We narrowed to 11,953 results for: SOM
-
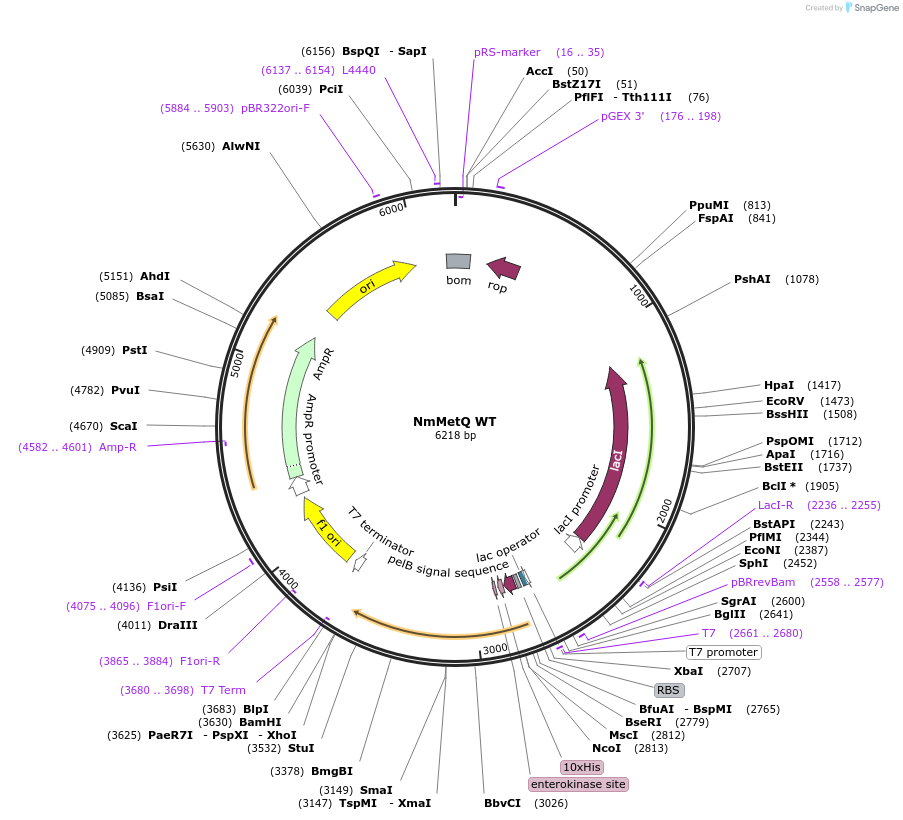
Plasmid#129647PurposeExpresses the mature metQ gene from Neisseria meningitides, without the signal sequenceDepositorInsertmetQ Gene
Tags10x Histidine Tag, Enterokinase, and pelB signal …ExpressionBacterialPromoterT7 PromoterAvailable SinceSept. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
pGEX-KG/Net1
Plasmid#28260DepositorAvailable SinceApril 11, 2011AvailabilityAcademic Institutions and Nonprofits only -
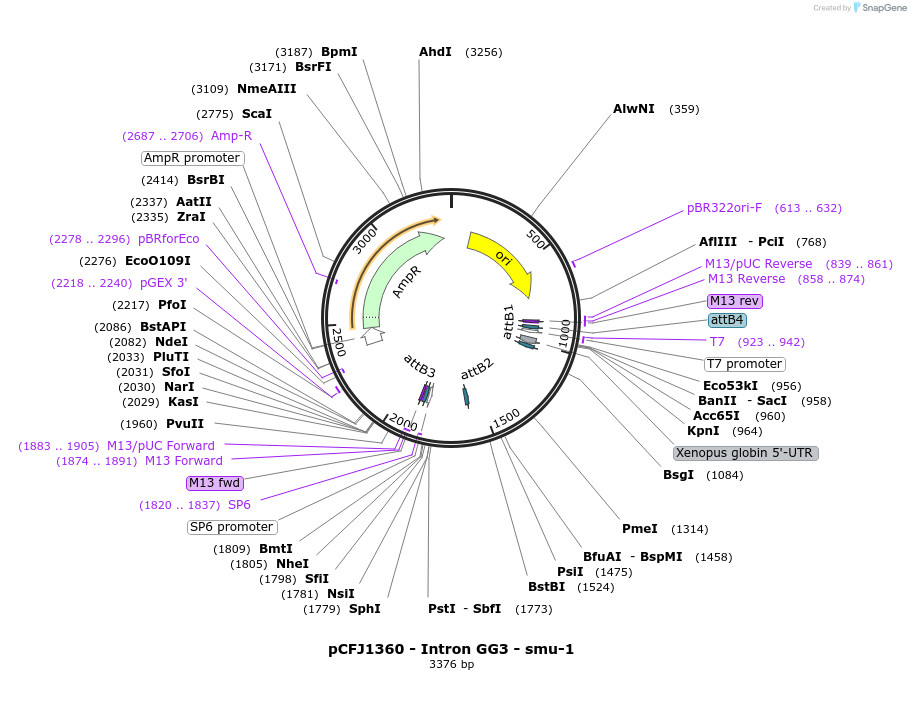
pCFJ1360 - Intron GG3 - smu-1
Plasmid#159806PurposeGolden Gate compatible intron with PATCs for reduced germline silencingDepositorInsertIntron GG3 - smu-1
ExpressionWormMutationNot applicableAvailable SinceDec. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
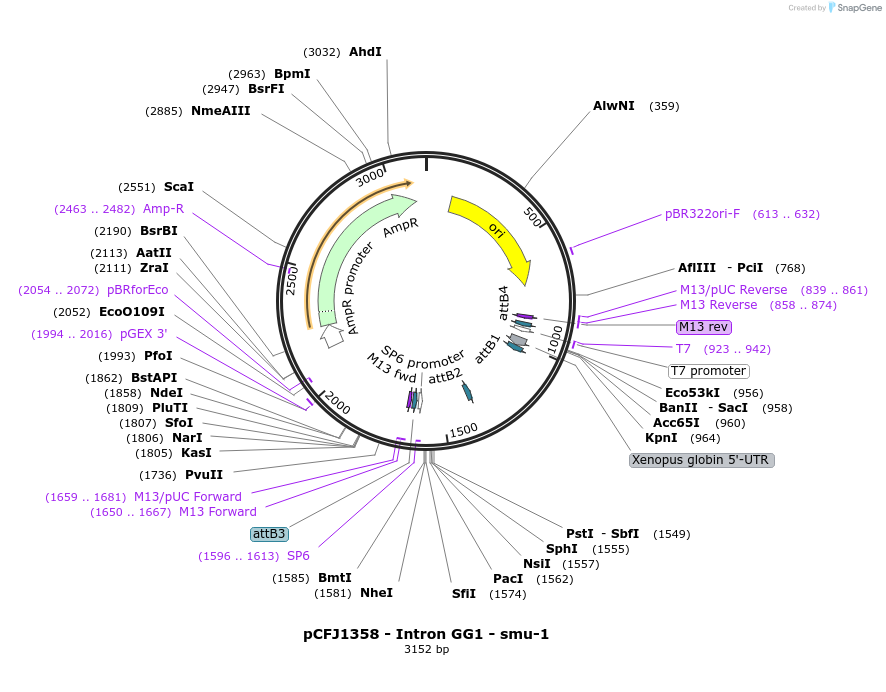
pCFJ1358 - Intron GG1 - smu-1
Plasmid#159804PurposeGolden Gate compatible intron with PATCs for reduced germline silencingDepositorInsertIntron GG1 - smu-1
ExpressionWormMutationNot applicableAvailable SinceDec. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
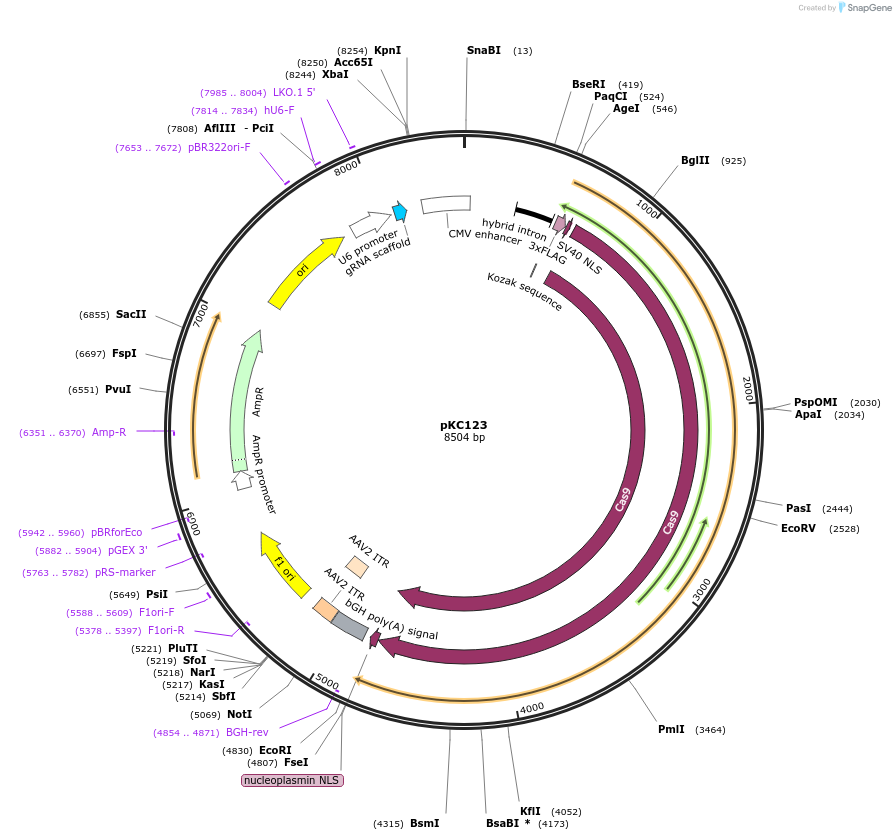
pKC123
Plasmid#134240Purposeexpresses gRNA targeting CENP A 3'DepositorInsertCENP A 3' CRISPR construct
UseCRISPRExpressionMammalianAvailable SinceFeb. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
-
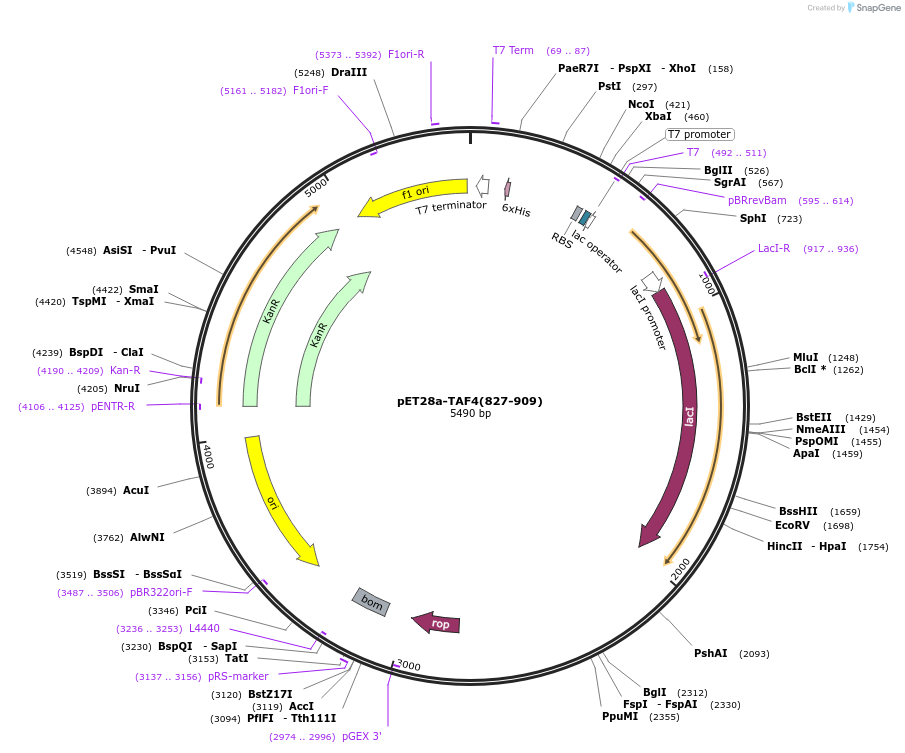
pET28a-TAF4(827-909)
Plasmid#105620Purposebacterially express TAF4 HFDDepositorInsertTAF4-HFD(827-909) (Taf4 Mouse)
ExpressionBacterialAvailable SinceFeb. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
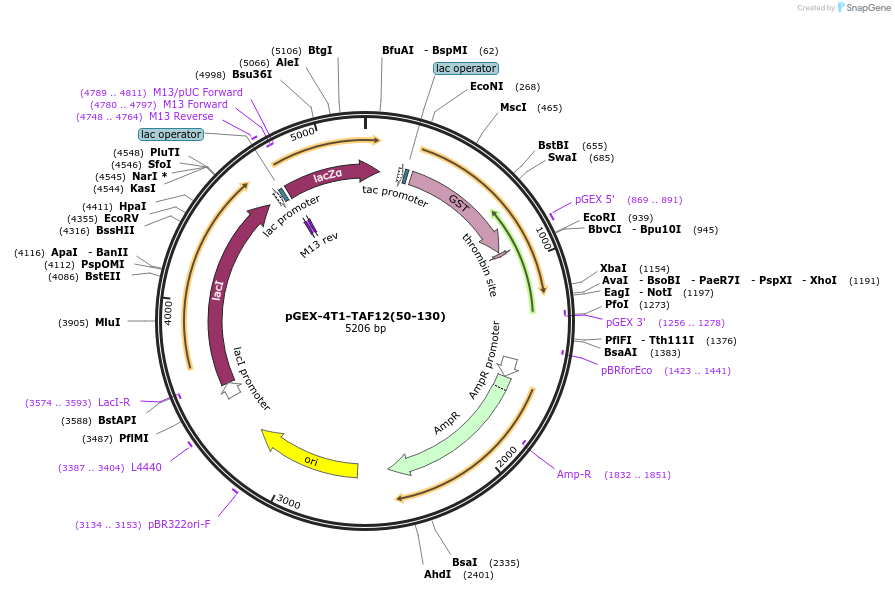
pGEX-4T1-TAF12(50-130)
Plasmid#105618Purposebacterially express GST tagged TAF12 HFDDepositorAvailable SinceFeb. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
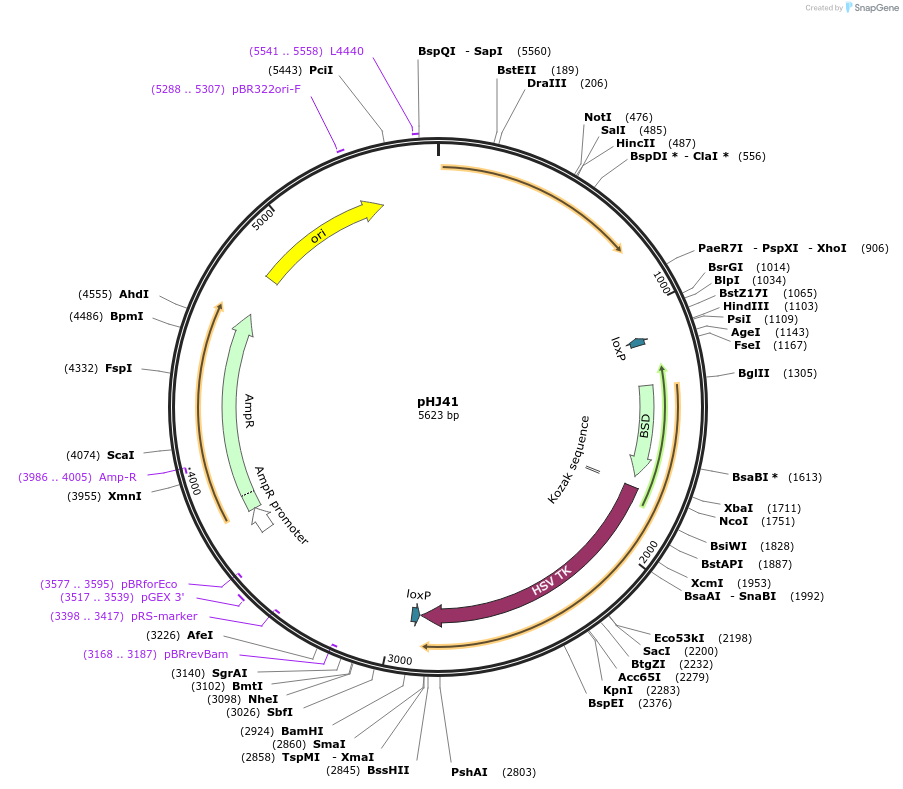
pHJ41
Plasmid#48360PurposeTargets beta-Tubulin for Cre-lox mediated knockout in T. brucei, Blasticidin/Gancyclovir selectableDepositorInsertPartial beta-TUB followed by 5' ALD UTR-loxP-SAS-BSD-Ty1-TK-loxP-3' ALD UTR
UseCre/Lox; Knockout in t. bruceiAvailable SinceJan. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
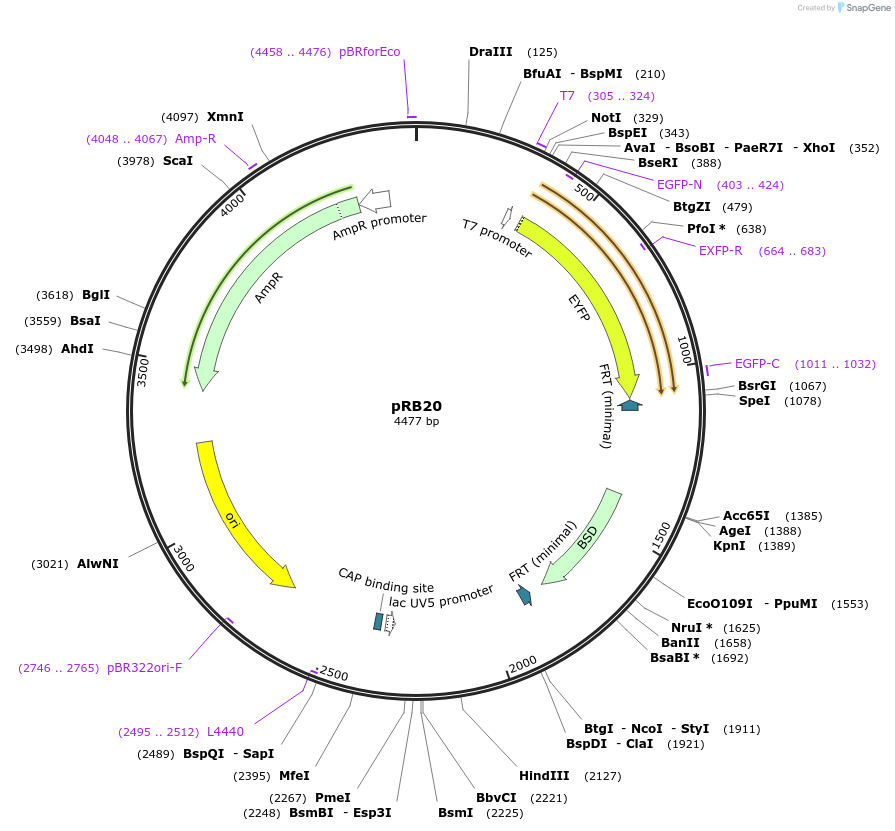
pRB20
Plasmid#52559Purposeserves as PCR-template to generate HR donors for C-terminal YFP-tagDepositorInserteYFP and Blasticigin resistance cassette
UseBlunt cloning kitAvailable SinceApril 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
RSPH1_pCSdest
Plasmid#53926DepositorAvailable SinceMarch 27, 2018AvailabilityIndustry, Academic Institutions, and Nonprofits -
-
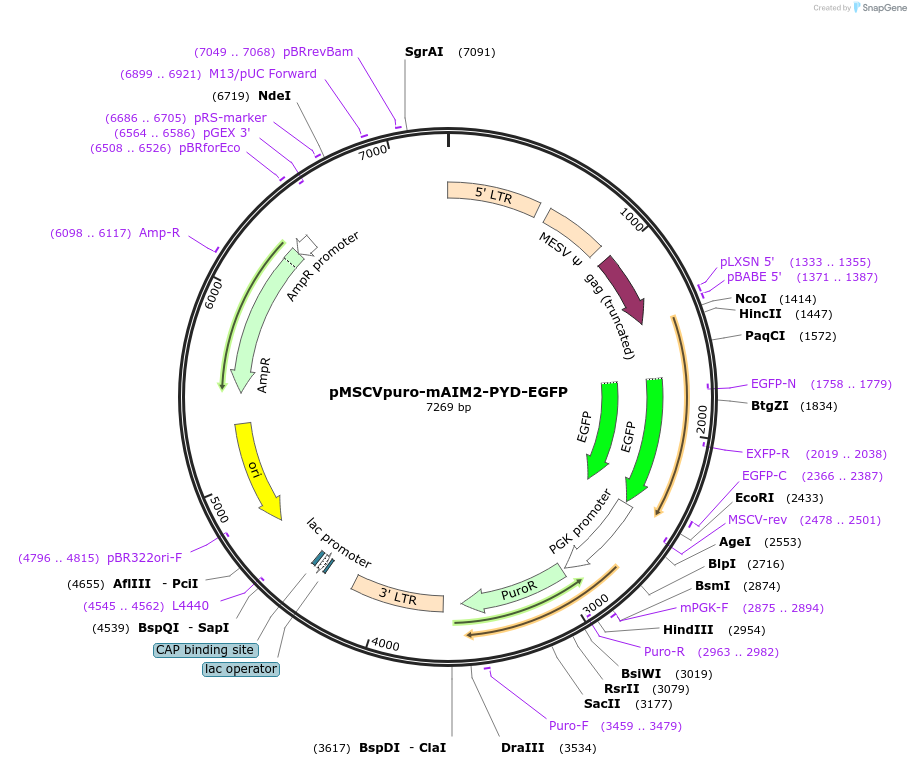
pMSCVpuro-mAIM2-PYD-EGFP
Plasmid#51610PurposeExpress GFP-tagged PYD of mAIM2 in mammalian cellsDepositorInsertAim2 (Aim2 Mouse)
UseRetroviralTagsEGFPExpressionMammalianMutationdeletion of HIN domainPromoterMESVAvailable SinceMarch 10, 2014AvailabilityAcademic Institutions and Nonprofits only -
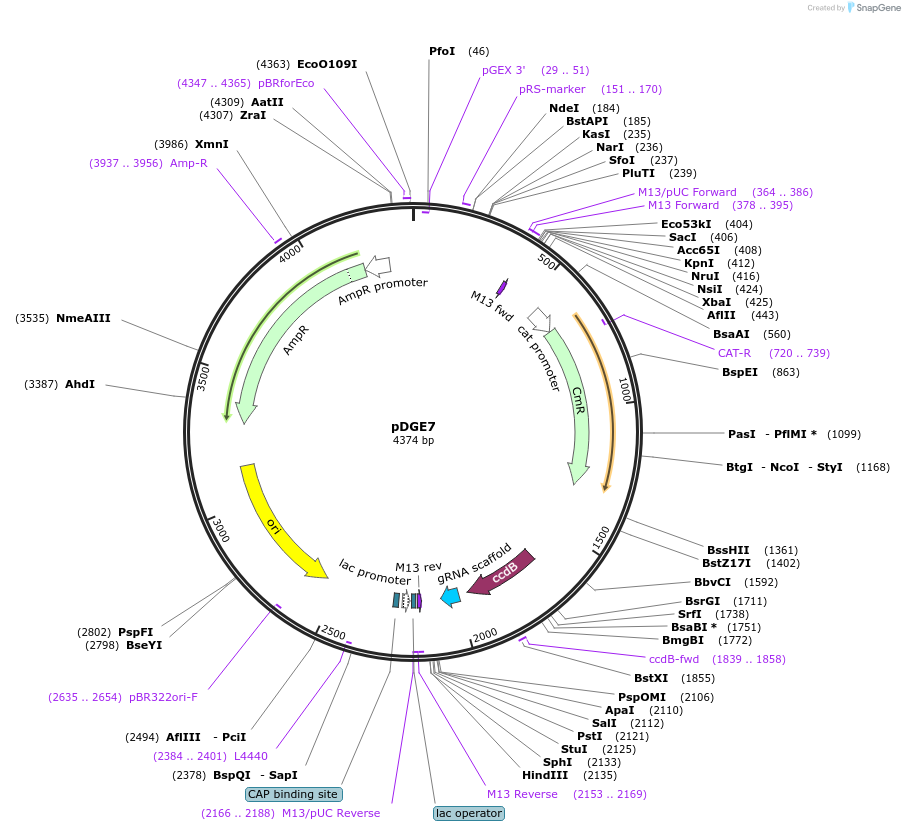
pDGE7
Plasmid#79462PurposesgRNA shuttle vector; module 2DepositorInsertpAtU6-ccdB-sgRNA (ccdB(letD) )
UseCRISPR and Synthetic BiologyMutationBsaI site eliminatedAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
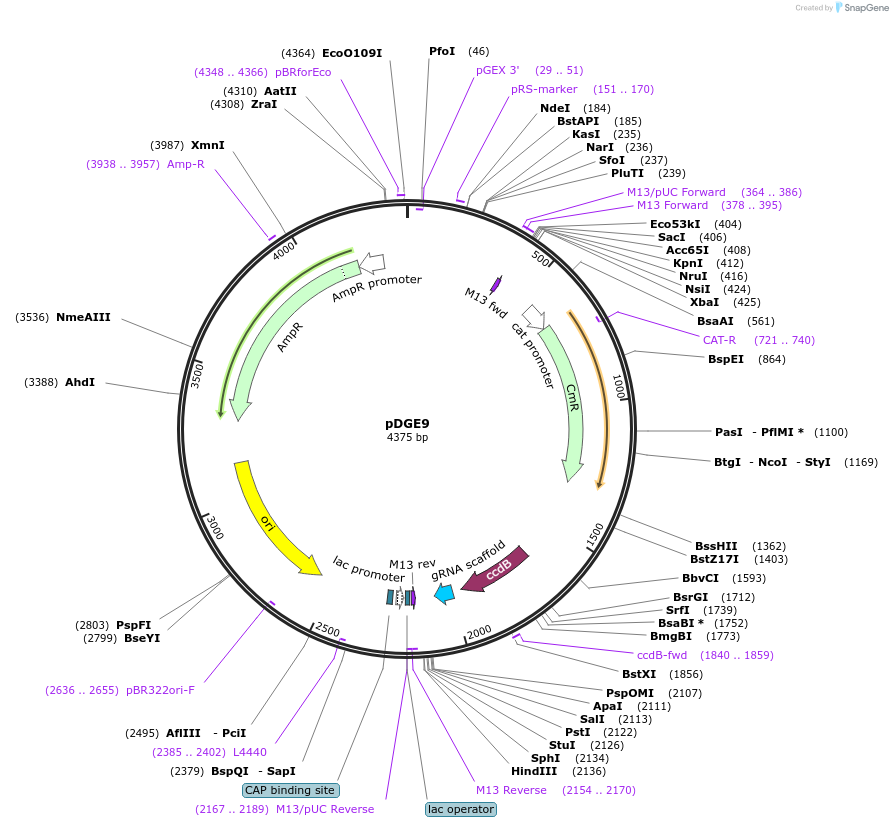
pDGE9
Plasmid#79465PurposesgRNA shuttle vector; module 3DepositorInsertpAtU6-ccdB-sgRNA (ccdB(letD) )
UseCRISPR and Synthetic BiologyMutationBsaI site eliminatedAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
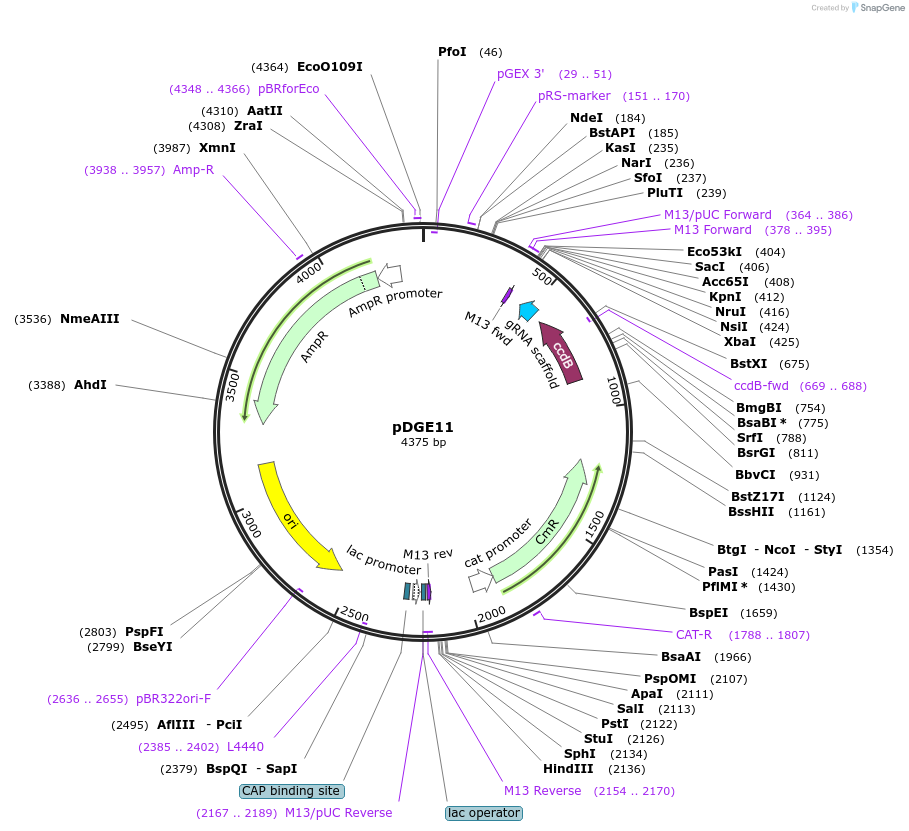
pDGE11
Plasmid#79467PurposesgRNA shuttle vector; module 4EDepositorInsertpAtU6-ccdB-sgRNA (ccdB(letD) )
UseCRISPR and Synthetic BiologyMutationBsaI site eliminatedAvailable SinceSept. 9, 2016AvailabilityAcademic Institutions and Nonprofits only -
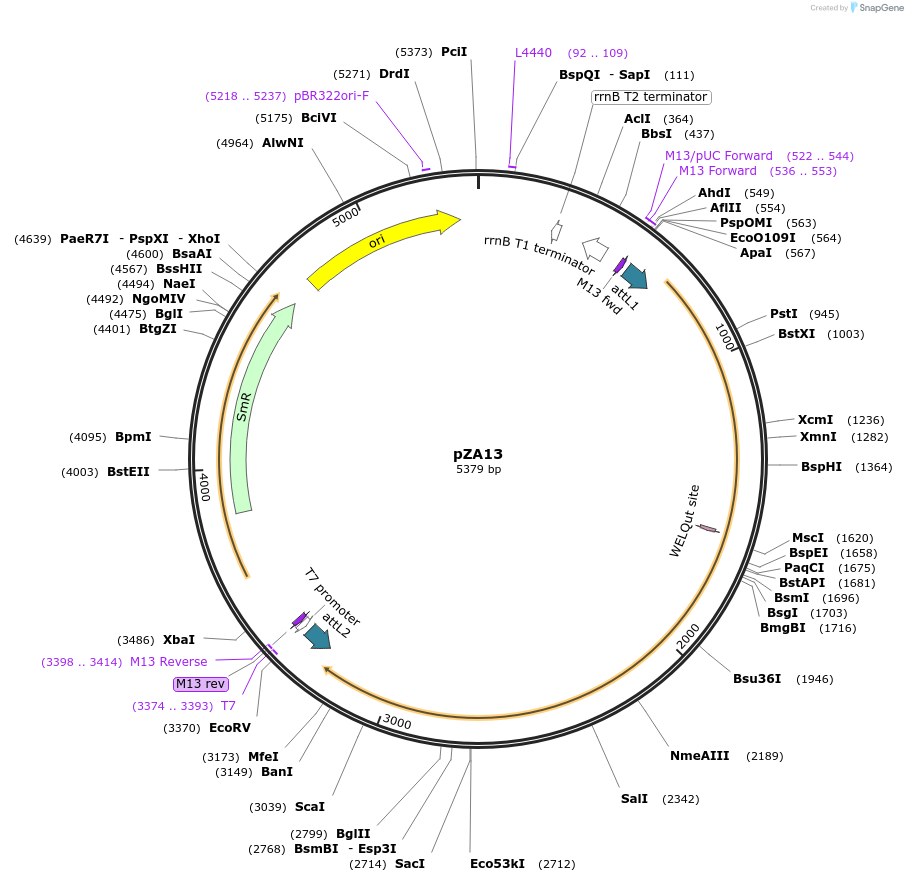
pZA13
Plasmid#158491PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 without STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationno STOP codonAvailable SinceSept. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
epiCRISPRe
Plasmid#135962PurposeExpresses SpCas9, and cloning backbone for sgRNADepositorTypeEmpty backboneExpressionMammalianAvailable SinceApril 30, 2020AvailabilityAcademic Institutions and Nonprofits only -
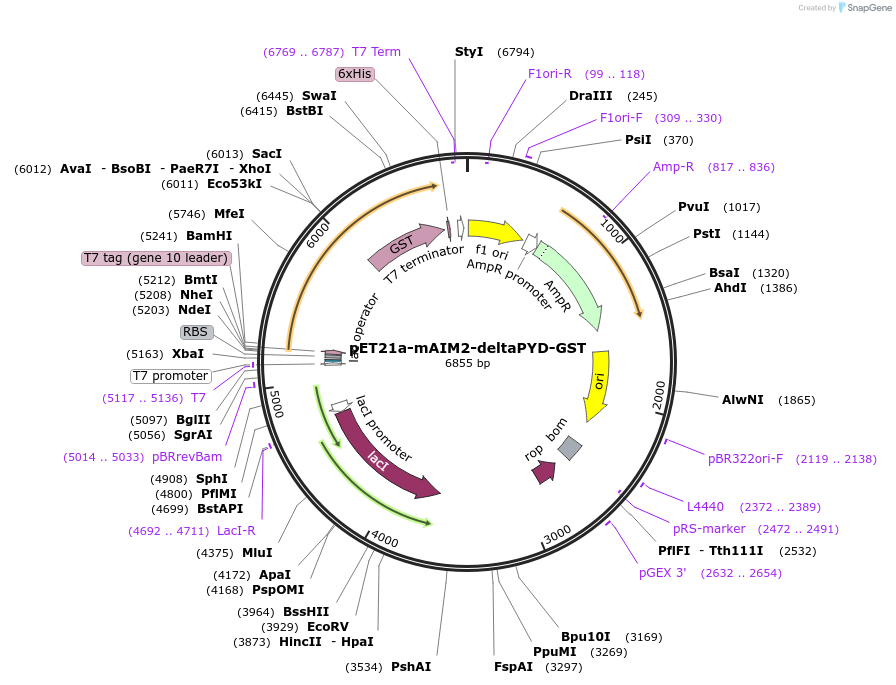
pET21a-mAIM2-deltaPYD-GST
Plasmid#51534PurposeExpress GST-tagged HIN domain of mAIM2 in bacteriaDepositorAvailable SinceMarch 11, 2014AvailabilityAcademic Institutions and Nonprofits only