We narrowed to 12,598 results for: 112
-
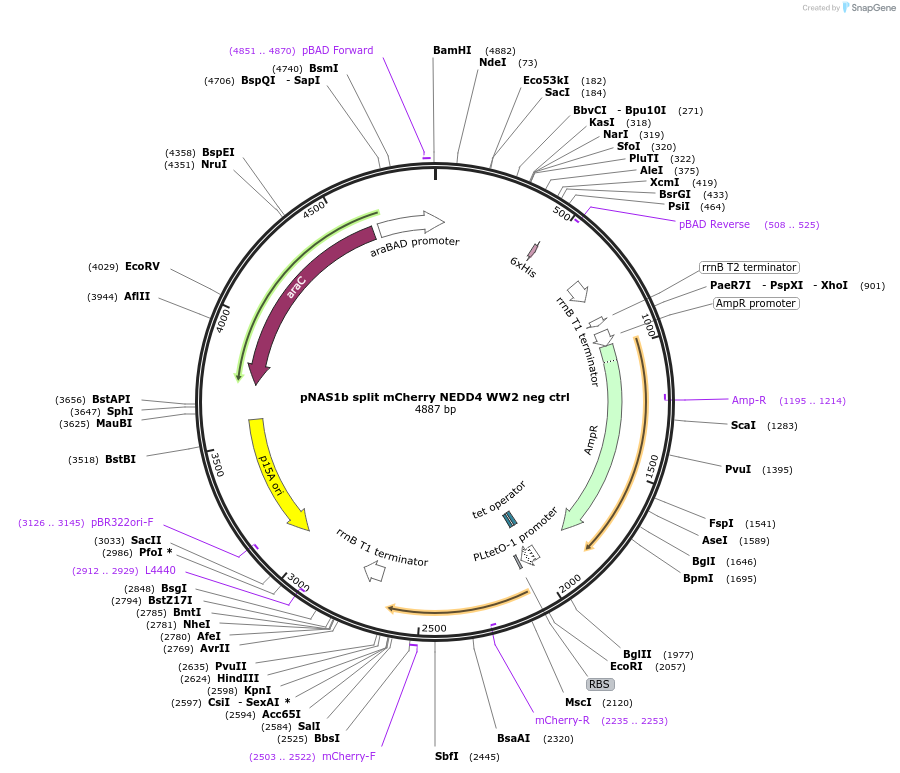
Plasmid#112030PurposeMode #2 phosphosite negative control (split mCherry, using NEDD4 WW2 binding domain paired with negative control peptide)DepositorInsertNEDD4 WW2 domain fused to C-terminal split mCherry, neg ctrl interacting peptide fused to N-terminal split mCherry
TagsSplit mCherryExpressionBacterialAvailable SinceAug. 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
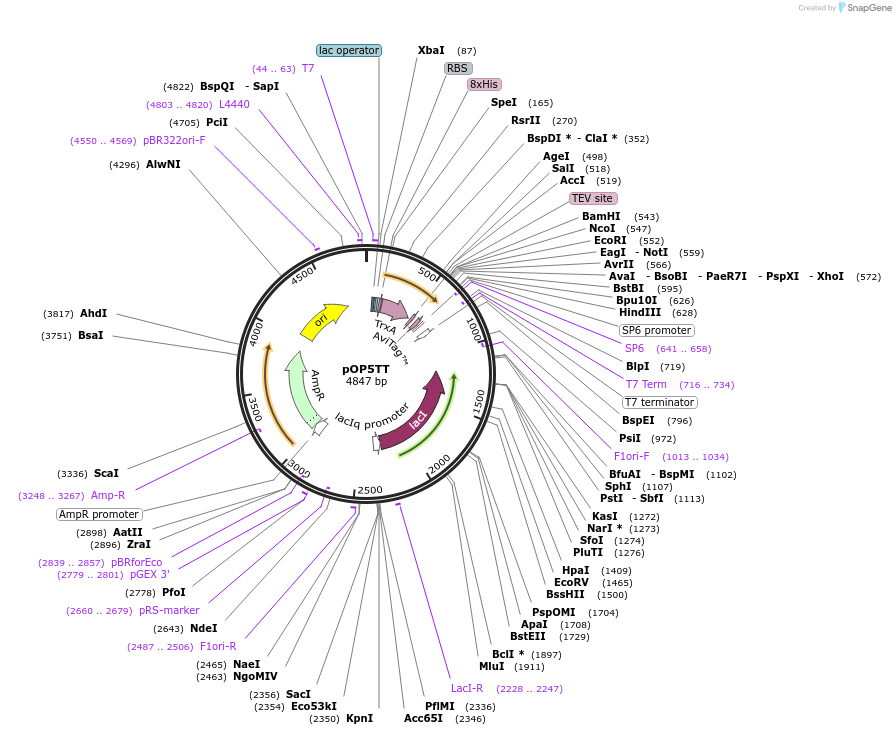
pOP5TT
Plasmid#112613PurposeUsed for expression of protein in E. coli as TEV cleavable N-terminual 8His and thioredoxin tag fusions and with C-terminal Avi-tagDepositorTypeEmpty backboneTags8His-thioredoxin and AviExpressionBacterialAvailable SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
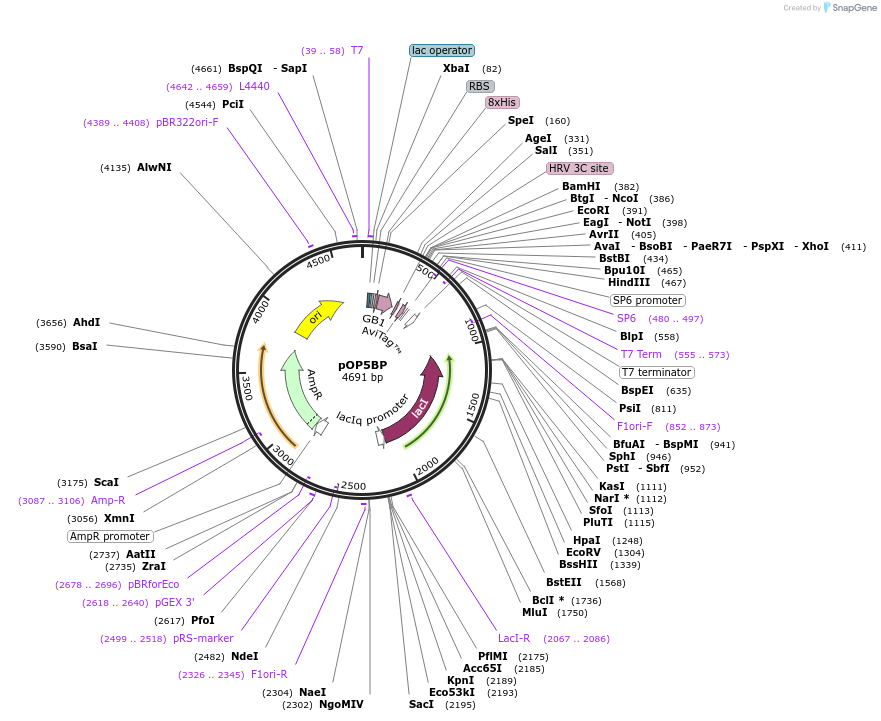
pOP5BP
Plasmid#112608PurposeUsed for expression of protein in E. coli as PreScission cleavable N-terminual 8His and GB1 tag fusions and with C-terminal Avi-tagDepositorTypeEmpty backboneTags8His-GB1 and AviExpressionBacterialAvailable SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
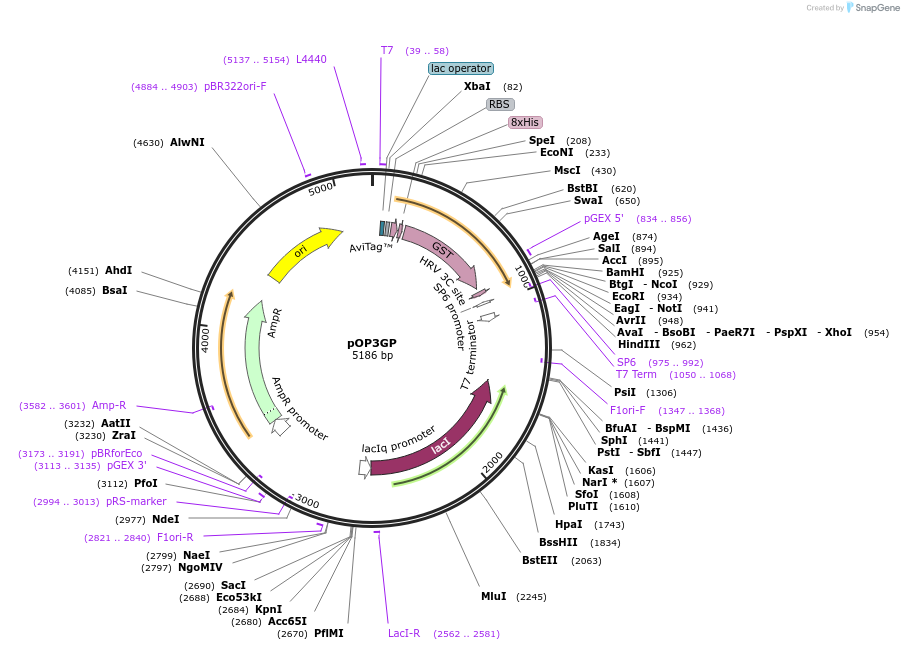
pOP3GP
Plasmid#112606PurposeUsed for expression of protein in E. coli as PreScission cleavable N-terminal Avi-His8-tag and GST fusionsDepositorTypeEmpty backboneTagsAvi - 8His-GSTExpressionBacterialAvailable SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
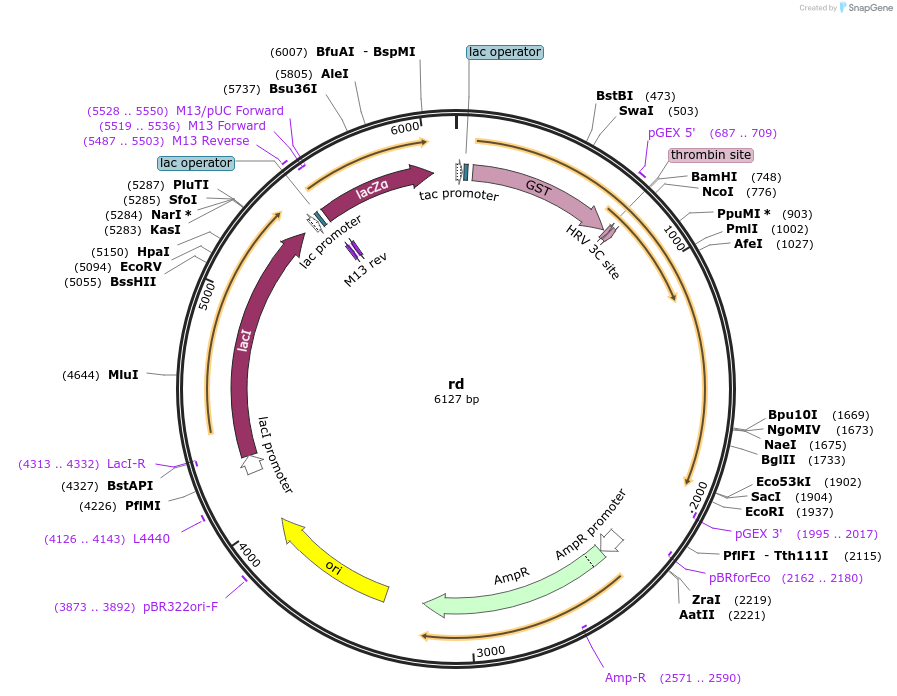
rd
Plasmid#112912PurposeExpress the PEST domain-replaced 48 kDa form of human dematin with a cleavable N-terminal GST tagDepositorInsertdematin (DMTN Human)
TagsGlutatione-S-Tranferase and precision protease si…ExpressionBacterialMutationPEST sequence near the N-terminal removed. 5′-GCG…PromoterT7Available SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
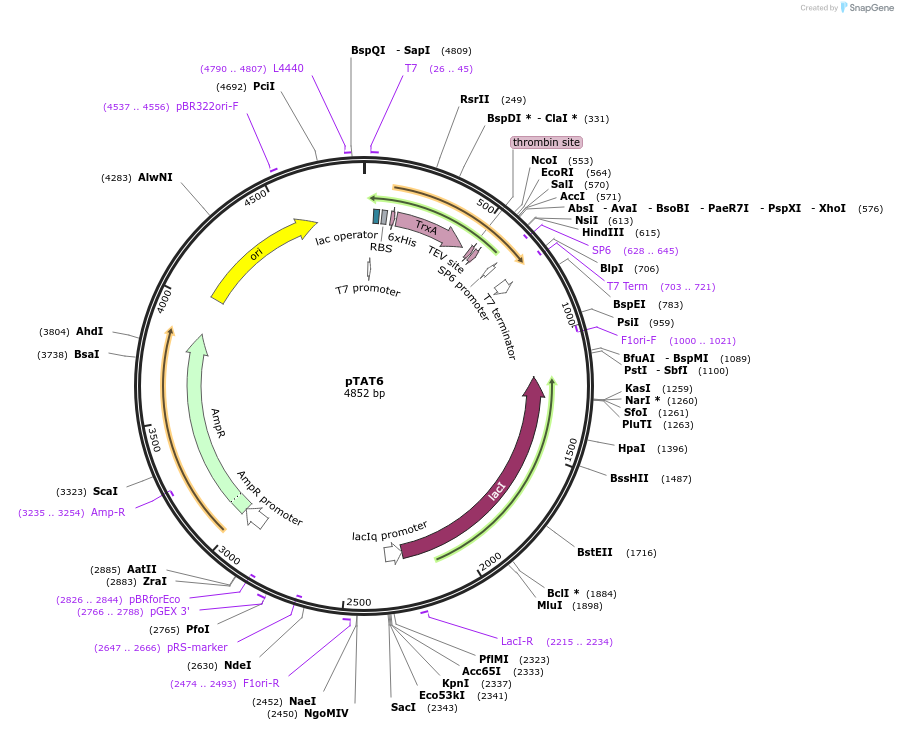
pTAT6
Plasmid#112594PurposeUsed for expression of protein in E. coli as Thrombin and TEV cleavable N-terminal 6His-tag-thioredoxin fusionsDepositorTypeEmpty backboneTags6xHis-thioredoxinExpressionBacterialAvailable SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
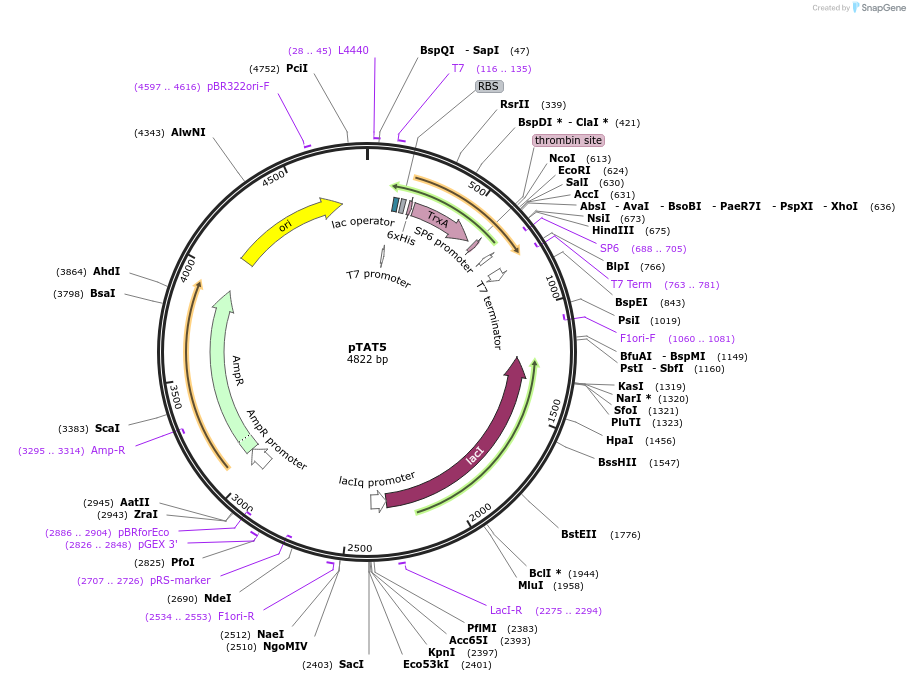
pTAT5
Plasmid#112593PurposeUsed for expression of protein in E. coli as Thrombin cleavable N-terminal 6His-tag-thioredoxin fusionsDepositorTypeEmpty backboneTags6xHis-thioredoxinExpressionBacterialAvailable SinceAug. 27, 2018AvailabilityAcademic Institutions and Nonprofits only -
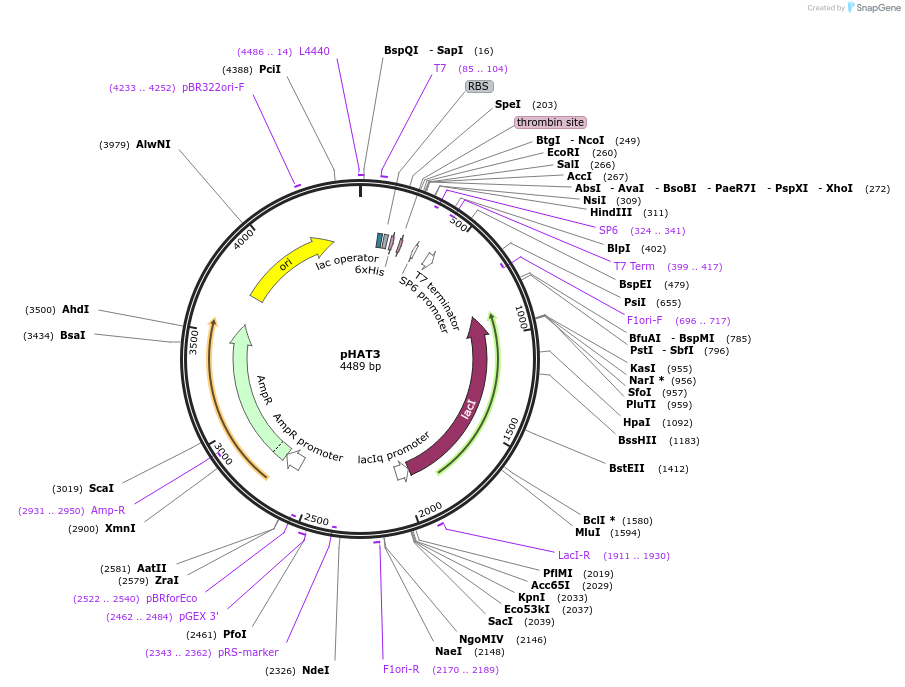
pHAT3
Plasmid#112584PurposeUsed for expression of protein in E. coli as Thrombin cleavable N-terminal 6His-tag fusionsDepositorTypeEmpty backboneTags6HisExpressionBacterialAvailable SinceAug. 24, 2018AvailabilityAcademic Institutions and Nonprofits only -
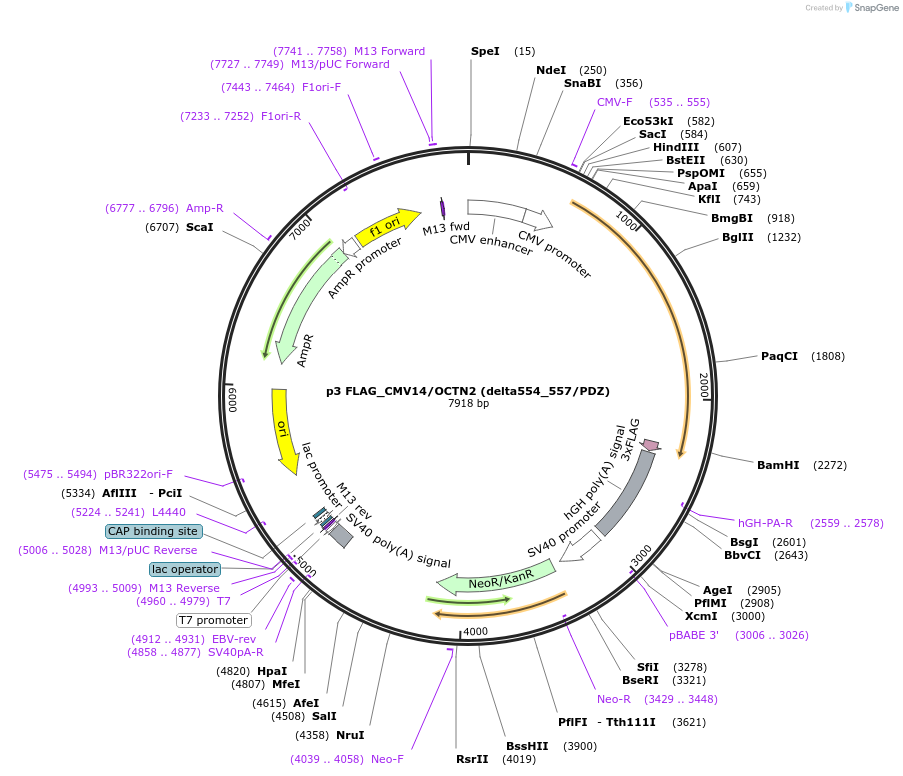
p3 FLAG_CMV14/OCTN2 (delta554_557/PDZ)
Plasmid#112803PurposeExpression of OCTN2 mutantDepositorInsertOCTN2/SLC22A5 (Slc22a5 Rat)
Tags3 x FlagExpressionMammalianMutationamino acids 554_557 deleted/PDZAvailable SinceAug. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
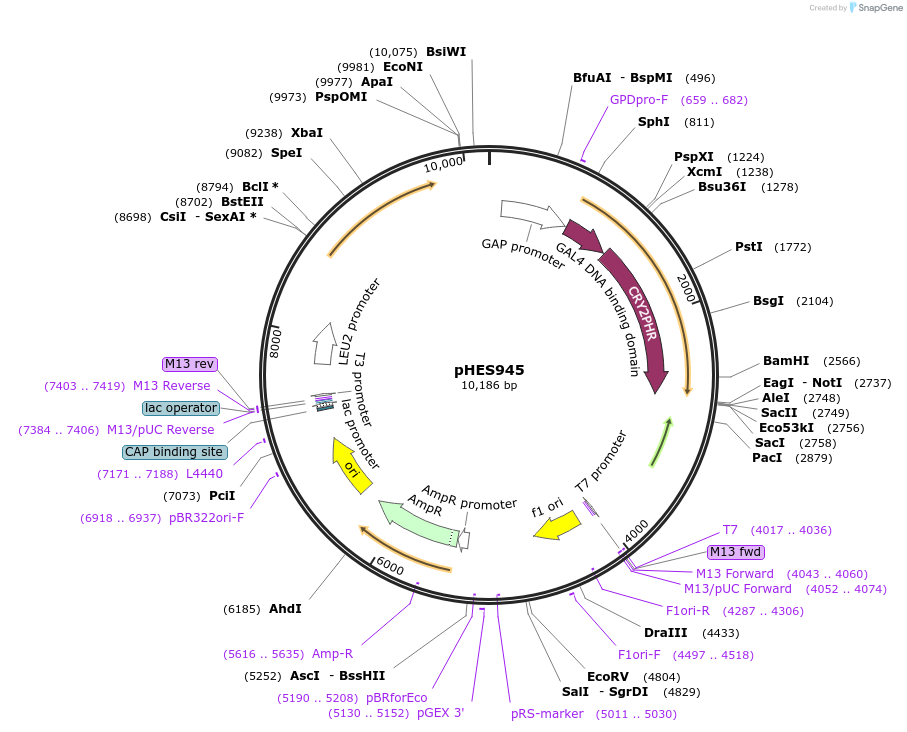
pHES945
Plasmid#112643Purposeblue light inducible cryptochrome based optogenetic expression systemDepositorInsertCRY2PHR-GAL4DBD
ExpressionYeastPromoterTDH3Available SinceAug. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
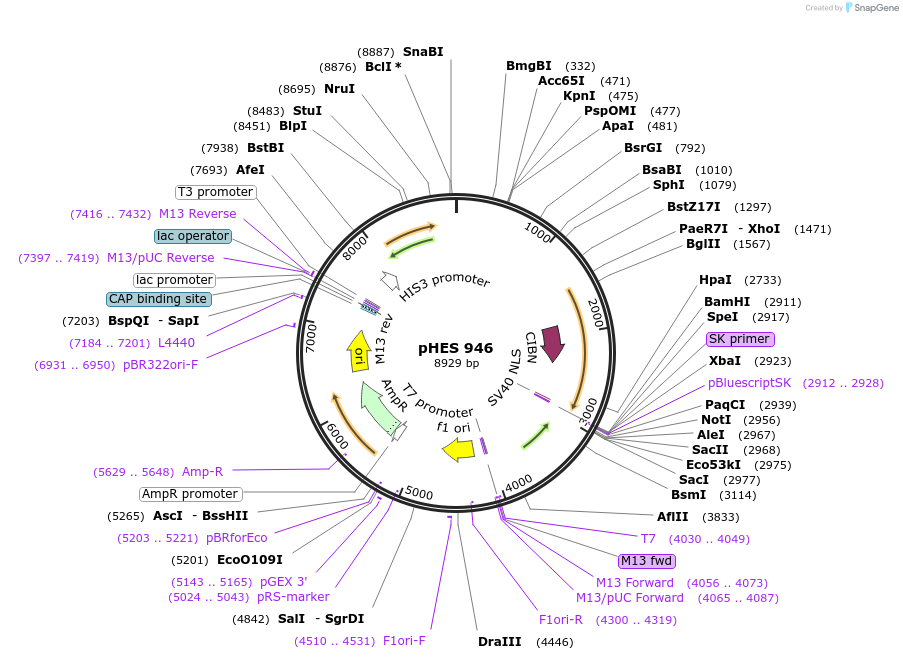
pHES 946
Plasmid#112644Purposeblue light inducible cryptochrome based optogenetic expression systemDepositorInsertRTG3AD-GSx7-CIB1-SV40NLS (CIB1 Synthetic, Mustard Weed, Budding Yeast)
ExpressionYeastPromoterADH1Available SinceAug. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
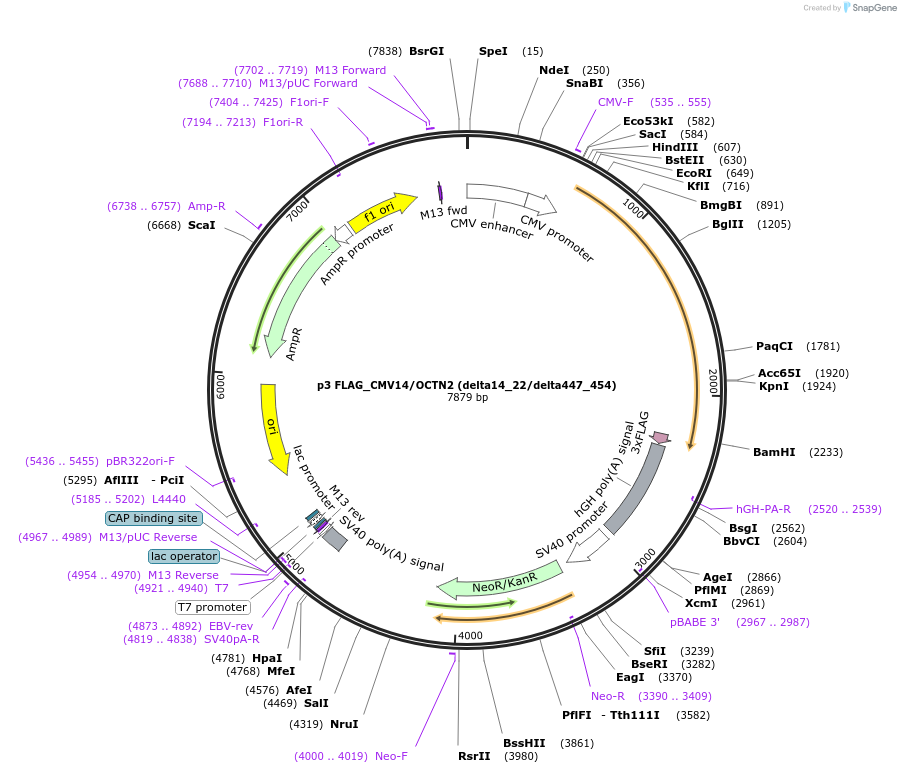
p3 FLAG_CMV14/OCTN2 (delta14_22/delta447_454)
Plasmid#112802PurposeExpression of OCTN2 mutantDepositorInsertOCTN2/SLC22A5 (Slc22a5 Rat)
Tags3 x FlagExpressionMammalianMutationamino acids 14_22 and 447_454 deletedAvailable SinceAug. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
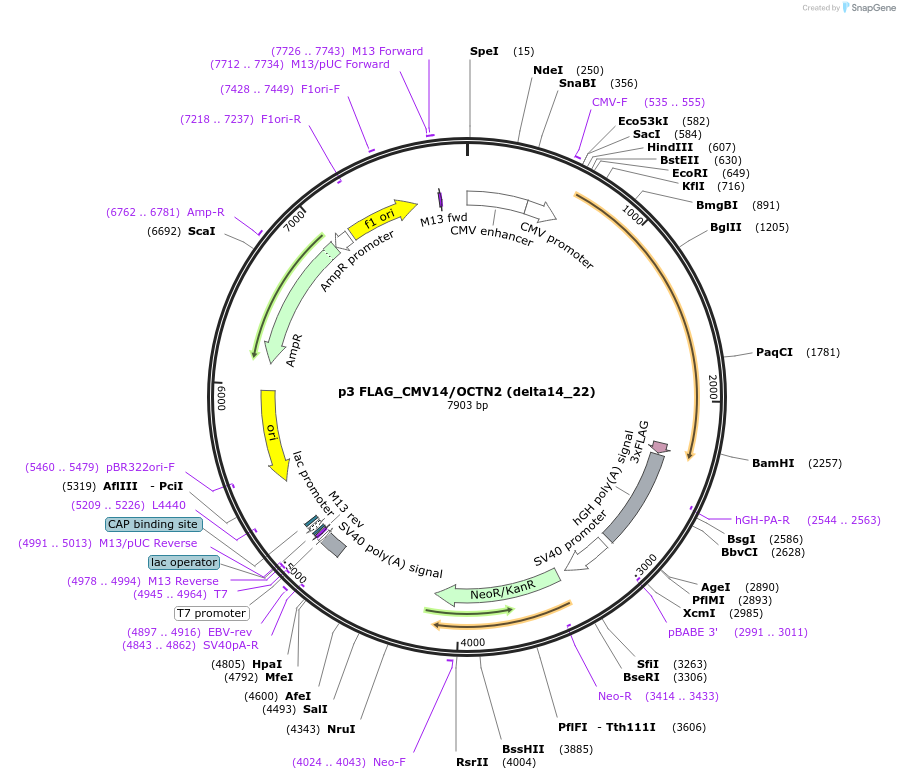
p3 FLAG_CMV14/OCTN2 (delta14_22)
Plasmid#112800PurposeExpression of OCTN2 mutantDepositorInsertOCTN2/SLC22A5 (Slc22a5 Rat)
Tags3 x FlagExpressionMammalianMutationamino acids 14_22 deletedAvailable SinceAug. 23, 2018AvailabilityAcademic Institutions and Nonprofits only -
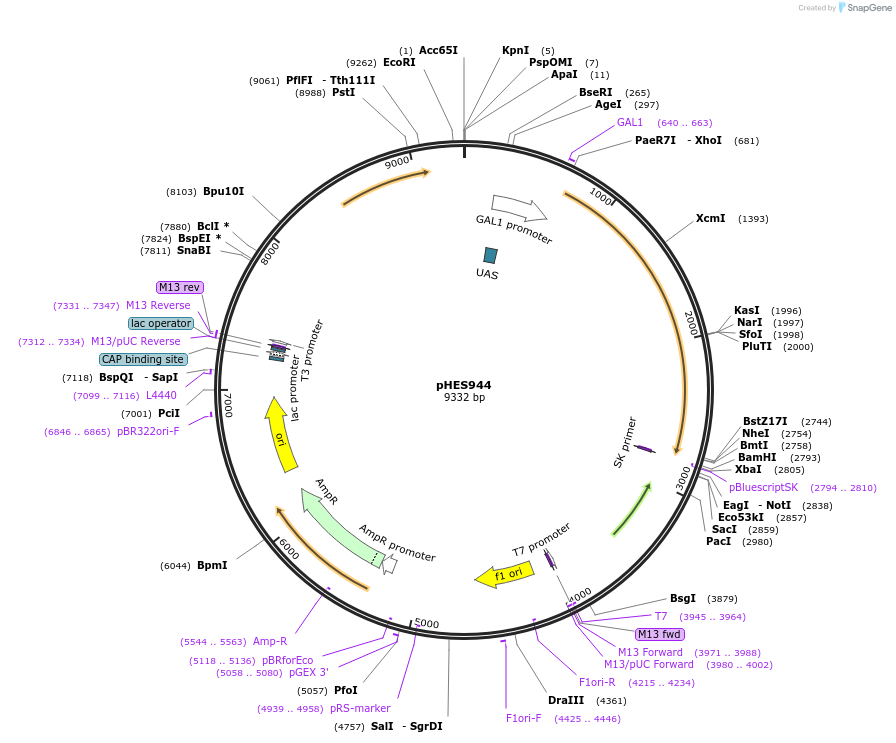
pHES944
Plasmid#112642Purposeexpress SST2 from the GAL1 promoterDepositorInsertSST2
ExpressionYeastPromoterGAL1Available SinceAug. 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
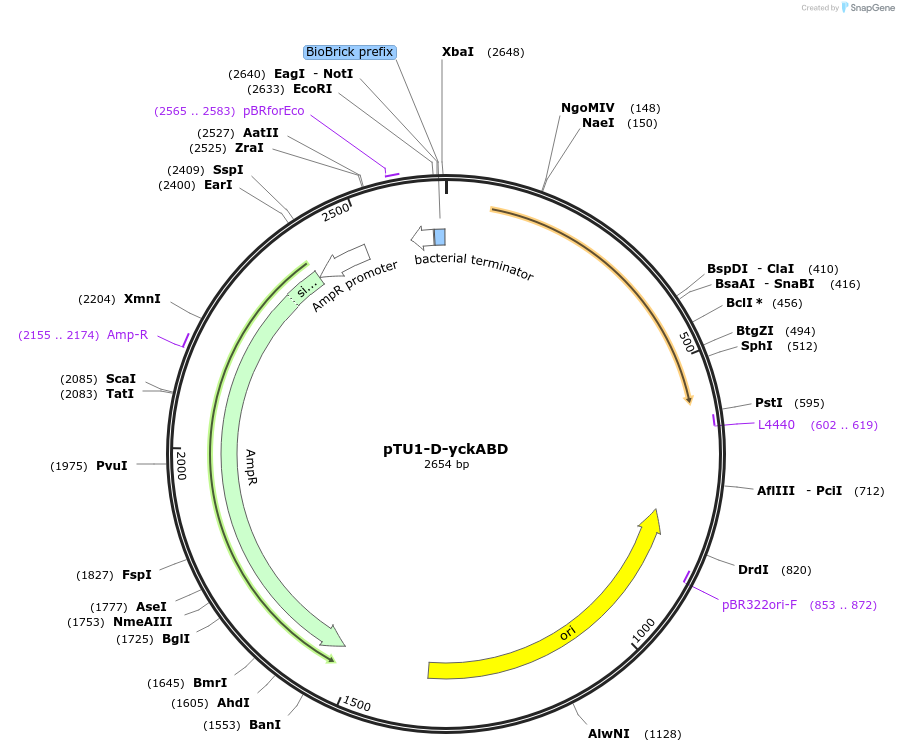
pTU1-D-yckABD
Plasmid#112790PurposeDownstream homology arm, Pair of HAs spans yckA and yckB, which share homology with ABC transporters in B. subtilisDepositorArticleInsertyckABD
ExpressionBacterialAvailable SinceAug. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
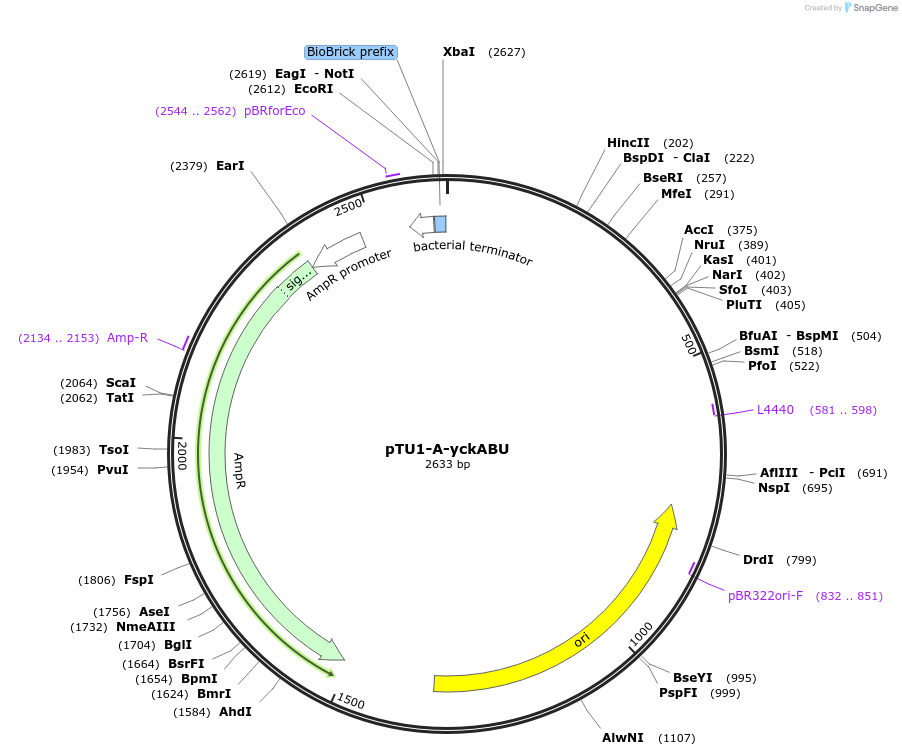
pTU1-A-yckABU
Plasmid#112789PurposeUpstream homology arm, Pair of HAs spans yckA and yckB, which share homology with ABC transporters in B. subtilisDepositorArticleInsertyckABU
ExpressionBacterialAvailable SinceAug. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
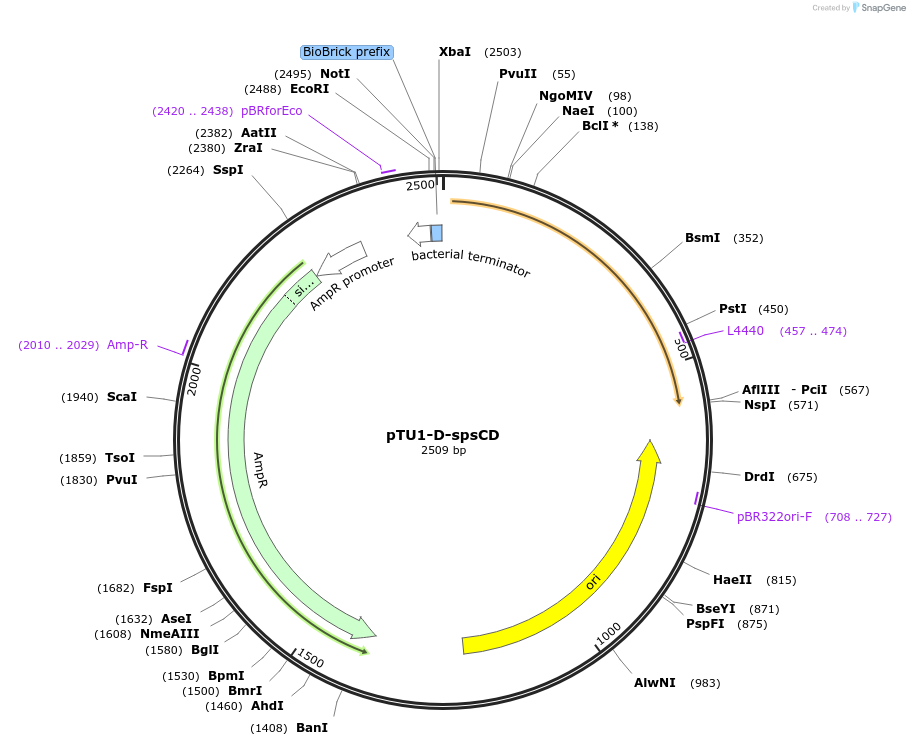
pTU1-D-spsCD
Plasmid#112788PurposeDownstream homology arm, SpsC is involved in spore coat polysaccharide synthesisDepositorArticleInsertspsCD
ExpressionBacterialAvailable SinceAug. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
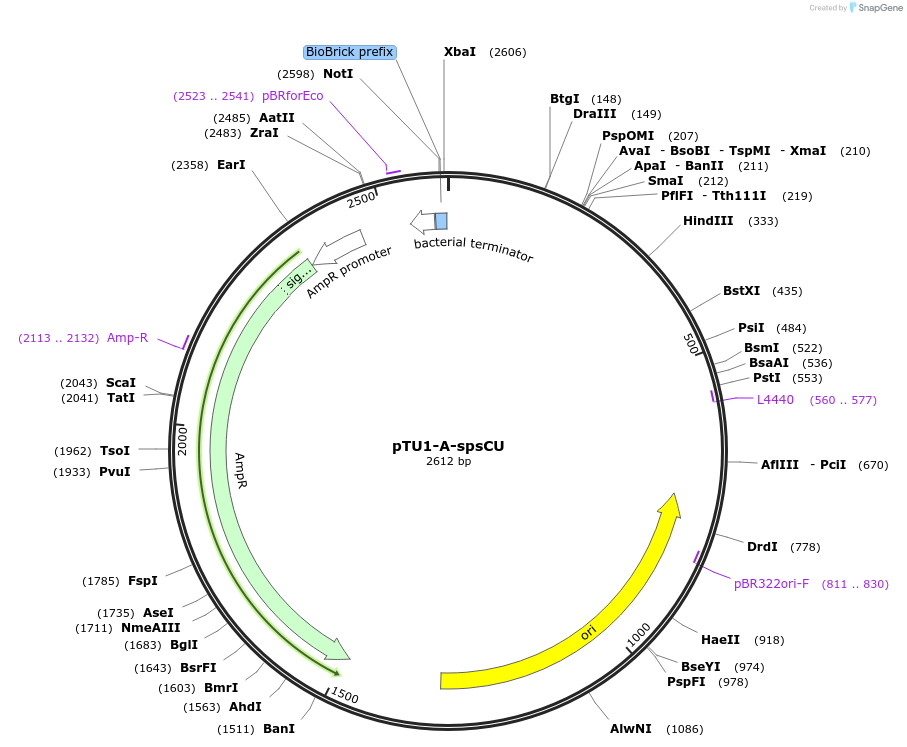
pTU1-A-spsCU
Plasmid#112787PurposeUpstream homology arm, SpsC is involved in spore coat polysaccharide synthesisDepositorArticleInsertspsCU
ExpressionBacterialAvailable SinceAug. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
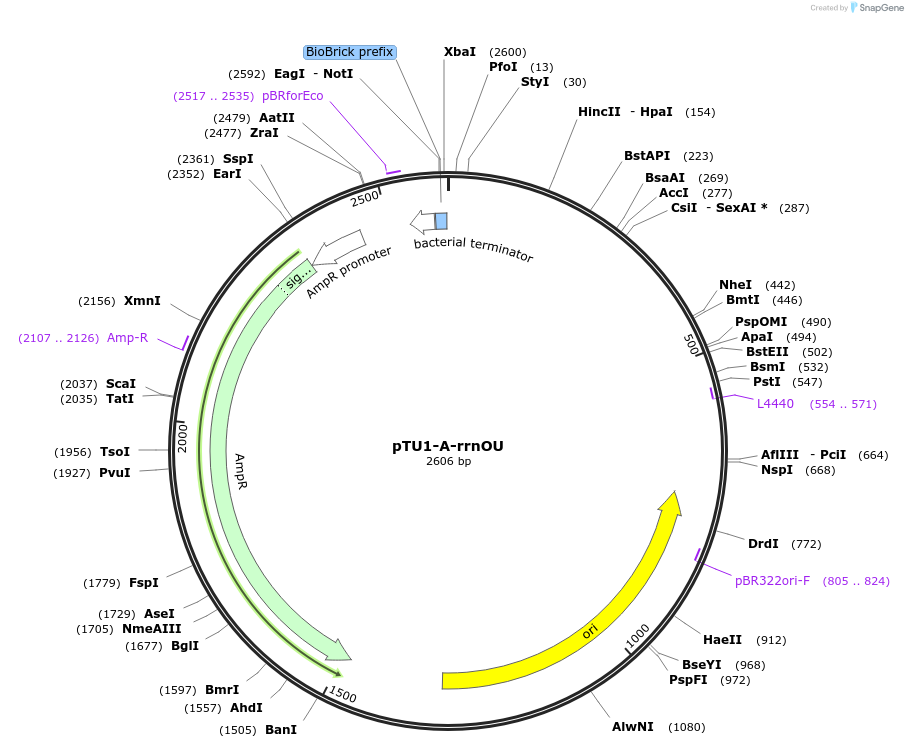
pTU1-A-rrnOU
Plasmid#112785PurposeUpstream homology arm, rrnO-23S (23S rRNA)DepositorArticleInsertrrnOU
ExpressionBacterialAvailable SinceAug. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
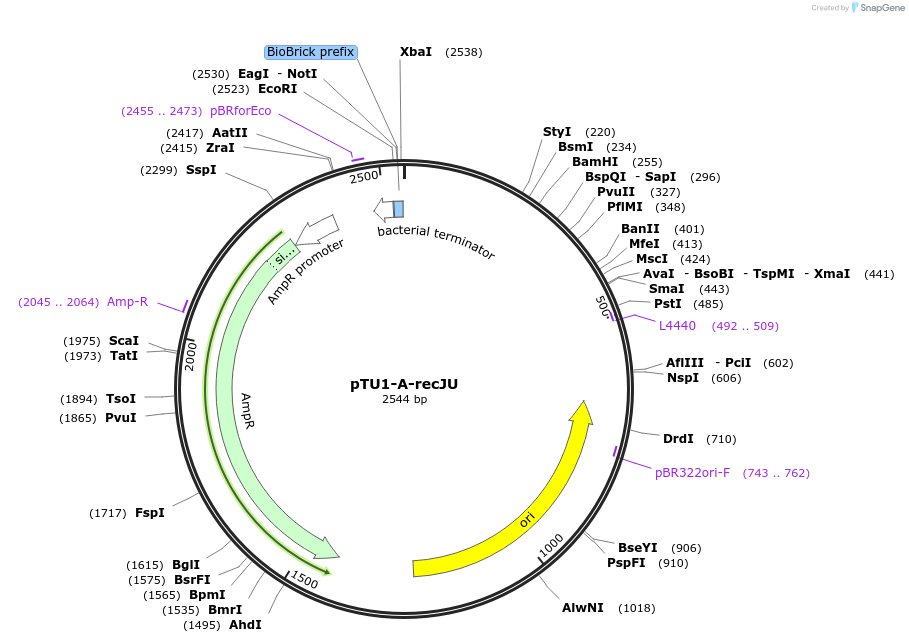
pTU1-A-recJU
Plasmid#112783PurposeUpstream homology arm, RecJ is a ssDNA specifiC exonuclease, involved in genome repair and maintenanceDepositorArticleInsertrecJU
ExpressionBacterialAvailable SinceAug. 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
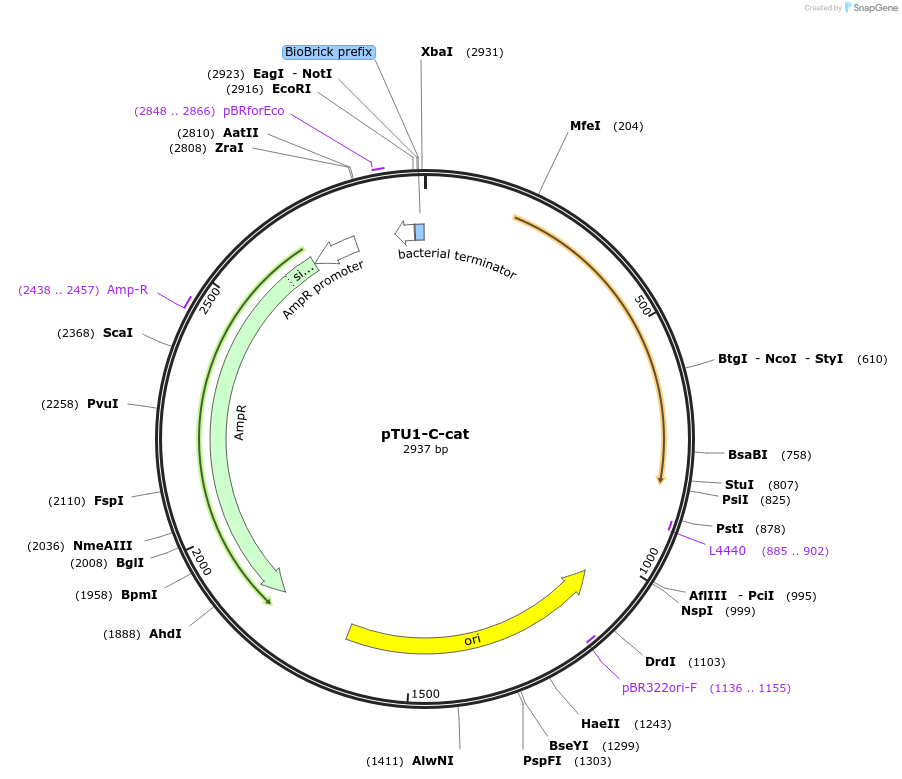
pTU1-C-cat
Plasmid#112791PurposeChloramphenicol resistance cassette (expressing chloramphenicol acetyltransferase)DepositorArticleInsertcat
ExpressionBacterialAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
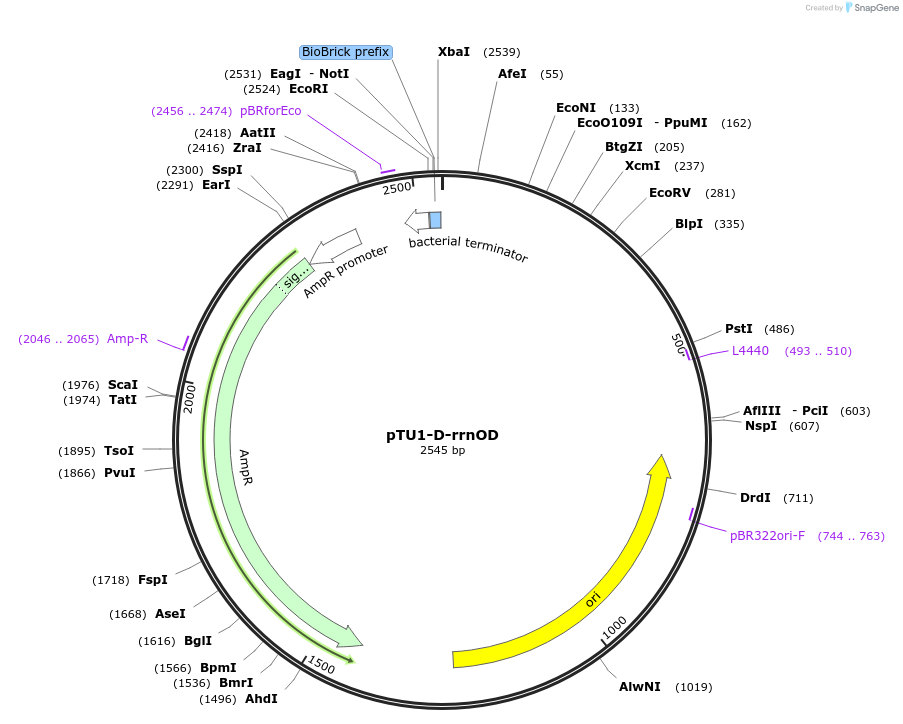
pTU1-D-rrnOD
Plasmid#112786PurposeDownstream homology arm, rrnO-23S (23S rRNA)DepositorArticleInsertrrnOD
ExpressionBacterialAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
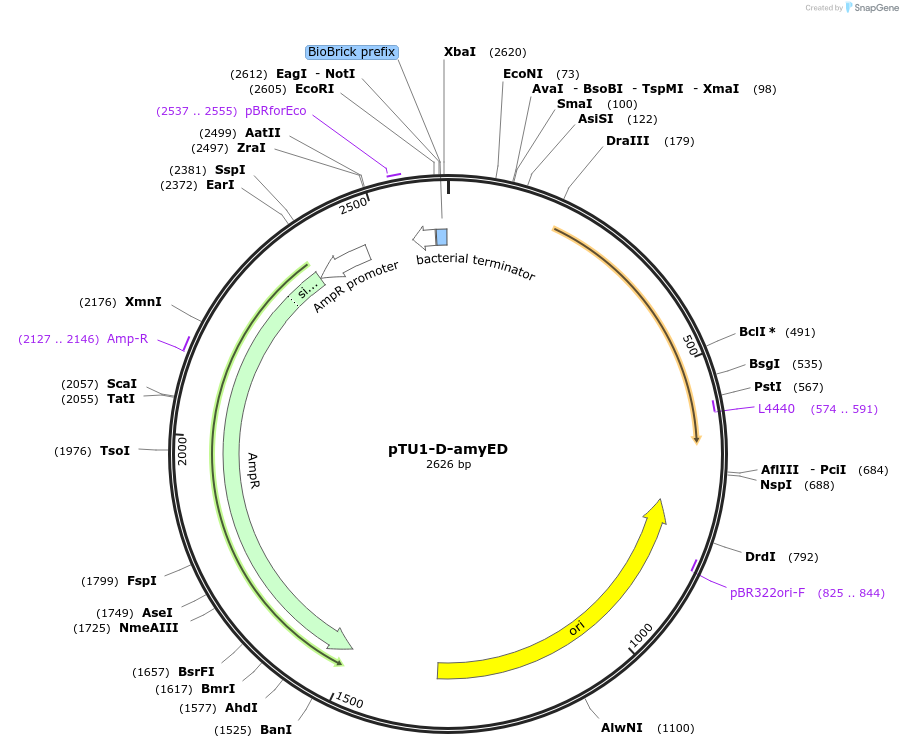
pTU1-D-amyED
Plasmid#112782PurposeDownstream homology arm, amyE encodes _-amylaseDepositorArticleInsertamyED
ExpressionBacterialAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
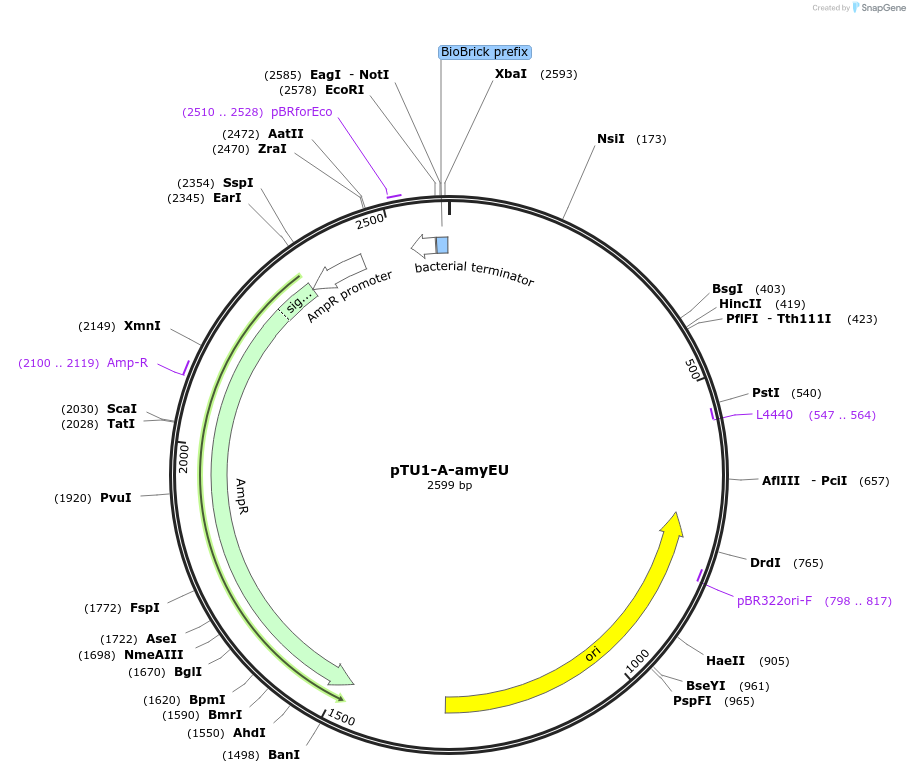
pTU1-A-amyEU
Plasmid#112781PurposeUpstream homology arm, amyE encodes _-amylaseDepositorArticleInsertamyEU
ExpressionBacterialAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
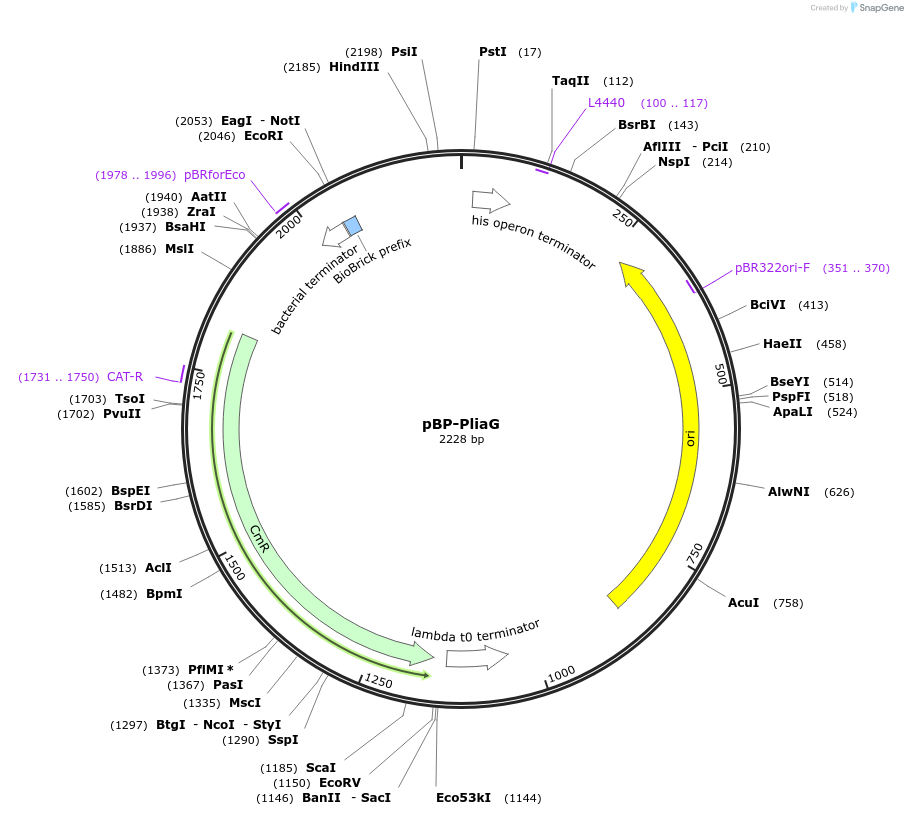
pBP-PliaG
Plasmid#112777PurposeWeak constitutive promoter of liaGFSR operon involved in detection of cell wall damageDepositorArticleInsertPliaG promoter
ExpressionBacterialAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
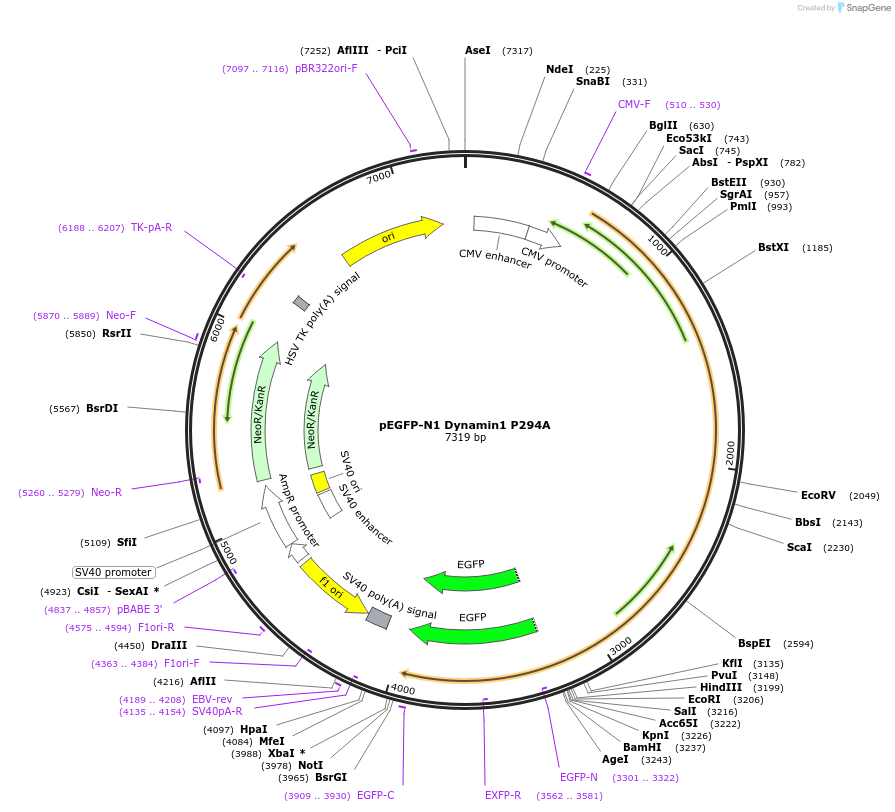
pEGFP-N1 Dynamin1 P294A
Plasmid#112102PurposeMammalian expression of GFP-tagged Dynamin protein.DepositorAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
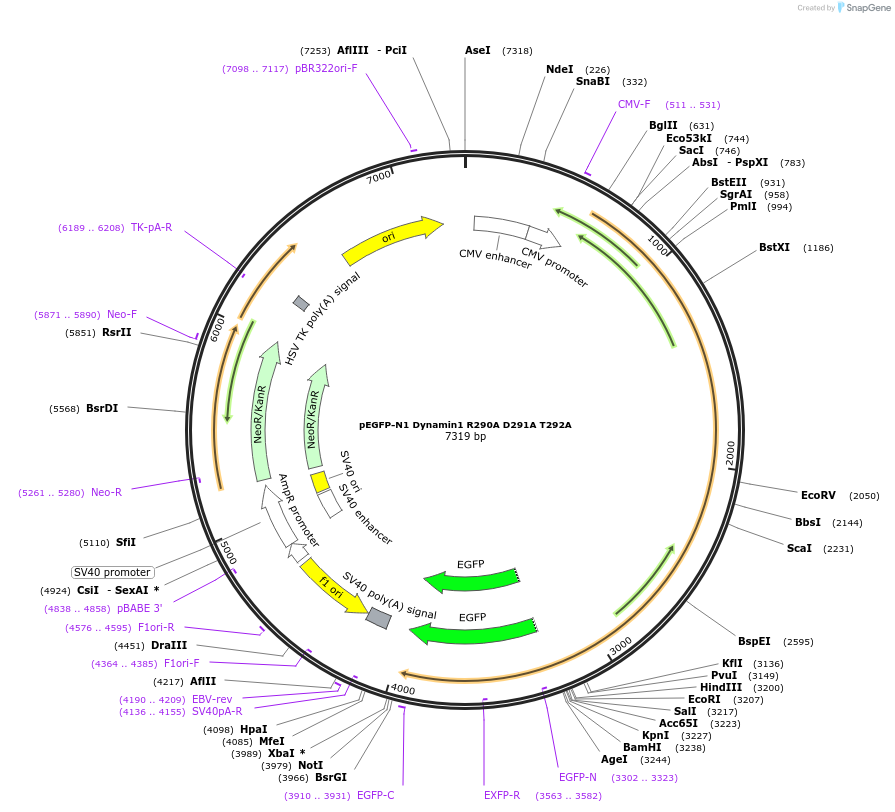
pEGFP-N1 Dynamin1 R290A D291A T292A
Plasmid#112103PurposeMammalian expression of GFP-tagged Dynamin protein.DepositorAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
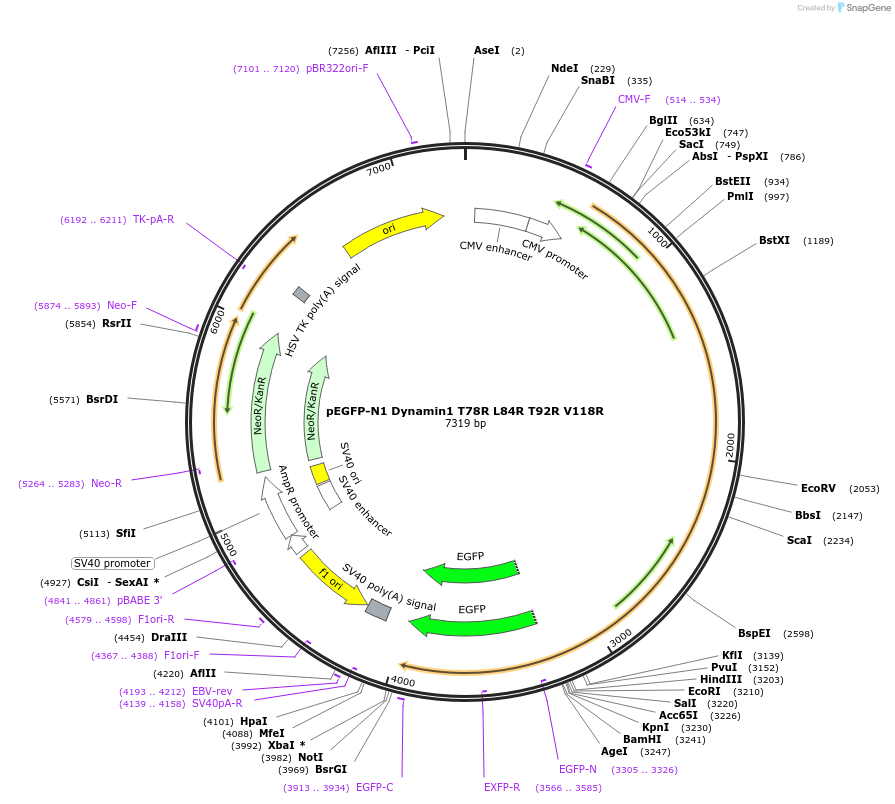
pEGFP-N1 Dynamin1 T78R L84R T92R V118R
Plasmid#112106PurposeMammalian expression plasmid of GFP-tagged Dynamin protein.DepositorInsertDynamin1 (DNM1 Human)
TagsEGFPExpressionMammalianMutationT78R L84R T92R V118RPromoterCMVAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
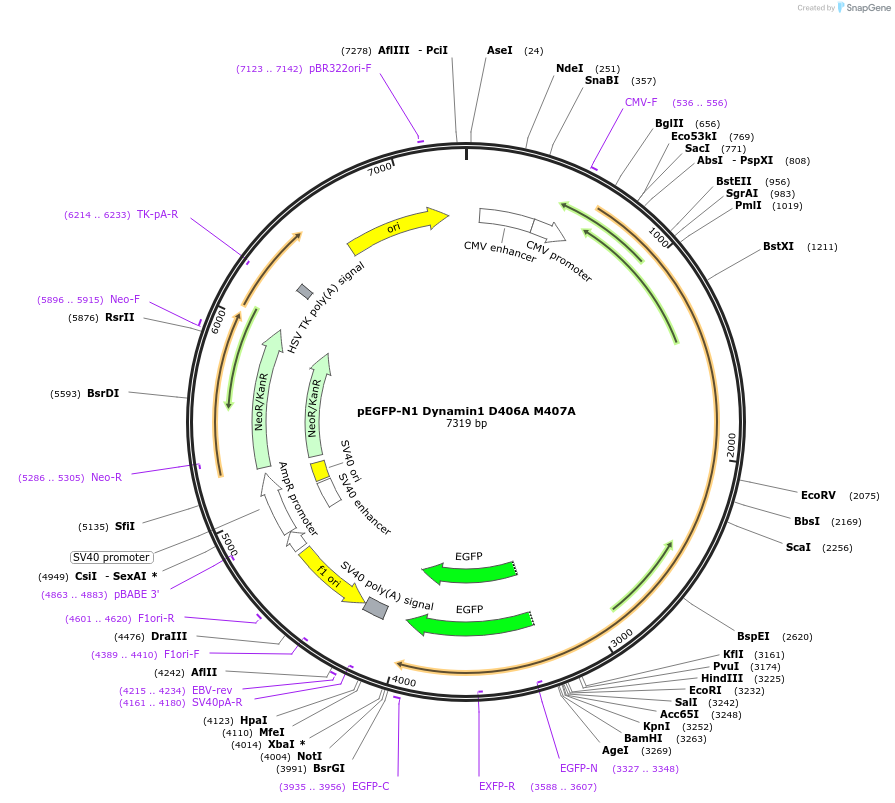
pEGFP-N1 Dynamin1 D406A M407A
Plasmid#112107PurposeMammalian expression plasmid of GFP-tagged Dynamin protein.DepositorAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
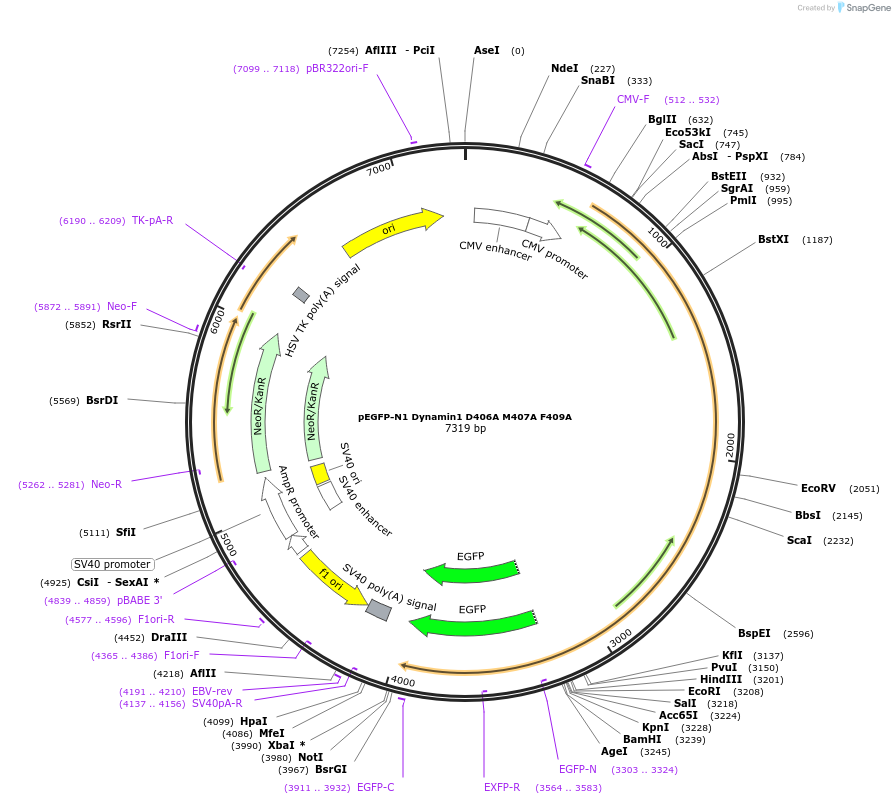
pEGFP-N1 Dynamin1 D406A M407A F409A
Plasmid#112108PurposeMammalian expression plasmid of GFP-tagged Dynamin protein.DepositorAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
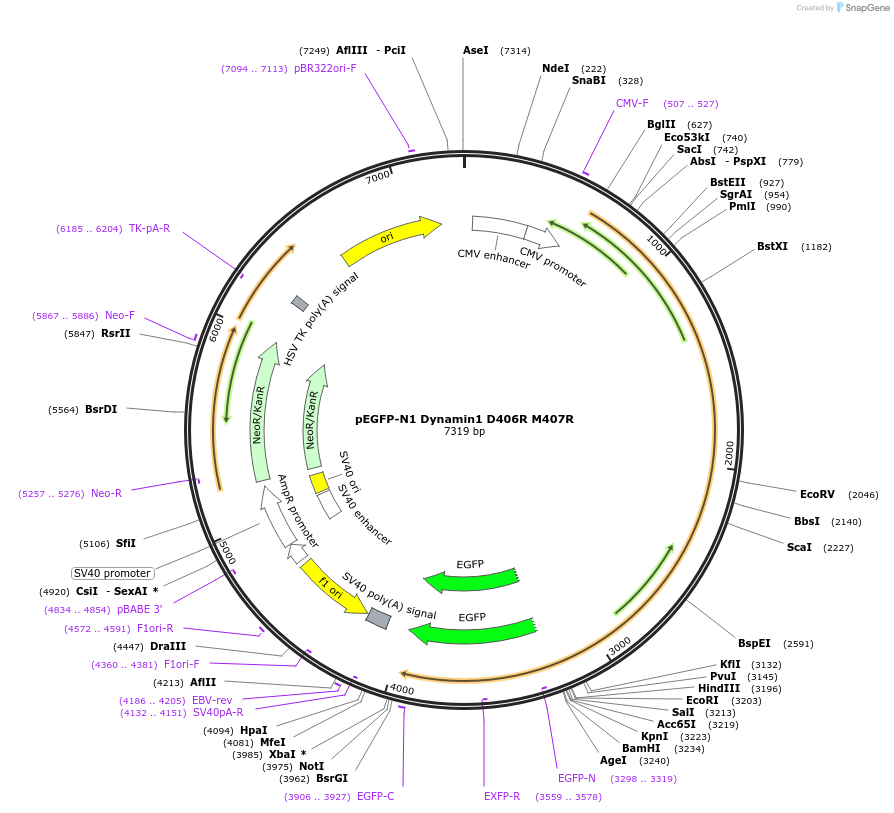
pEGFP-N1 Dynamin1 D406R M407R
Plasmid#112109PurposeMammalian expression plasmid of GFP-tagged Dynamin protein.DepositorAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
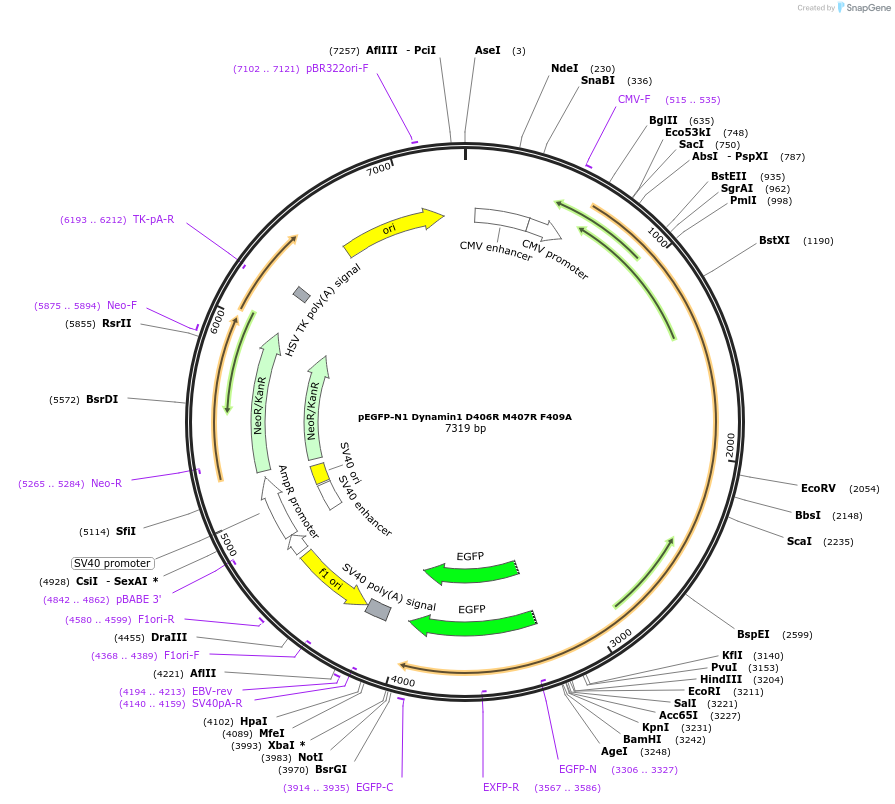
pEGFP-N1 Dynamin1 D406R M407R F409A
Plasmid#112110PurposeMammalian expression plasmid of GFP-tagged Dynamin protein.DepositorAvailable SinceAug. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
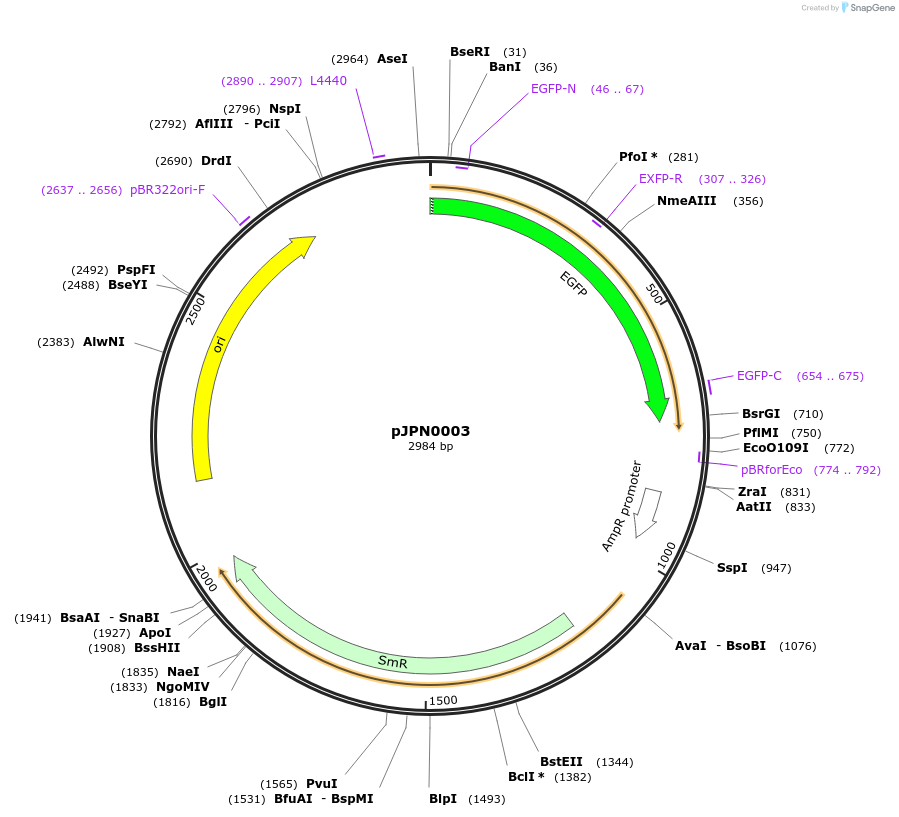
pJPN0003
Plasmid#112080PurposeGFP module NT2DepositorInsertGFP
ExpressionPlantMutationBsaI and BpiI restriction sites have been remove…Available SinceAug. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
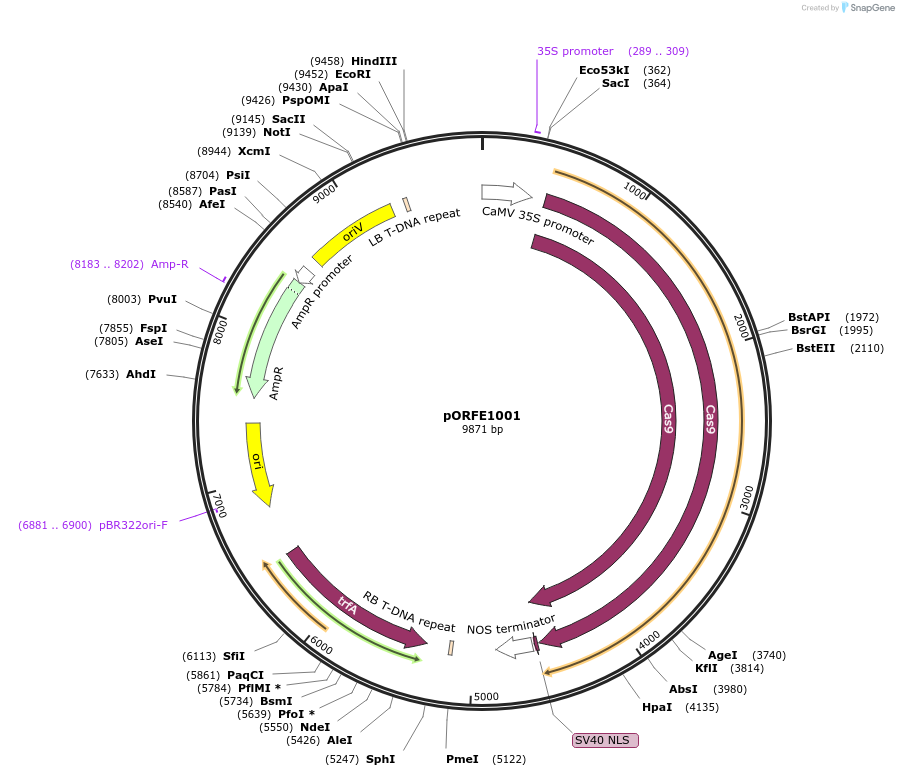
pORFE1001
Plasmid#112079PurposeOverexpression of the AtCAS9 in plant under 2x35S promoterDepositorInsertAtCAS9
UseCRISPRExpressionPlantMutationPlant codon optimized Cas9 S. pyogenesPromoter2x35SAvailable SinceAug. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
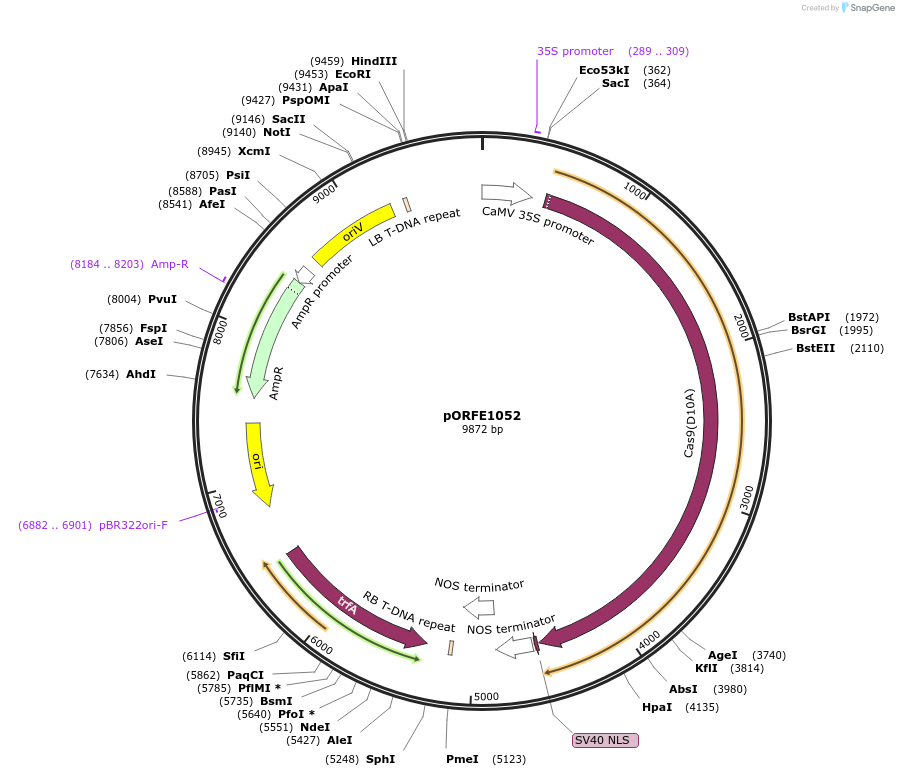
pORFE1052
Plasmid#112078PurposeOverexpression of the AtCAS9 nickase in plant under 2x35S promoterDepositorInsertAtCAS9 D10A
UseCRISPRExpressionPlantMutationPlant codon optimized nCas9 D10A S. pyogenesPromoter2x35SAvailable SinceAug. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
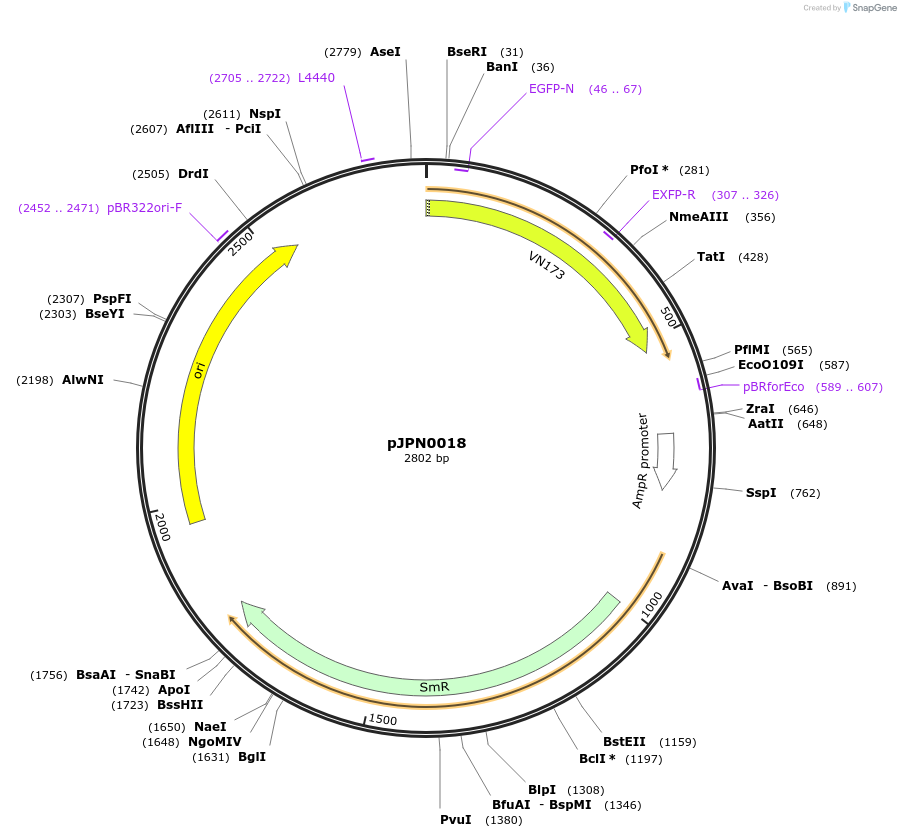
pJPN0018
Plasmid#112083PurposeeCFP module CTDepositorInserteCFP
ExpressionPlantMutationBsaI and BpiI restriction sites have been remove…Available SinceAug. 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
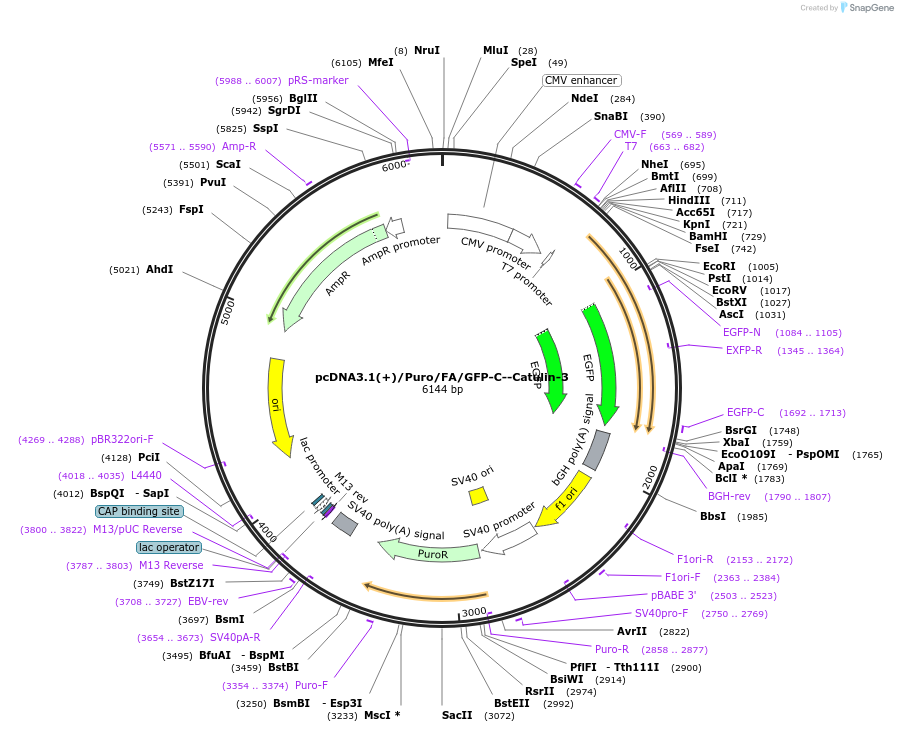
pcDNA3.1(+)/Puro/FA/GFP-C--Catulin-3
Plasmid#112843Purposetransient/stable overexpression of the C-terminal domain of Catulin.DepositorAvailable SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
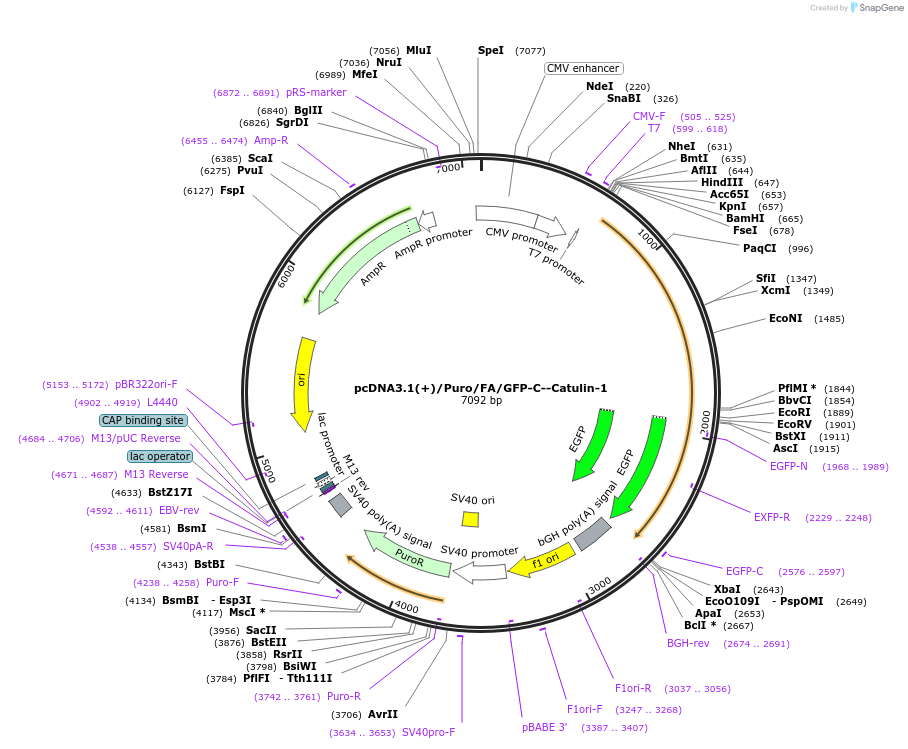
pcDNA3.1(+)/Puro/FA/GFP-C--Catulin-1
Plasmid#112841Purposetransient/stable overexpression of the N-terminal domain of Catulin.DepositorAvailable SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
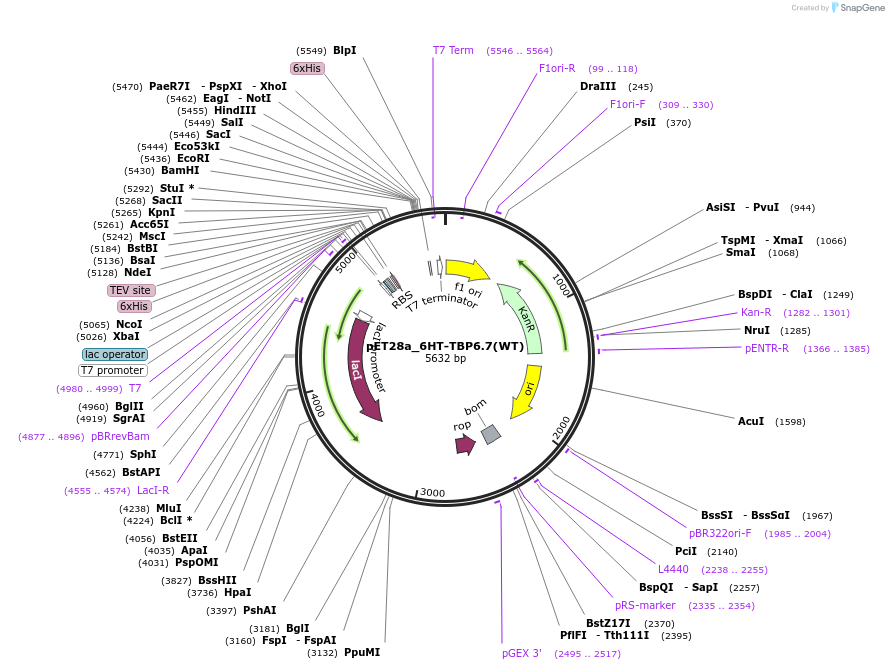
pET28a_6HT-TBP6.7(WT)
Plasmid#112720PurposeWild-type TBP6.7 was one of the first evolved TAR-binding proteins to bind with exceptional affinity. It was this protein that was used to obtain co-crystals with WT TAR RNA.DepositorInsert6His-TEV-TBP6.7(WT)
Tags6His-TEVExpressionBacterialPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
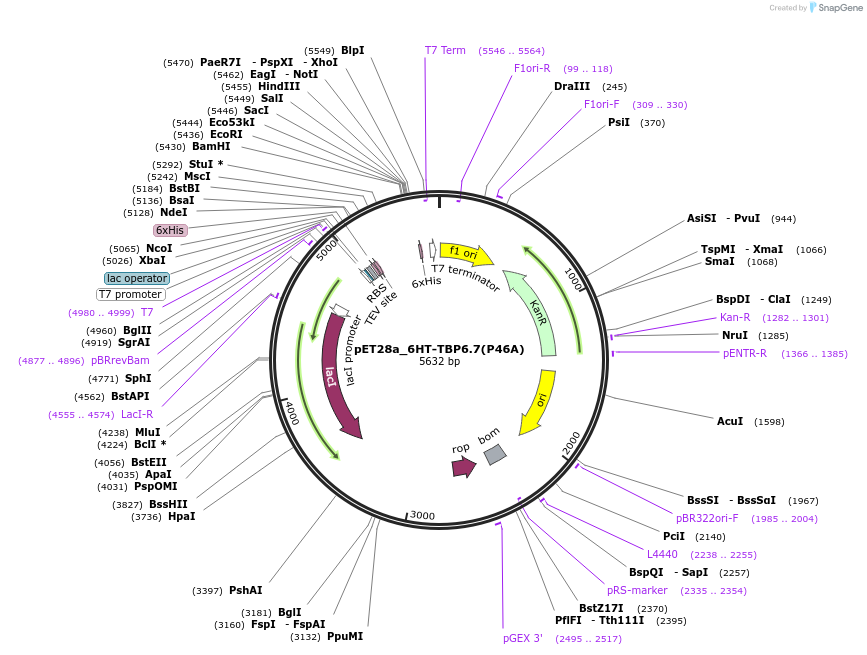
pET28a_6HT-TBP6.7(P46A)
Plasmid#112721PurposeThis mutation (P46A) marks the first residue within the evolved region of U1A.DepositorInsert6His-TEV-TBP6.7(P46A)
Tags6His-TEVExpressionBacterialMutationP46APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
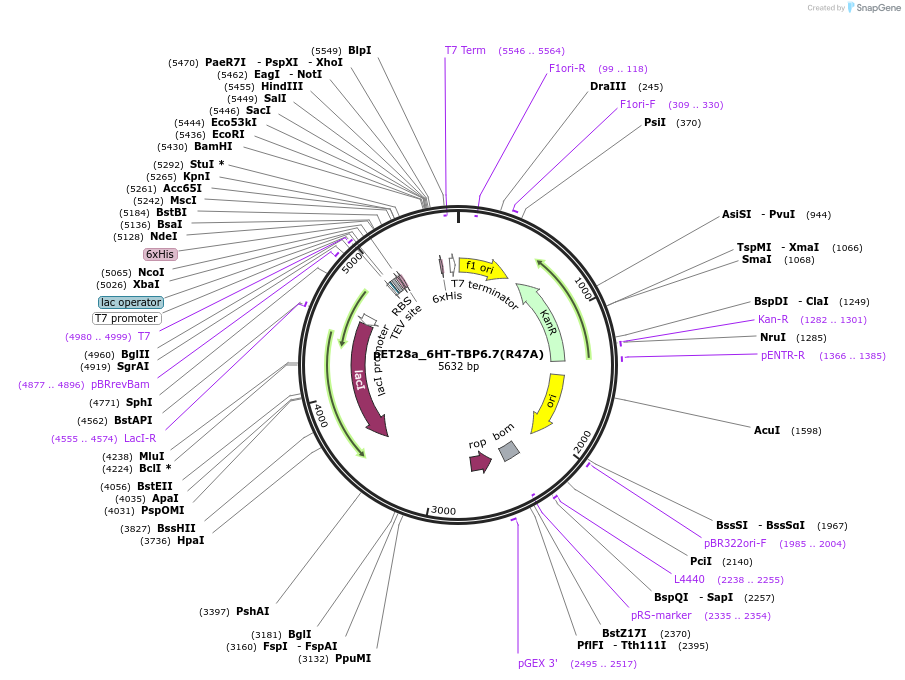
pET28a_6HT-TBP6.7(R47A)
Plasmid#112722PurposeArg47 within TBP6.7 is the most critical residue in terms of forming interactions with WT TAR. Mutating this amino acid to Ala results in ~600-fold reduction in Kd.DepositorInsert6His-TEV-TBP6.7(R47A)
Tags6His-TEVExpressionBacterialMutationR47APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
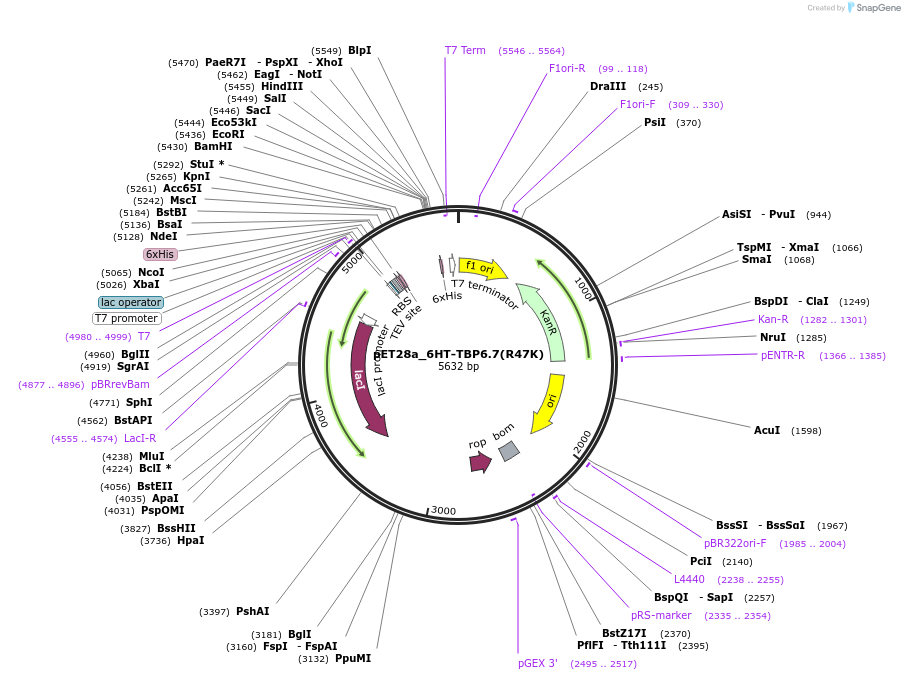
pET28a_6HT-TBP6.7(R47K)
Plasmid#112723PurposeArg47 of TBP6.7 interacts with Hoogsteen edge of TAR's Gua26. Mutating this amino acid to Lys reduces binding to the WT TAR RNA ~330-fold.DepositorInsert6His-TEV-TBP6.7(R47K)
Tags6His-TEVExpressionBacterialMutationR47KPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
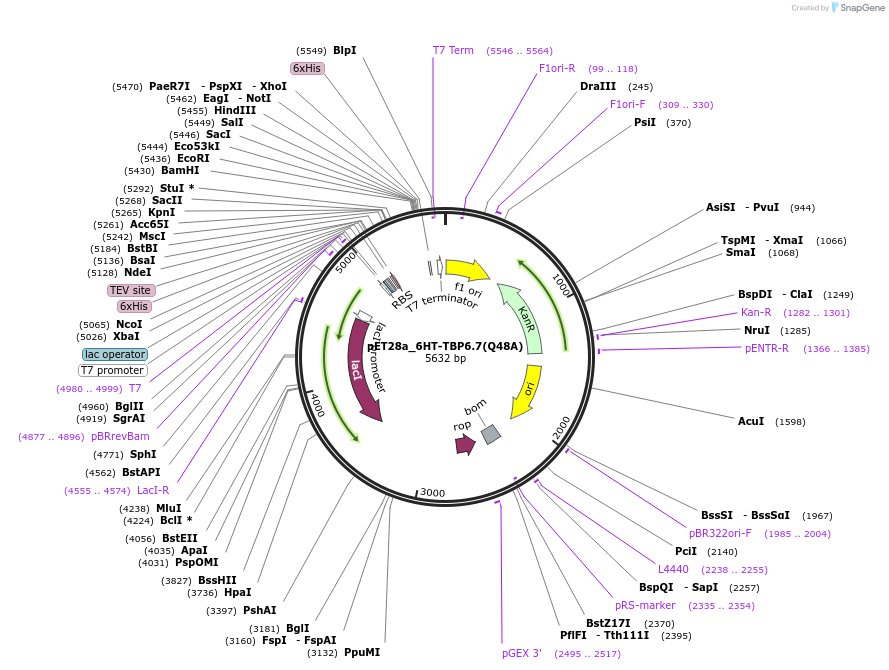
pET28a_6HT-TBP6.7(Q48A)
Plasmid#112724PurposeGln48 contacts the backbone of TAR RNA as well as mediates intramoecular interaction to stabilize the conformation of beta2-beta3 loop within TBP6.7.DepositorInsert6His-TEV-TBP6.7(Q48A)
Tags6His-TEVExpressionBacterialMutationQ48APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
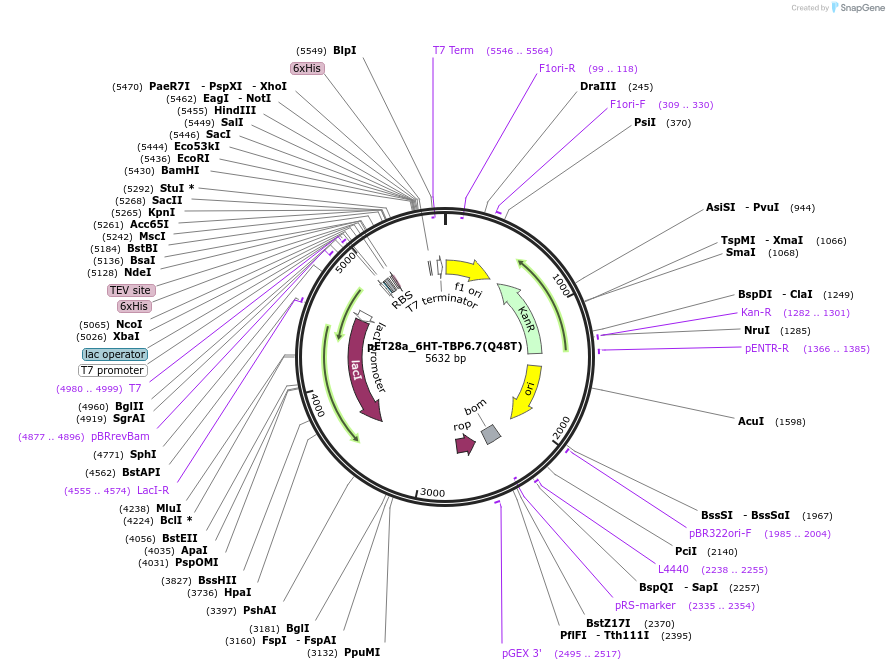
pET28a_6HT-TBP6.7(Q48T)
Plasmid#112725PurposeThis mutant of TBP6.7 binds to TAR very similarly to TBP6.7(WT), with only 1.4-fold reduced Kd.DepositorInsert6His-TEV-TBP6.7(Q48T)
Tags6His-TEVExpressionBacterialMutationQ48TPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
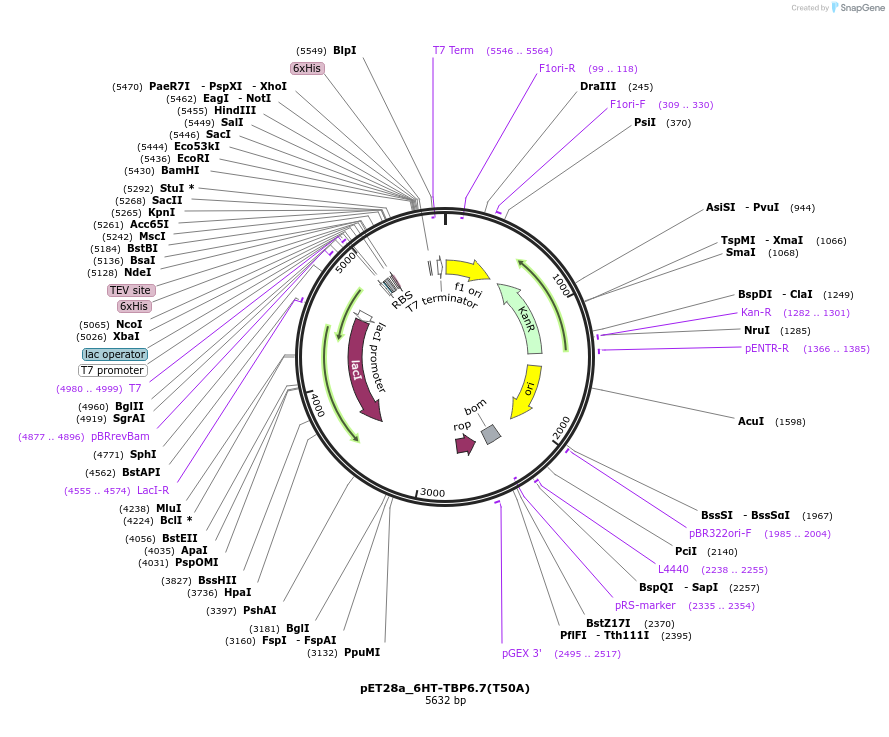
pET28a_6HT-TBP6.7(T50A)
Plasmid#112727PurposeLav-evolved amino acid T50 within TBPs is important in engaging intramolecularly to steer also lab-evolved R49 into conformation compatible binding to RNA.DepositorInsert6His-TEV-TBP6.7(T50A)
Tags6His-TEVExpressionBacterialMutationT50APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
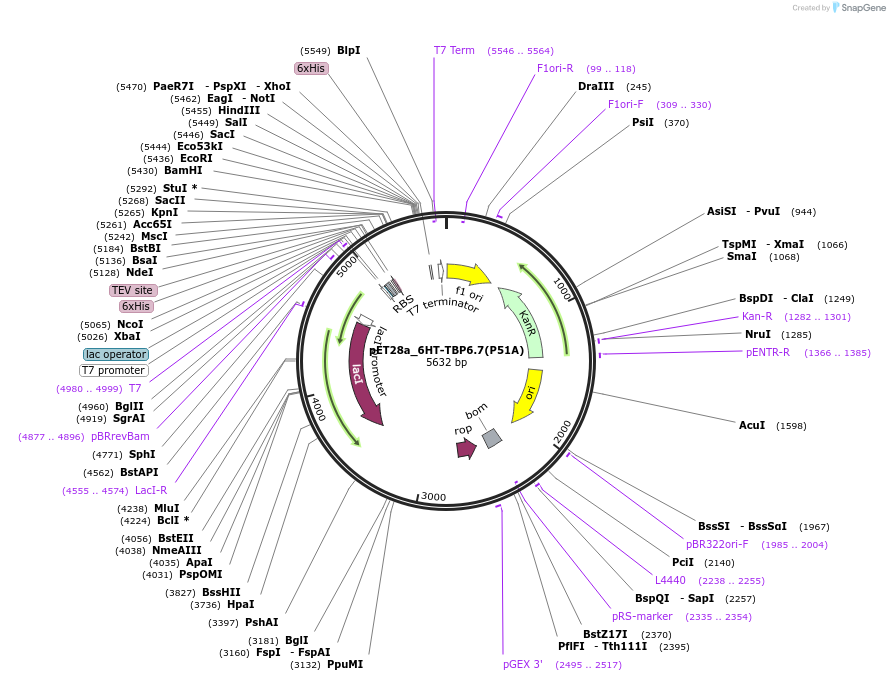
pET28a_6HT-TBP6.7(P51A)
Plasmid#112728PurposeSimilar to P46, P51 was selected and is present in all evolved TBPs. Mutating this position into Ala has a modest effect on binding.DepositorInsert6His-TEV-TBP6.7(P51A)
Tags6His-TEVExpressionBacterialMutationP51APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
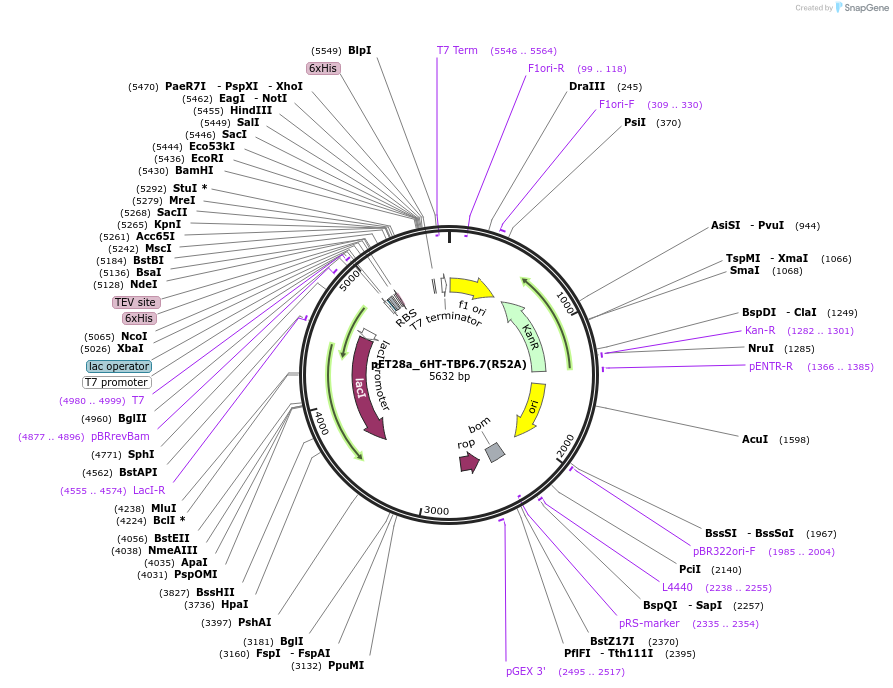
pET28a_6HT-TBP6.7(R52A)
Plasmid#112729Purpose52nd position is Arg in both wild-type U1A and TBPs, but the RNA recognition by this residue in both types of proteins is fundamentally different.DepositorInsert6His-TEV-TBP6.7(R52A)
Tags6His-TEVExpressionBacterialMutationR52APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
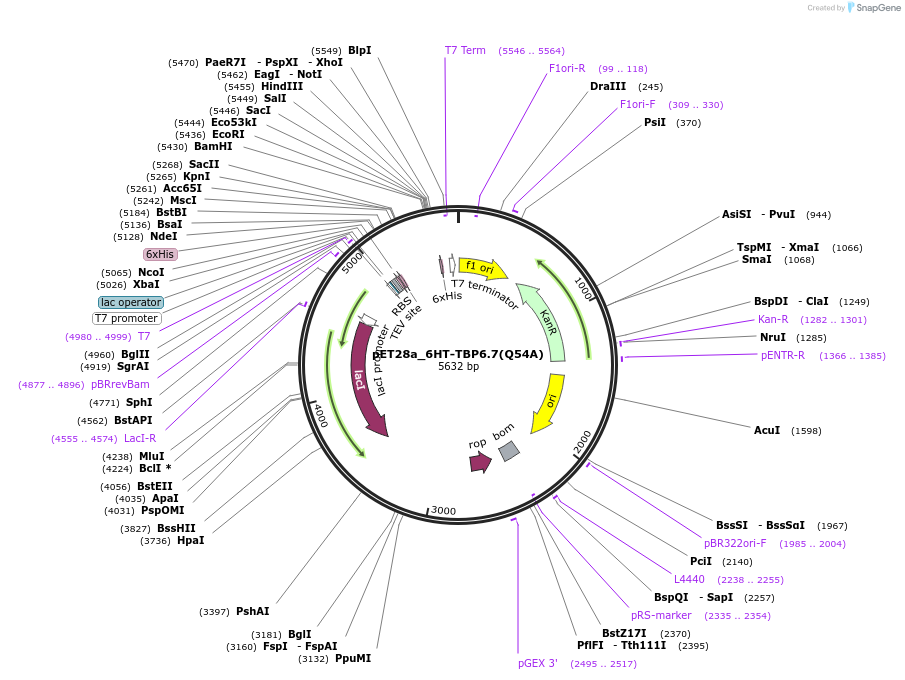
pET28a_6HT-TBP6.7(Q54A)
Plasmid#112730PurposeAlthough not evolved, this residue within TBPs participates in stabilizing the polypeptide loop responsible for RNA recognition.DepositorInsert6His-TEV-TBP6.7(Q54A)
Tags6His-TEVExpressionBacterialMutationQ54APromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
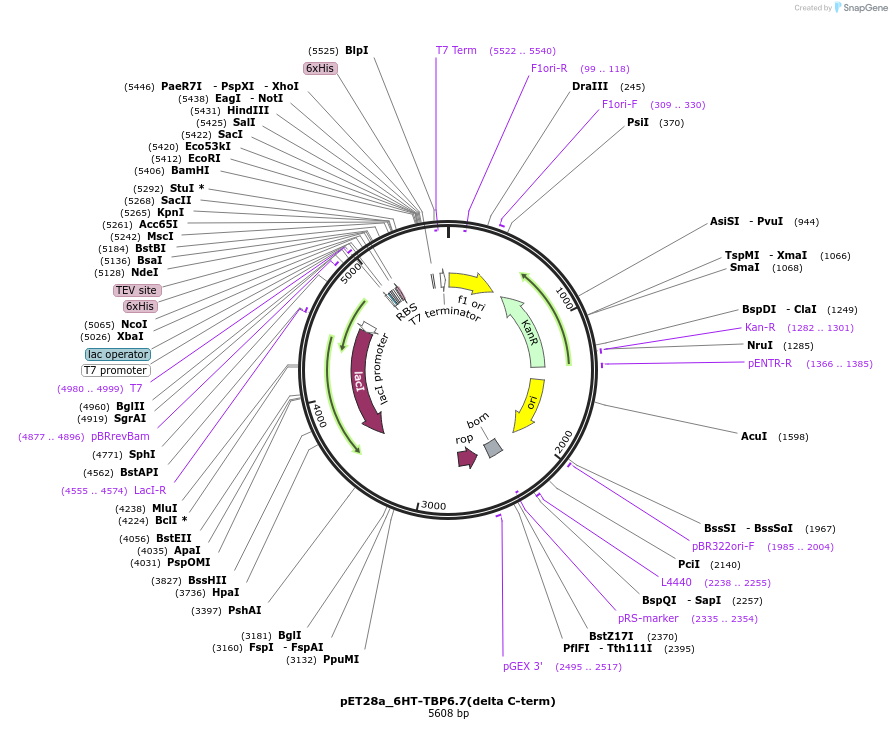
pET28a_6HT-TBP6.7(delta C-term)
Plasmid#112731PurposeThis mutant of TBP6.7 is lacking residues 91-98 at C-terminus, and was used to test the involvement of those residues in TAR recognition.DepositorInsert6His-TEV-TBP6.7(delta C-term)
Tags6His-TEVExpressionBacterialMutationdelta C-termPromoterT7Available SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only -
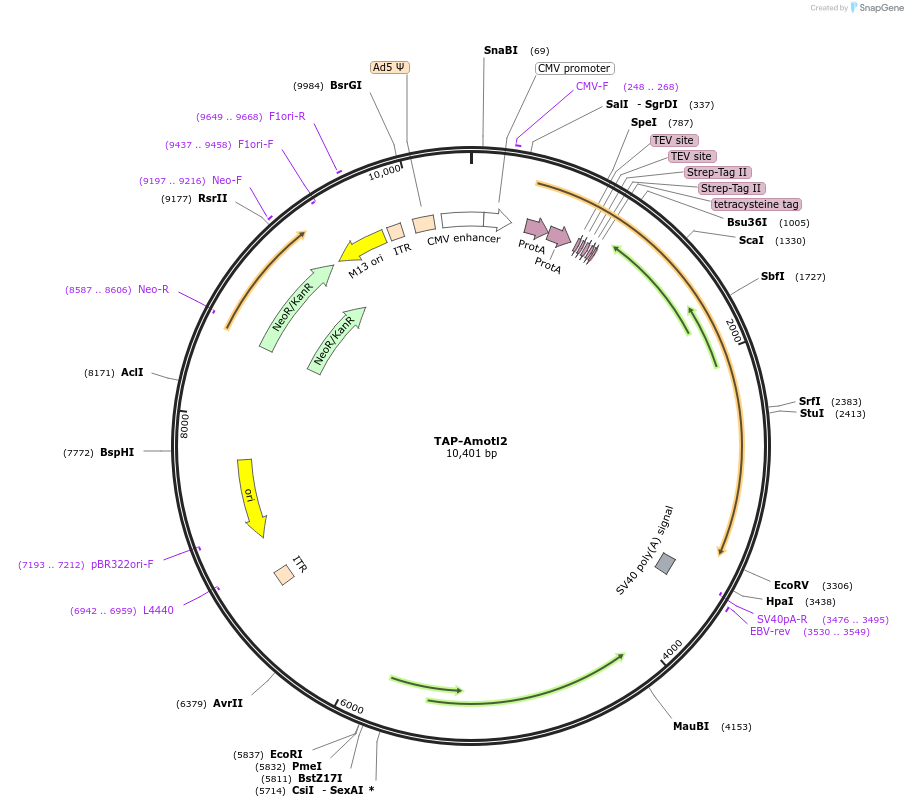
TAP-Amotl2
Plasmid#112834Purposetransient/stable overexpressionDepositorAvailable SinceAug. 8, 2018AvailabilityAcademic Institutions and Nonprofits only