We narrowed to 51,827 results for: ANG
-
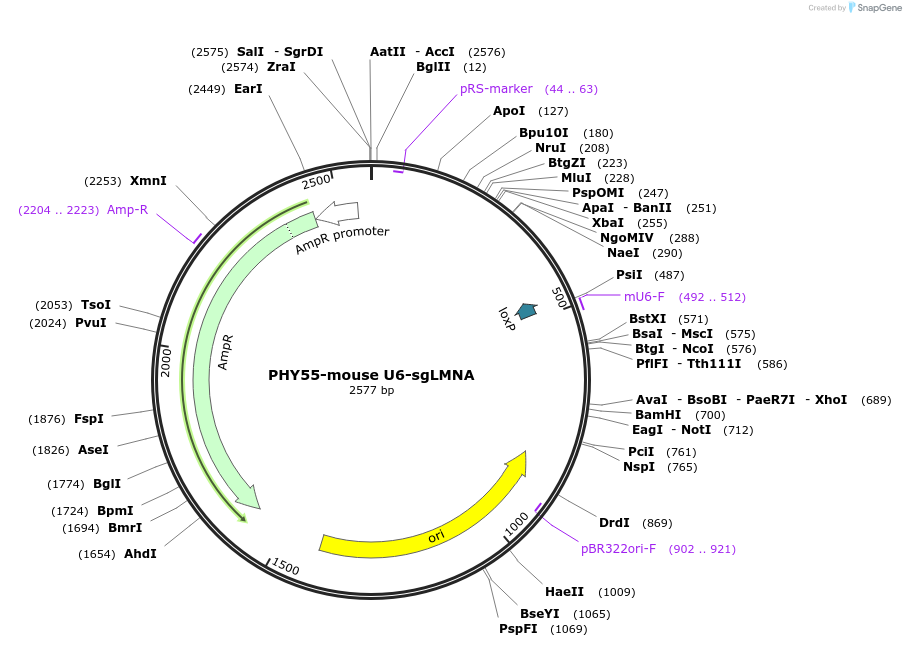
Plasmid#164045PurposeU6-driven sgRNA targeting LMNADepositorInsertLMNA targeting sgRNA
UseCRISPRPromotermouse U6Available SinceApril 9, 2021AvailabilityAcademic Institutions and Nonprofits only -
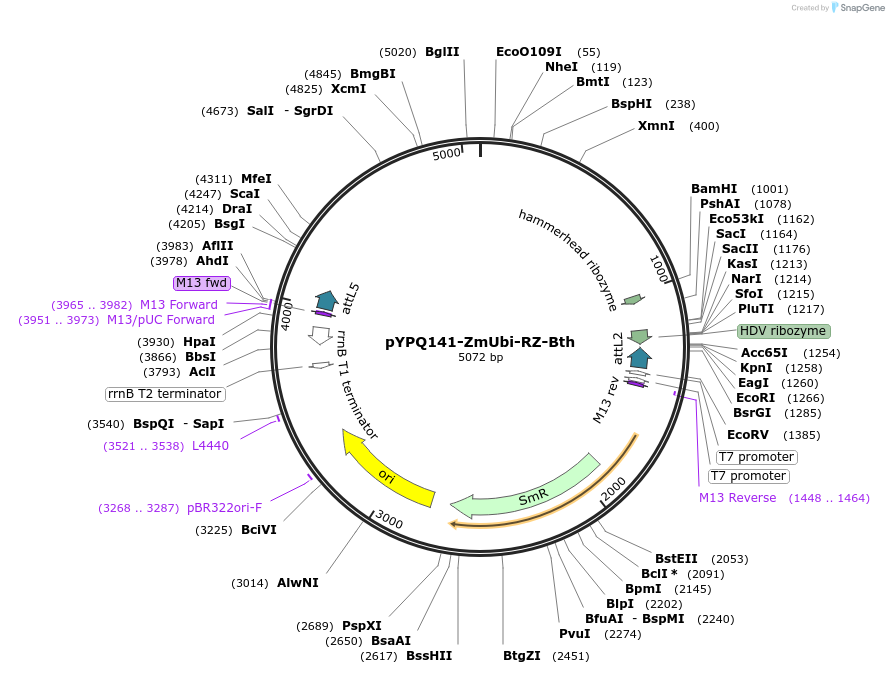
pYPQ141-ZmUbi-RZ-Bth
Plasmid#129686Purpose(Empty backbone) Gateway entry clone with sgRNA under ZmUbi promoterDepositorInsertEmpty Backbone
UseCRISPRExpressionPlantAvailable SinceMarch 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
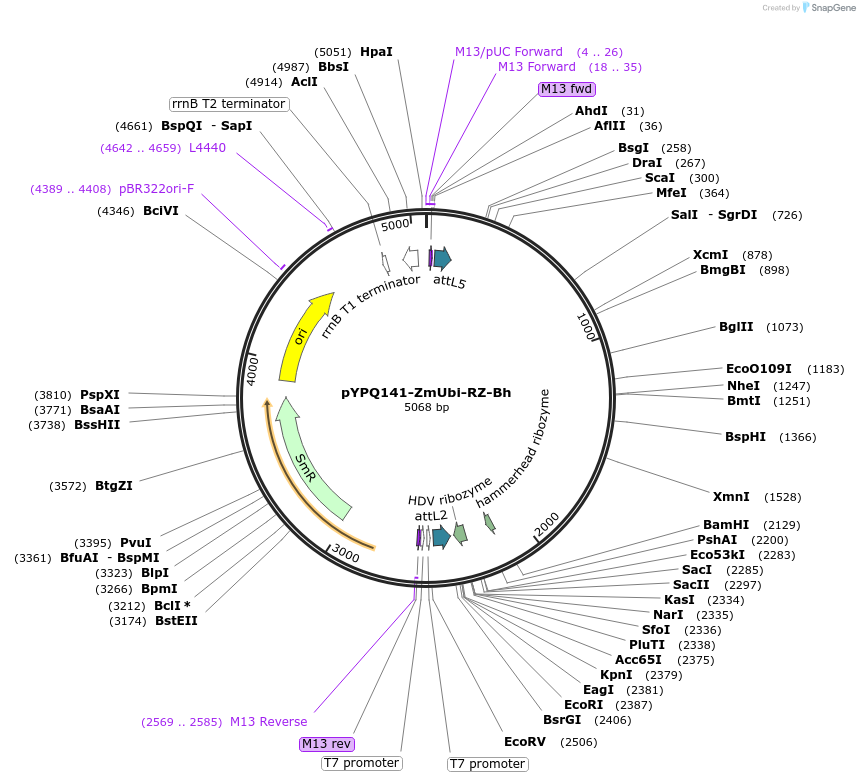
pYPQ141-ZmUbi-RZ-Bh
Plasmid#136378PurposeGateway entry clone for BhCas12b sgRNA expression under ZmUbi promoter with ribozyme processingDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceFeb. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
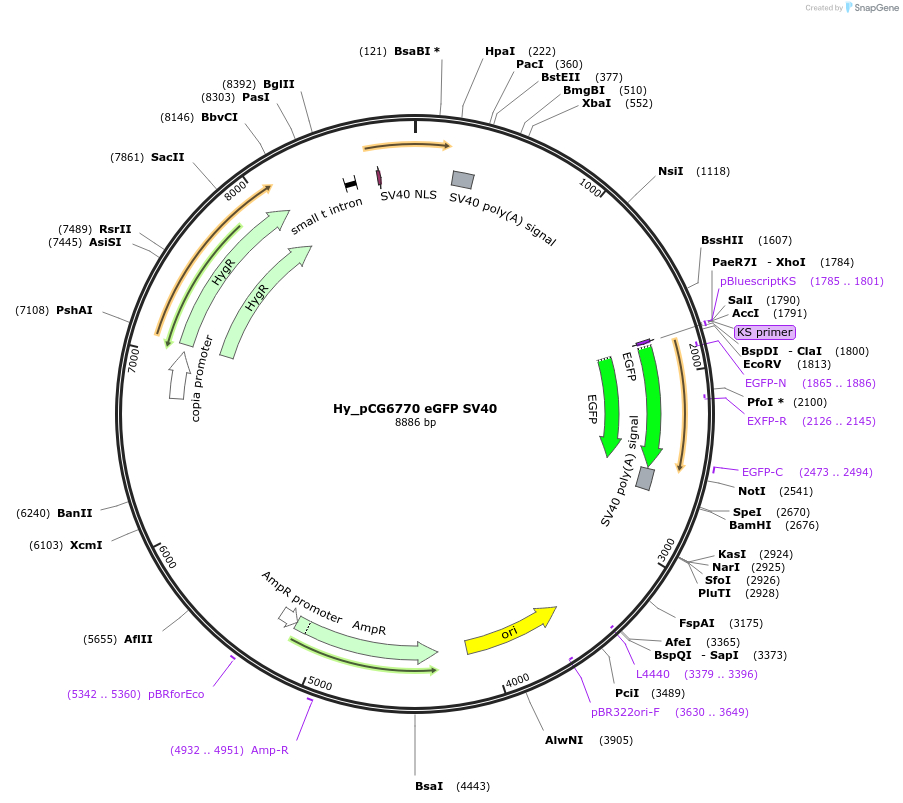
Hy_pCG6770 eGFP SV40
Plasmid#132649PurposeExpresses eGFP mRNA from the Drosophila CG6770 promoterDepositorInserteGFP
ExpressionInsectPromoterCG6770Available SinceOct. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
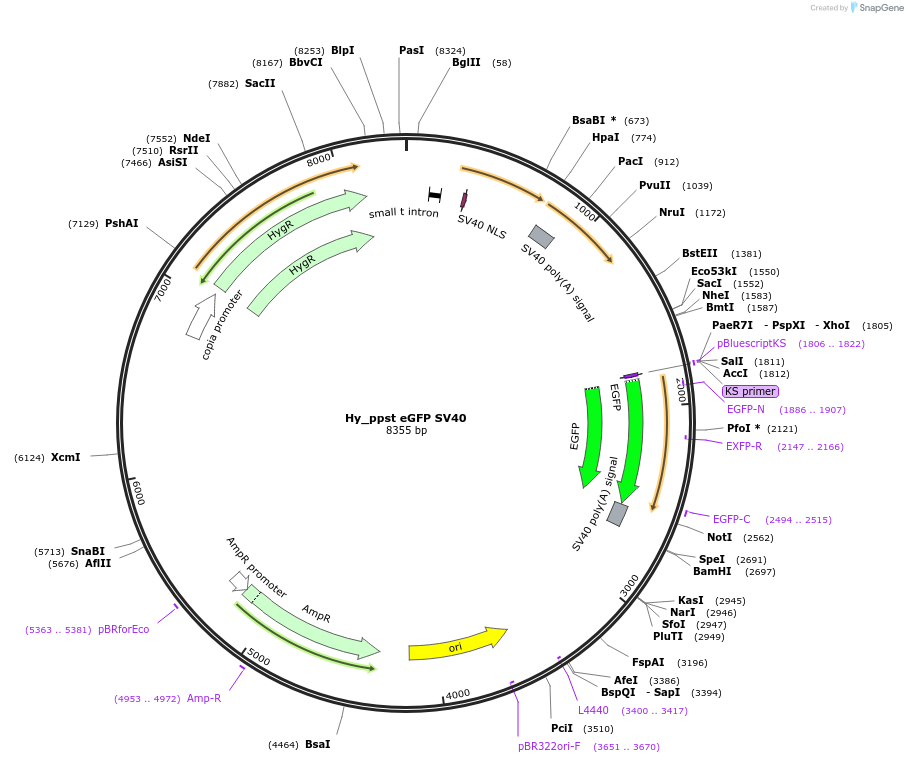
Hy_ppst eGFP SV40
Plasmid#132648PurposeExpresses eGFP mRNA from the Drosophila pst promoterDepositorInserteGFP
ExpressionInsectPromoterpstAvailable SinceOct. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
Hy_pSirup eGFP SV40
Plasmid#132647PurposeExpresses eGFP mRNA from the Drosophila Sirup promoterDepositorInserteGFP
ExpressionInsectPromoterSirupAvailable SinceOct. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
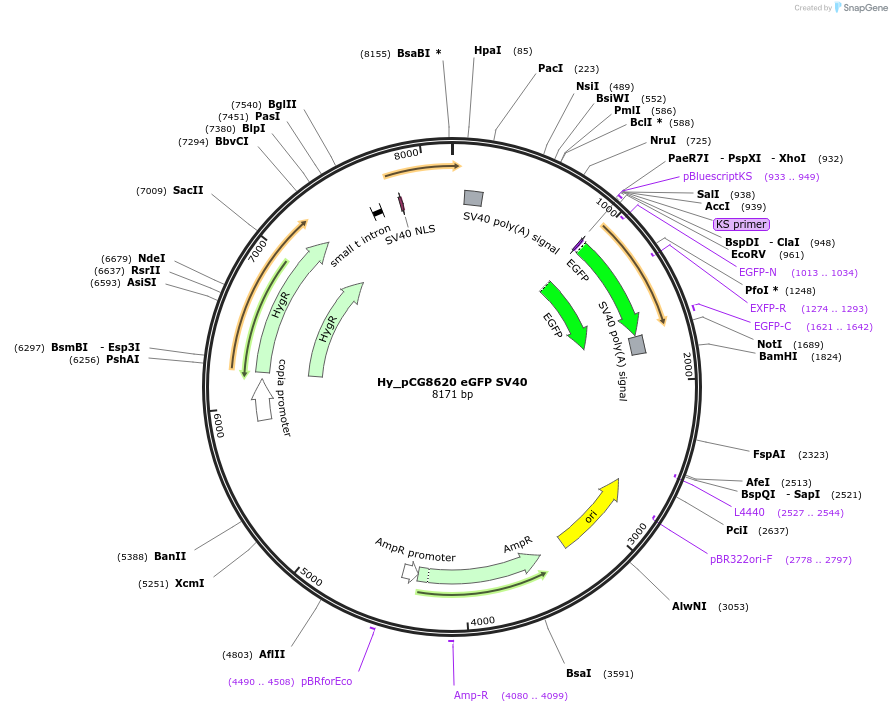
Hy_pCG8620 eGFP SV40
Plasmid#132646PurposeExpresses eGFP mRNA from the Drosophila CG8620 promoterDepositorInserteGFP
ExpressionInsectPromoterHmlAvailable SinceOct. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
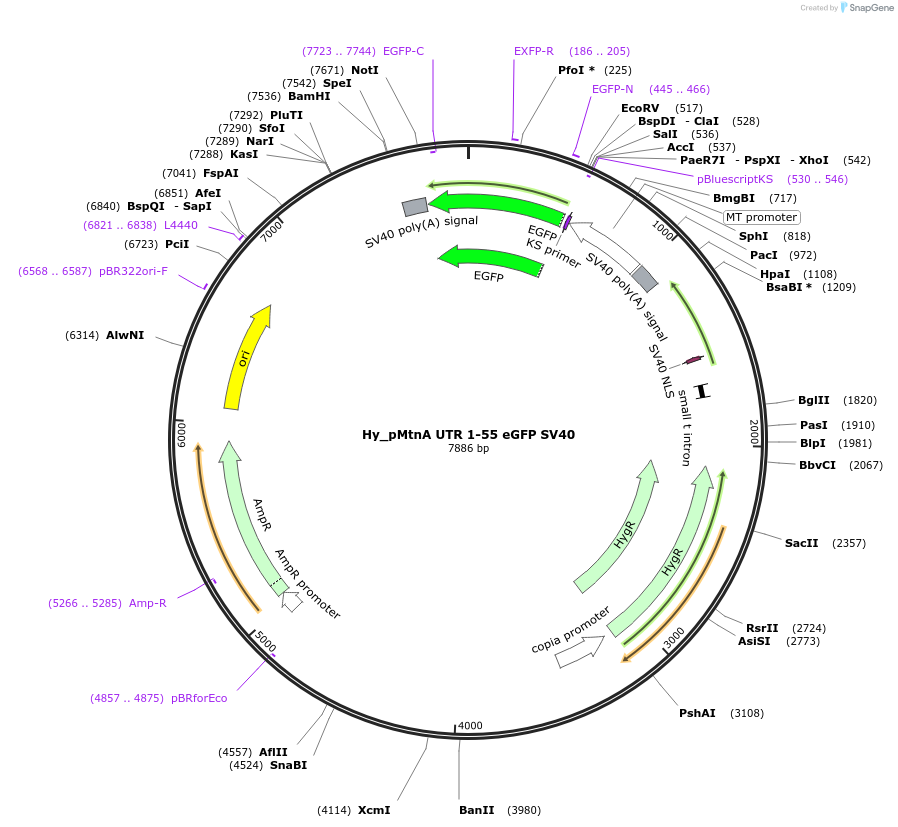
Hy_pMtnA UTR 1-55 eGFP SV40
Plasmid#132651PurposeExpresses an eGFP mRNA from the Drosophila Metallothionein A promoter that has 55 nt of the MtnA 5' UTRDepositorInserteGFP
ExpressionInsectPromoterMtnAAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
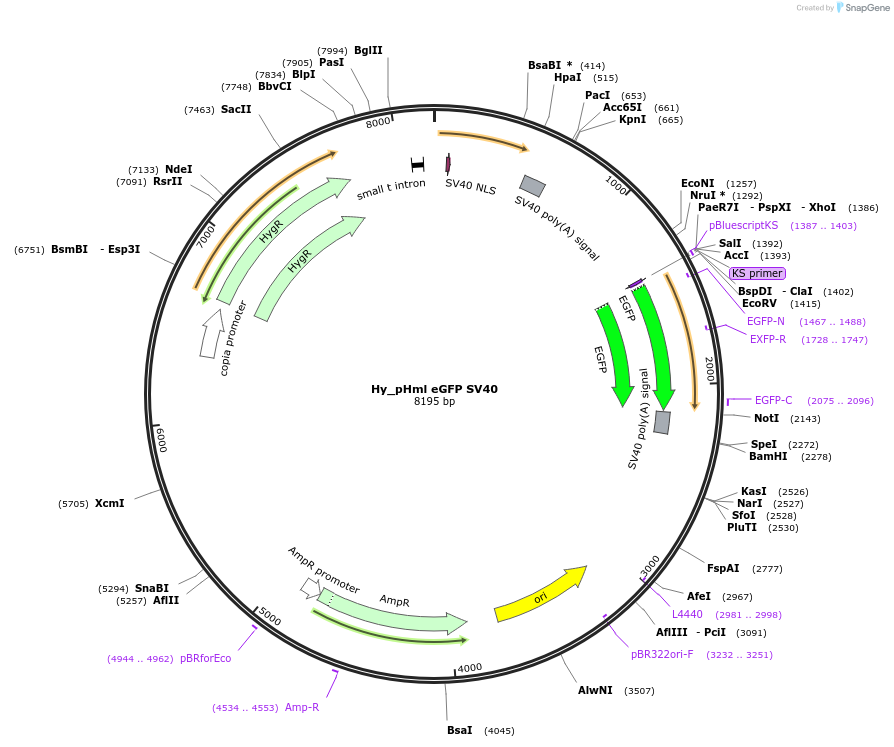
Hy_pHml eGFP SV40
Plasmid#132645PurposeExpresses eGFP mRNA from the Drosophila Hml promoterDepositorInserteGFP
ExpressionInsectPromoterHmlAvailable SinceOct. 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
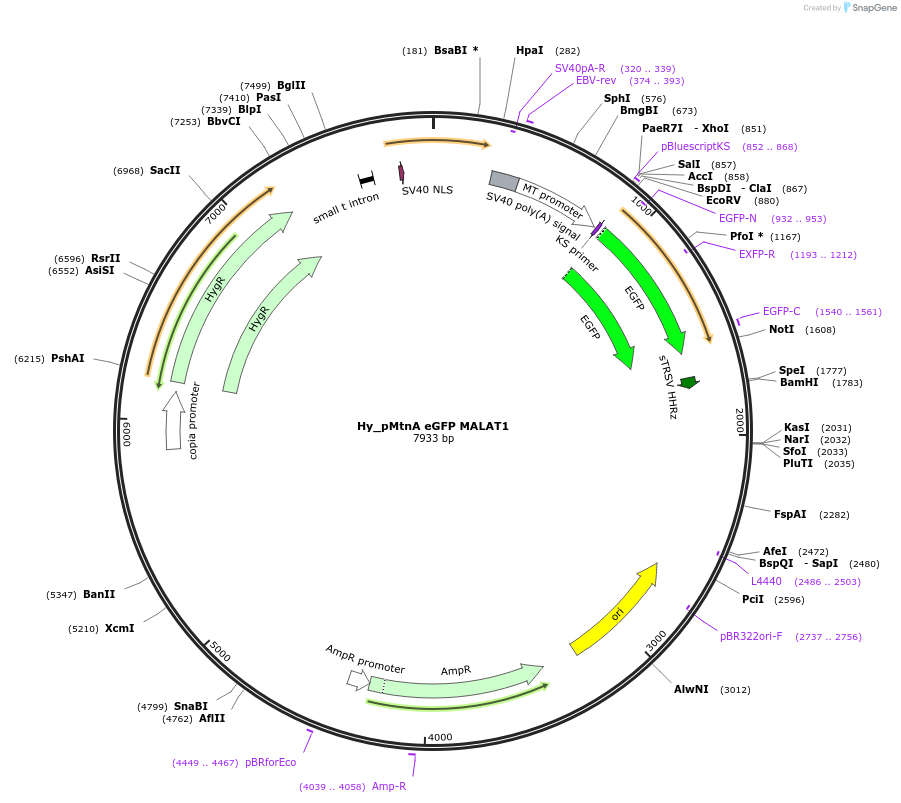
Hy_pMtnA eGFP MALAT1
Plasmid#132643PurposeExpresses an eGFP mRNA from the Drosophila Metallothionein A promoter that ends in the MALAT1 triple helixDepositorInserteGFP
ExpressionInsectPromoterMtnAAvailable SinceSept. 24, 2019AvailabilityAcademic Institutions and Nonprofits only -
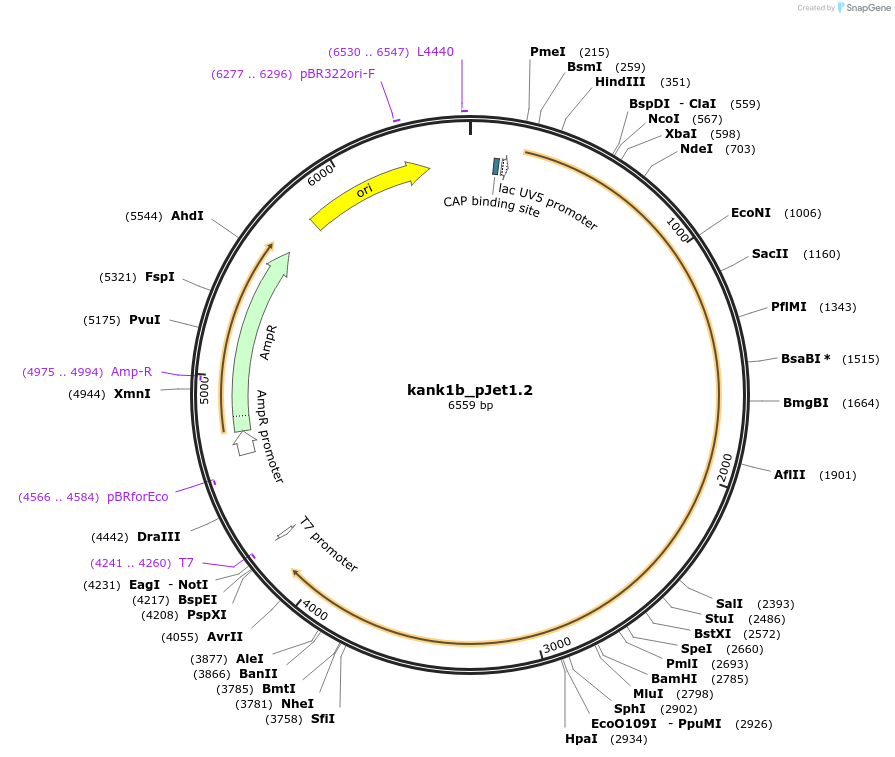
kank1b_pJet1.2
Plasmid#121979Purposefor generating kank1b riboprobes for WISH in zebrafishDepositorAvailable SinceFeb. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
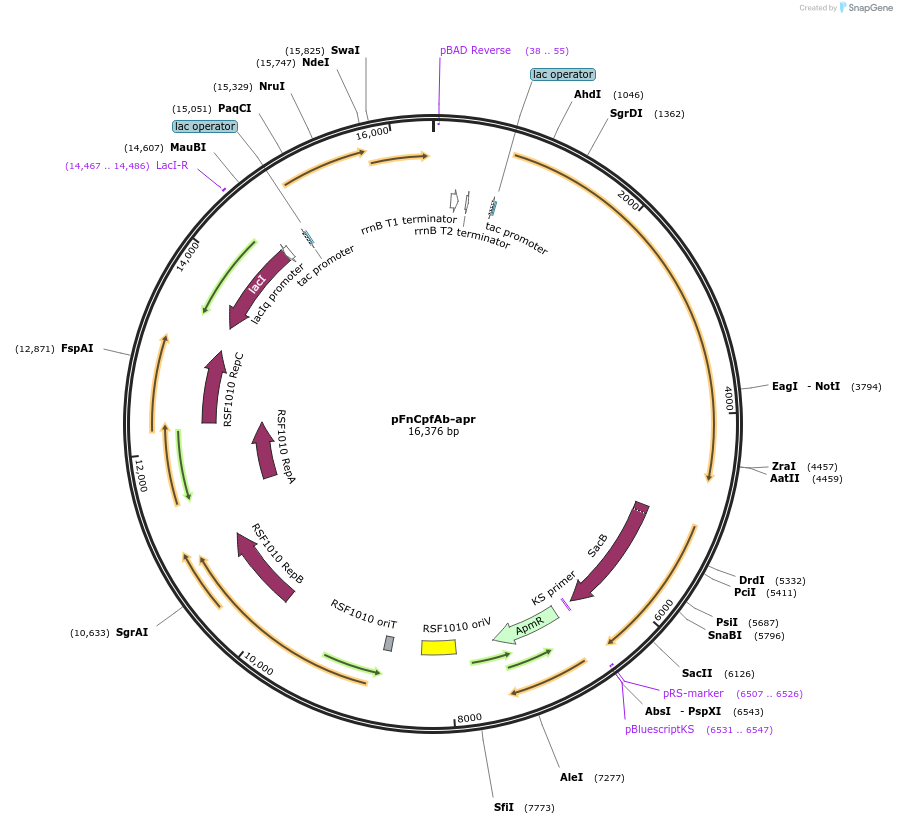
pFnCpfAb-apr
Plasmid#214198PurposeExpresses FnCpf1 and RecAb recombination system for genome editing in Acinetobacter baumanniiDepositorInsertFnCpf1
UseCRISPRPromotertac promoterAvailable SinceJune 11, 2024AvailabilityAcademic Institutions and Nonprofits only -
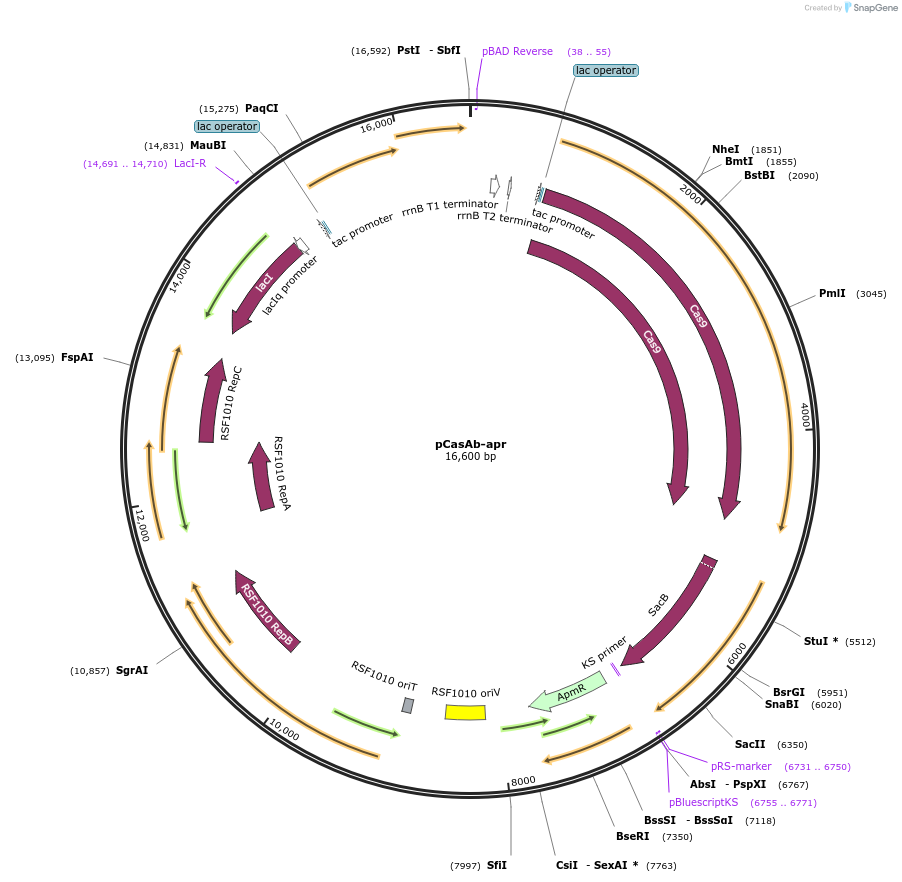
pCasAb-apr
Plasmid#121998PurposeBacterial expression of Cas9 nuclease and RecAb recombination system in Acinetobacter baumanniiDepositorTypeEmpty backboneUseCRISPRExpressionBacterialAvailable SinceSept. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
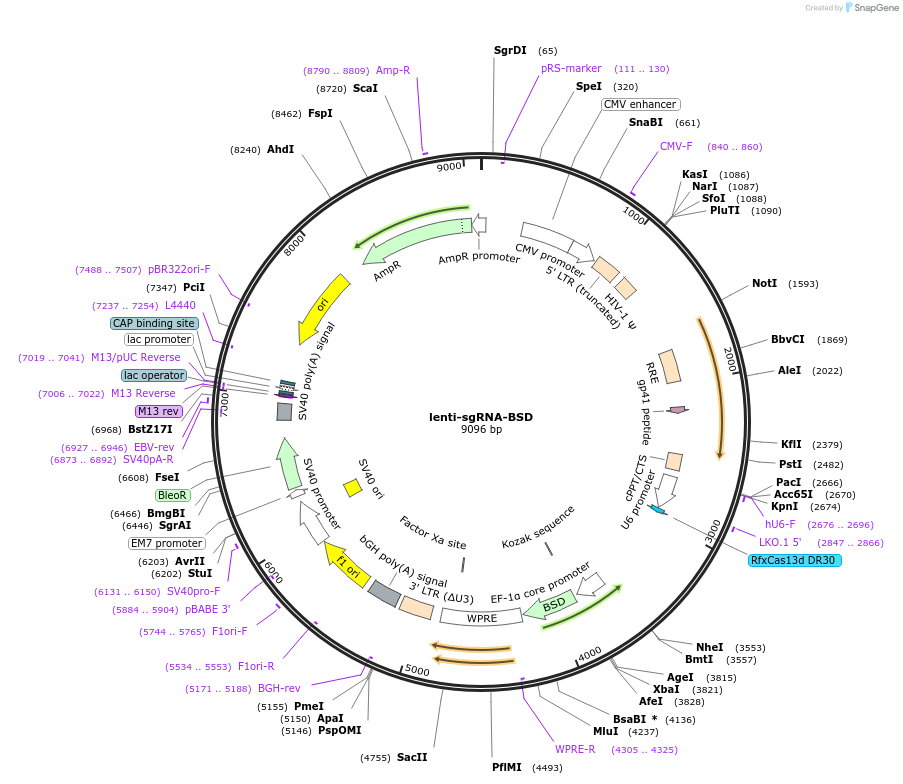
lenti-sgRNA-BSD
Plasmid#175583PurposeExpresses sgRNA for CasRx in mammalian cellDepositorInsertsgRNA
UseCRISPR and LentiviralExpressionMammalianPromoterU6Available SinceNov. 5, 2021AvailabilityAcademic Institutions and Nonprofits only -
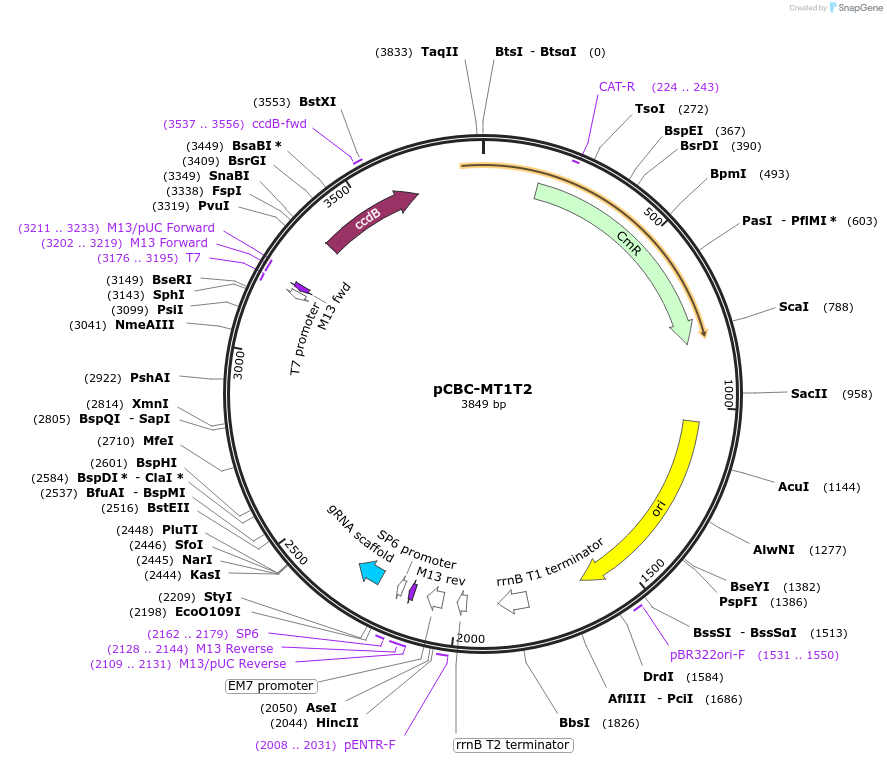
pCBC-MT1T2
Plasmid#50593PurposeCRISPR/Cas based plant genome editing and gene regulation; used as template for making expression cassette with multiple gRNA target sitesDepositorInsertT1, gRNA scaffold, OsU3t plus, TaU3p, T2
UseCRISPR; Pcr templateExpressionPlantAvailable SinceDec. 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
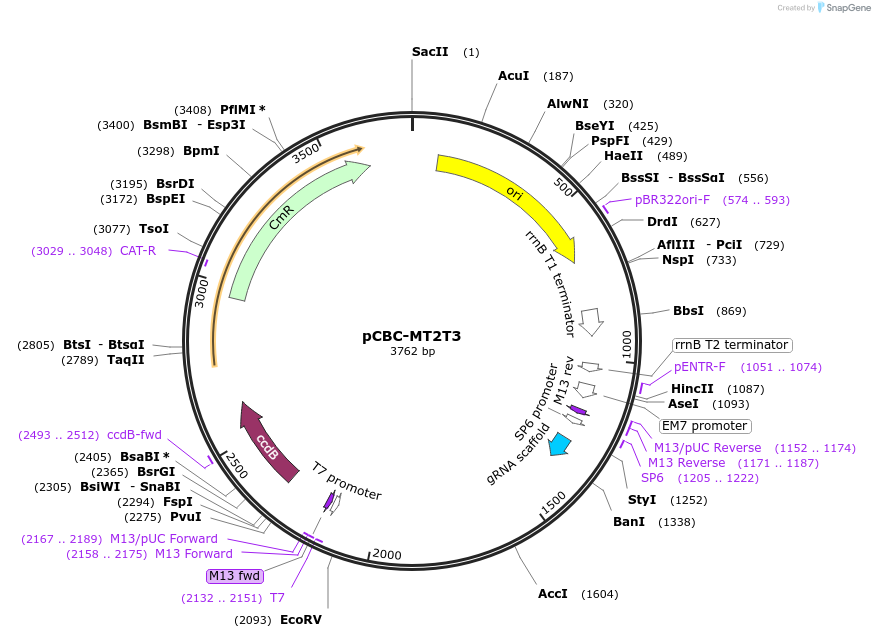
pCBC-MT2T3
Plasmid#50594PurposeCRISPR/Cas based plant genome editing and gene regulation; used as template for making expression cassette with multiple gRNA target sitesDepositorInsertT2, gRNA scaffold, TaU3t plus, U6-26p, T3
UseCRISPR; Pcr templateExpressionPlantAvailable SinceDec. 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
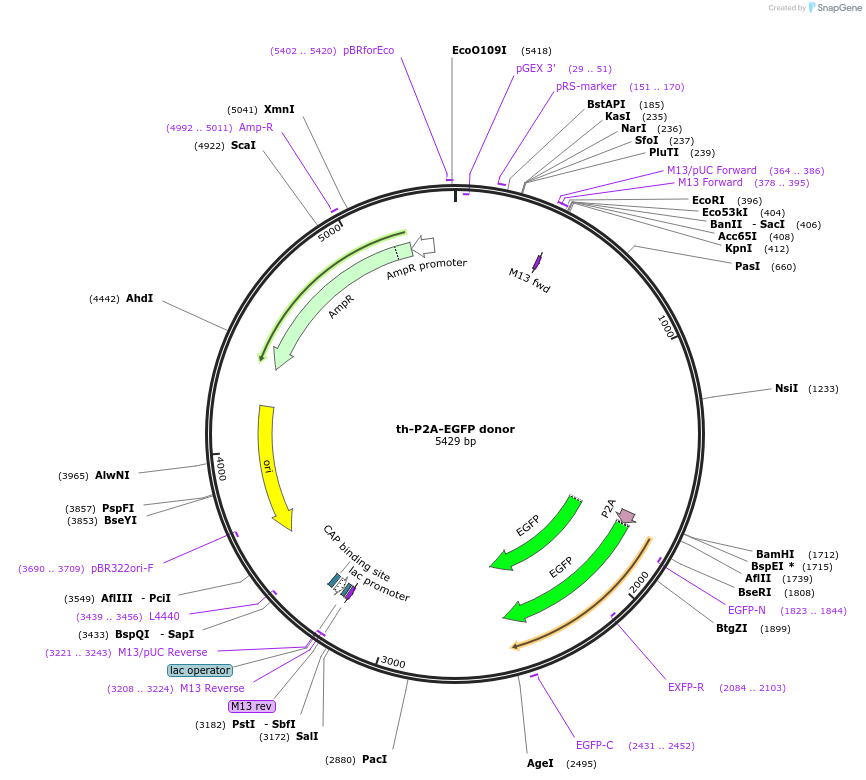
th-P2A-EGFP donor
Plasmid#65562PurposeIntron targeting-mediated EGFP knockin at the zebrafish th locus.DepositorAvailable SinceJune 3, 2015AvailabilityAcademic Institutions and Nonprofits only -
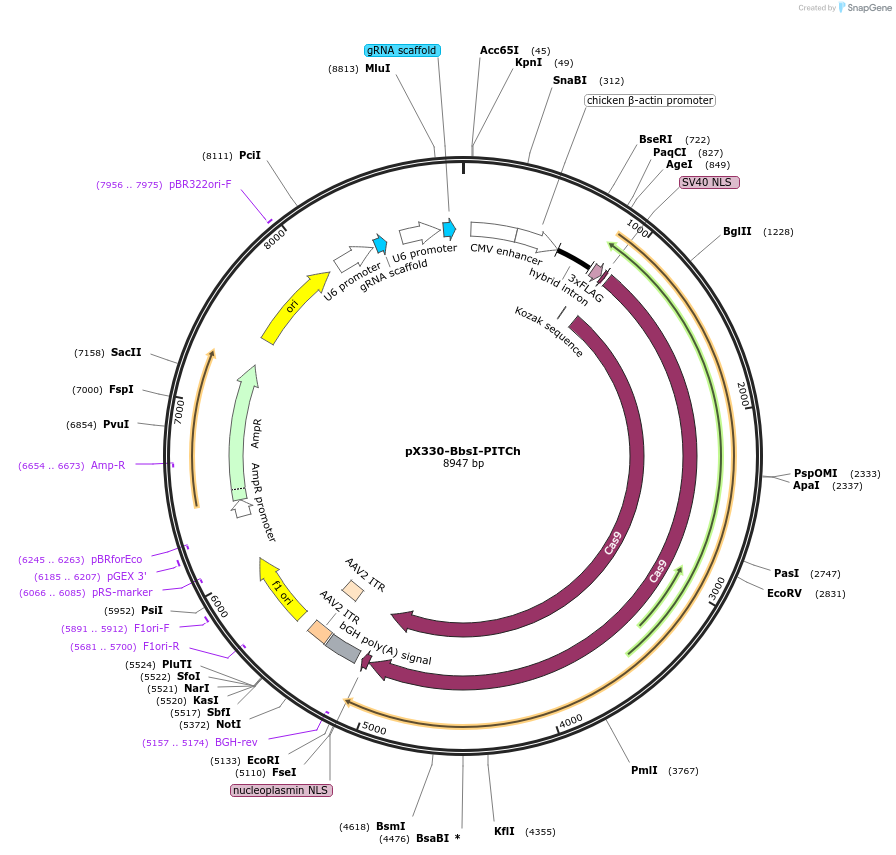
pX330-BbsI-PITCh
Plasmid#127875PurposeFor user to clone their gRNA oligos to BbsI site. PITCh gRNA expression cassette is already build in so there is no need to perform Golden Gate assemblyDepositorTypeEmpty backboneUseCRISPRExpressionMammalianAvailable SinceJuly 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
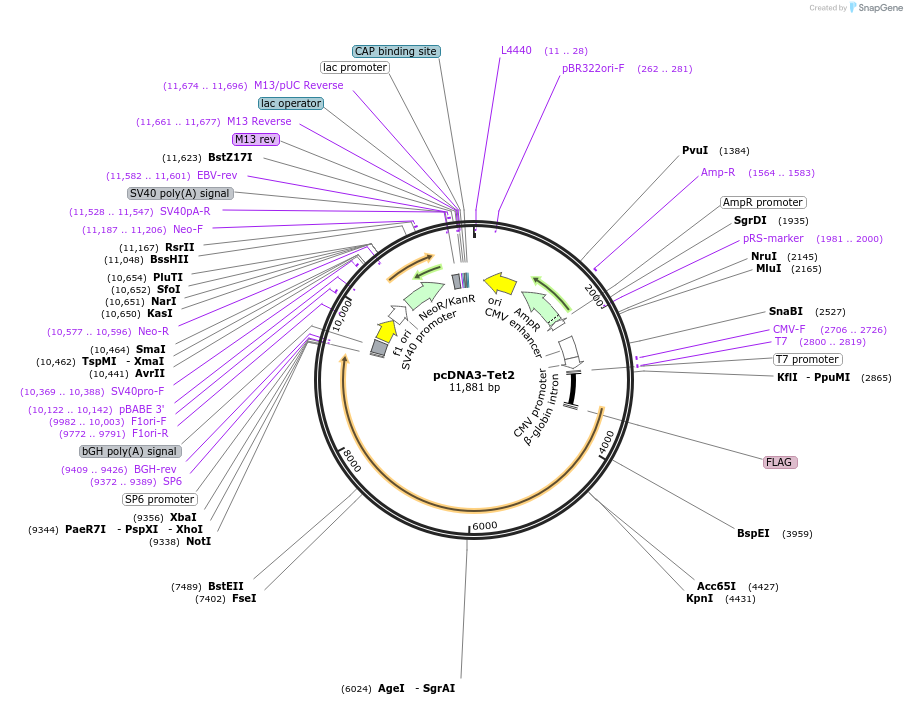
pcDNA3-Tet2
Plasmid#60939PurposeExpresses Tet2 in mammalian cellsDepositorAvailable SinceNov. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
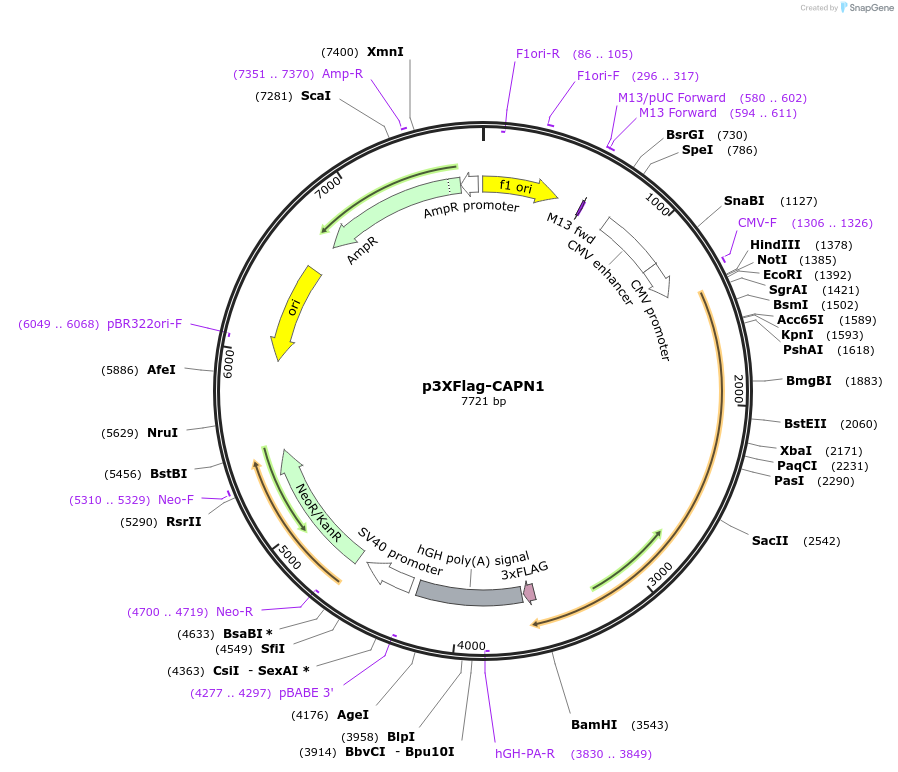
p3XFlag-CAPN1
Plasmid#60941PurposeExpresses calpain1 in mammalian cellsDepositorAvailable SinceDec. 22, 2014AvailabilityAcademic Institutions and Nonprofits only