We narrowed to 81,061 results for: Mycs;
-
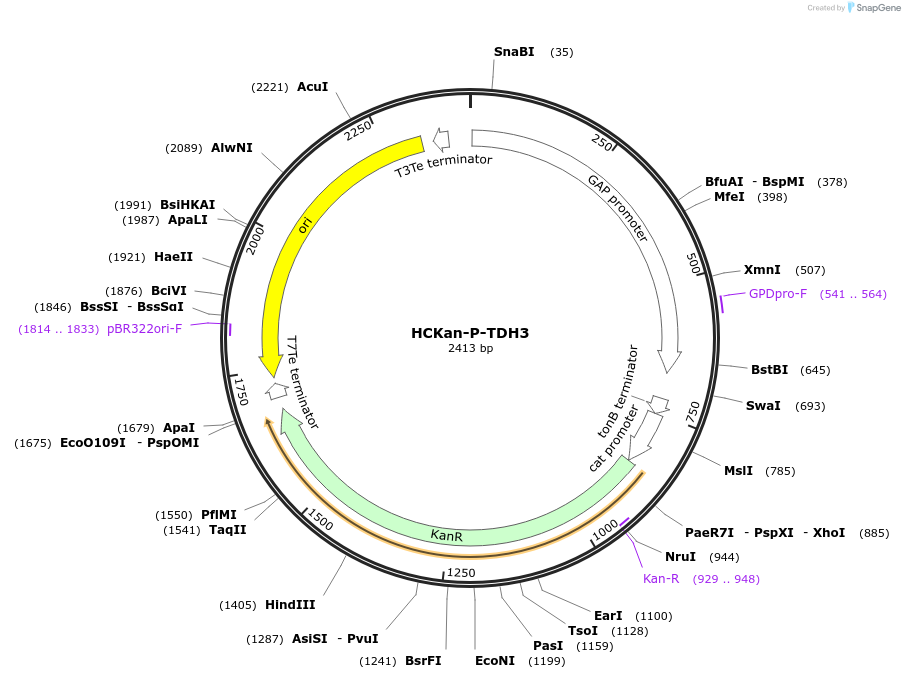
Plasmid#65336Purposeconstruction of TDH3 promoter in HCKan-P receiving vectorDepositorInsertTDH3 promoter
UseSynthetic BiologyAvailable SinceFeb. 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
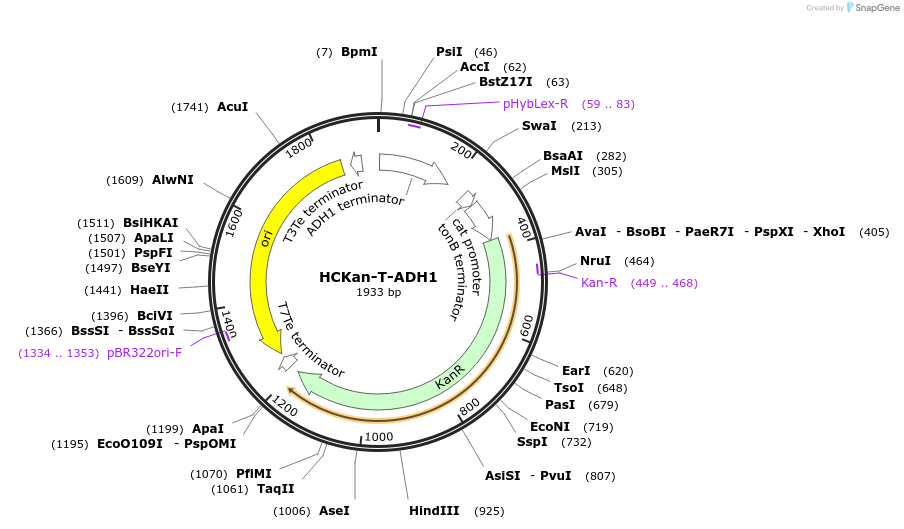
HCKan-T-ADH1
Plasmid#65339Purposeconstruction of ADH1 terminator in HCKan-P receiving vectorDepositorInsertADH1 terminator
UseSynthetic BiologyAvailable SinceFeb. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
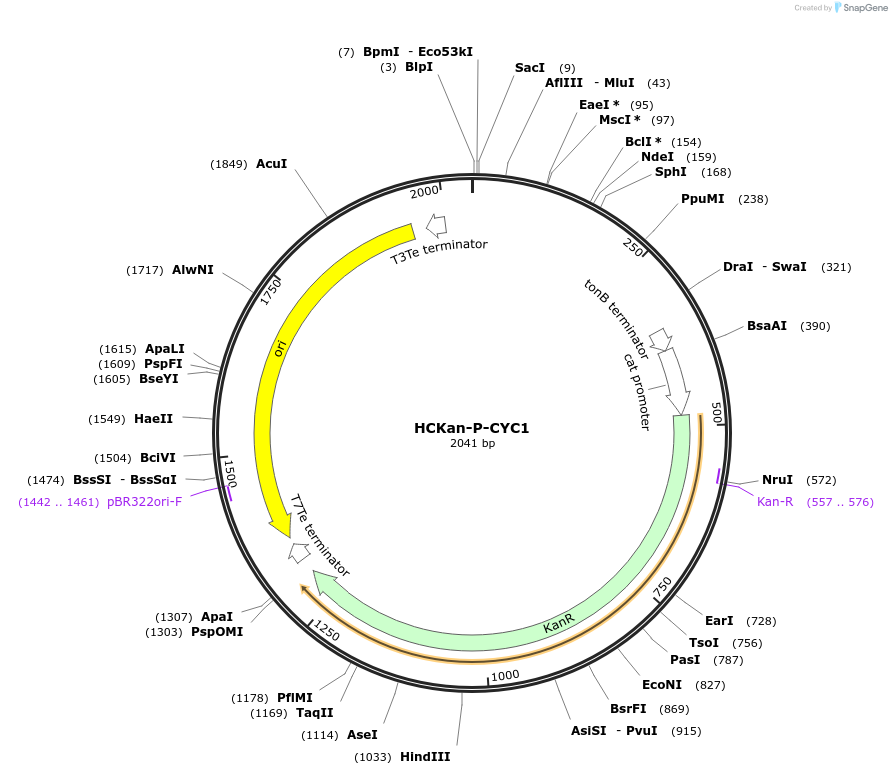
HCKan-P-CYC1
Plasmid#65338Purposeconstruction of CYC1 promoter in HCKan-P receiving vectorDepositorInsertCYC1 promoter
UseSynthetic BiologyAvailable SinceSept. 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
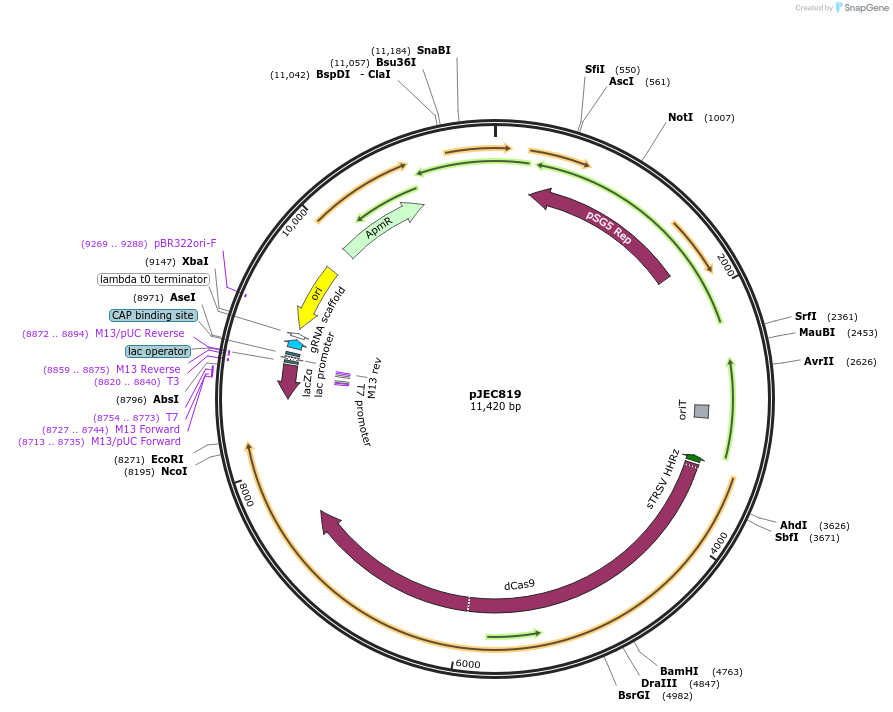
pJEC819
Plasmid#190815PurposeCRISPRa backbone plasmid harboring BbsI sites for easy cloning of sgRNA targeting regions via Golden gate.DepositorInsertdCas9-XTEN-rpoA
UseCRISPR and Synthetic BiologyTagsXTEN (linker), followed by the N-terminal domain …PromoterSP30Available SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
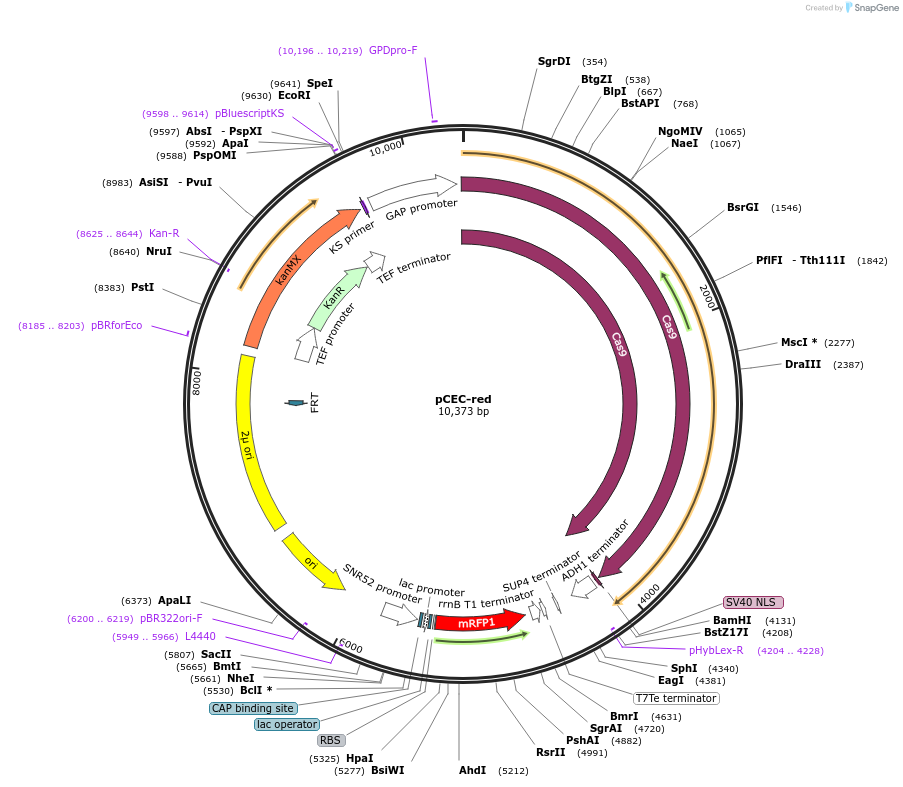
pCEC-red
Plasmid#196040PurposeAllows the expression of Cas9 in Saccharomyces cerevisiae and contains a gRNA expression unit with a mRFP1 placeholder for user-defined gRNA insertion using BsaI-mediated Golden Gate AssemblyDepositorTypeEmpty backboneUseCRISPR and Synthetic Biology; Grna cloningExpressionYeastAvailable SinceMarch 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
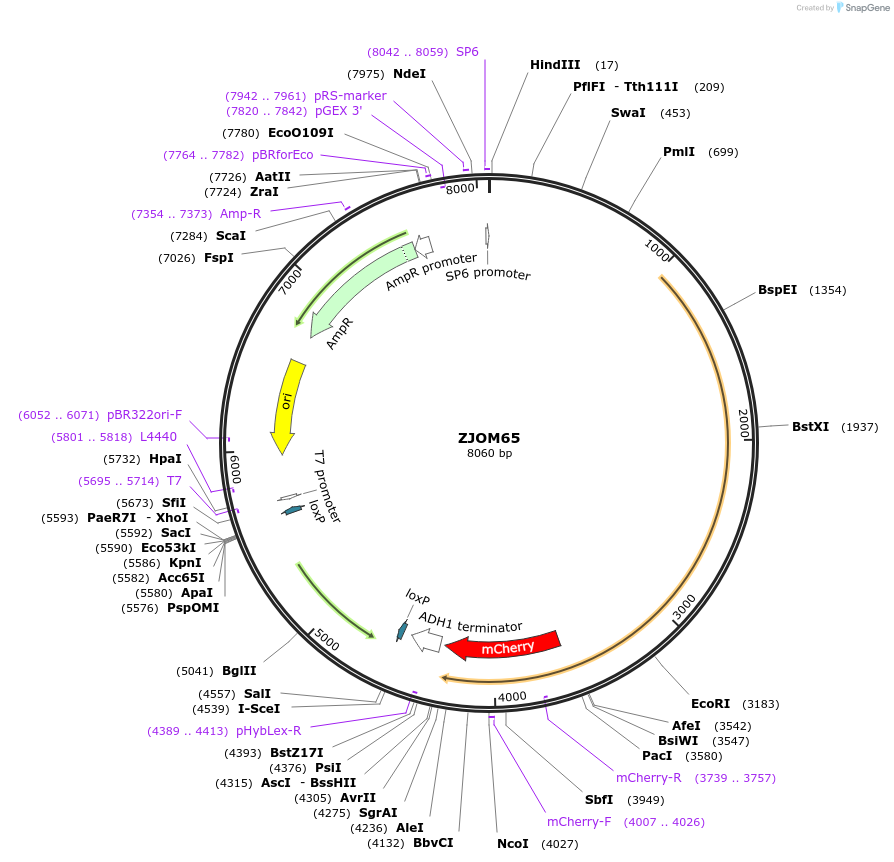
ZJOM65
Plasmid#133654PurposeIntegration of VPH1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
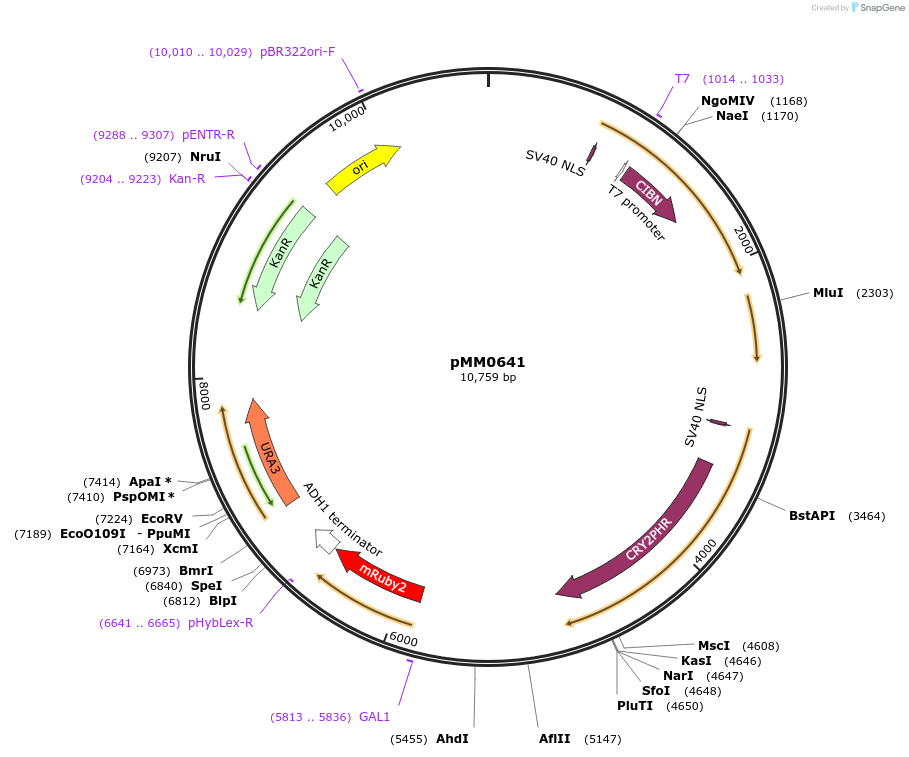
pMM0641
Plasmid#129006PurposeIntegration vector for optogenetic machinery (pMEDIUM-VP16-CIB1/pMEDIUM-ZCRY2PHR) and pZF-mRUBY2DepositorInsert5' URA3-pRPL18B-SV40NLS-VP16-CIB1-tENO1-pRPL18B-SV40NLS-ZiF268DBD-CRY2PHR-tSSA1-pZF(BS)-mRUBY2-tADH1-URA3 URA3 3'-KanR-ColE1
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
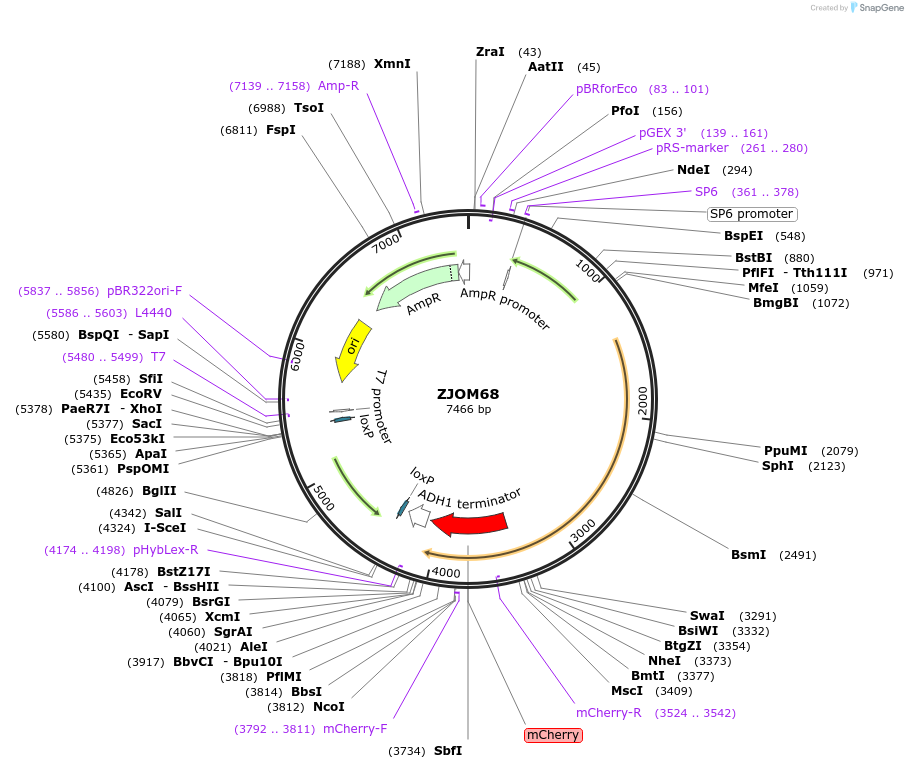
ZJOM68
Plasmid#133657PurposeIntegration of TGL3-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
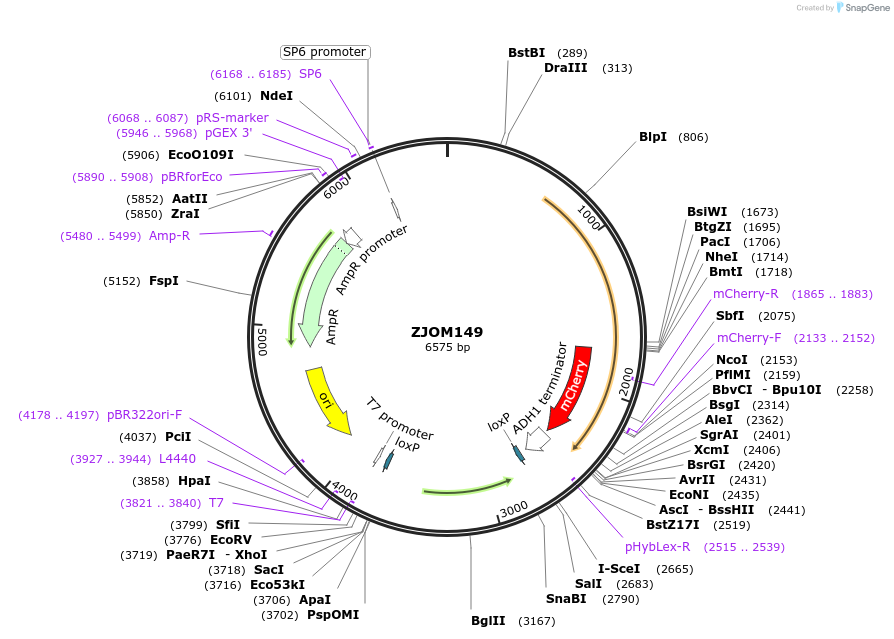
ZJOM149
Plasmid#133646PurposeIntegration of ELO3-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
pTiGc
Plasmid#78314PurposeMycobacterial dual reporter plasmid for fluorescence dilution. Description: hsp60-gfp, hsp60(ribo)-turboFP635 (inducible TurboFP635 under control of theophylline - inducible riboswitch).DepositorInsertsTurboFP635
GFP
UseSynthetic Biology; Mycobacterial replicating vect…ExpressionBacterialPromoterhsp60 and hsp60(ribo)Available SinceJuly 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
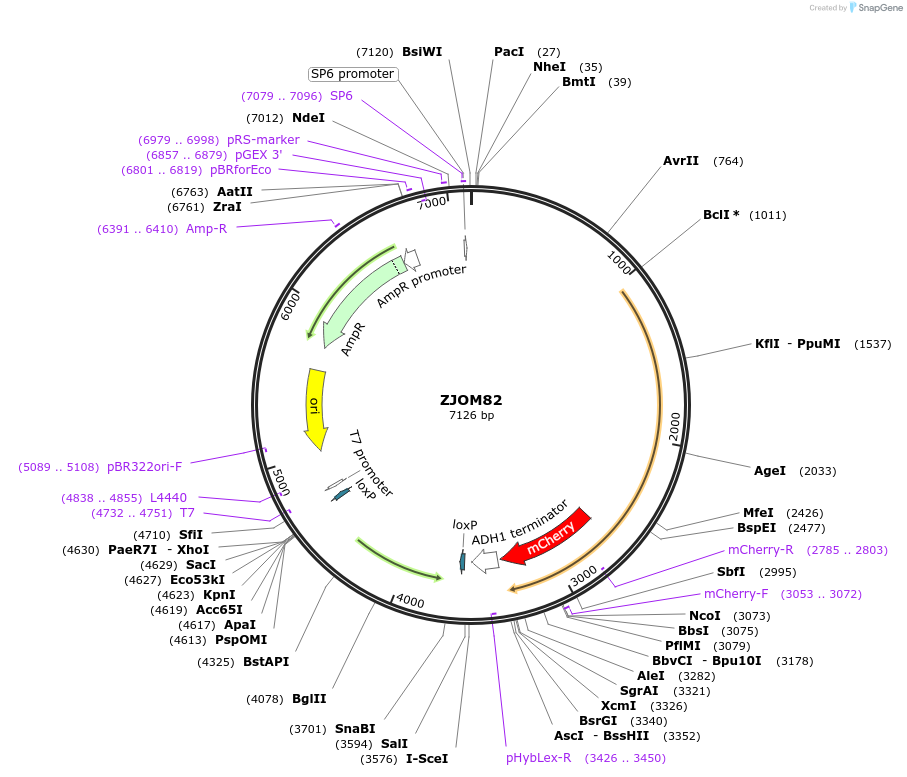
ZJOM82
Plasmid#133648PurposeIntegration of NAB2-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceJan. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
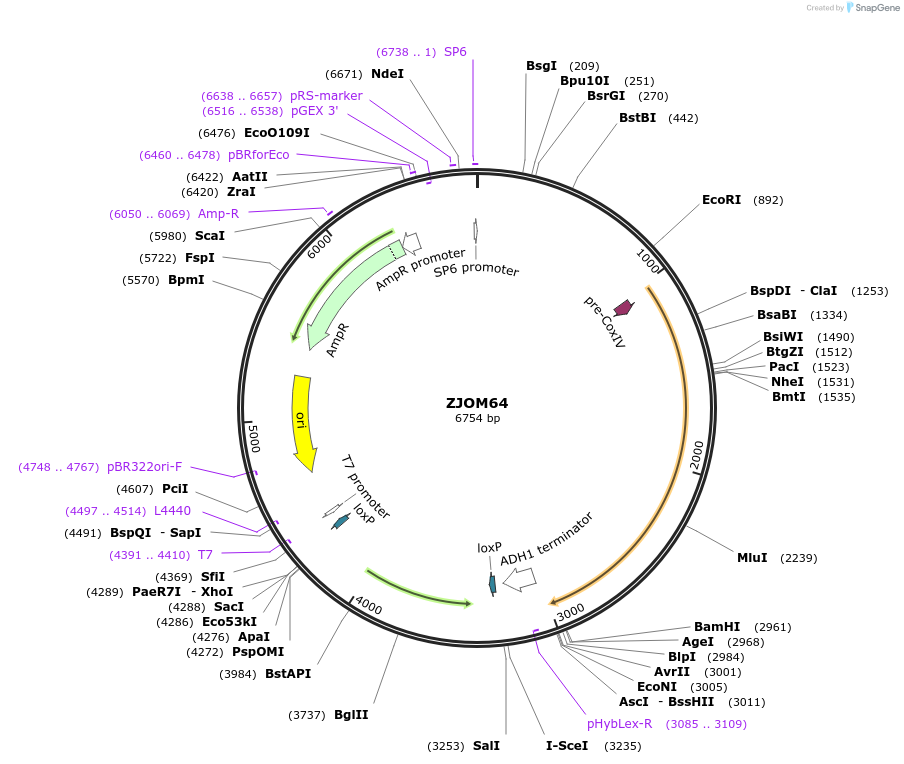
ZJOM64
Plasmid#133655PurposeIntegration of COX4-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
ZJOM135
Plasmid#133659PurposeIntegration of SEC63-2mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
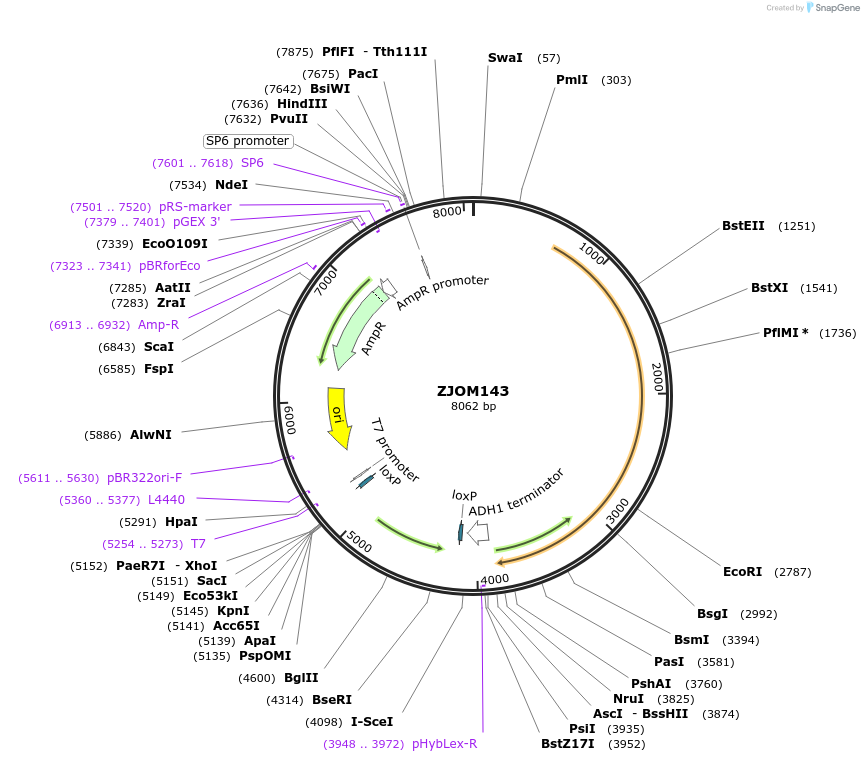
ZJOM143
Plasmid#133667PurposeIntegration of VPH1-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
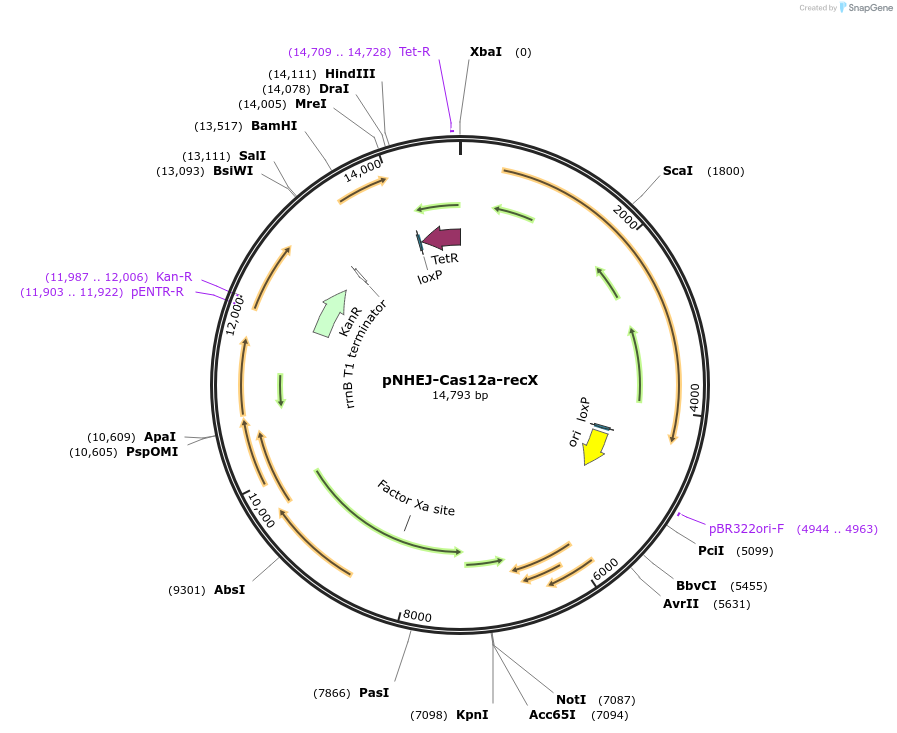
pNHEJ-Cas12a-recX
Plasmid#158714PurposeCRISPR-assisted-NHEJ system used for genome editing in Mycobacterium smegmatis. Helper plasmid expresses FnCpf1, NHEJ machinery of M. marinum and recXDepositorInsertrecX
UseCRISPRExpressionBacterialPromoterPmycAvailable SinceJan. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
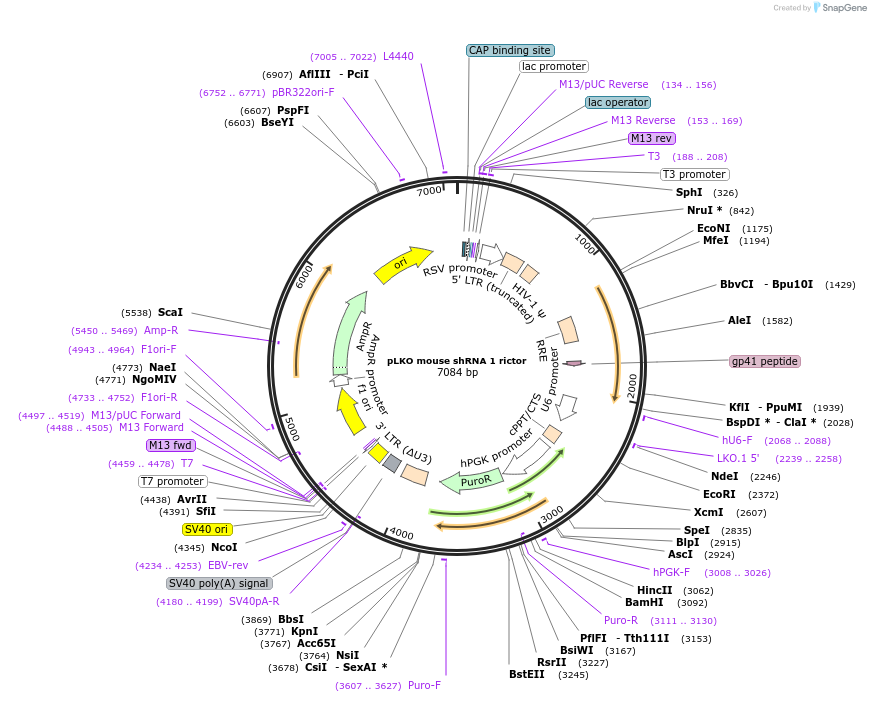
pLKO mouse shRNA 1 rictor
Plasmid#21341DepositorAvailable SinceJuly 6, 2009AvailabilityAcademic Institutions and Nonprofits only -
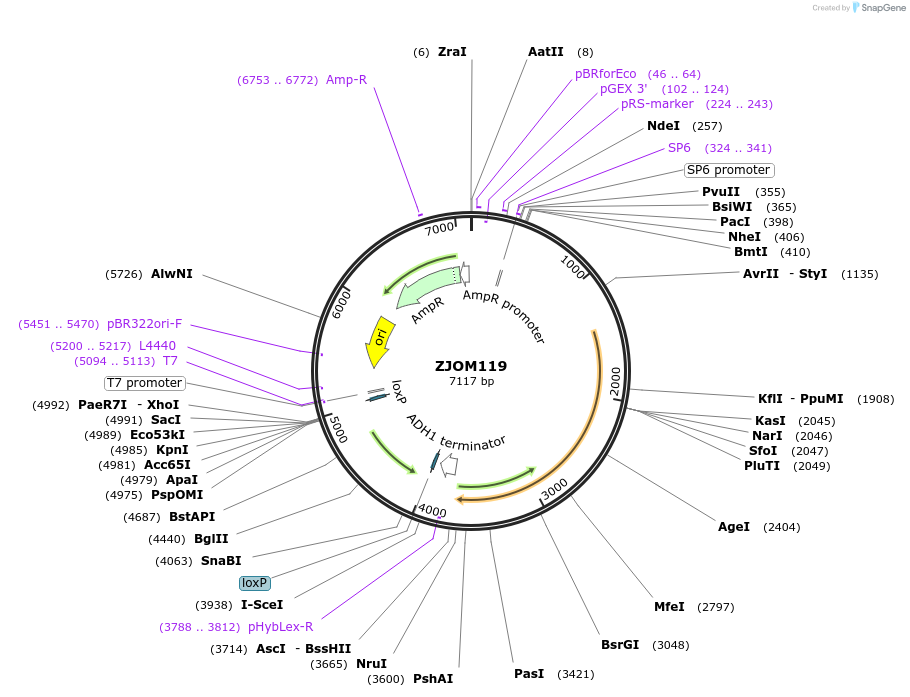
ZJOM119
Plasmid#133661PurposeIntegration of NAB2-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceDec. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
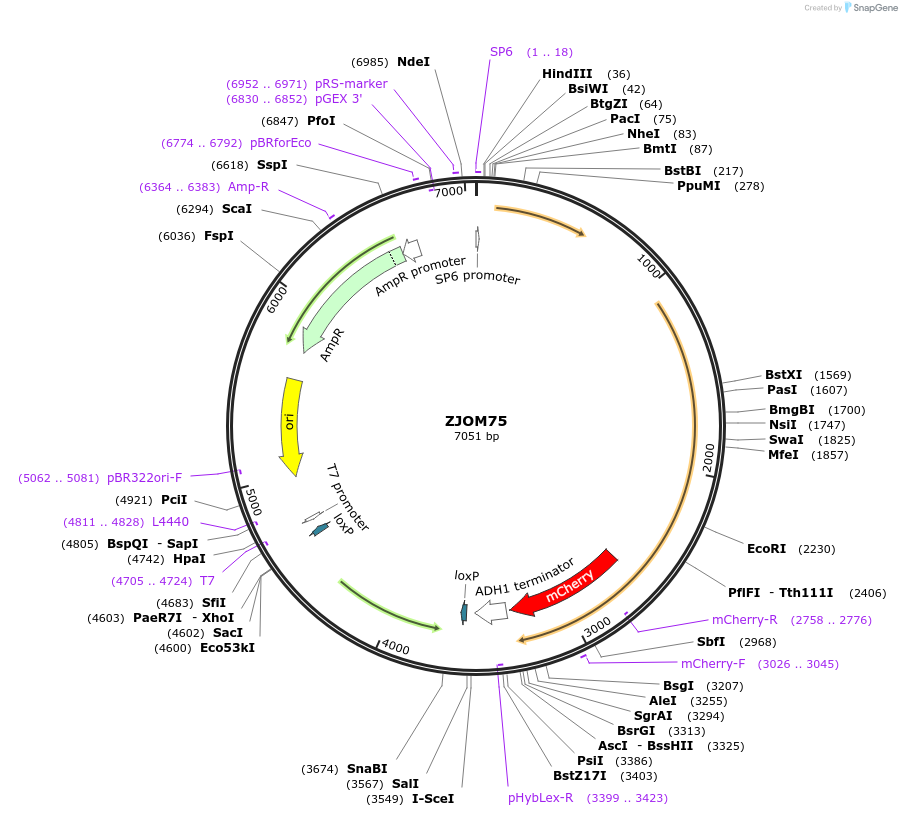
ZJOM75
Plasmid#133649PurposeIntegration of ANP1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceJan. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
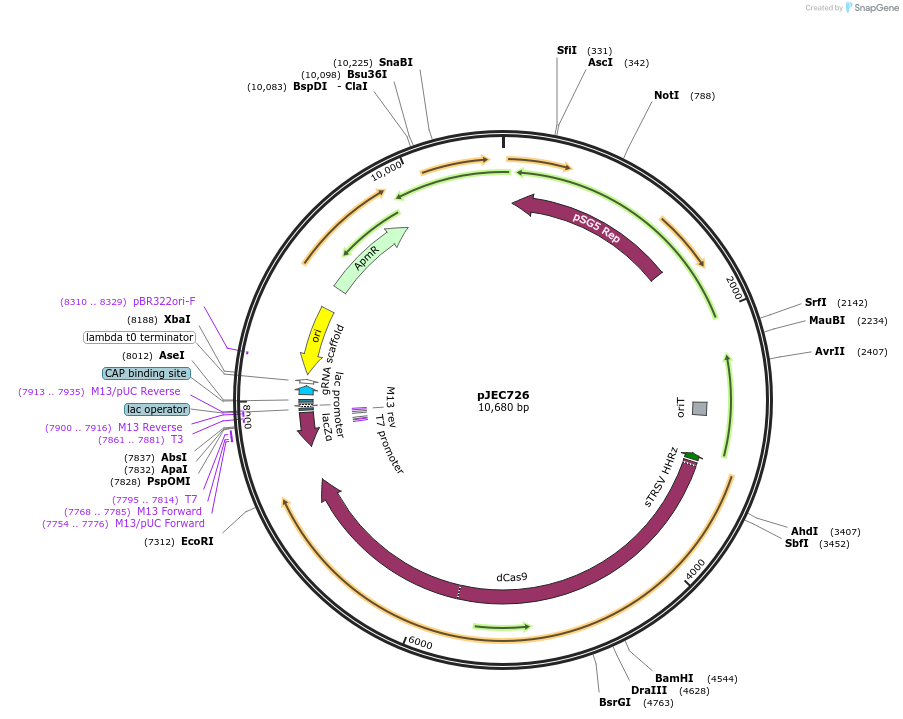
pJEC726
Plasmid#190814PurposeCRISPRi backbone plasmid harboring BbsI sites for easy cloning of sgRNA targeting regions via Golden gate.DepositorInsertdCas9
UseCRISPR and Synthetic BiologyMutationS. pyogenes dCas9 codon optimized for Streptomyce…PromoterSP30Available SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLKO mouse shRNA 1 raptor
Plasmid#21339DepositorAvailable SinceJuly 7, 2009AvailabilityAcademic Institutions and Nonprofits only