We narrowed to 40,628 results for: ANT
-
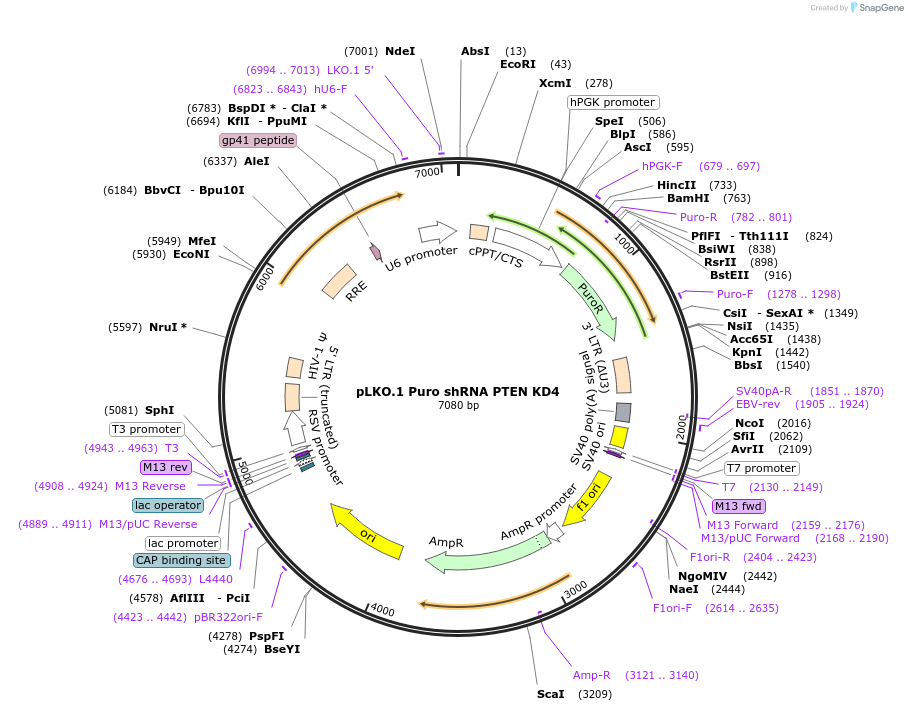
Plasmid#162010PurposePTEN shRNADepositorInsertPTEN (PTEN Canis Lupus)
UseLentiviralAvailable SinceDec. 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
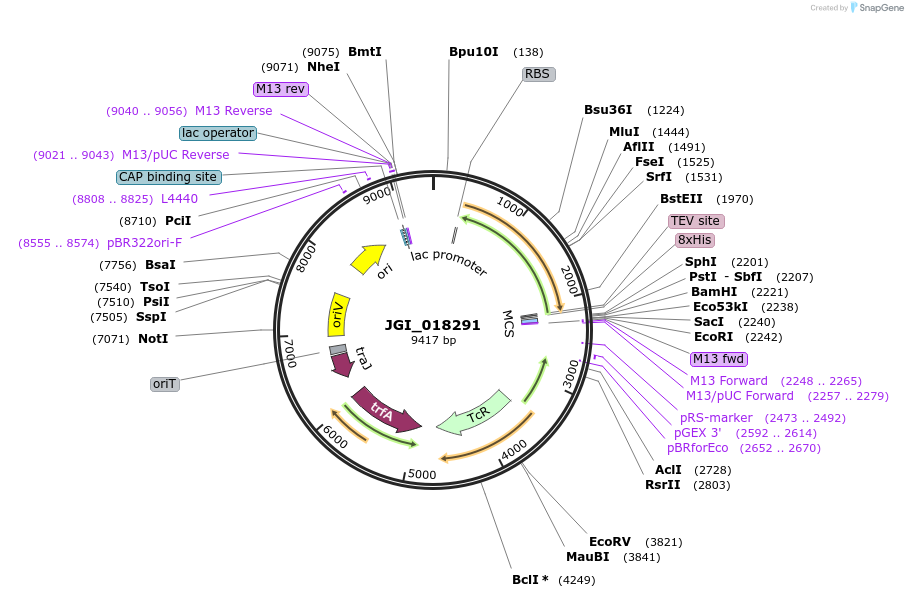
JGI_018291
Plasmid#158823PurposeRare earth element alcohol dehydrogenasesDepositorInsertJanthino_4798
UseSynthetic BiologyAvailable SinceNov. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
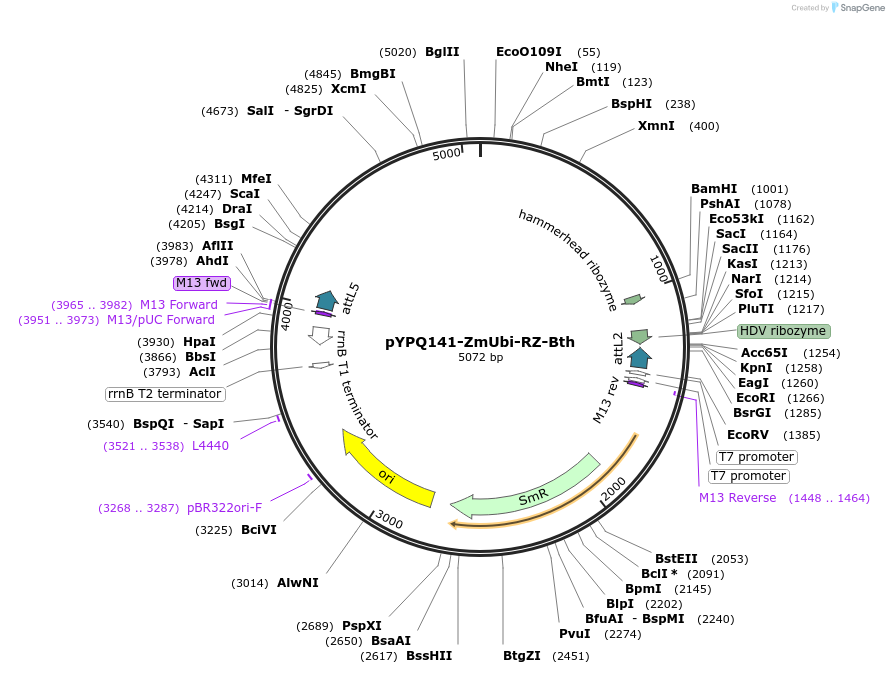
pYPQ141-ZmUbi-RZ-Bth
Plasmid#129686Purpose(Empty backbone) Gateway entry clone with sgRNA under ZmUbi promoterDepositorInsertEmpty Backbone
UseCRISPRExpressionPlantAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
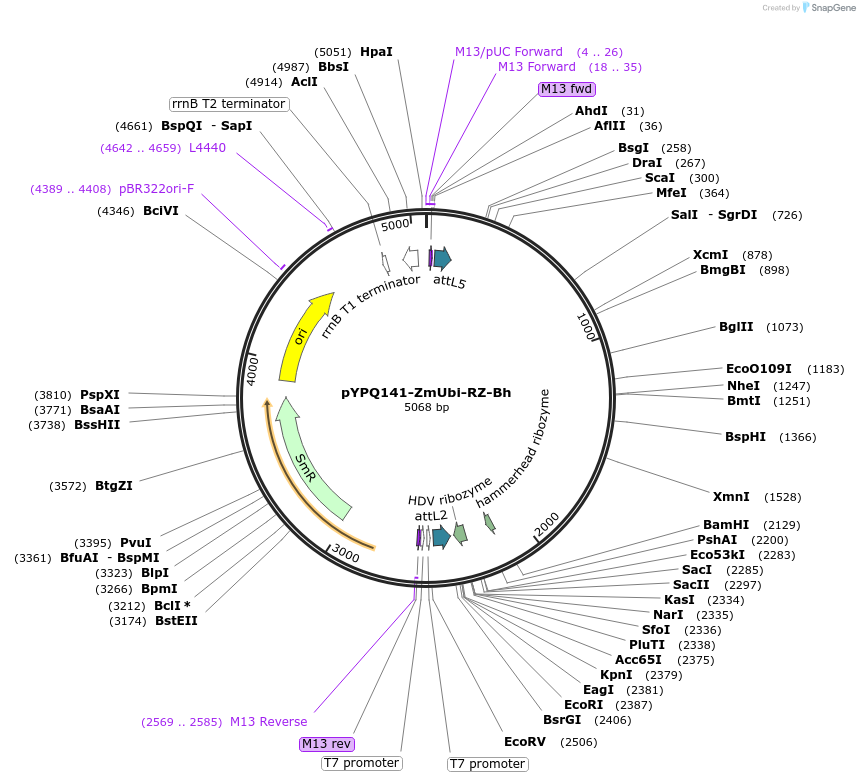
pYPQ141-ZmUbi-RZ-Bh
Plasmid#136378PurposeGateway entry clone for BhCas12b sgRNA expression under ZmUbi promoter with ribozyme processingDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
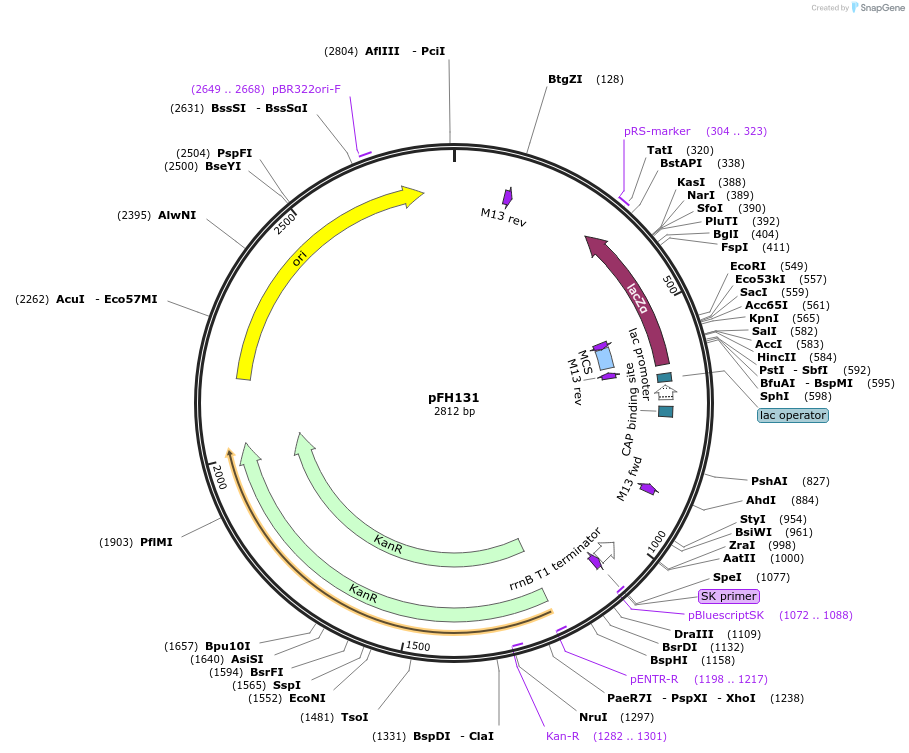
pFH131
Plasmid#136550PurposecrRNA backbone for FnCas12a/CmsI nucleases, combine with OsU3 promoter module (pFH39)DepositorInsertcrRNA backbone for FnCas12a/CmsI nucleases, combine with OsU3 promoter module (pFH39)
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
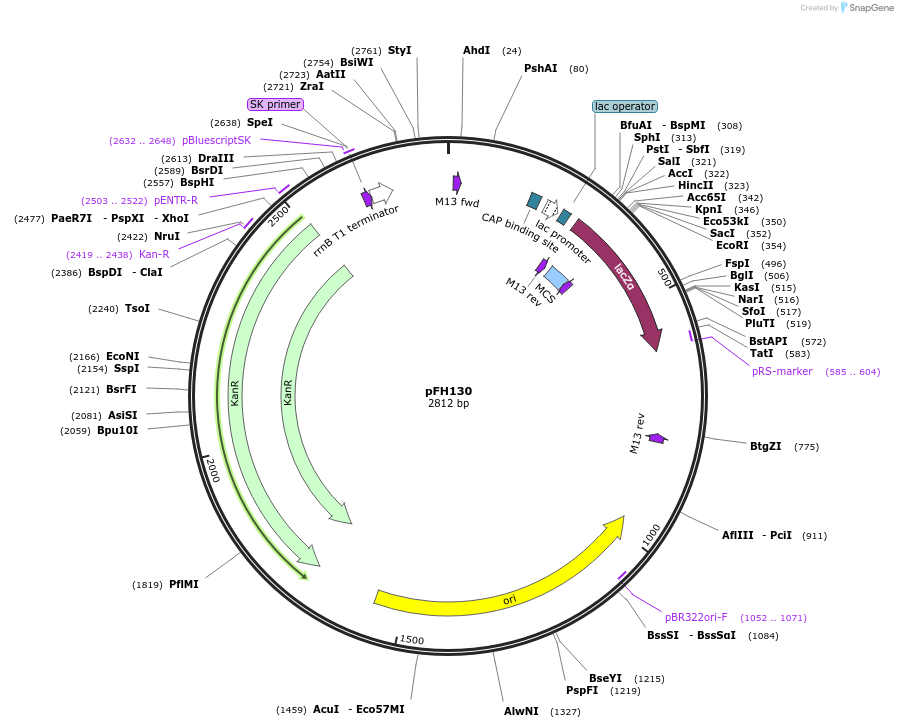
pFH130
Plasmid#136549PurposecrRNA backbone for FnCas12a/CmsI nucleases, combine with OsU6-2 promoter module (pFH37)DepositorInsertcrRNA backbone for FnCas12a/CmsI nucleases, combine with OsU6-2 promoter module (pFH37)
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
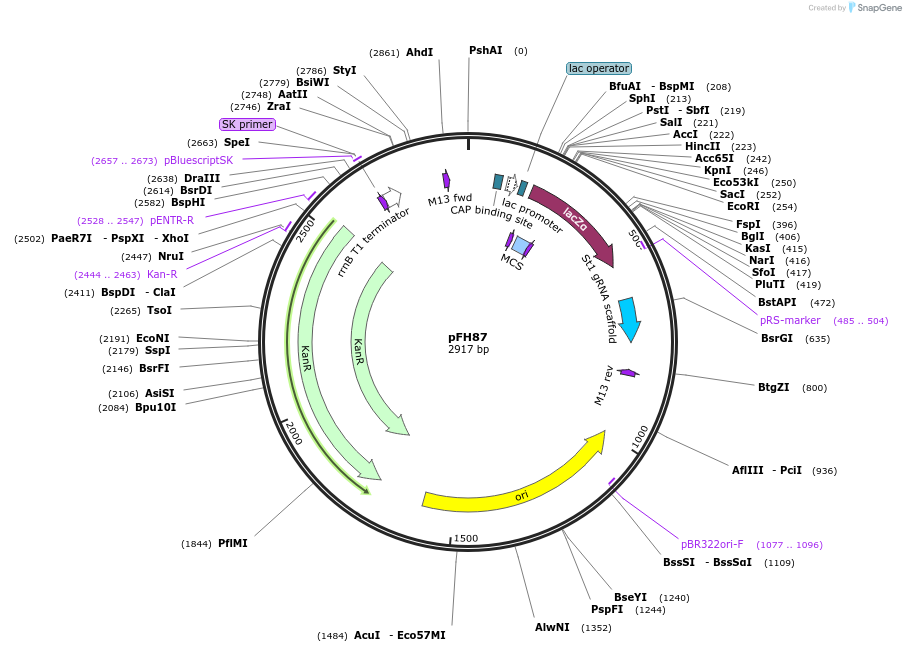
pFH87
Plasmid#128200PurposesgRNA backbone for StCas9, combine with TaU3 promoter module (pFH33)DepositorInsertsgRNA backbone for StCas9, combine with TaU3 promoter module (pFH33)
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
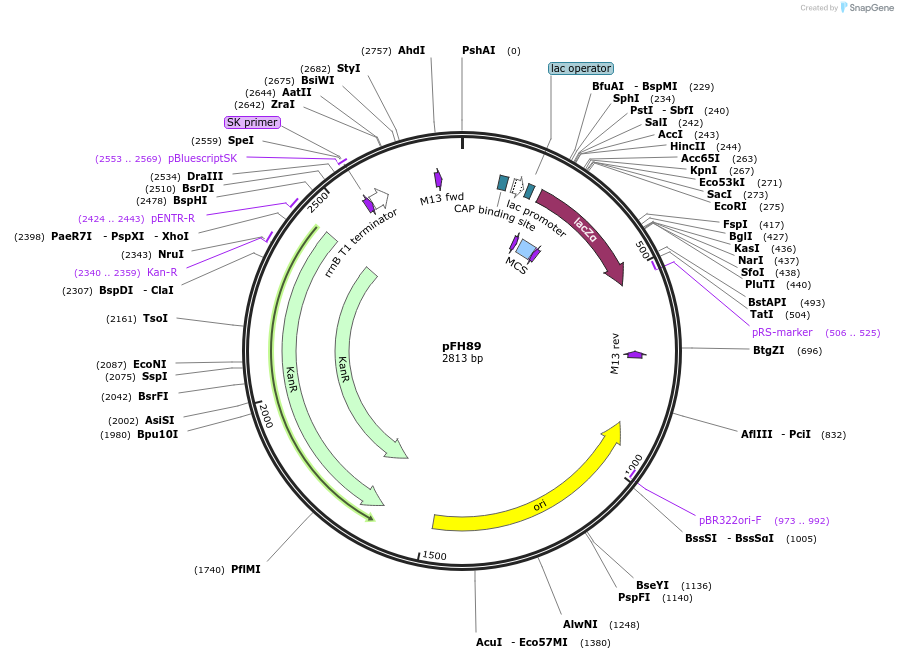
pFH89
Plasmid#128202PurposecrRNA backbone for LbCas12a, combine with TaU3 promoter module (pFH33)DepositorInsertcrRNA backbone for LbCas12a, combine with TaU3 promoter module (pFH33)
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
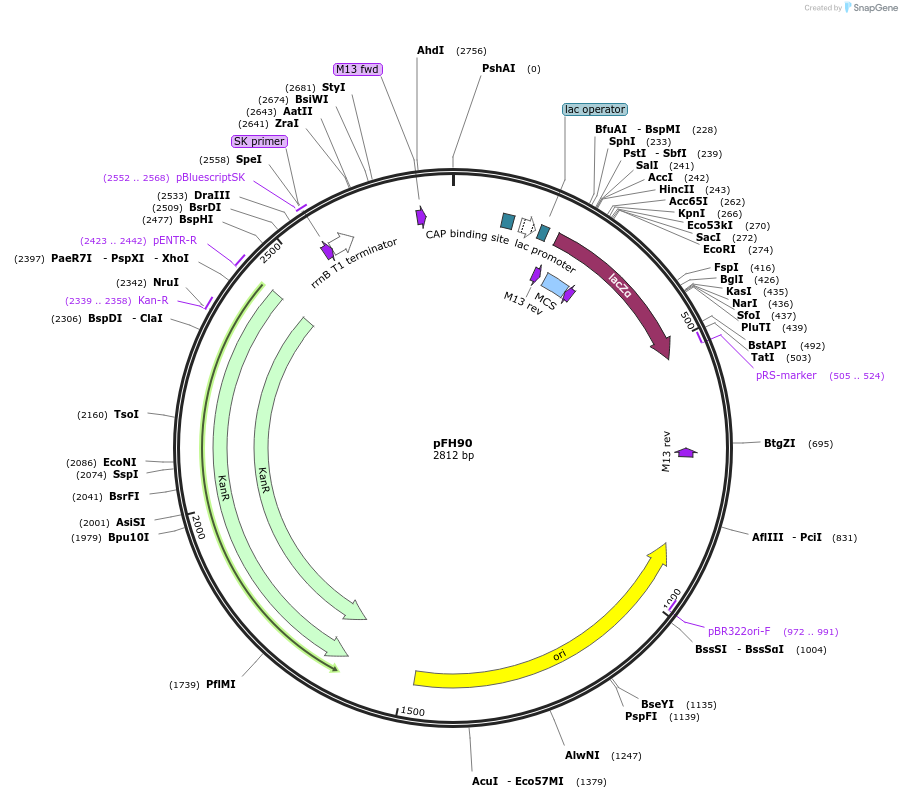
pFH90
Plasmid#128203PurposecrRNA backbone for FnCas12a/CmsI nucleases, combine with TaU3 promoter module (pFH33)DepositorInsertcrRNA backbone for FnCas12a/CmsI nucleases, combine with TaU3 promoter module (pFH33)
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
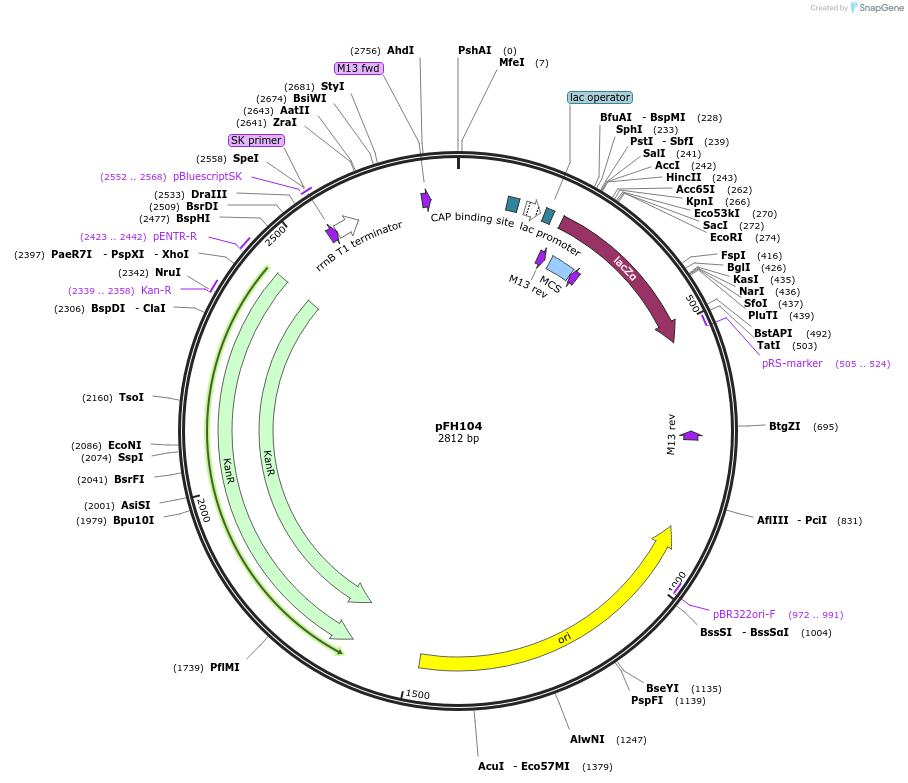
pFH104
Plasmid#128209PurposecrRNA backbone for FnCas12a/CmsI nucleases, combine with AtU6-26 promoter module (pFH35)DepositorInsertcrRNA backbone for FnCas12a/CmsI nucleases, combine with AtU6-26 promoter module (pFH35)
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
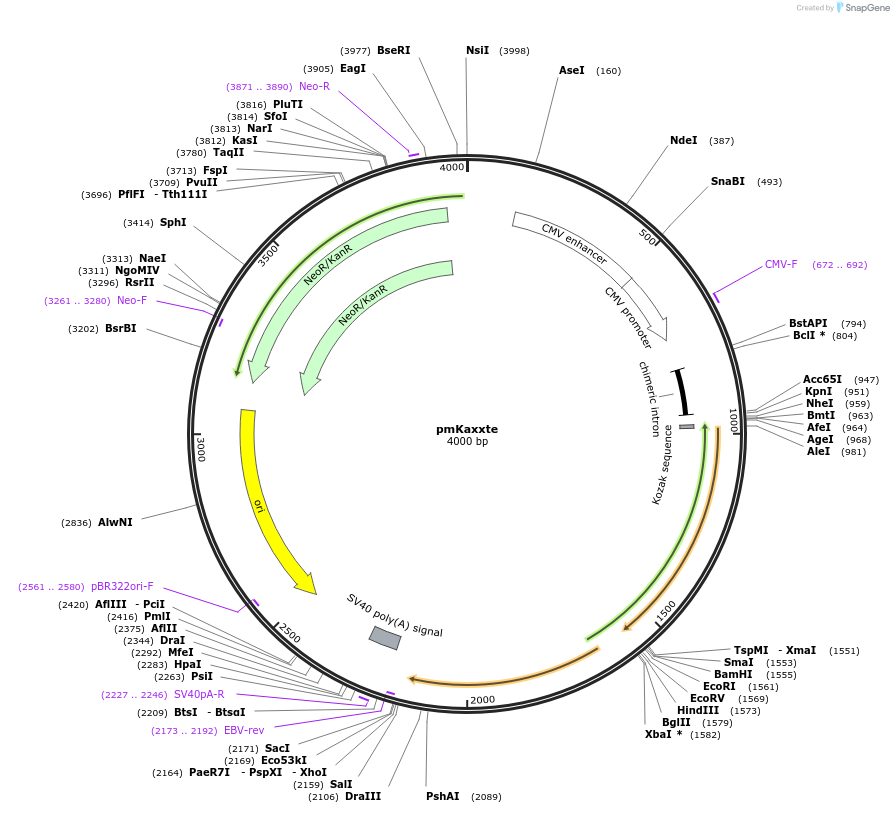
pmKaxxte
Plasmid#113630Purposeto test Cas9 gRNA efficiency or HR-capacityDepositorInsertmKaxxte2.5
ExpressionMammalianPromoterCMVAvailable SinceFeb. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
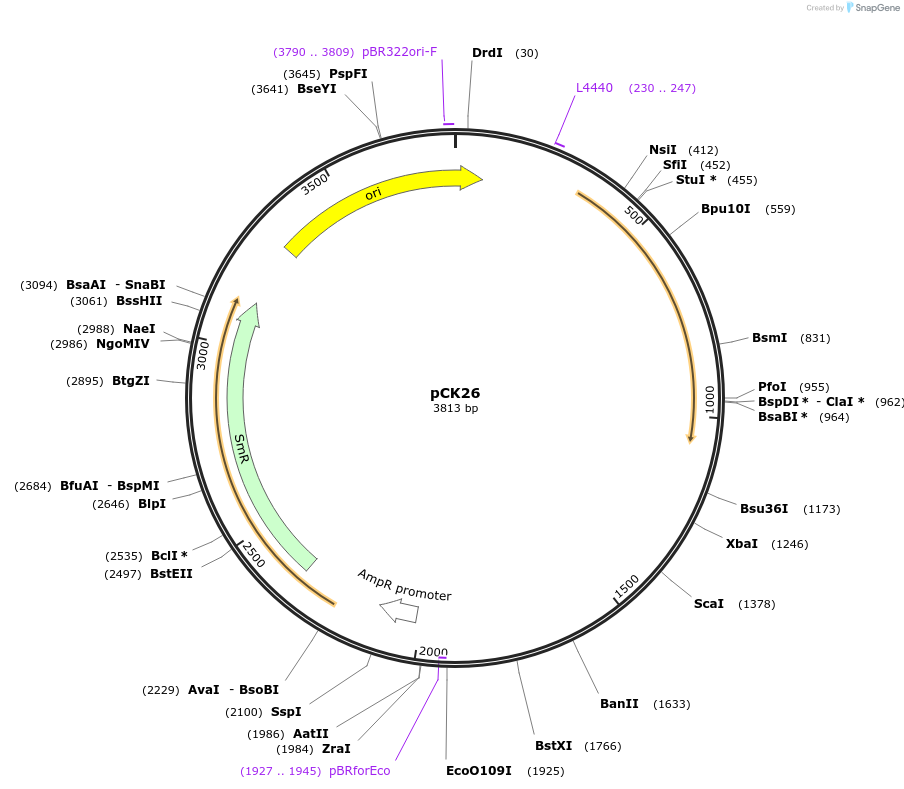
pCK26
Plasmid#105429PurposeSynthetic biologyDepositorInsertBs3
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
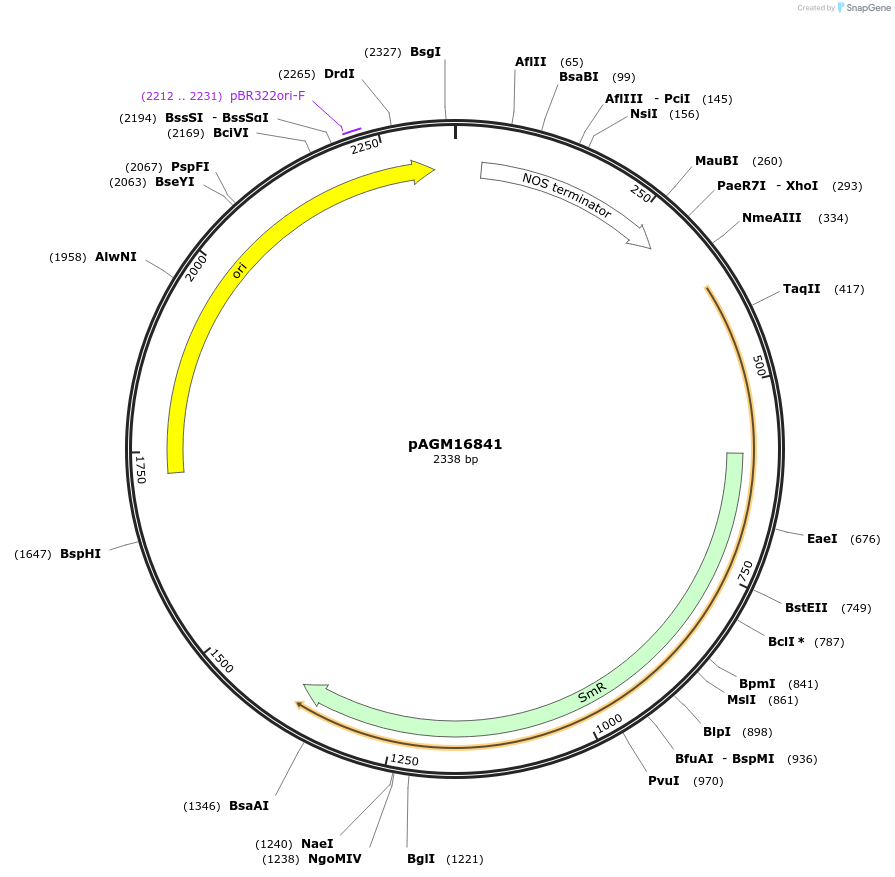
pAGM16841
Plasmid#105416PurposeSynthetic biologyDepositorInsertSTOP codon and nos terminator
UseSynthetic BiologyAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pJOG021
Plasmid#105420PurposeSynthetic biologyDepositorInserthpt (hygromycin phosphotransferase)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
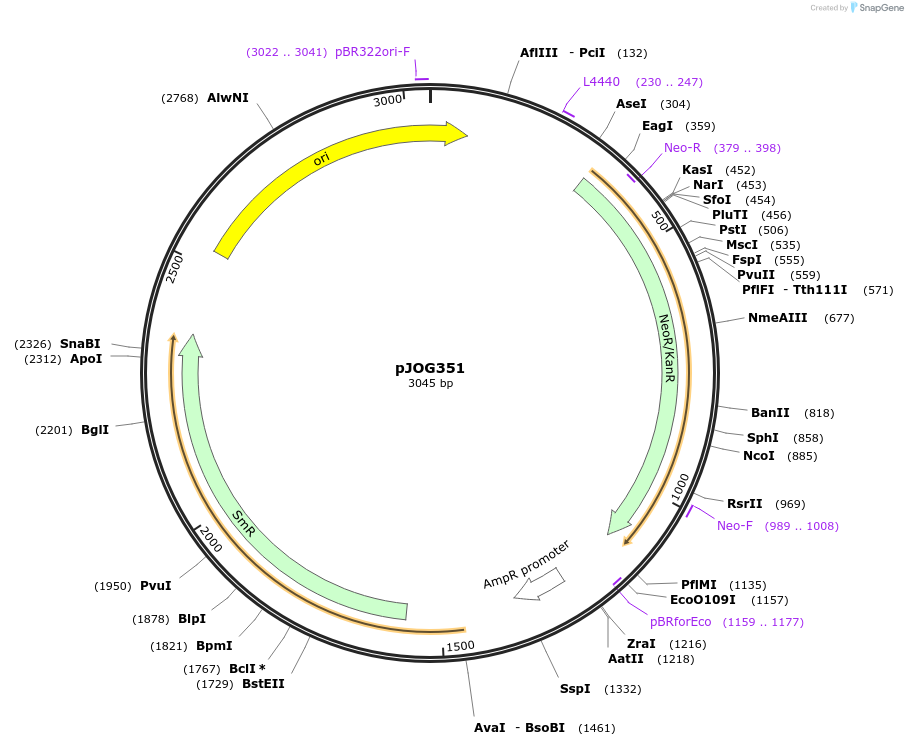
pJOG351
Plasmid#105421PurposeSynthetic biologyDepositorInsertnptII (neomycin phosphotransferase II)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
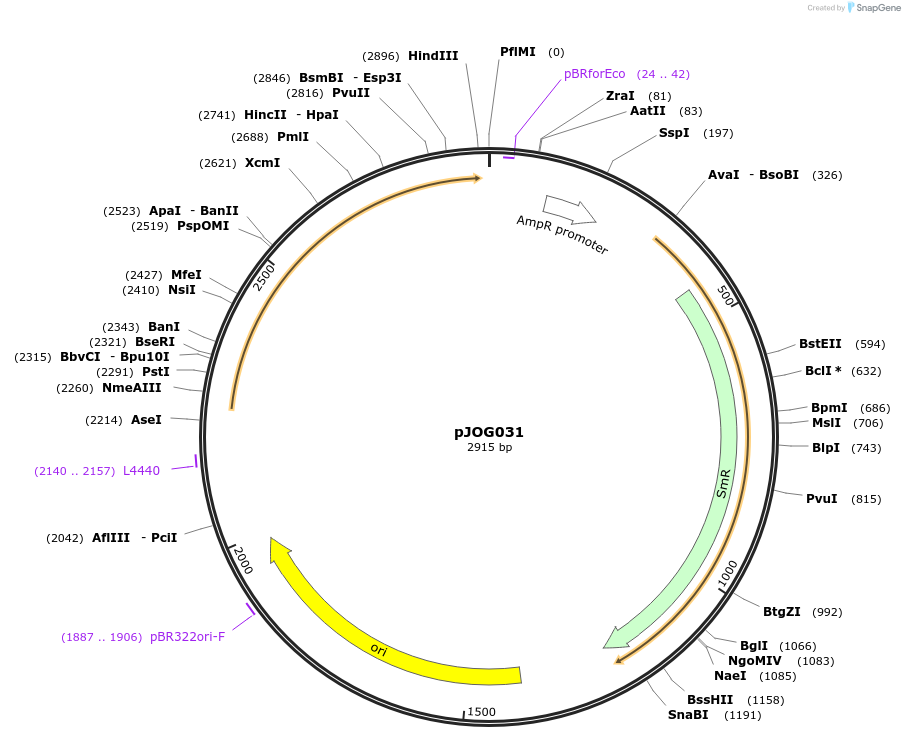
pJOG031
Plasmid#105422PurposeSynthetic biologyDepositorInsertAvrRps4 Pseudomonas syringae pv. pisi)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
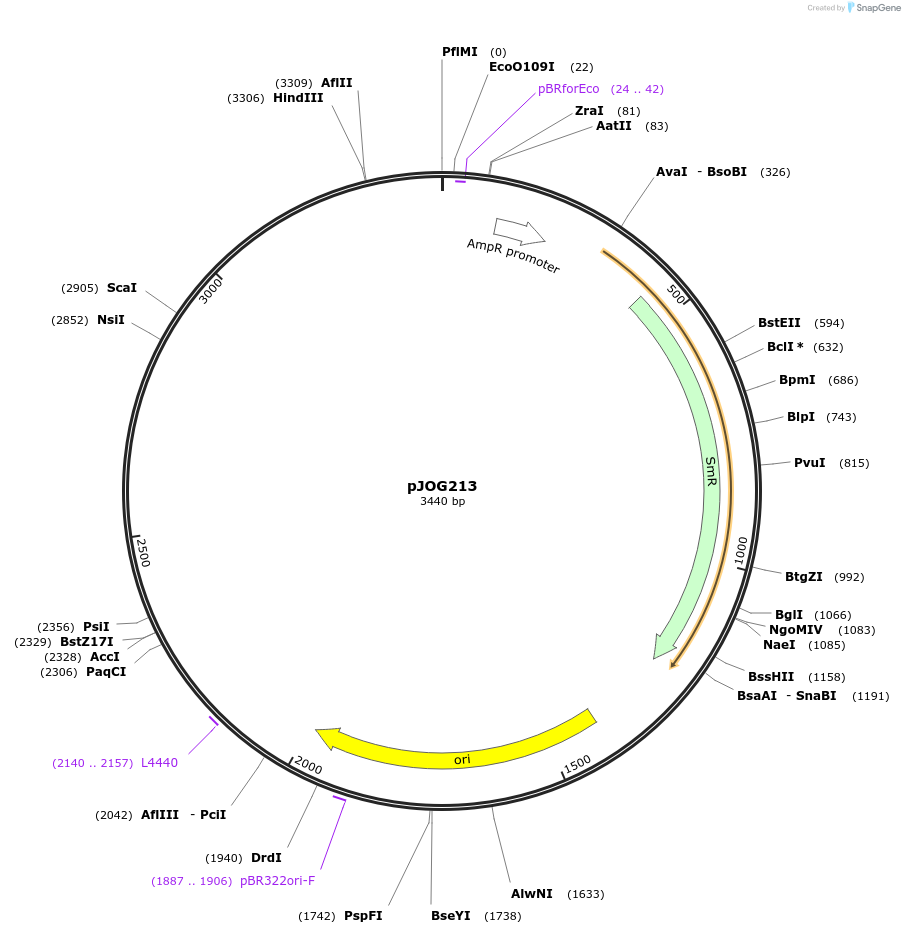
pJOG213
Plasmid#105385PurposeSynthetic biologyDepositorInsertpromotor SlEDS1 (Solyc060g071280; 1200bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
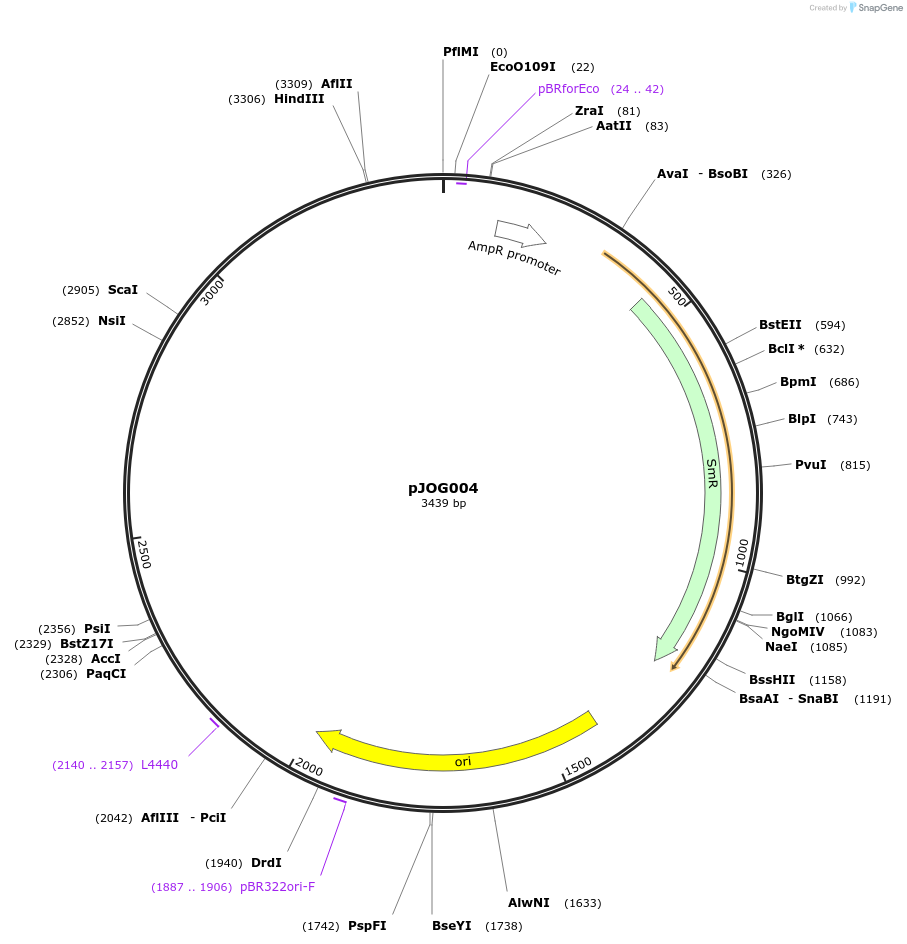
pJOG004
Plasmid#105386PurposeSynthetic biologyDepositorInsertpromotor SlEDS1 (Solyc060g071280; 1200bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
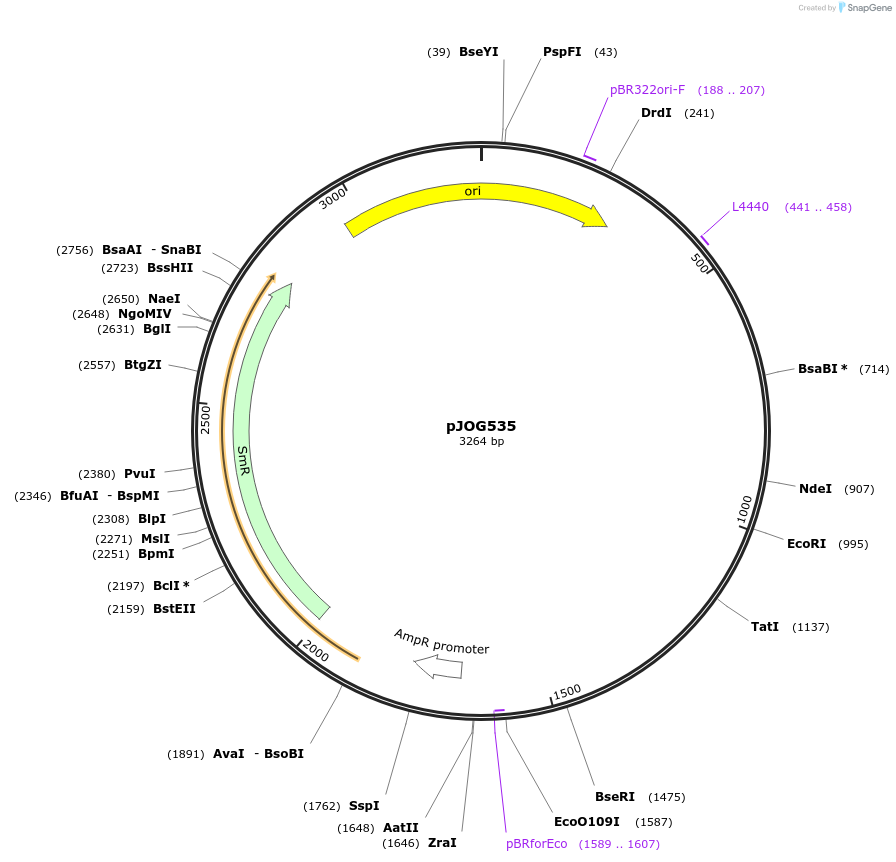
pJOG535
Plasmid#105387PurposeSynthetic biologyDepositorInsertpromotor CaBs3 (1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
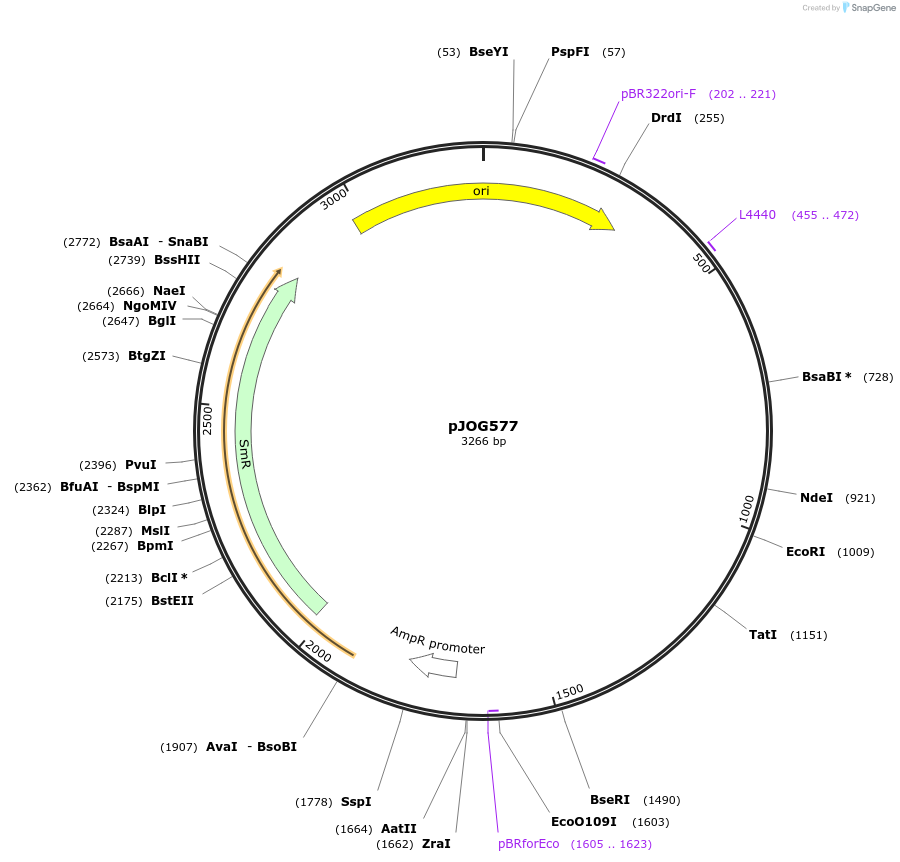
pJOG577
Plasmid#105388PurposeSynthetic biologyDepositorInsertpromotor CaBs3 (1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
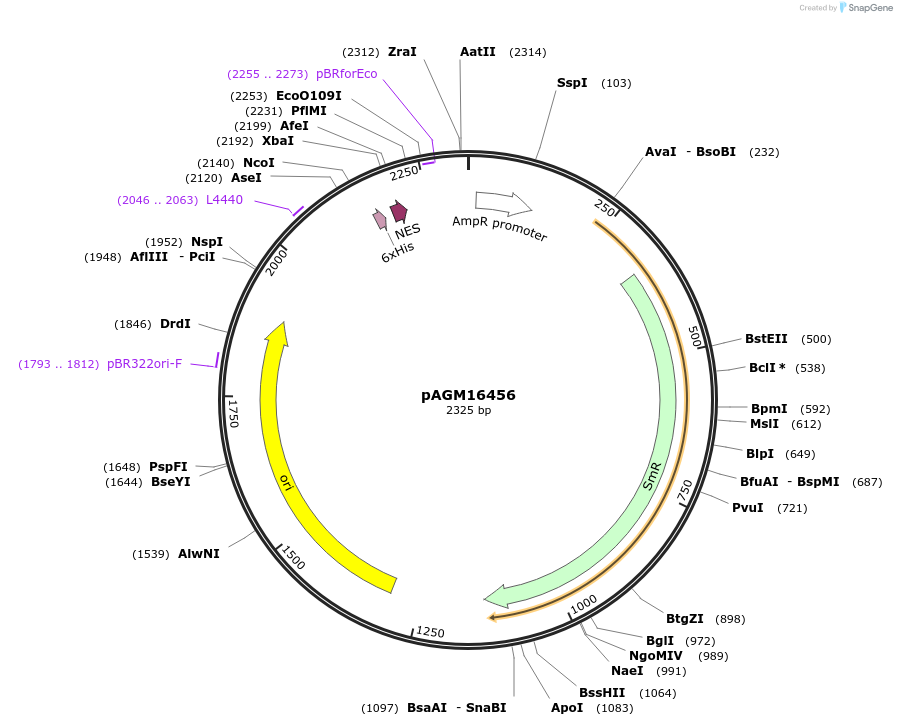
pAGM16456
Plasmid#105402PurposeSynthetic biologyDepositorInsert6xHis-NES(Rev)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
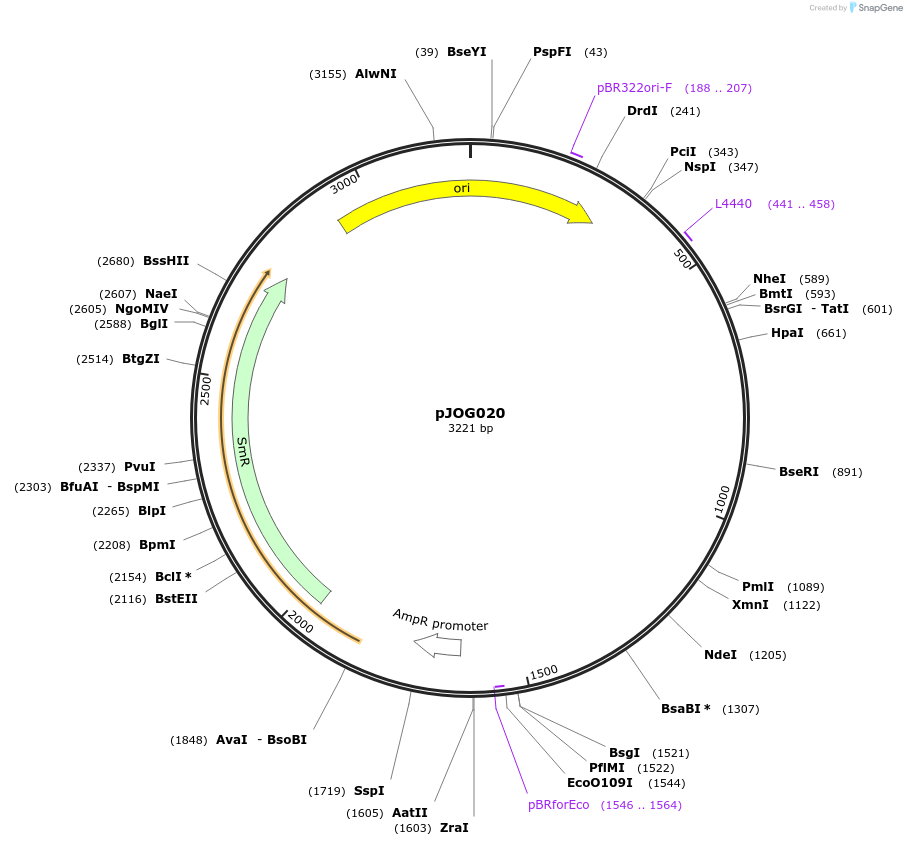
pJOG020
Plasmid#105382PurposeSynthetic biologyDepositorInsertpromotor Ubi4-2 (1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
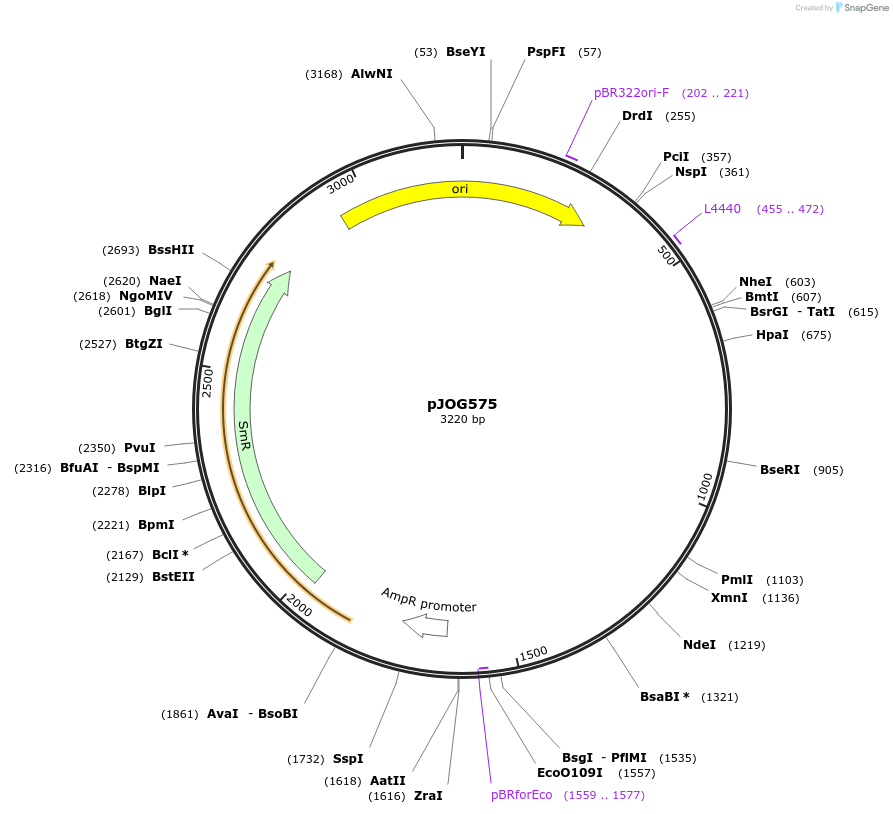
pJOG575
Plasmid#105383PurposeSynthetic biologyDepositorInsertpromotor Ubi4-2 (1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
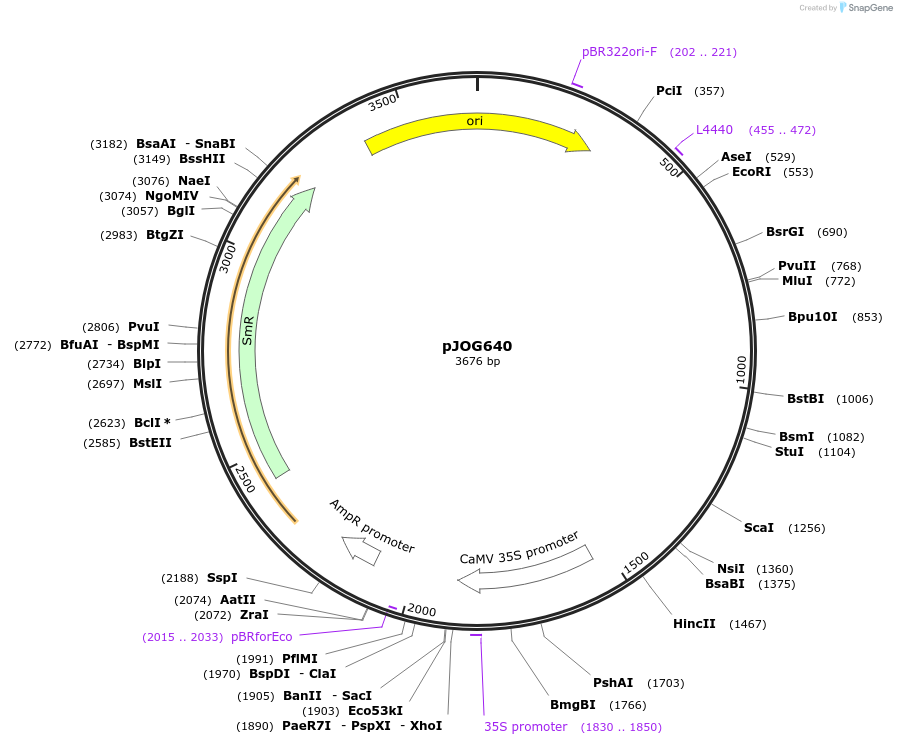
pJOG640
Plasmid#105354PurposeSynthetic biologyDepositorInsertpromotor 35S (1300bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
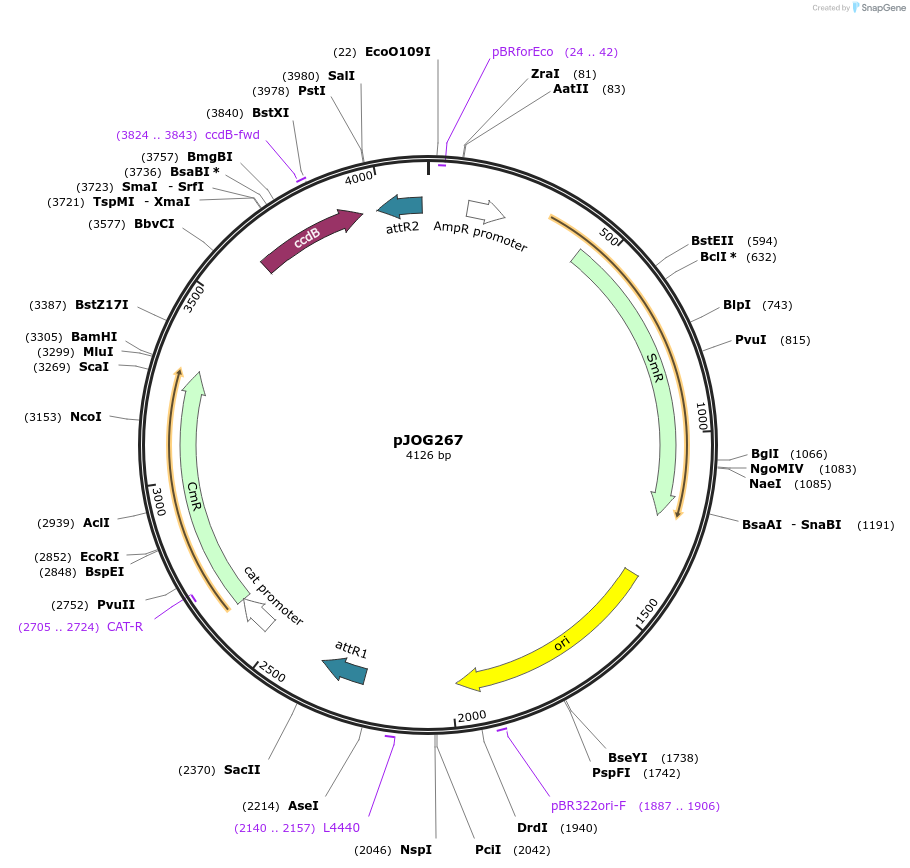
pJOG267
Plasmid#105339PurposeSynthetic biology; gateway destination vector assemblyDepositorTypeEmpty backboneUseSynthetic BiologyAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pRK5-FLAG-dSestrin (CG11299-PD)
Plasmid#73675PurposeoverexpressionDepositorAvailable SinceMarch 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
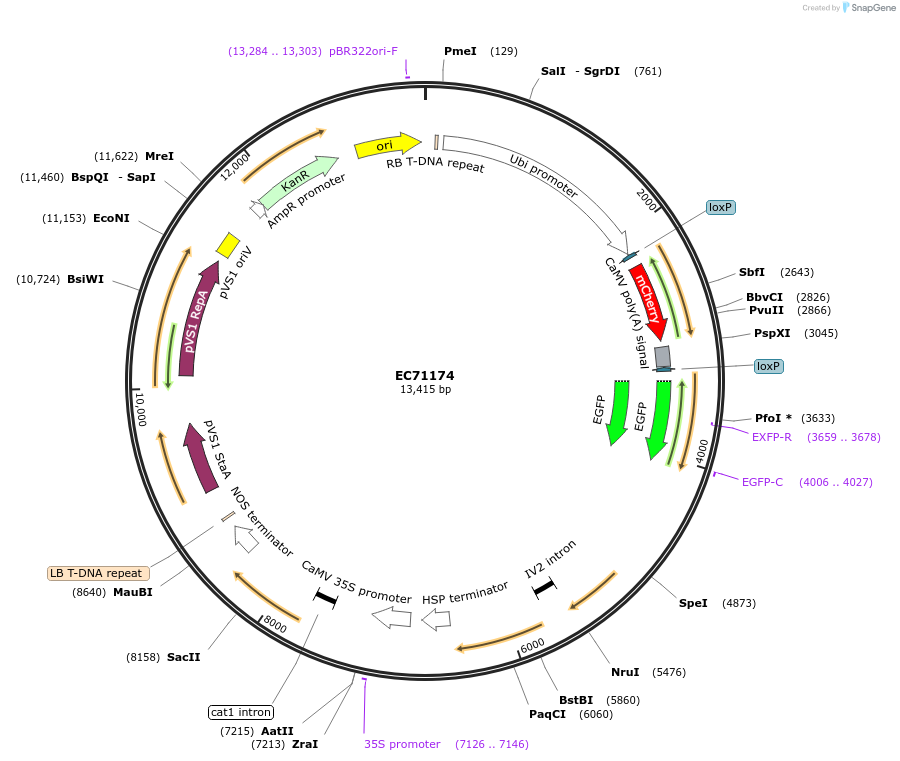
EC71174
Plasmid#154067PurposeLevel 2 Golden Gate vector, pL2M-HvHSP17-CREU5-UBQ-loxmCHERRYER-eGFPERDepositorInsertsHS Cre recombinase
Ubiquitin promoter driving mCherry with the 35S terminator, flanked by lox sites, followed by eGFP with the Actin terminator
HYGROMYCIN Resistance Cassette
UseCre/Lox and Synthetic Biology; Golden gateExpressionBacterial and PlantMutationThe Arabidopsis thaliana U5 small nuclear ribonuc…Promoter35S promoter, Hordeum vulgare HSP 17 promoter use…Available SinceSept. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
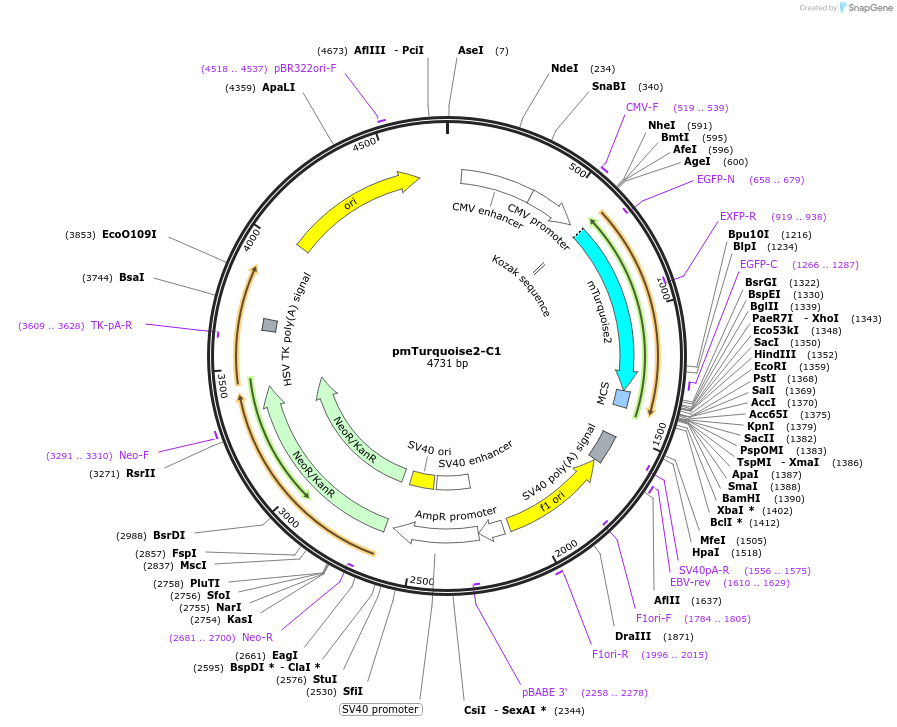
pmTurquoise2-C1
Plasmid#60560PurposeExpresses the cyan fluorescent protein variant mTurquoise2, can be used to create fusion proteinsDepositorInsertmTurquoise2
ExpressionMammalianPromoterCMVAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
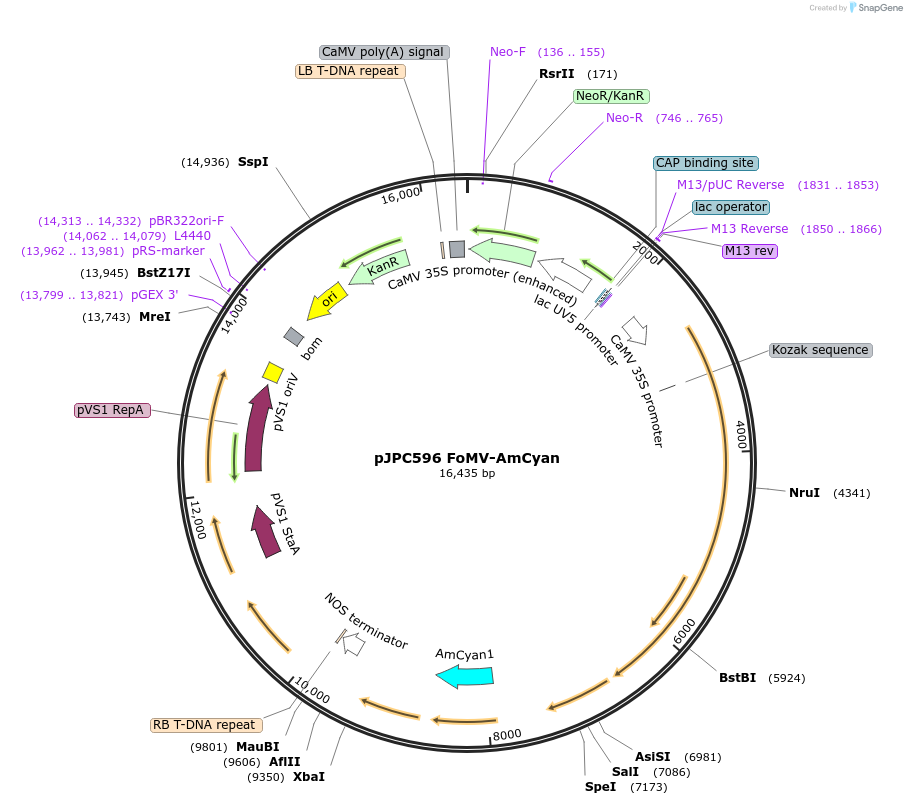
pJPC596 FoMV-AmCyan
Plasmid#234812PurposeTo express flourescent protein AmCyan from the Foxtail Mosaic Virus Vector (T-DNA)DepositorInsertAmCyan
ExpressionPlantAvailable SinceMay 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
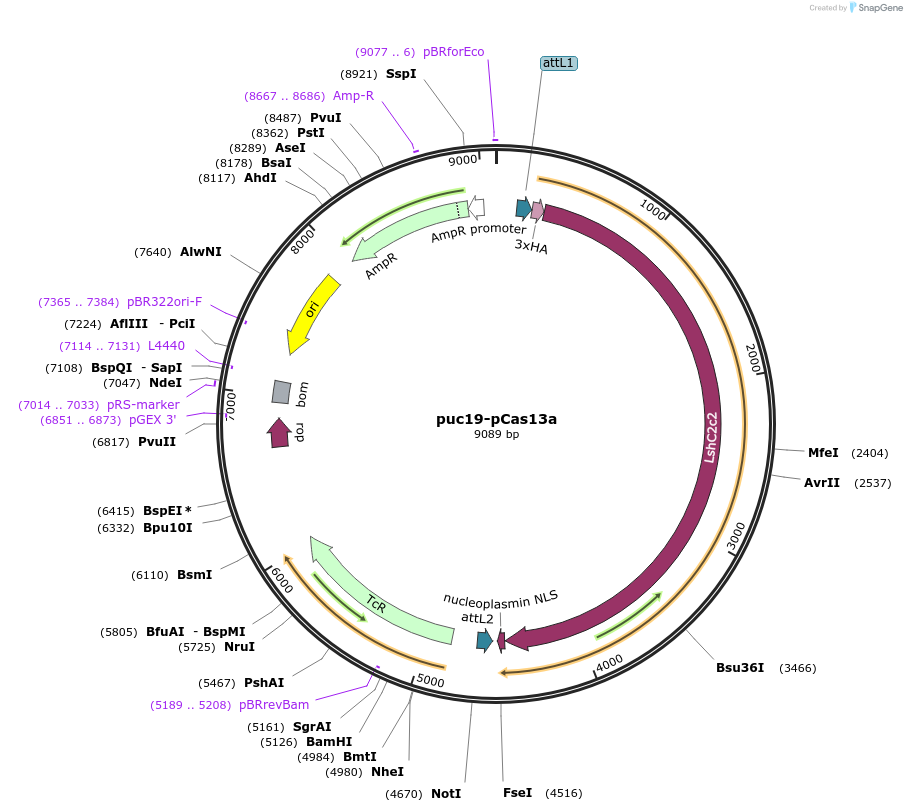
puc19-pCas13a
Plasmid#118963PurposeIntermediate/cloning vector that can be used for sub-cloning into final destination vectors based on one's requirements.DepositorInsertCas13a
ExpressionPlantAvailable SinceFeb. 6, 2019AvailabilityAcademic Institutions and Nonprofits only -
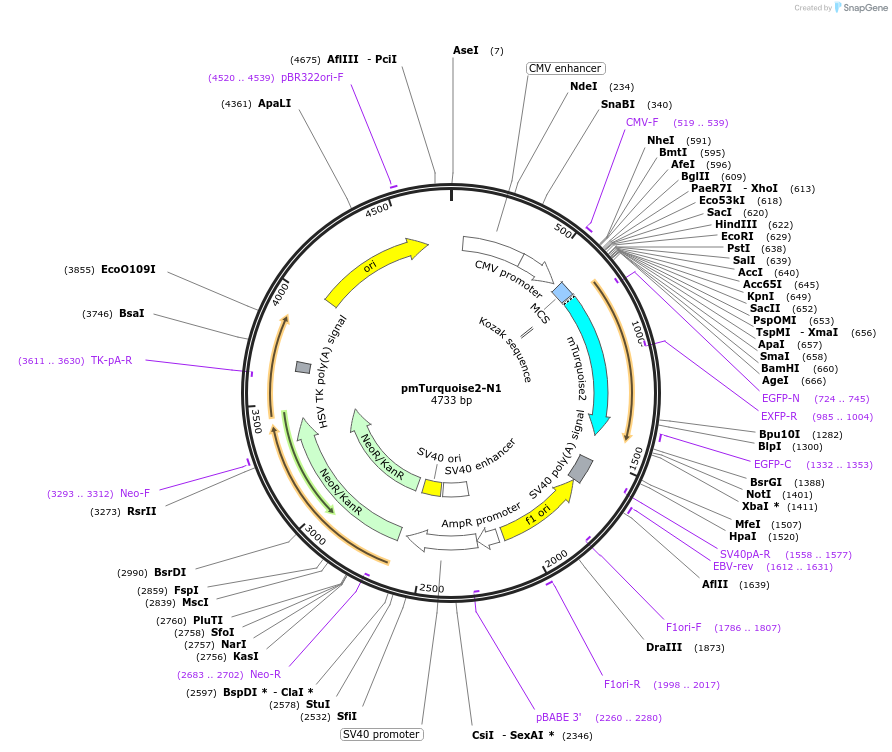
pmTurquoise2-N1
Plasmid#60561PurposeExpresses the cyan fluorescent protein variant mTurquoise2, can be used to create fusion proteinsDepositorInsertmTurquoise2
ExpressionMammalianPromoterCMVAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
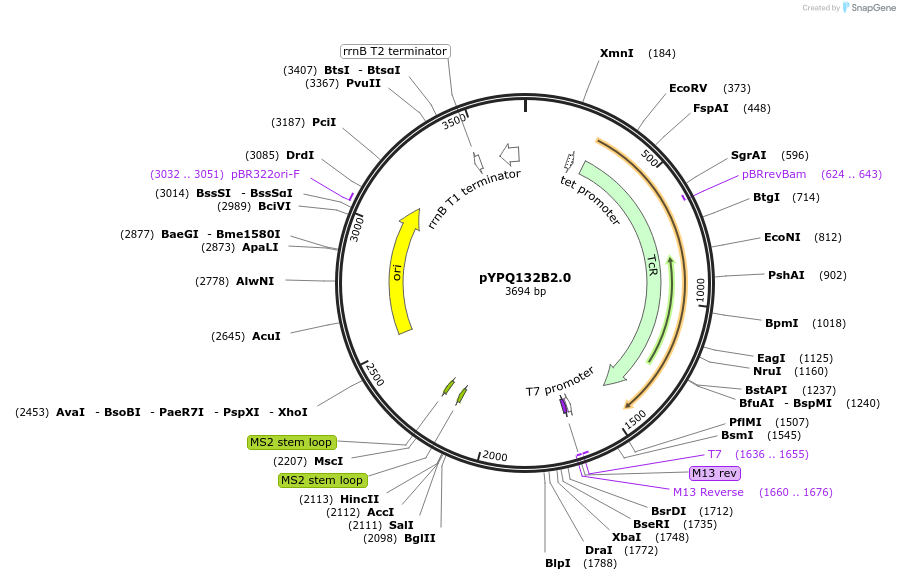
pYPQ132B2.0
Plasmid#99888PurposeGolden Gate entry vector to express the 2nd gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceMarch 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
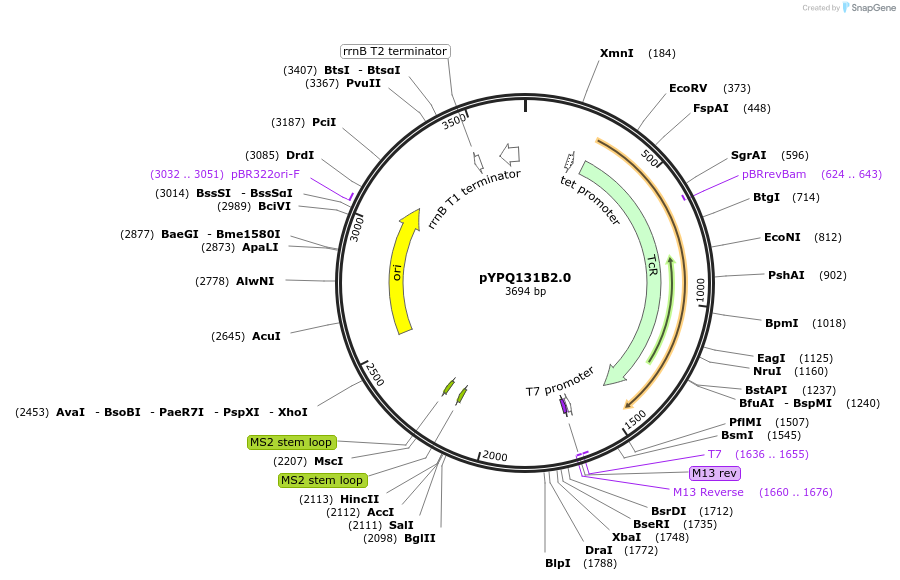
pYPQ131B2.0
Plasmid#99885PurposeGolden Gate entry vector to express the 1st gRNA with gRNA2.0 scaffold (with four MS2 binding sites) under AtU3 promoterDepositorTypeEmpty backboneUseCRISPRExpressionPlantPromoterAtU3Available SinceApril 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
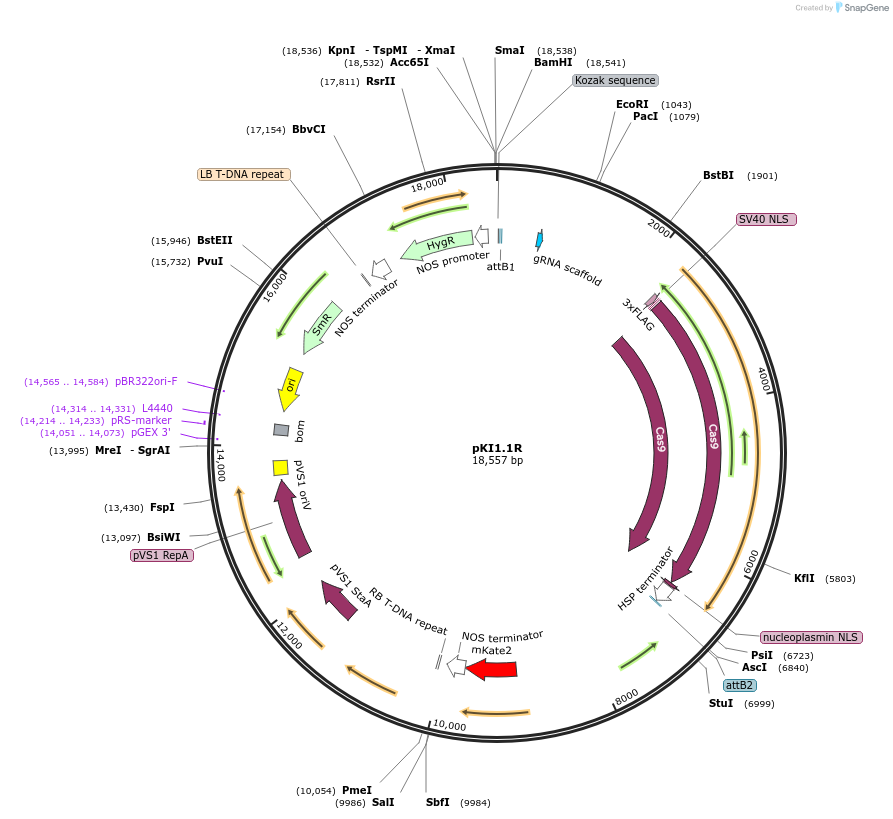
pKI1.1R
Plasmid#85808PurposeCRISPR/Cas9 in Arabidopsis with seed a fluorescent reporter.DepositorInserthuman-codon-optimized SpCas9
UseCRISPRTagsFLAG and NLSExpressionPlantPromoterAtRPS5AAvailable SinceJan. 11, 2017AvailabilityAcademic Institutions and Nonprofits only -
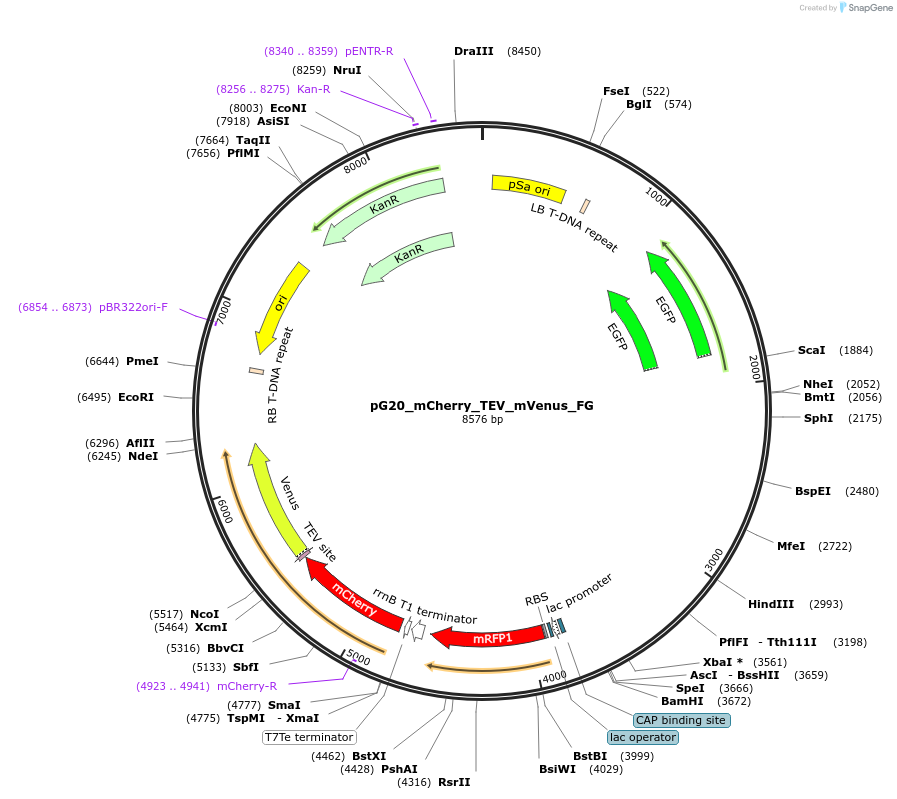
pG20_mCherry_TEV_mVenus_FG
Plasmid#240673PurposeExpression plasmid for mCherry-TEV-mVenus-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsmCherry-TEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
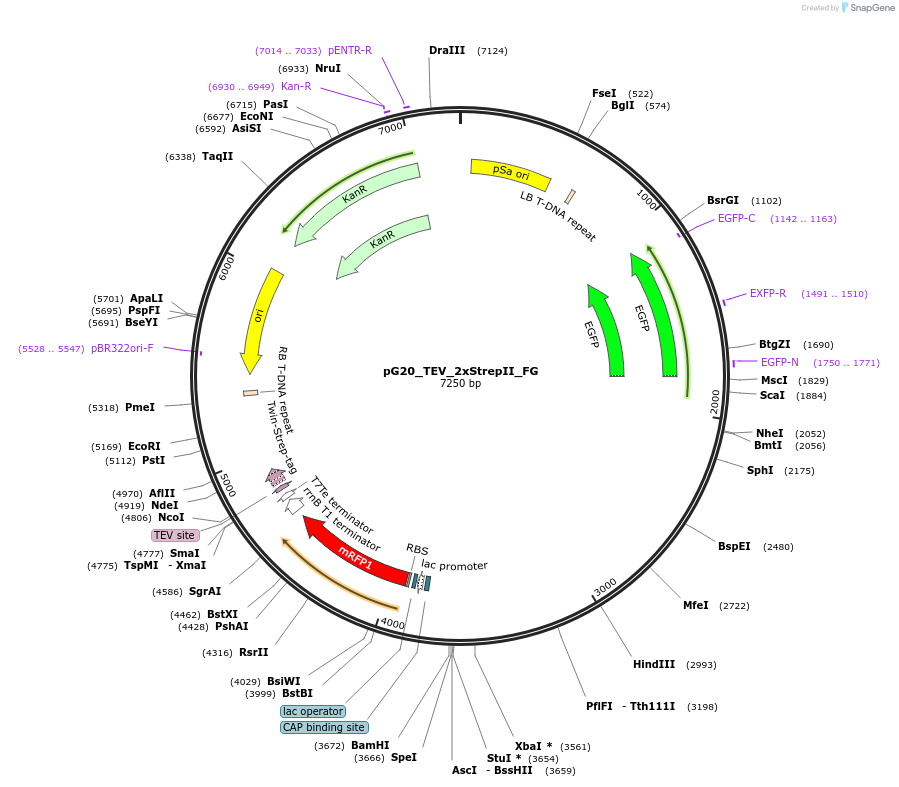
pG20_TEV_2xStrepII_FG
Plasmid#240675PurposeExpression plasmid for TEV-2xStrepII-tagged proteins with FAST-Green transgenic seed selectionDepositorTypeEmpty backboneTagsTEV-2xStrepIIExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
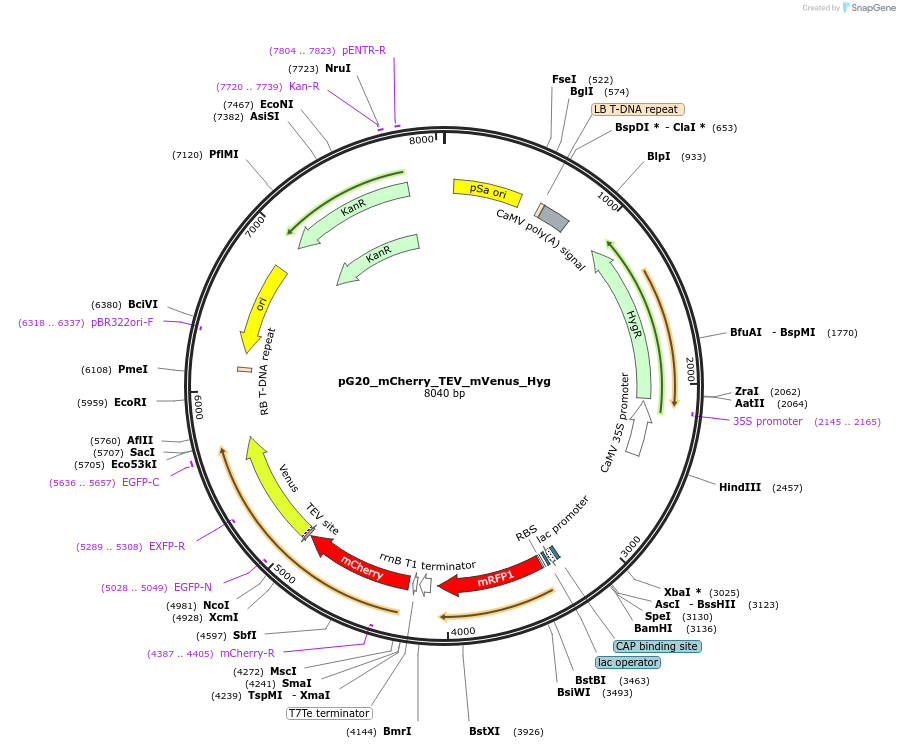
pG20_mCherry_TEV_mVenus_Hyg
Plasmid#240653PurposeExpression plasmid for mCherry-TEV-mVenus-tagged proteins with Hygromycin transgenic seed selectionDepositorTypeEmpty backboneTagsmCherry-TEV-mVenusExpressionPlantAvailable SinceNov. 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
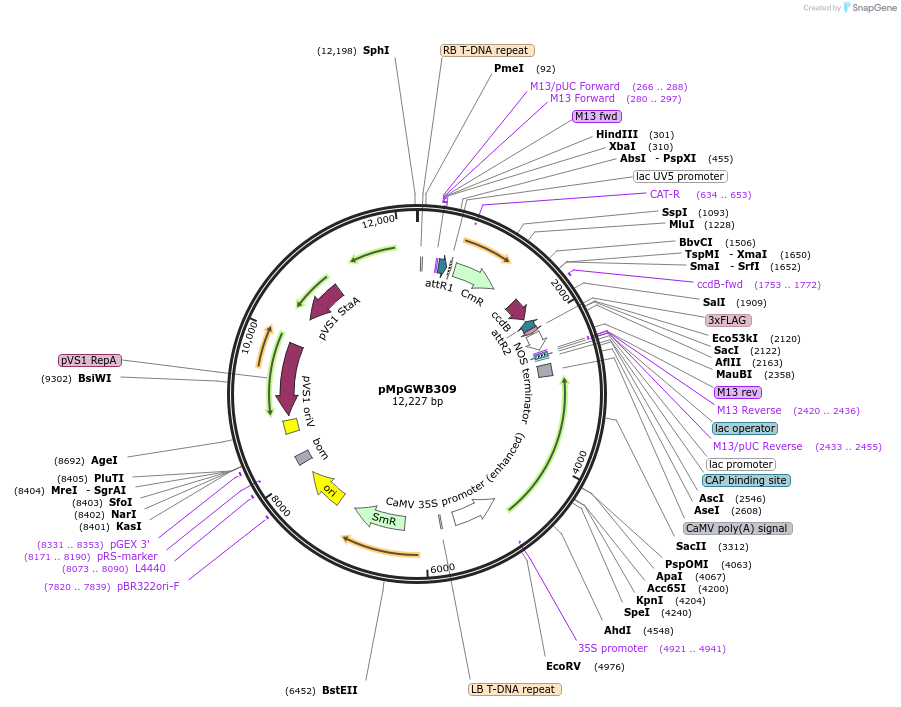
pMpGWB309
Plasmid#68637PurposeGateway binary vector designed for transgenic research with Marchantia polymorpha as well as other plantsDepositorTypeEmpty backboneTags3xFLAGExpressionPlantAvailable SinceJune 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
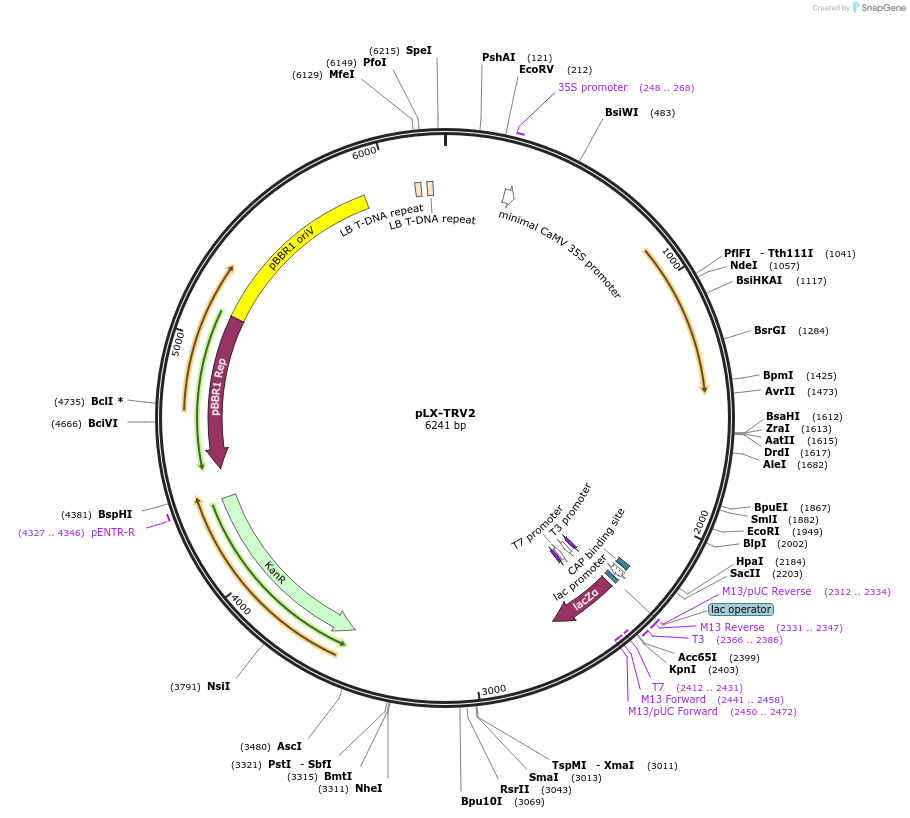
pLX-TRV2
Plasmid#180516PurposeAgrobacterium tumefaciens-based expression of TRV2 including a PEBV heterologous promoter and a polylinkerDepositorInsertTobacco rattle virus RNA2 with a PEBV heterologous promoter and a polylinker
ExpressionPlantPromoter35S CaMVAvailable SinceApril 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
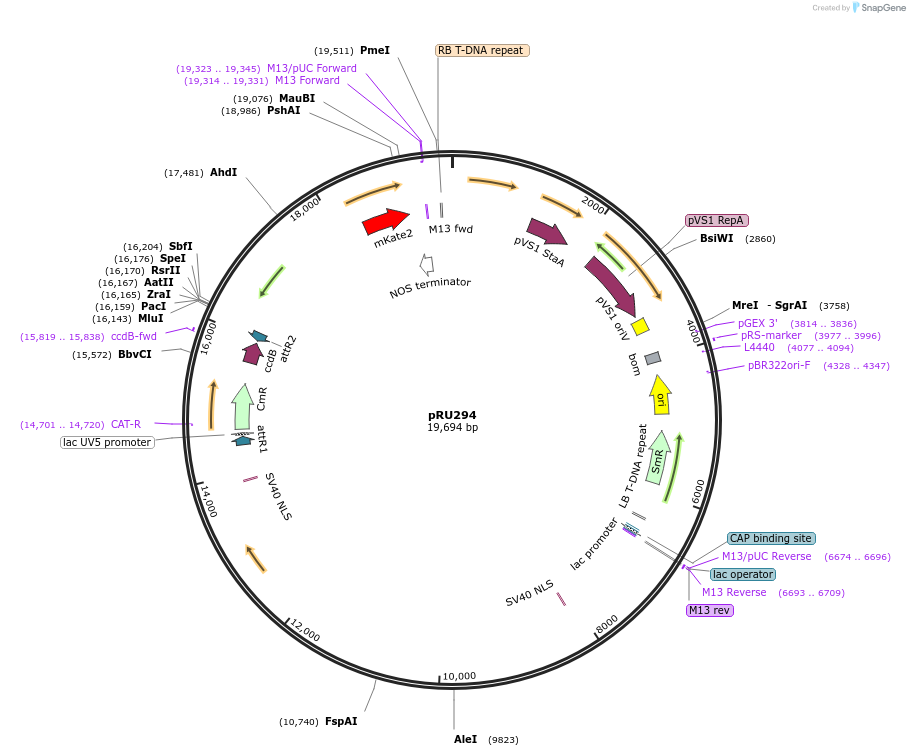
pRU294
Plasmid#167689PurposeFinal destination vector for single Gateway LR reactionDepositorInsertpEC1.2::Cas9i
UseCRISPRAvailable SinceJuly 20, 2021AvailabilityAcademic Institutions and Nonprofits only