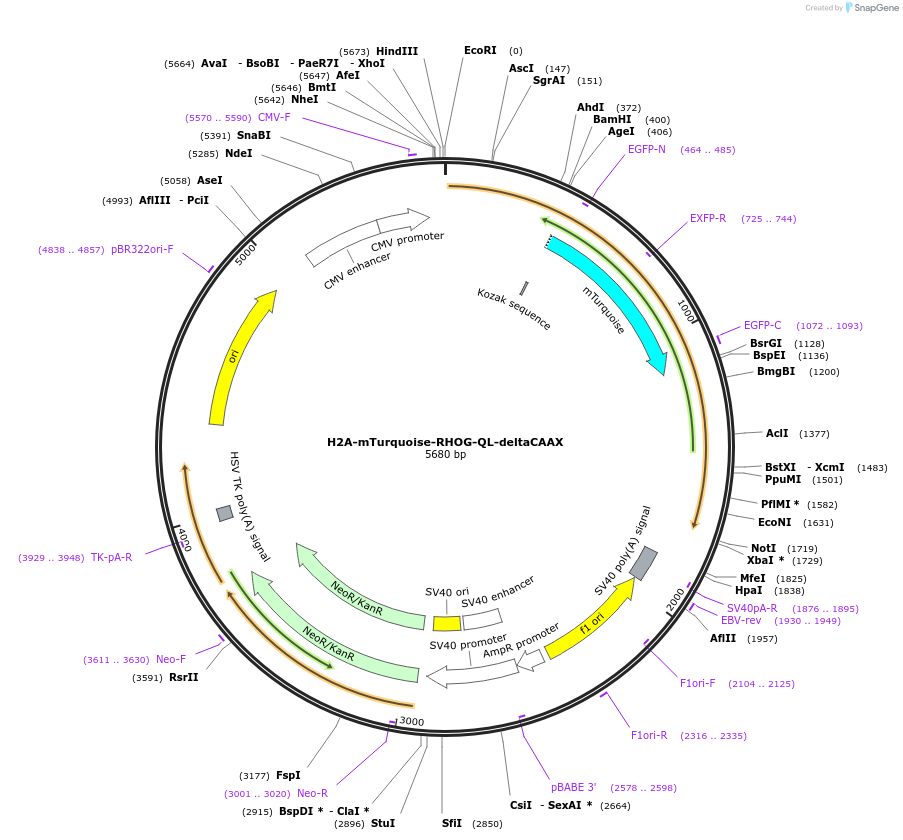
H2A-mTurquoise-RHOG-QL-deltaCAAX
Plasmid#176096PurposeConstitutively active, nuclear localized RhoGDepositorAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
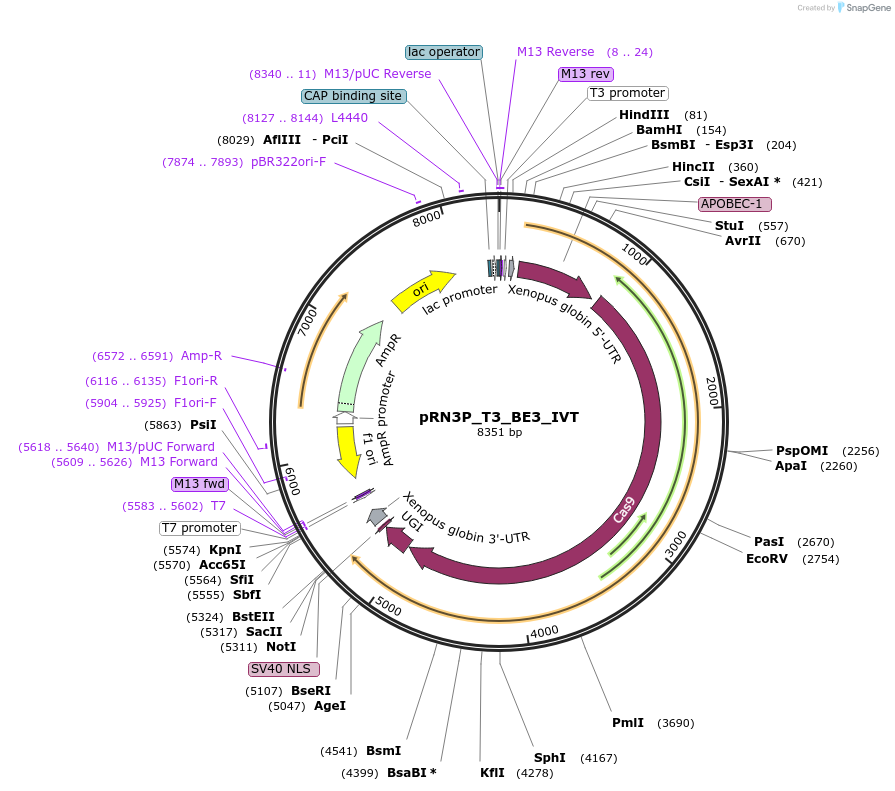
pRN3P_T3_BE3_IVT
Plasmid#177015PurposePlasmid to be used as DNA template for in-vitro RNA transcription of the BE3 base editor (C to T) by T3 RNA polymeraseDepositorInsertBE3
UseCRISPR; Vector for in-vitro transcriptionPromoterT3 promoterAvailable SinceDec. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
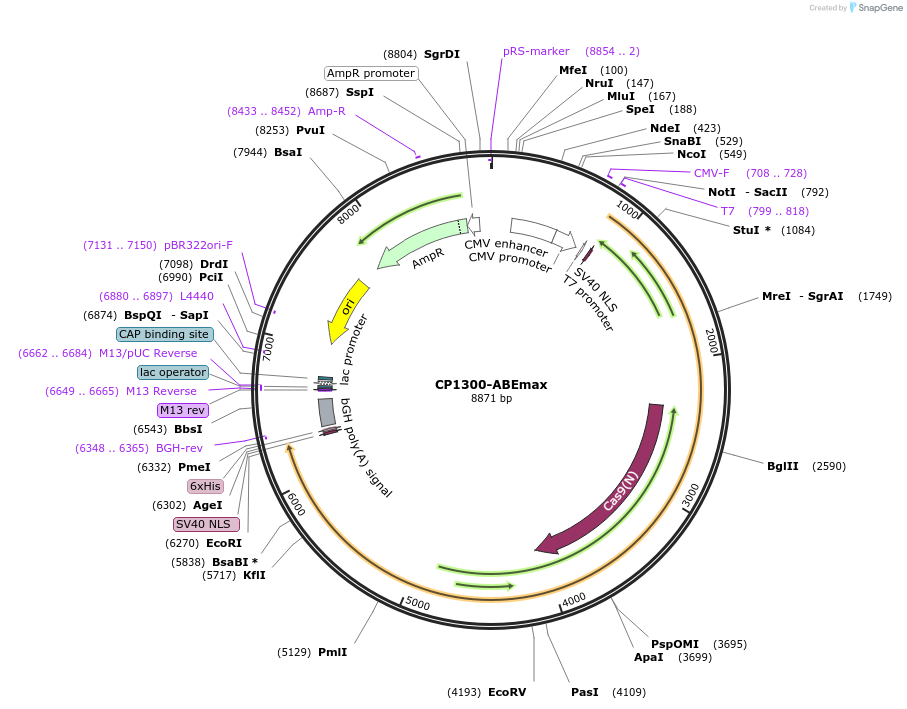
CP1300-ABEmax
Plasmid#119810PurposeA-to-G base editorDepositorInsertecTadA-ecTadA*(7.10)-nSpCas9 (CP1300)
ExpressionMammalianAvailable SinceMay 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
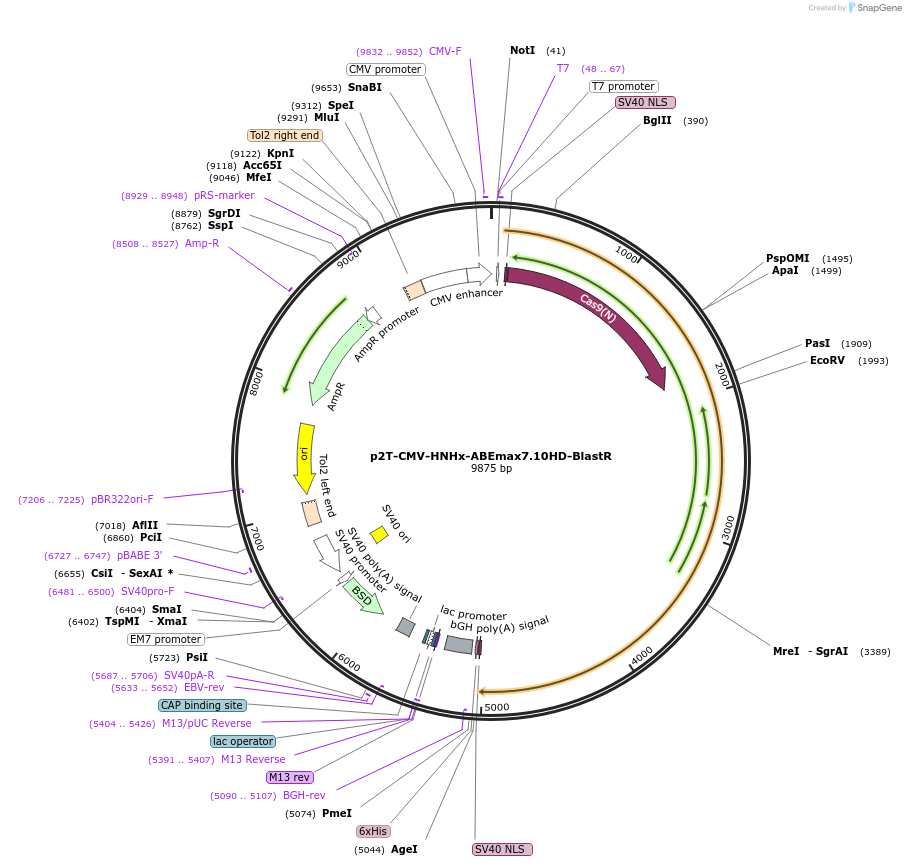
p2T-CMV-HNHx-ABEmax7.10HD-BlastR
Plasmid#176477PurposeA-to-G base editor with alternative targeting scopeDepositorInsertHNHx-ABEmax7.10HD
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceNov. 17, 2021AvailabilityAcademic Institutions and Nonprofits only -
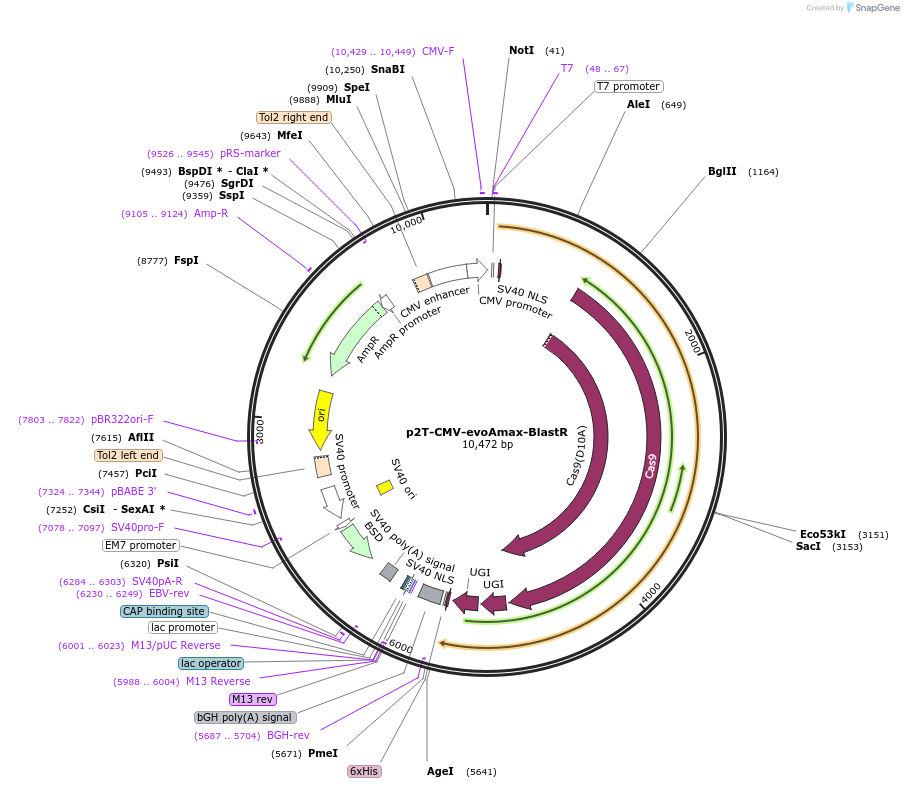
p2T-CMV-evoAmax-BlastR
Plasmid#152996PurposeC-to-T base editorDepositorInsertevoA-nSpCas9-UGI-UGI
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceAug. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
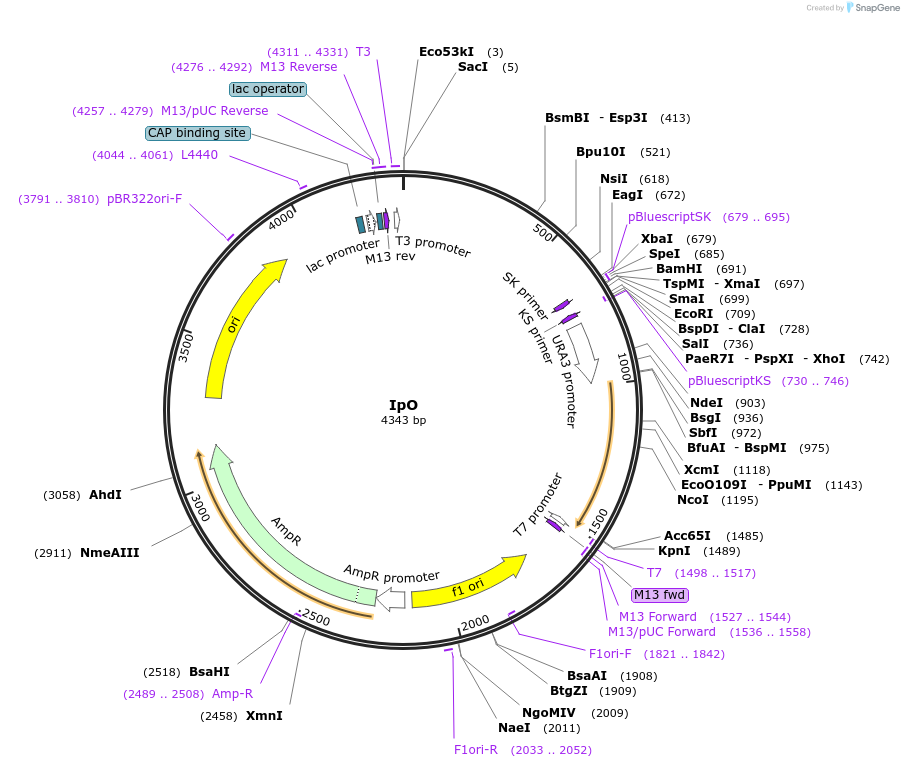
IpO
Plasmid#48233PurposeBase vector for PCR-based DNA transplant in yeast. DNA segments to be transplanted into the yeast genome are cloned into the MCS.DepositorInsertsURA3 3' fragment
URA3 5' fragment
UseRoutine cloning vectorMutation-242 upstream to URA3 ORF +495 and URA3 ORF from …PromoterURA3 promoter and noneAvailable SinceFeb. 10, 2014AvailabilityAcademic Institutions and Nonprofits only -
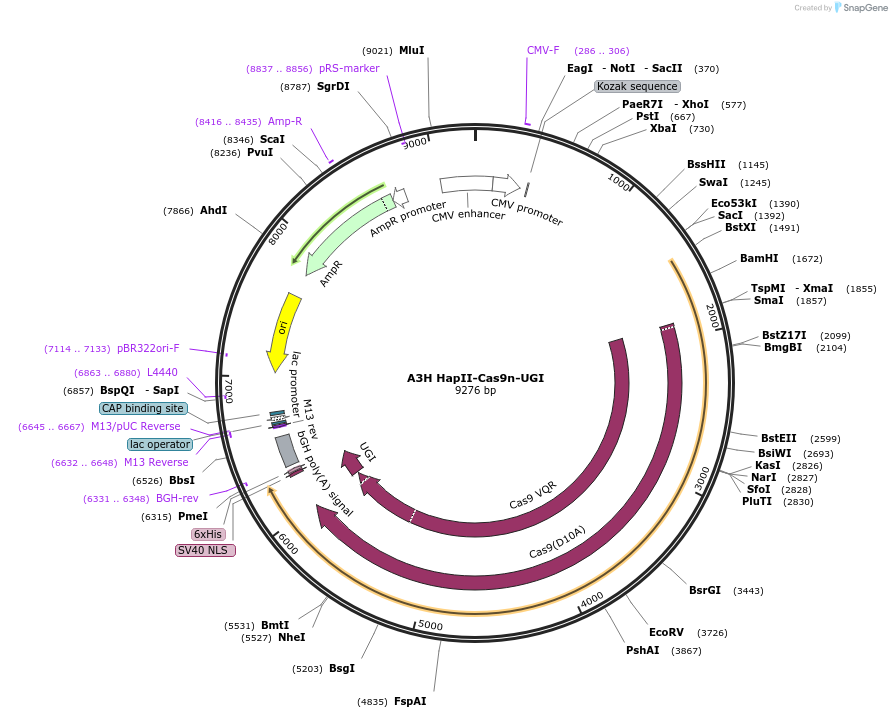
A3H HapII-Cas9n-UGI
Plasmid#119141PurposeBase editor made from APOBEC3H Haplotype IIDepositorInsertAPOBEC3H Haplotype II-Cas9 nickase-UGI
UseCRISPRMutationNonePromoterCMVAvailable SinceJune 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
A3H HapI-Cas9n-UGI
Plasmid#119140PurposeBase editor made from APOBEC3H Haplotype IDepositorInsertAPOBEC3H Haplotype I-Cas9 nickase-UGI
UseCRISPRMutationNonePromoterCMVAvailable SinceJune 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
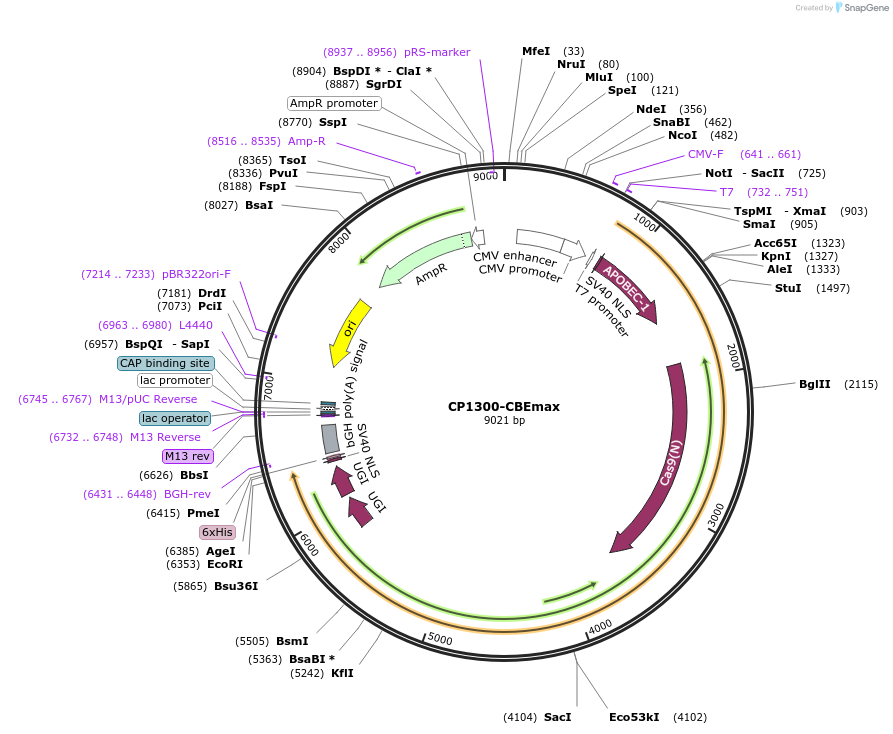
CP1300-CBEmax
Plasmid#119805PurposeC-to-T base editorDepositorInsertrApobec1-nSpCas9 (CP1300) -UGI-UGI
ExpressionMammalianAvailable SinceMay 22, 2019AvailabilityAcademic Institutions and Nonprofits only -
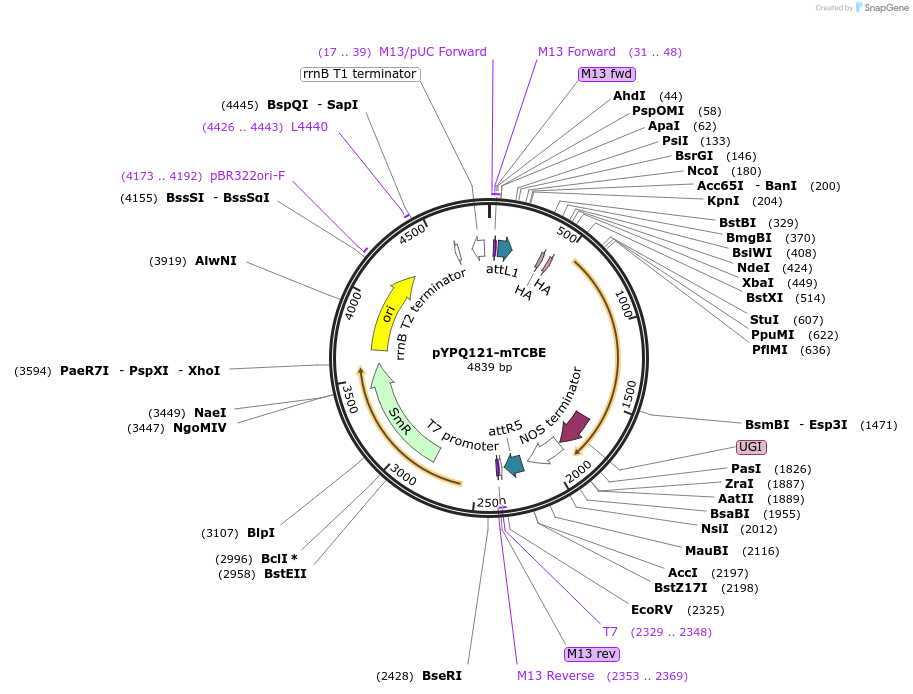
pYPQ121-mTCBE
Plasmid#218172PurposeExpress plastid monomeric TALE-linked CBE, composed of TALE, a catalytically impaired, full length DddA variant (E1347A), a human APOBEC3A variant (hA3A-Y130F), and UGIDepositorInsertTALE-Intron-hA3A-Y130F-DddA full (E1347A)-UGI
UsePlastid base editingExpressionPlantAvailable SinceJune 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
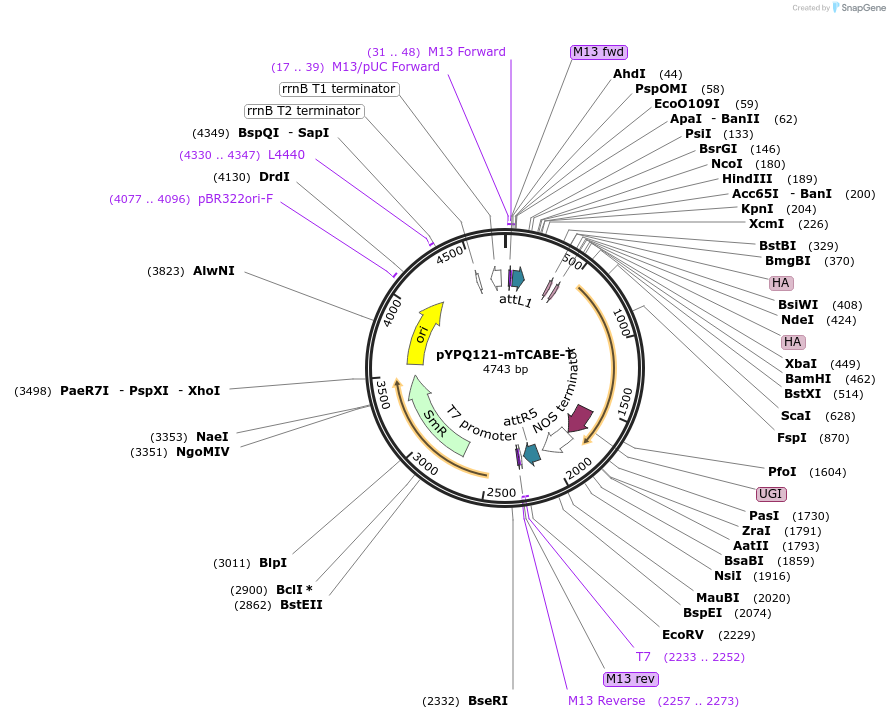
pYPQ121-mTCABE-T
Plasmid#218173PurposeExpress plastid monomeric TALE-linked CABE-T, composed of TALE, a catalytically impaired, full length DddA variant (E1347A), a TadA-derived deaminase, and UGIDepositorInsertTALE-Intron-TadAC-DddA full (E1347A)-UGI
UsePlastid base editingExpressionPlantAvailable SinceJune 26, 2024AvailabilityAcademic Institutions and Nonprofits only -
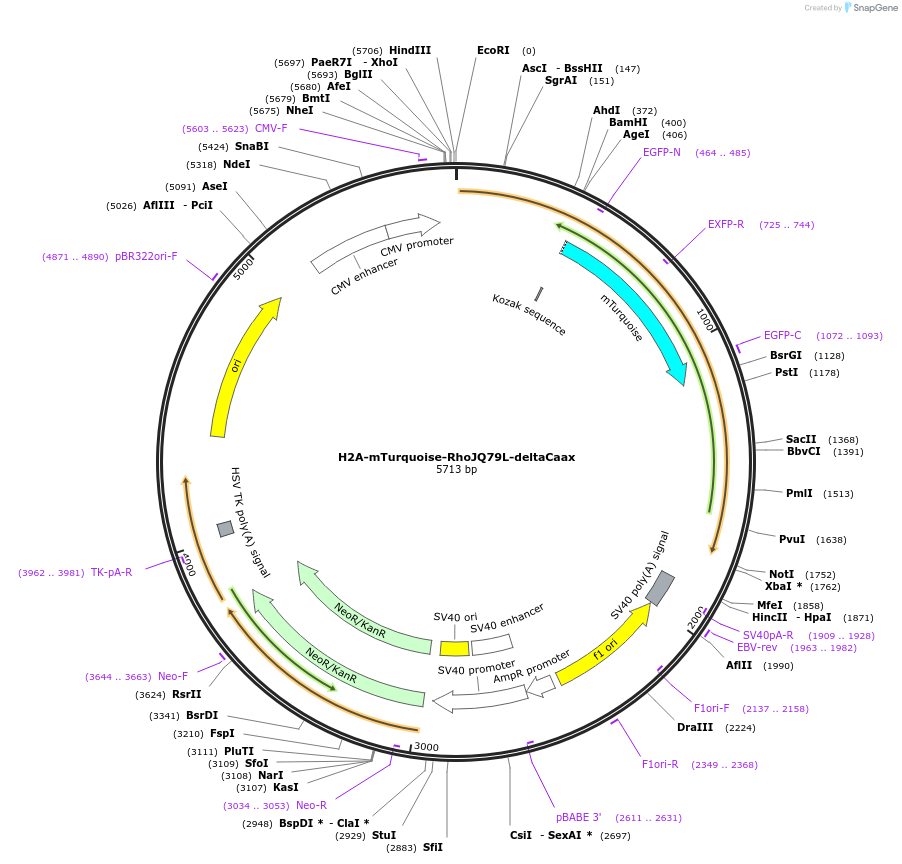
H2A-mTurquoise-RhoJQ79L-deltaCaax
Plasmid#176110PurposeConstitutively active, nucelar localized RhoJDepositorAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
TOMM20-mTurquoise-RhoG-QL-deltaCaaX
Plasmid#176122PurposeConstitutively active, mitochondiral localized RhoGDepositorAvailable SinceOct. 29, 2021AvailabilityAcademic Institutions and Nonprofits only -
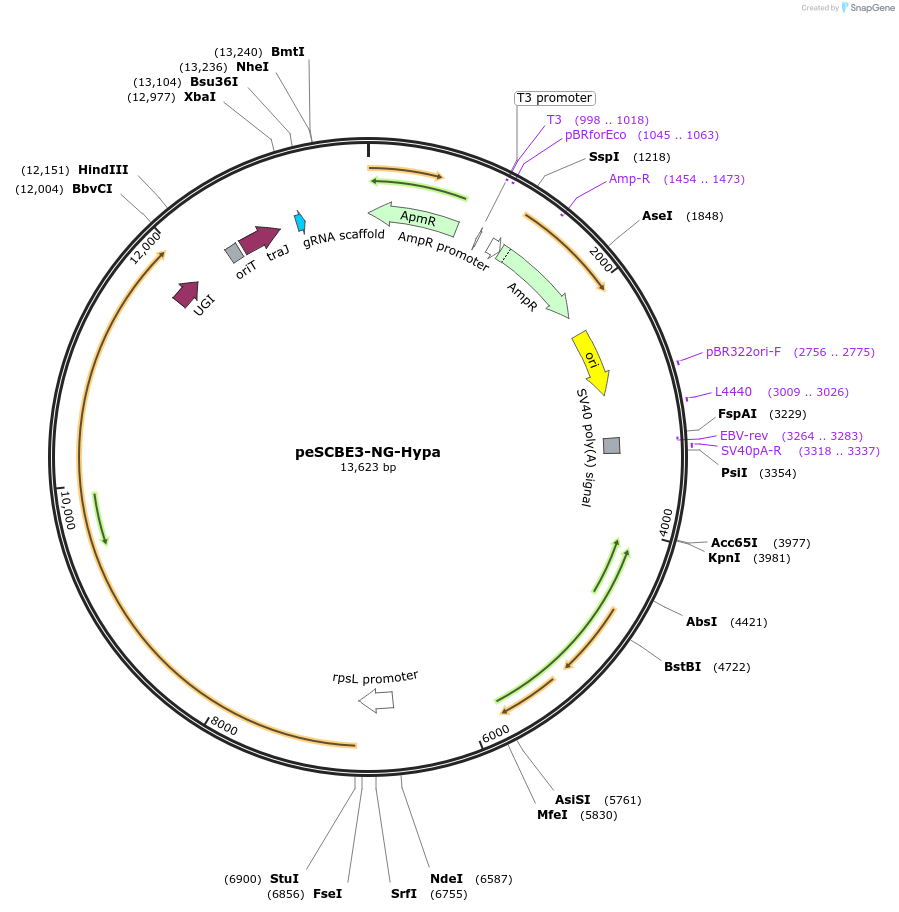
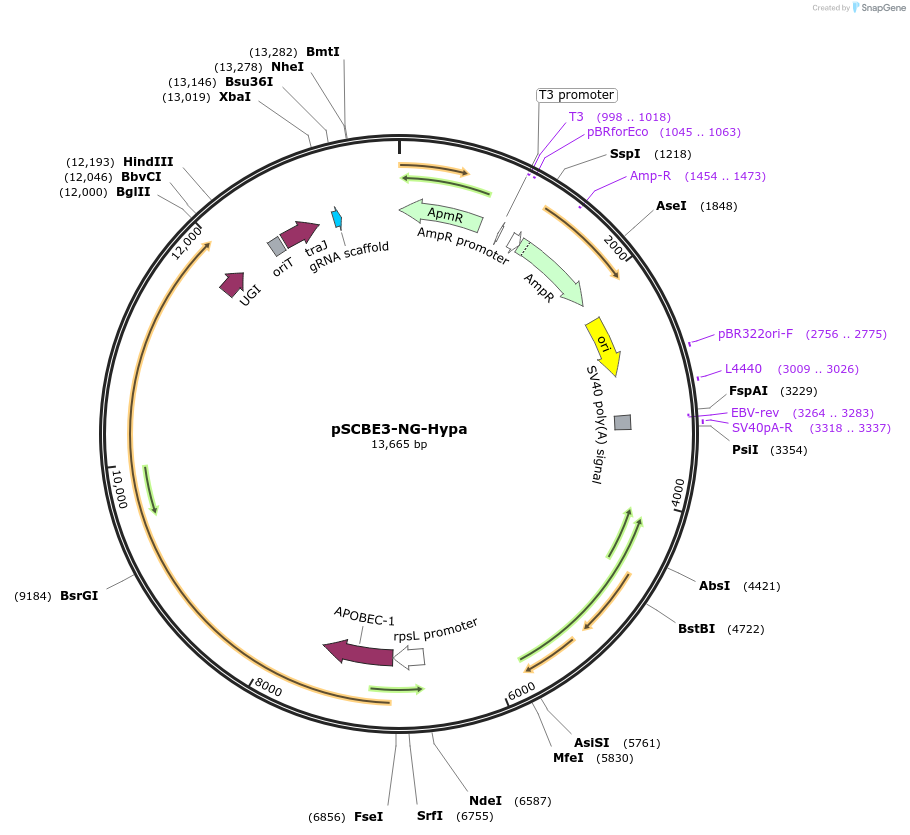
peSCBE3-NG-Hypa
Plasmid#218165PurposeThis plasmid harbors the base editor eSCBE3-NG-Hypa along with an sgRNA cloning cassette, facilitating high-efficiency and high-fidelity cytosine base editing at targets bearing NG PAM in StreptomycesDepositorInsertsescbe3-NG-Hypa
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutation in the portion of deaminase hAPOBEC3A : …PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
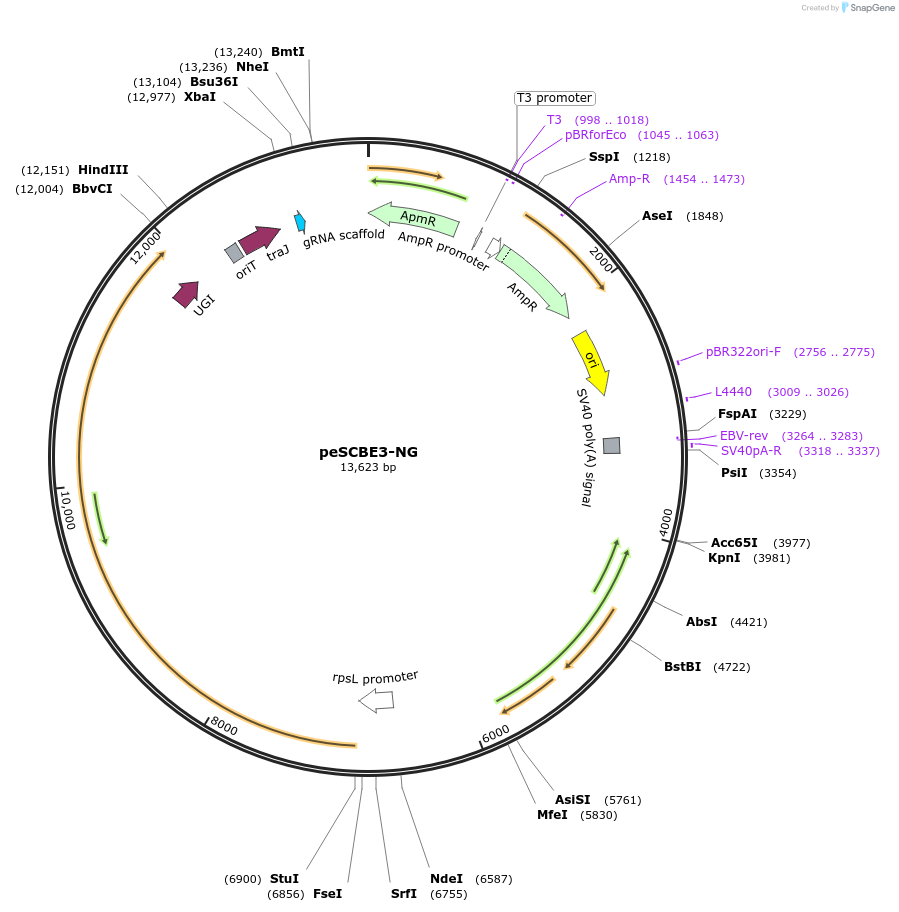
peSCBE3-NG
Plasmid#218163PurposeThis plasmid harbors the base editor eSCBE3-NG along with an sgRNA cloning cassette, facilitating high-efficiency cytosine base editing at targets bearing NG PAM in Streptomyces.DepositorInsertsescbe3-NG
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutation in the portion of deaminase hAPOBEC3A : …PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
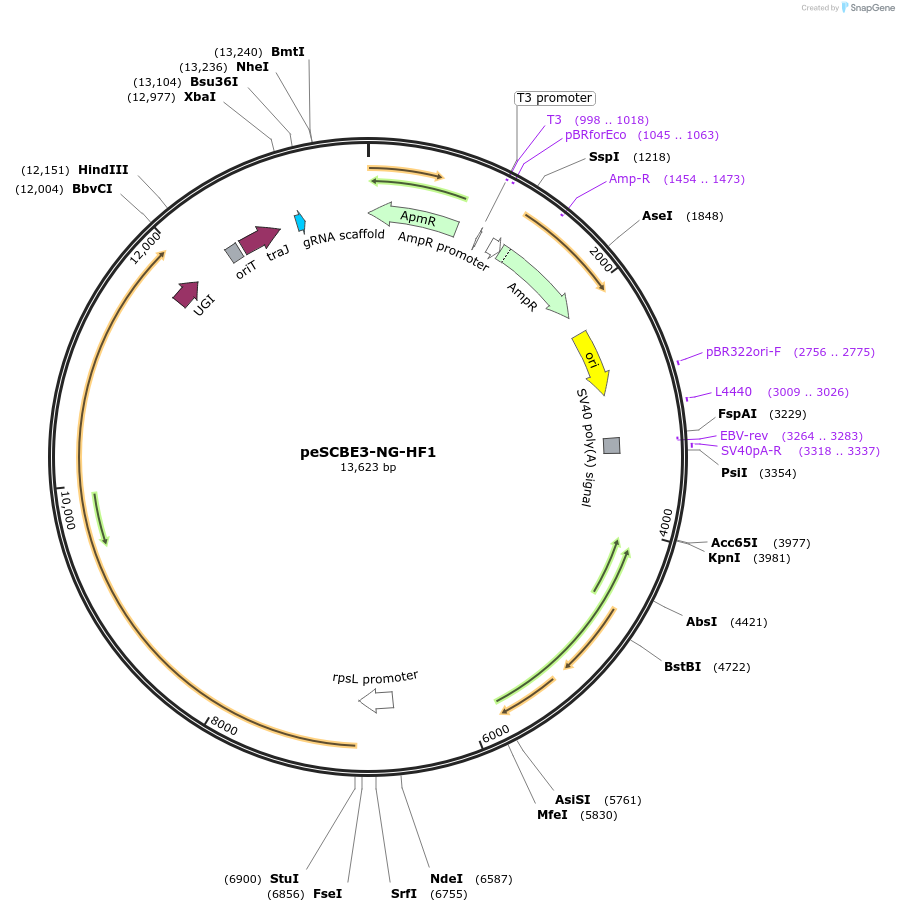
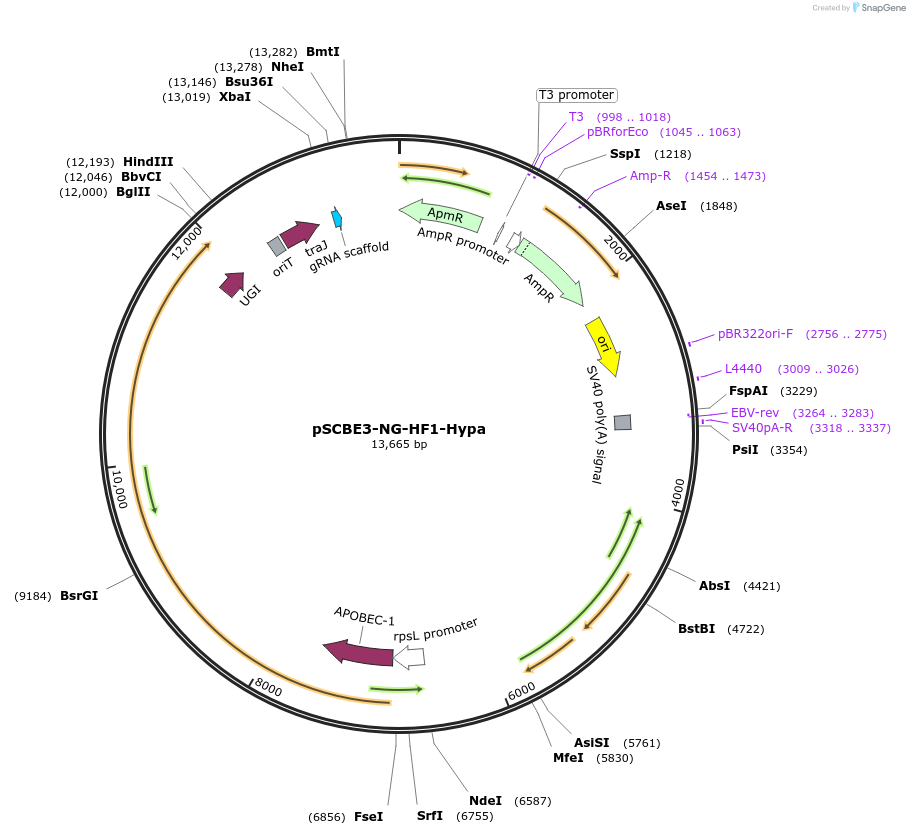
peSCBE3-NG-HF1
Plasmid#218164PurposeThis plasmid harbors the base editor eSCBE3-NG-HF1 along with an sgRNA cloning cassette, facilitating high-efficiency and high-fidelity cytosine base editing at targets bearing NG PAM in Streptomyces.DepositorInsertsescbe3-NG-HF1
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutation in the portion of deaminase hAPOBEC3A : …PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
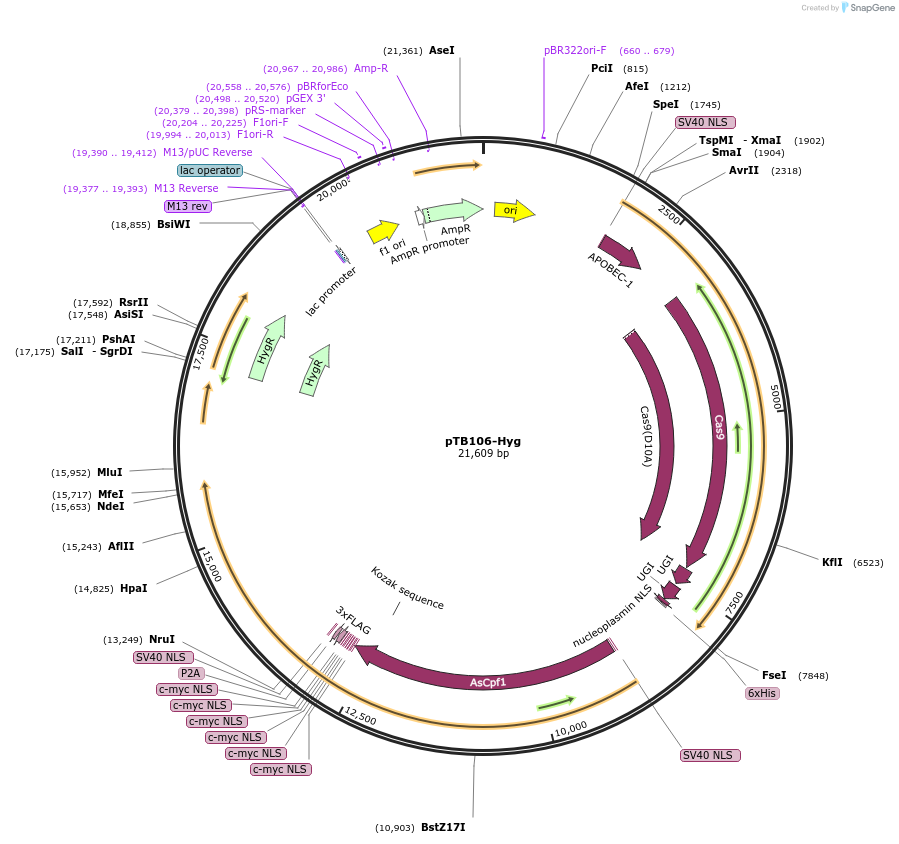
pTB106-Hyg
Plasmid#236779PurposeExpression of Cas9 cytosine base editor (CBE), T7 RNAP, Cas12a and hygromycin selection marker. The CBE contains a ssDNA-DBD from the L. major RAD51 protein. This plasmid is transfected as episome.DepositorInserthyBE4max (CBE); Cas12a (Acidaminococcus sp. BV3L6)-P2A-T7 RNAP
UseCRISPRAvailable SinceDec. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
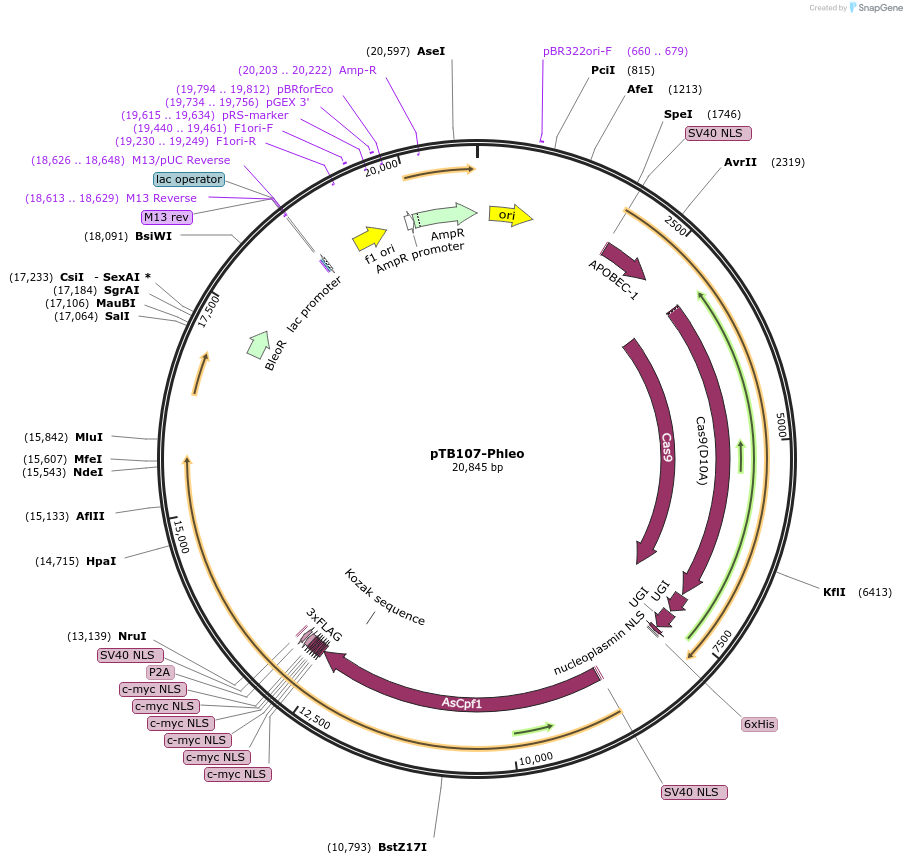
pTB107-Phleo
Plasmid#236782PurposeExpression of Cas9 cytosine base editor (CBE), T7 RNAP, Cas12a and phleomycin selection marker. The CBE contains a ssDNA-DBD from the H. sapiens RAD51 protein. This plasmid is transfected as episome.DepositorInserthyBE4max (CBE); Cas12a (Acidaminococcus sp. BV3L6)-P2A-T7 RNAP
UseCRISPRAvailable SinceDec. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
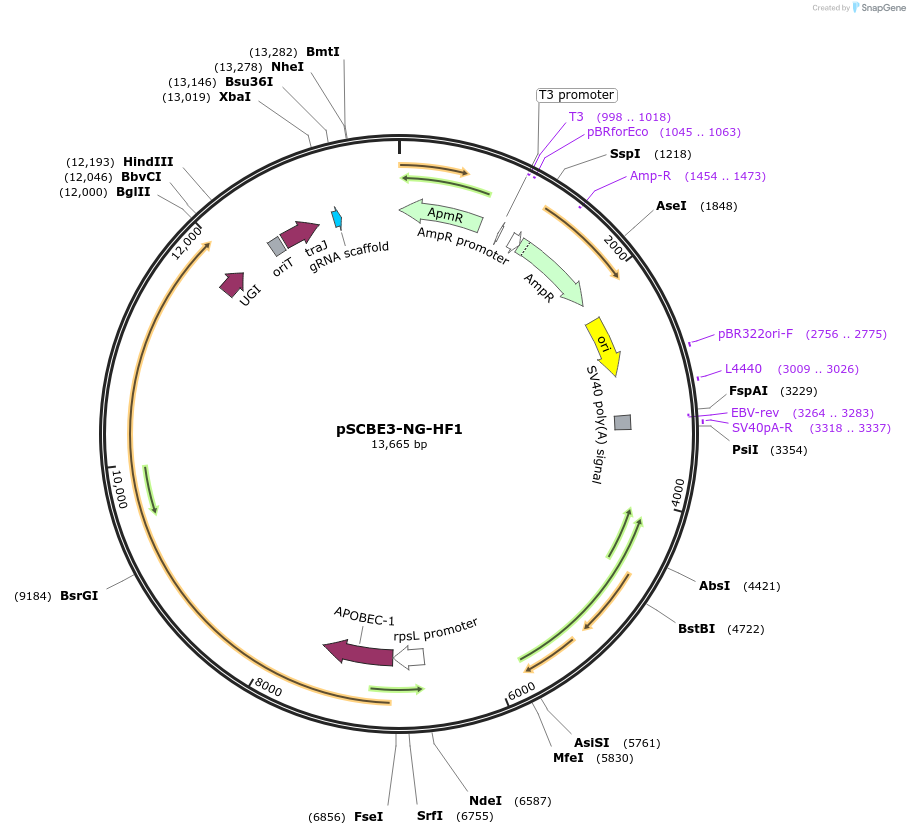
pSCBE3-NG-HF1
Plasmid#218160PurposeThis plasmid harbors the base editor SCBE3-NG-HF1 along with an sgRNA cloning cassette, facilitating high-fidelity cytosine base editing at targets bearing NG PAM in Streptomyces.DepositorInsertsscbe3-NG-HF1
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutations in the portion of SpCas9 : D10A / N497A…PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
pSCBE3-NG-Hypa
Plasmid#218161PurposeThis plasmid harbors the base editor SCBE3-NG-Hypa along with an sgRNA cloning cassette, facilitating high-fidelity cytosine base editing at targets bearing NG PAM in Streptomyces.DepositorInsertsscbe3-NG-Hypa
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutations in the portion of SpCas9 : D10A / N692A…PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
pSCBE3-NG-HF1-Hypa
Plasmid#218162PurposeThis plasmid harbors the base editor SCBE3-NG-HF1-Hypa along with an sgRNA cloning cassette, facilitating high-fidelity cytosine base editing at targets bearing NG PAM in Streptomyces.DepositorInsertsscbe3-NG-HF1-Hypa
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutations in the portion of SpCas9 : D10A / N497A…PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
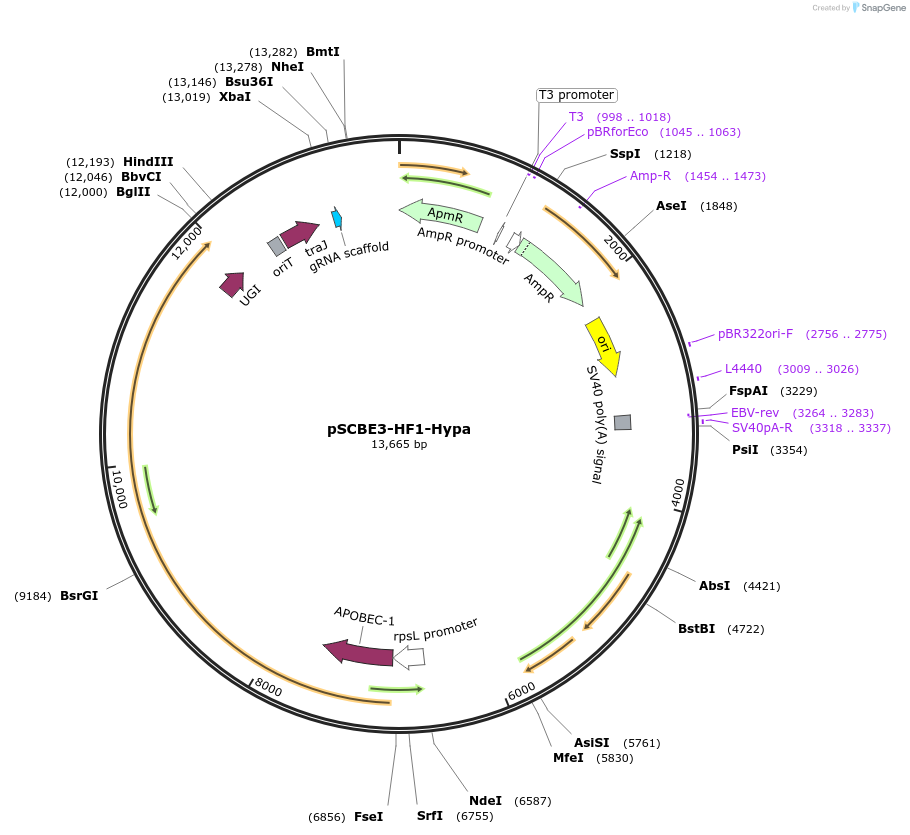
pSCBE3-HF1-Hypa
Plasmid#218158PurposeThis plasmid harbors the base editor SCBE3-HF1-Hypa along with an sgRNA cloning cassette, facilitating high-fidelity cytosine base editing in Streptomyces.DepositorInsertsscbe3-HF1-Hypa
a programmable sgRNA cloning cassette
UseCRISPR; Sgrna transcriptionExpressionBacterialMutationMutations in the portion of SpCas9 : D10A / N497A…PromoterrpsL promoterAvailable SinceMay 13, 2024AvailabilityAcademic Institutions and Nonprofits only -
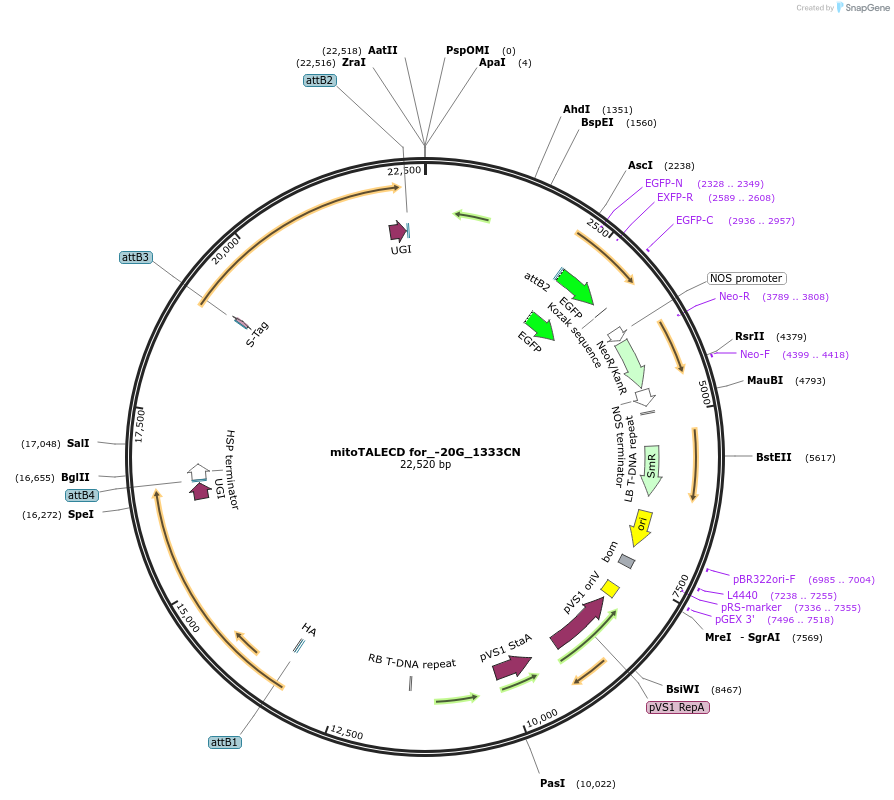
mitoTALECD for_-20G_1333CN
Plasmid#186200PurposeFor genome editing (base editing) of Arabidopsis mitochondria, targeted 20th G to A of RNA editing site of ATP1 by OTP87DepositorInsertsmitochondrial targeted TALE left hand with 1333C of Cytidine Deaminase (mitoTALECD)
mitochondrial targeted TALE right hand with 1333N of Cytidine Deaminase (mitoTALECD)
UseSynthetic Biology; Ti plasmid for plant transform…Available SinceAug. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
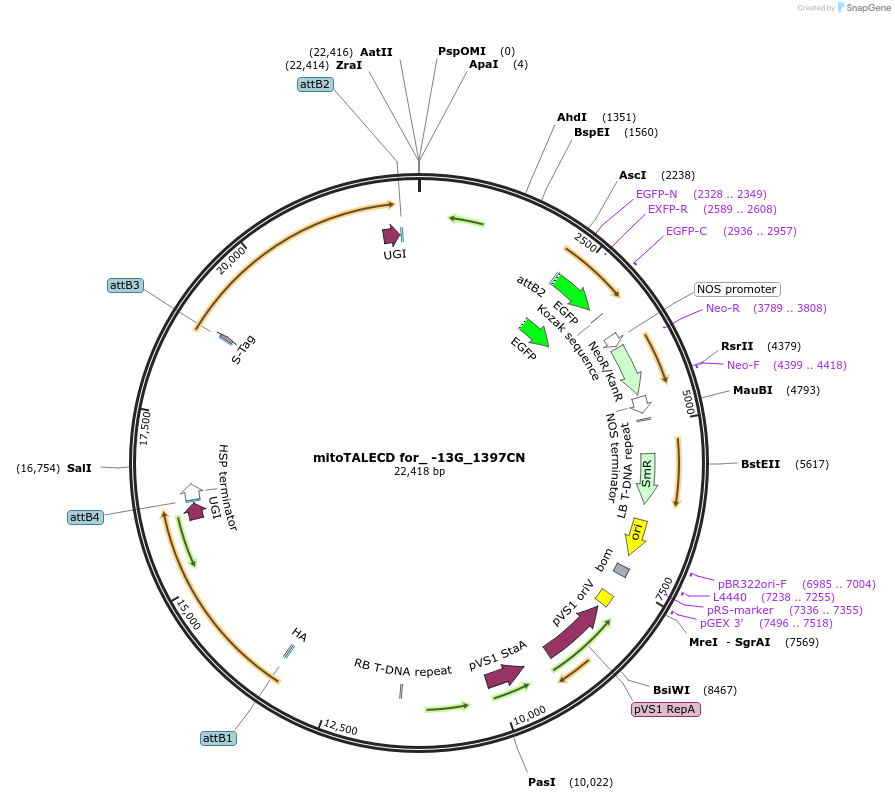
mitoTALECD for_ -13G_1397CN
Plasmid#186199PurposeFor genome editing (base editing) of Arabidopsis mitochondria, targeted -13th G to A of RNA editing site of ATP1 by OTP87DepositorInsertsmitochondrial targeted TALE left hand with 1397C of Cytidine Deaminase (mitoTALECD)
mitochondrial targeted TALE right hand with 1397N of Cytidine Deaminase (mitoTALECD)
UseSynthetic Biology; Ti plasmid for plant transform…Available SinceAug. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
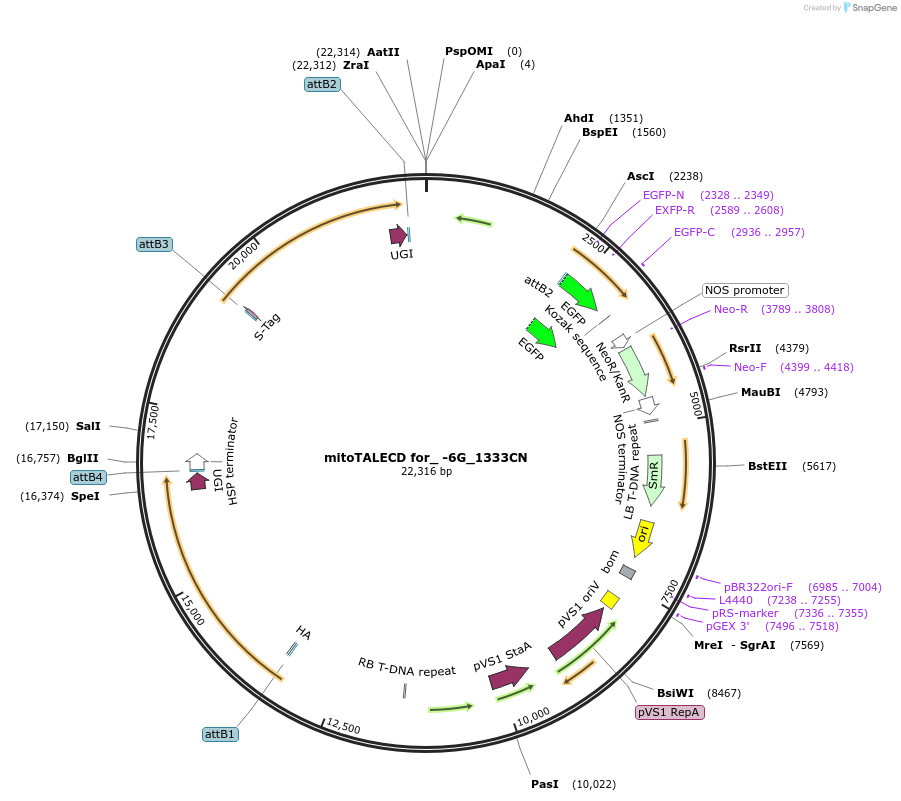
mitoTALECD for_ -6G_1333CN
Plasmid#186198PurposeFor genome editing (base editing) of Arabidopsis mitochondria, targeted -6th G to A of RNA editing site of ATP1 by OTP87DepositorInsertsmitochondrial targeted TALE left hand with 1333C of Cytidine Deaminase (mitoTALECD)
mitochondrial targeted TALE right hand with 1333N of Cytidine Deaminase (mitoTALECD)
UseSynthetic Biology; Ti plasmid for plant transform…Available SinceAug. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
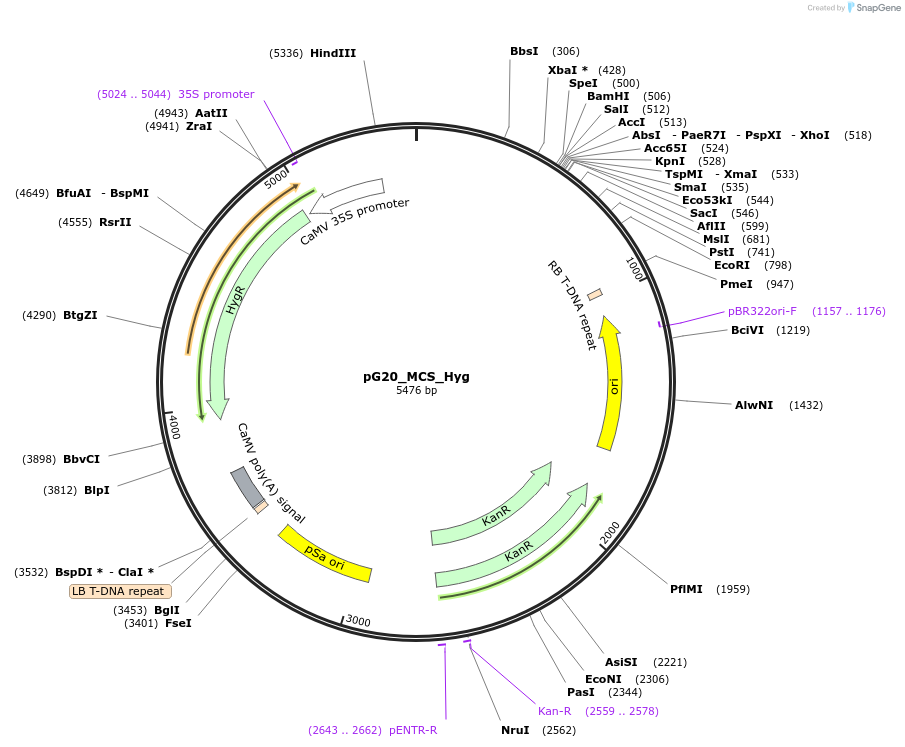
pG20_MCS_Hyg
Plasmid#159709PurposepGREEN-based cloning construct for expression in plantaDepositorTypeEmpty backboneExpressionPlantAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
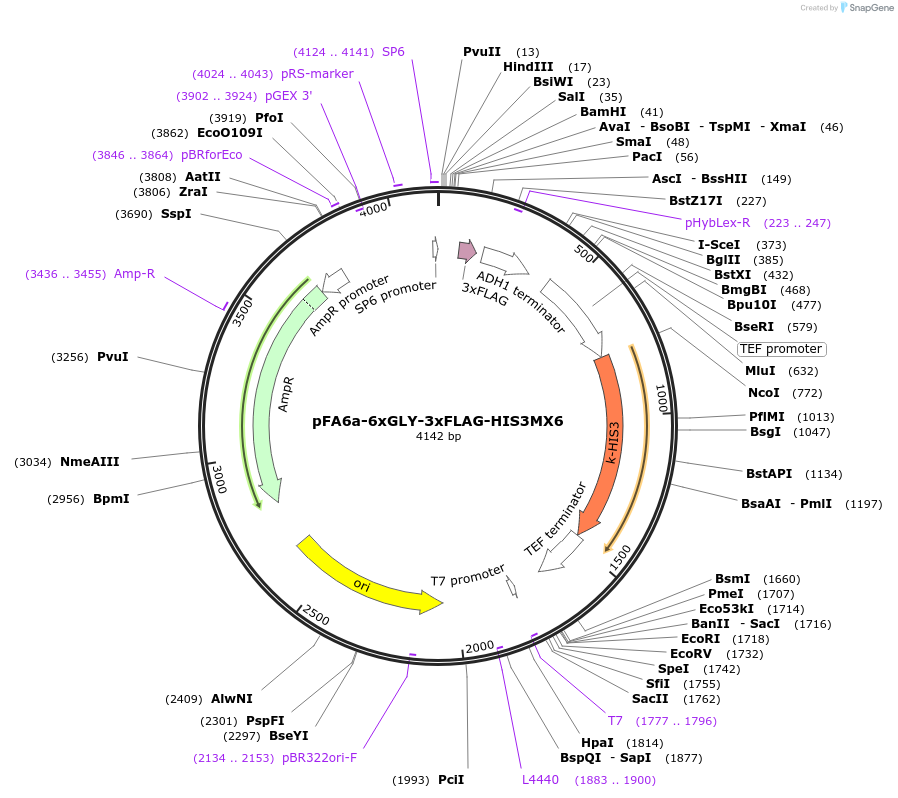
pFA6a-6xGLY-3xFLAG-HIS3MX6
Plasmid#20753DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
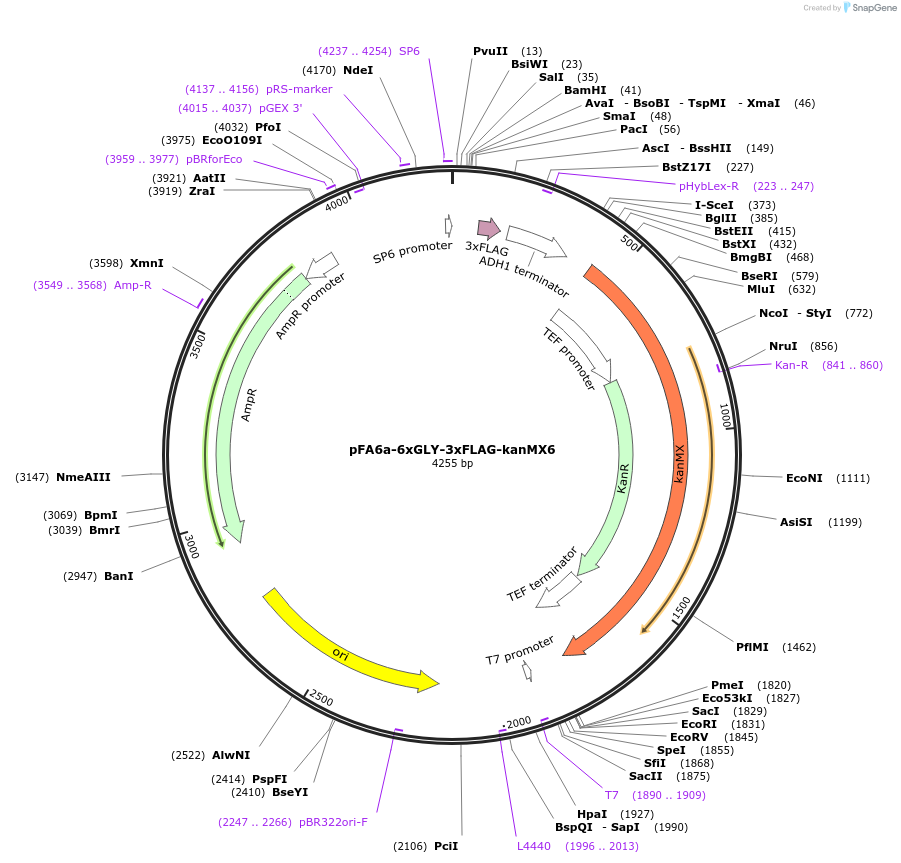
pFA6a-6xGLY-3xFLAG-kanMX6
Plasmid#20754DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
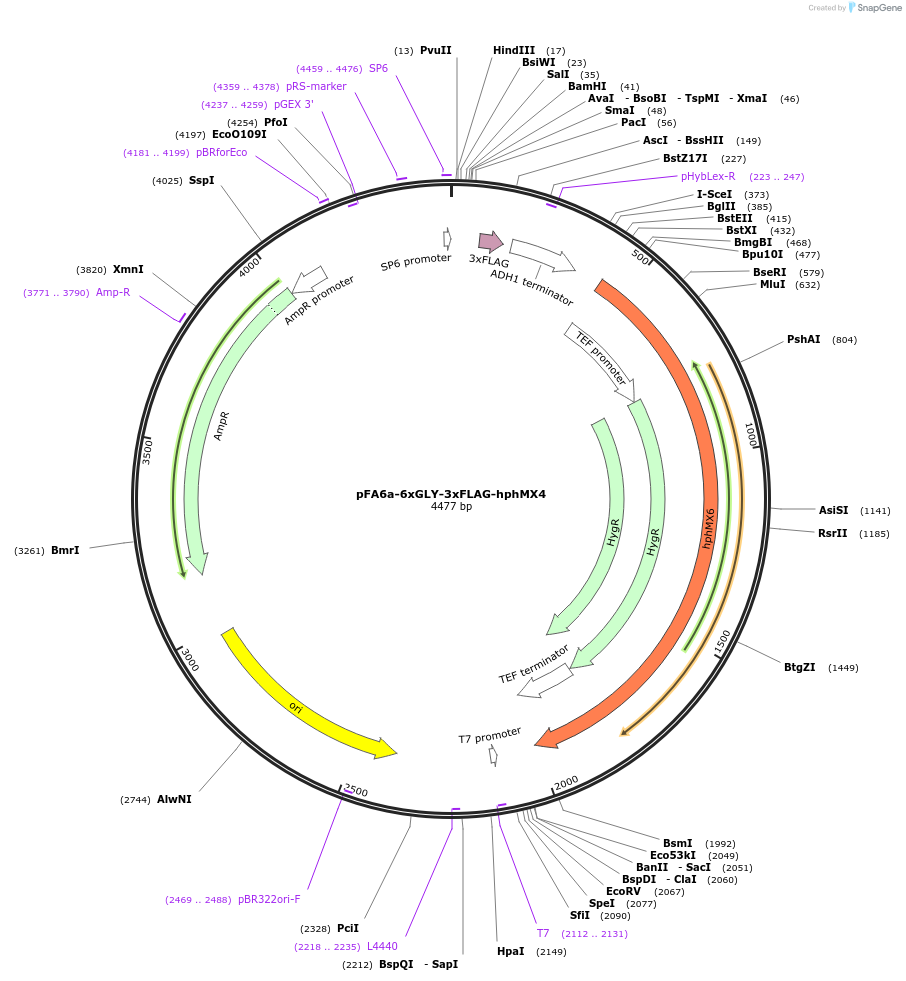
pFA6a-6xGLY-3xFLAG-hphMX4
Plasmid#20755DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
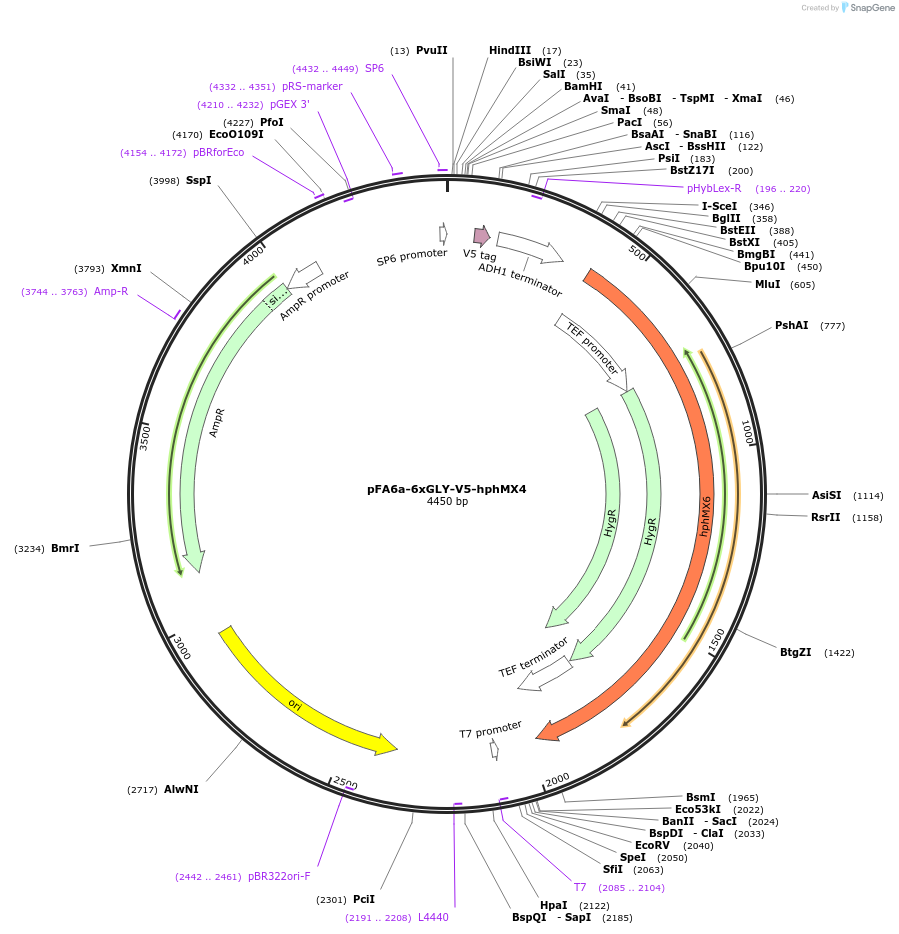
pFA6a-6xGLY-V5-hphMX4
Plasmid#20779DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
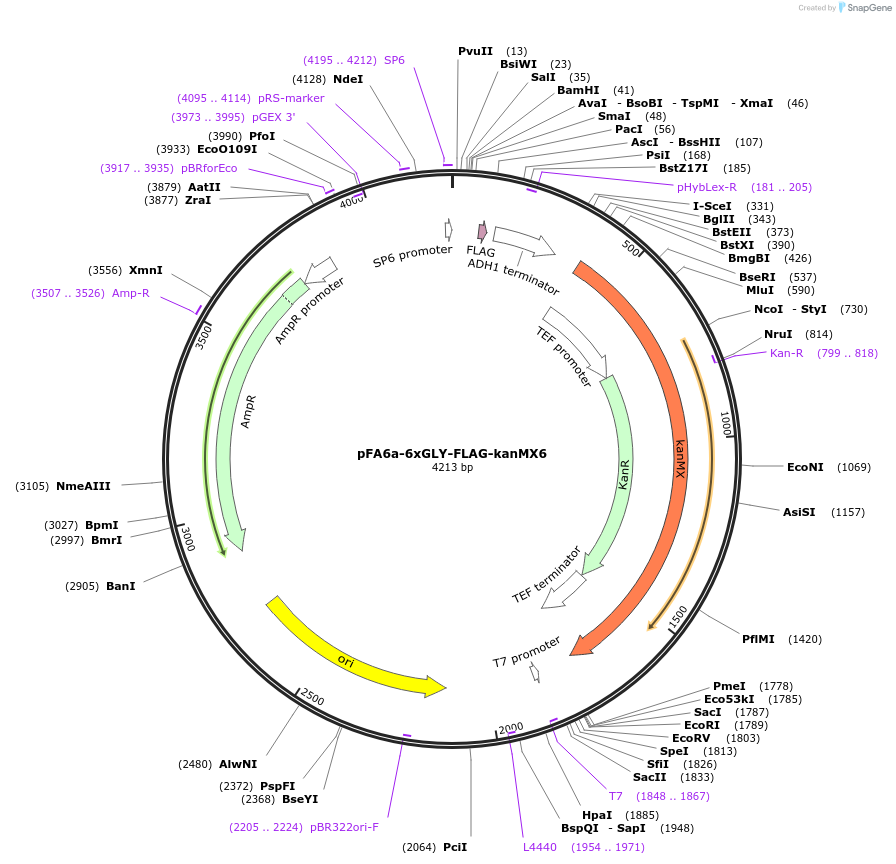
pFA6a-6xGLY-FLAG-kanMX6
Plasmid#20751DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
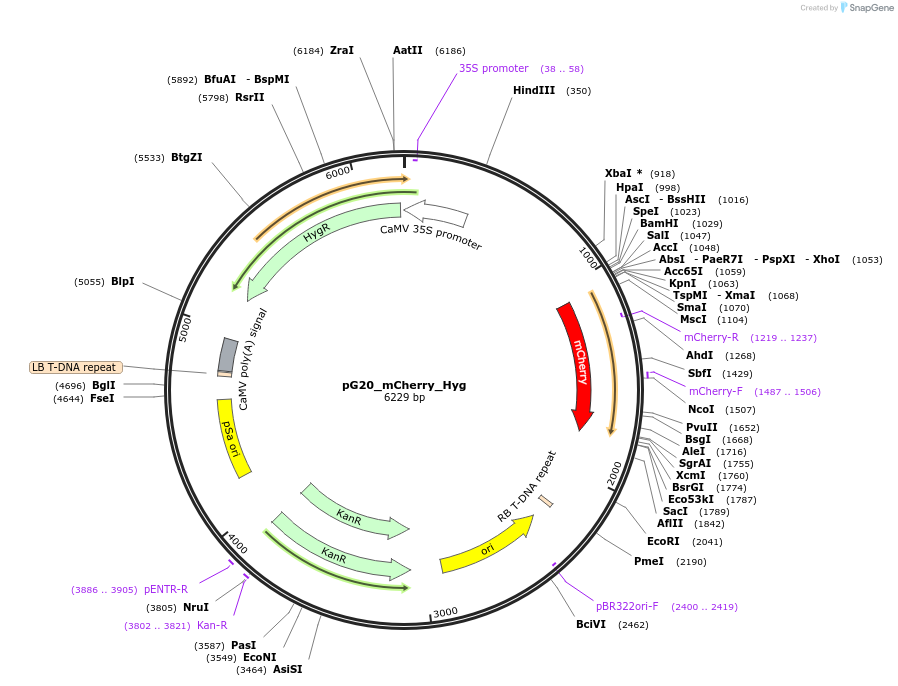
pG20_mCherry_Hyg
Plasmid#159701PurposepGREEN-based cloning construct for expression in plantaDepositorTypeEmpty backboneTagsmCherryExpressionPlantAvailable SinceOct. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
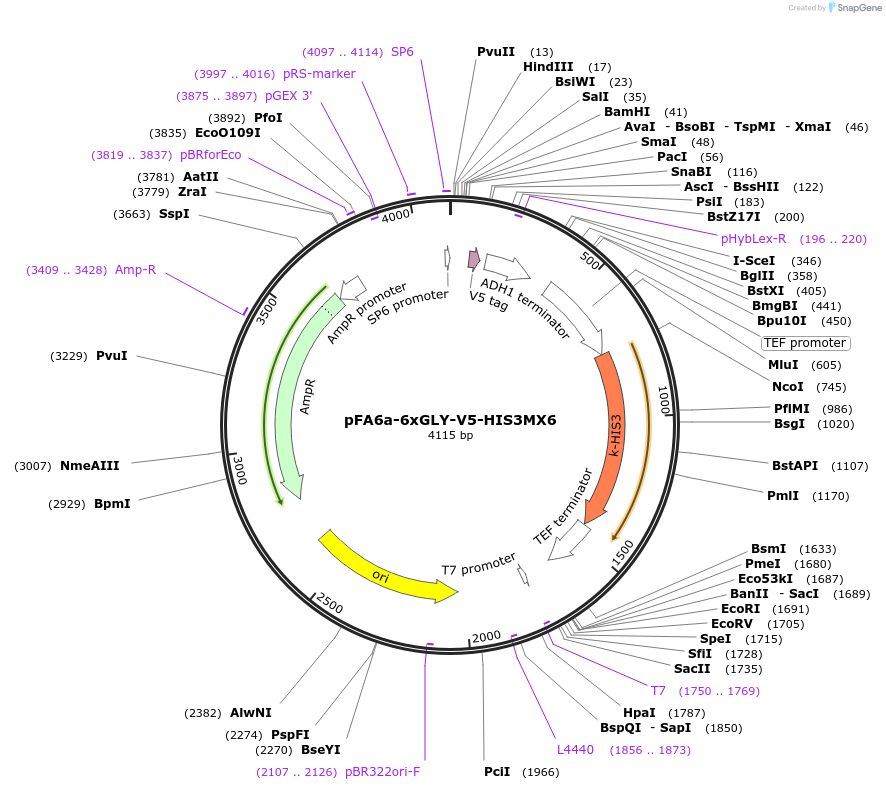
pFA6a-6xGLY-V5-HIS3MX6
Plasmid#20777DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 27, 2009AvailabilityAcademic Institutions and Nonprofits only -
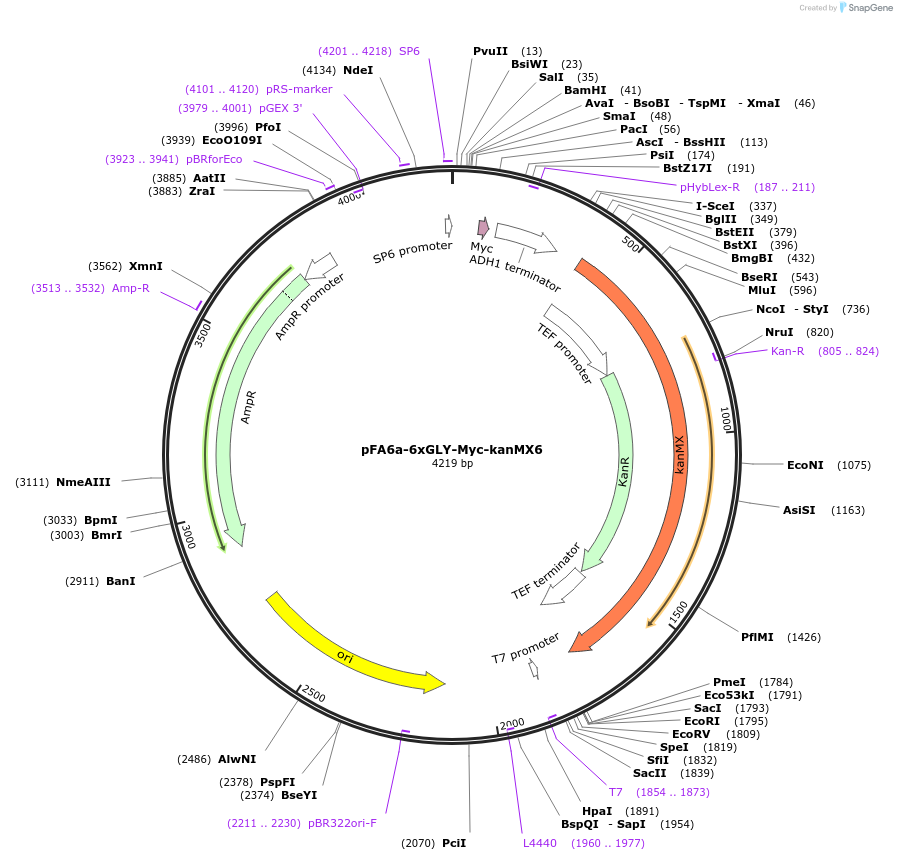
pFA6a-6xGLY-Myc-kanMX6
Plasmid#20769DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
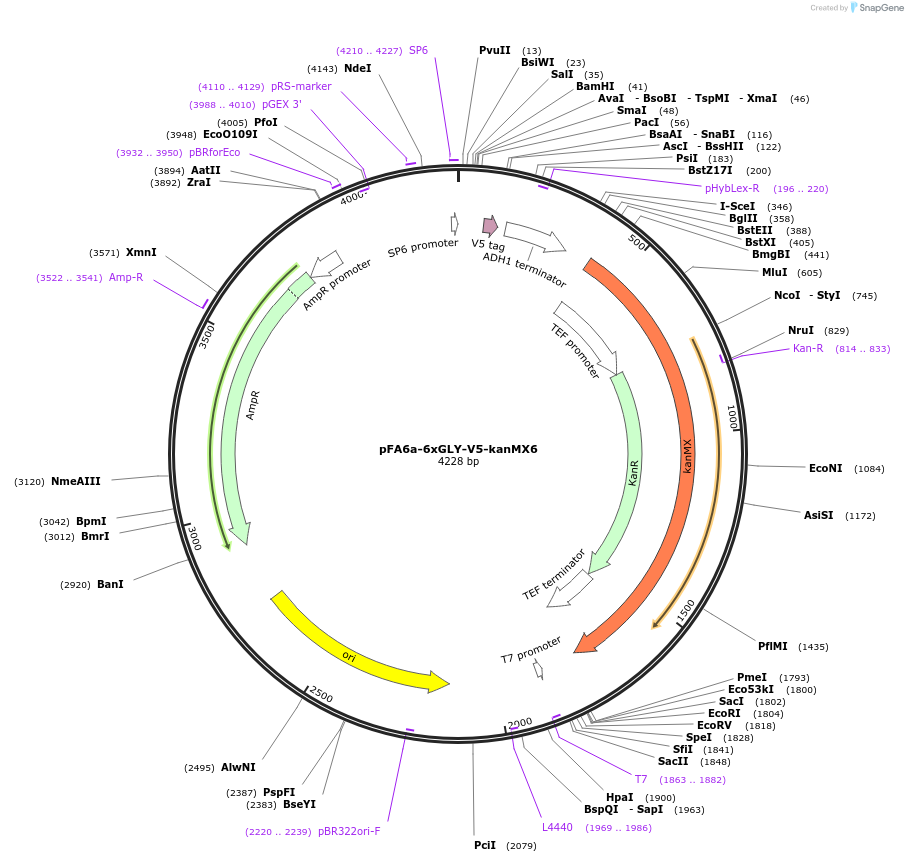
pFA6a-6xGLY-V5-kanMX6
Plasmid#20780DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
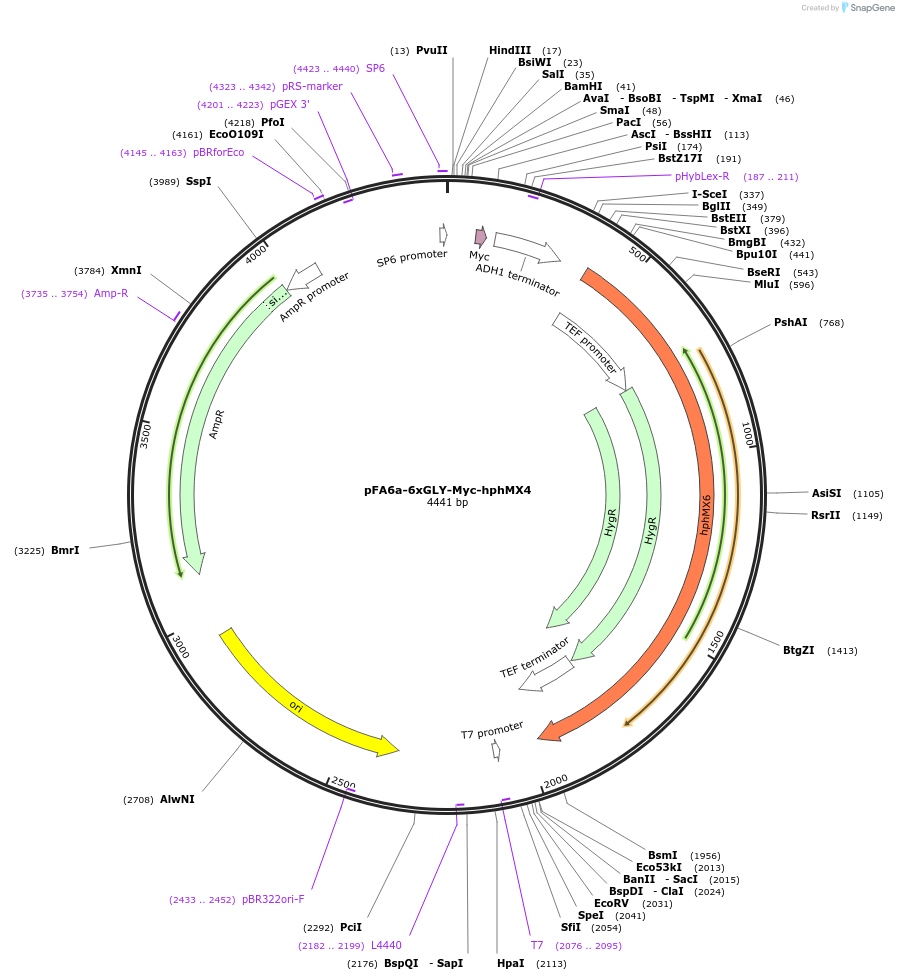
pFA6a-6xGLY-Myc-hphMX4
Plasmid#20770DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only -
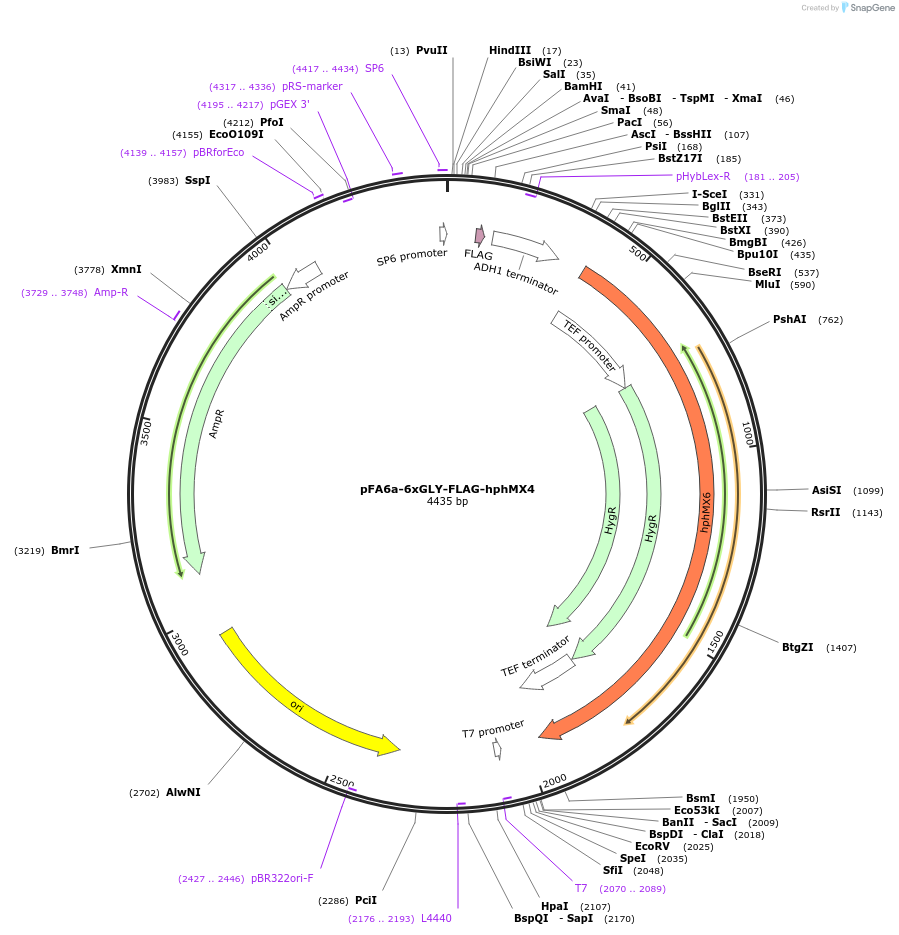
pFA6a-6xGLY-FLAG-hphMX4
Plasmid#20752DepositorTypeEmpty backboneUsePcr-based c-terminal taggingExpressionYeastAvailable SinceMarch 23, 2009AvailabilityAcademic Institutions and Nonprofits only