We narrowed to 9,878 results for: EPO
-
Plasmid#71178PurposeLuciferase reporterDepositorInsert757_S2-1_GATA-to-HEY
UseLuciferaseTagsluciferaseExpressionInsectMutationPromoterDSCP Drosophila synthetic core promoterAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pSGE_722_S2-1_CGCG-to-HEY
Plasmid#71177PurposeLuciferase reporterDepositorInsert722_S2-1_CGCG-to-HEY
UseLuciferaseTagsluciferaseExpressionInsectMutationPromoterDSCP Drosophila synthetic core promoterAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pSGE_720_S2-1_CGCG-to-ETS21C
Plasmid#71176PurposeLuciferase reporterDepositorInsert720_S2-1_CGCG-to-ETS21C
UseLuciferaseTagsluciferaseExpressionInsectMutationPromoterDSCP Drosophila synthetic core promoterAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
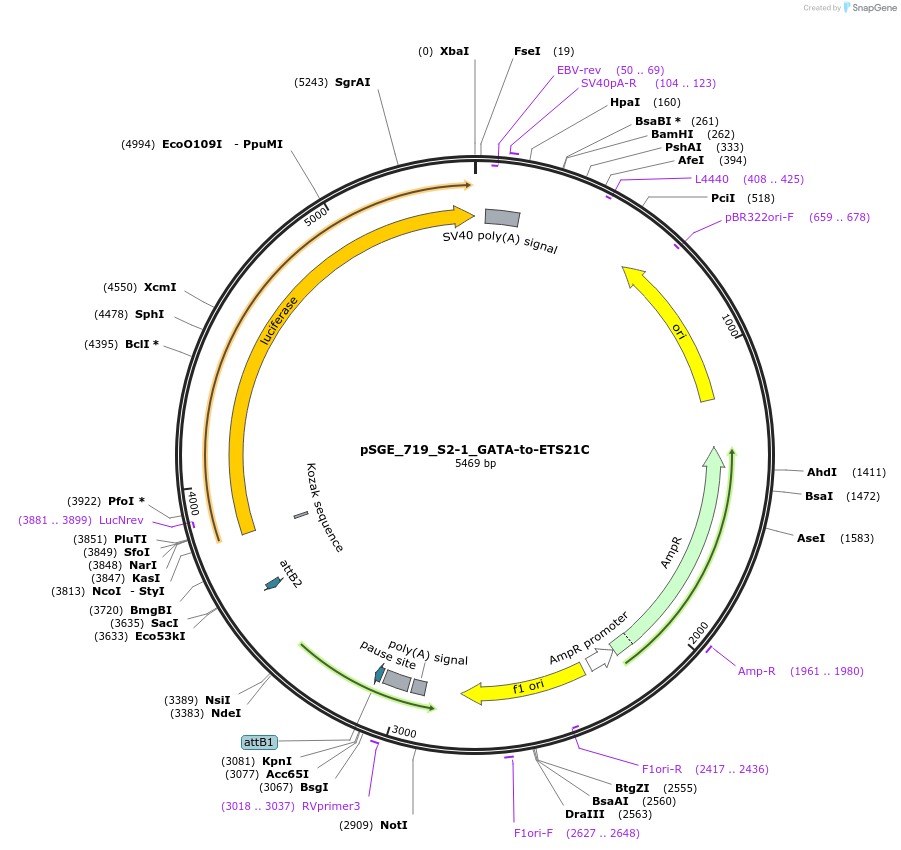
pSGE_719_S2-1_GATA-to-ETS21C
Plasmid#71175PurposeLuciferase reporterDepositorInsert719_S2-1_GATA-to-ETS21C
UseLuciferaseTagsluciferaseExpressionInsectMutationPromoterDSCP Drosophila synthetic core promoterAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAS-7 (mm12-21)
Plasmid#40842PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm12-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-6 (mm17-21)
Plasmid#40841PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm17-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-5 (mm18-21)
Plasmid#40840PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm18-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-4 (mm19-21)
Plasmid#40839PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm19-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-3 (mm20-21)
Plasmid#40838PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm20-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-2 (mm21)
Plasmid#40837PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-16 (mm14-21)
Plasmid#40851PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm14-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-15 (mm15-21)
Plasmid#40850PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm15-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS14 (mm16-21)
Plasmid#40849PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm16-21
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-13
Plasmid#40848PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–Bartel
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-12 (mm1-8)
Plasmid#40847PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–mm1-8
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-11 (bulge 9-15)
Plasmid#40846PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge9-15
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-10 (bulge 9-13)
Plasmid#40845PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-13
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pAS-9 (bulge 9-11)
Plasmid#40844PurposeTarget RNAs used to determine complementarity requirements for in vitro tailing and degradationDepositorInsertlet-7–bulge 9-11
UseMirna reporterTagsEGFPExpressionInsectMutationPromoterActin5CAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
pG6(3'Pro) (P#1478)
Plasmid#14660DepositorInsertUAS repeats
UseTagsExpressionBacterialMutationPromoterAvailable SinceJune 30, 2008AvailabilityAcademic Institutions and Nonprofits only -
pKB234.1
Plasmid#170022PurposeEncodes LacI-controlled expression of surface-displayed hBD3 alpha-helix fragment and PvirK-mNeonGreen reporterDepositorArticleInsertsmNeonGreen
hBD3 alpha-helix
UseSynthetic BiologyTagsLpp, OmpA, and TetherExpressionBacterialMutationPromoterPtac and PvirKAvailable SinceAvailabilityAcademic Institutions and Nonprofits only -
pKB235.4
Plasmid#170026PurposeEncodes LacI-controlled expression of surface-displayed CCL20 fragment lacking alpha-helix sequence and PvirK-mNeonGreen reporterDepositorArticleInsertsmNeonGreen
CCL20 Δ alpha-helix
UseSynthetic BiologyTagsLpp, OmpA, and TetherExpressionBacterialMutationPromoterPtac and PvirKAvailable SinceAvailabilityAcademic Institutions and Nonprofits only -
pKB236.2
Plasmid#170029PurposeEncodes LacI-controlled expression of surface-displayed TCP C-terminal alpha-helix fragment and PvirK-mNeonGreen reporterDepositorArticleInsertsmNeonGreen
TCP C-terminal alpha-helix
UseSynthetic BiologyTagsLpp, OmpA, and TetherExpressionBacterialMutationPromoterPtac and PvirKAvailable SinceAvailabilityAcademic Institutions and Nonprofits only -
pmGFP-P2A-K0-P2A-RFP
Plasmid#105686PurposeDouble fluorescent stalling reporter K0DepositorInsertsP2A-3xFLAG-VHPbeta
K0
P2A-mCherry
UseTagsExpressionMammalianMutationPromoterCMV (from vector)Available SinceFeb. 6, 2018AvailabilityAcademic Institutions and Nonprofits only -
pLminP_Luc2P_RE17
Plasmid#90359PurposeNFAT - PKC/Ca++ gene reporterDepositorInsertFirefly luciferase
UseLentiviralTagsExpressionMutationPromoterNFATAvailable SinceAug. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLminP_Luc2P_RE1
Plasmid#90335PurposeNFkB - p50/p65 pathway gene reporterDepositorInsertFirefly luciferase
UseLentiviralTagsExpressionMutationPromoterNFKBAvailable SinceAug. 22, 2022AvailabilityAcademic Institutions and Nonprofits only -
CMV-mCherry-(F30-2xPepper)10
Plasmid#129401PurposemCherry mRNA reporter tagged with (F30-2xPepper)10DepositorInsertmCherry-(F30-2xPepper)10
UseTagsExpressionMammalianMutationPromoterCMVAvailable SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
PNCA-GFP-dYY1
Plasmid#67791PurposeUse together with pCMV-intron and VSV-G to package MMLV reporter virusDepositorInsertMMLV-LTR-GFP
UseTagsExpressionMutationDeletion of LTR U3 YY1 binding domainPromoterAvailable SinceSept. 1, 2015AvailabilityAcademic Institutions and Nonprofits only