We narrowed to 102 results for: cryptococcus
-
Plasmid#128370PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 6, a small gene-free region. Contains the nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseUnspecifiedPromoterACT1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only
-
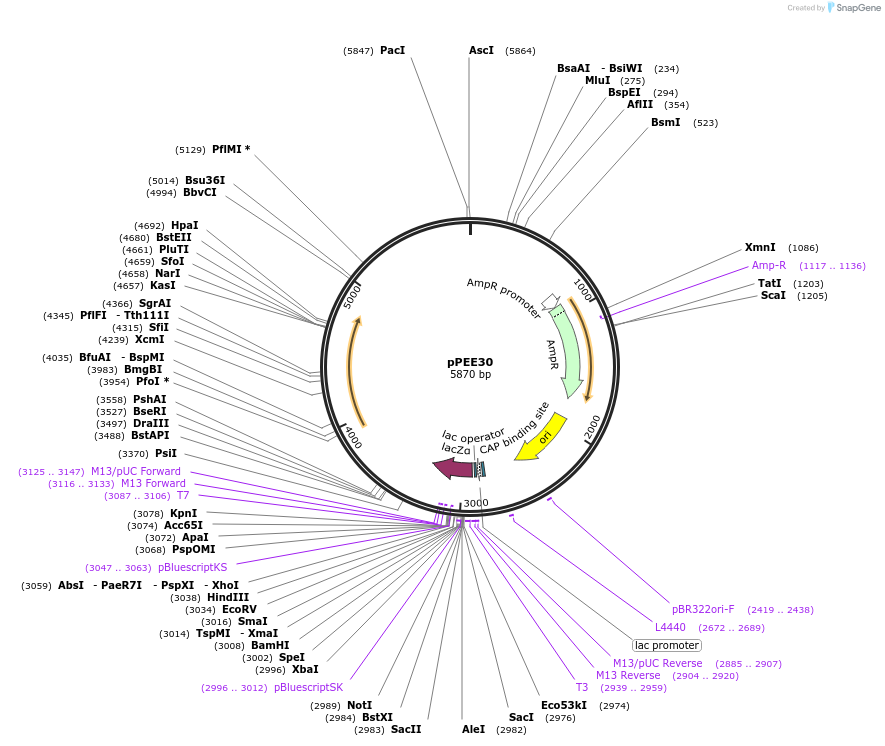
pPEE19
Plasmid#128362PurposeDominant marker in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site 3, a small gene-free region. Contains the amdS2 counterselectable marker.DepositorTypeEmpty backboneUseUnspecifiedPromoterTEF1Available SinceAug. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
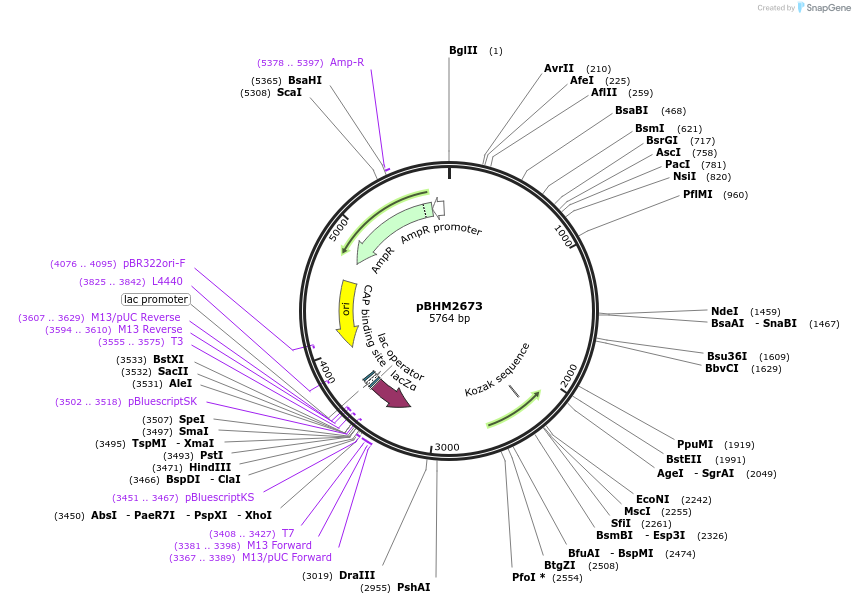
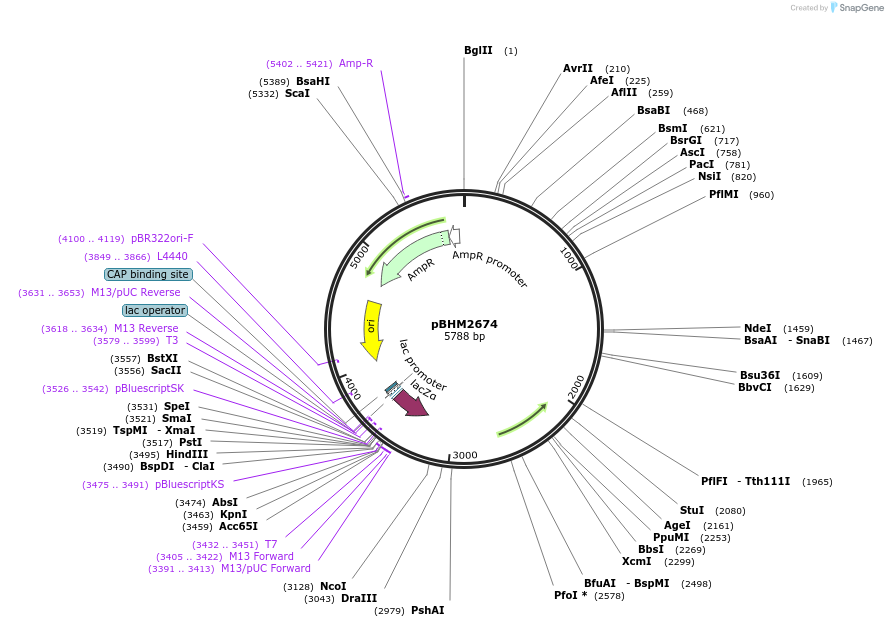
pBHM2673
Plasmid#241720PurposeIntegration of DNA into Cryptococcus neoformans Safe Haven 1 locus, contains BSD marker for selection on blasticidin SDepositorTypeEmpty backboneExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
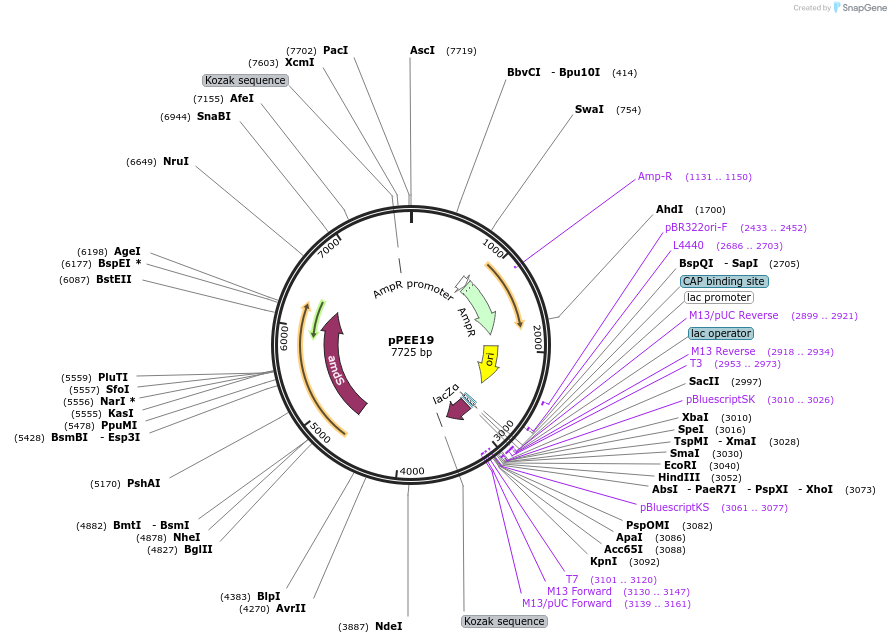
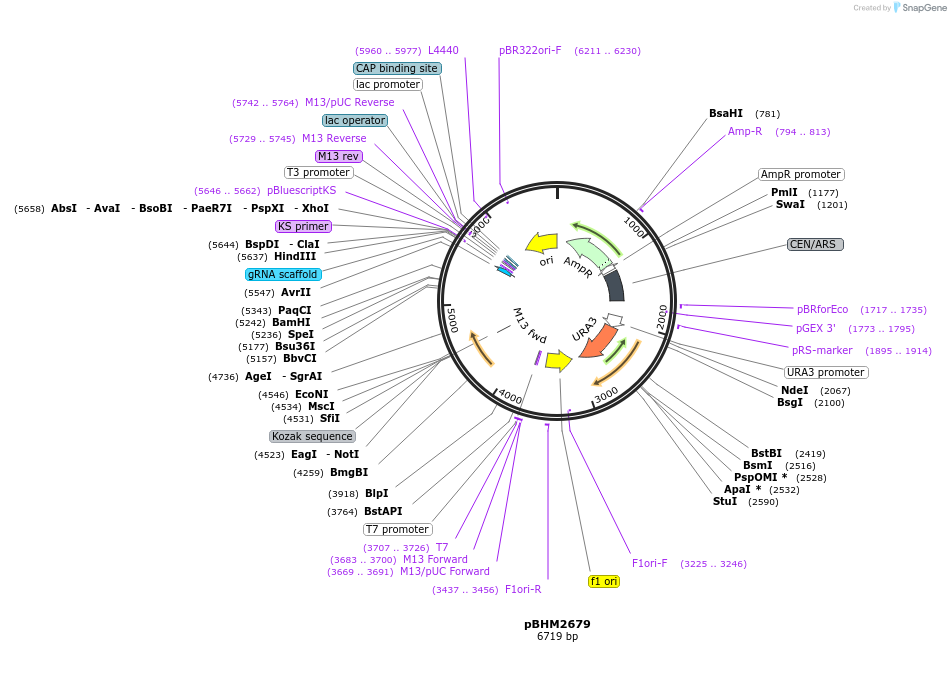
pBHM2679
Plasmid#241724PurposeConstruction of fused marker-sgRNA inserts for Cryptococcus neoformans genome editing, contains BSD marker for selection on blasticidin SDepositorTypeEmpty backboneUseCRISPRExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
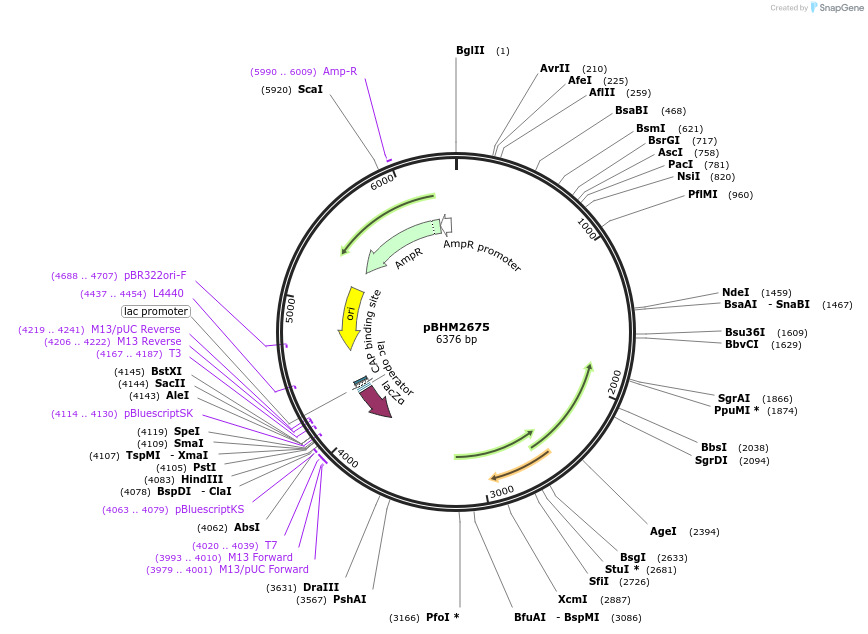
pBHM2675
Plasmid#241722PurposeIntegration of DNA into Cryptococcus neoformans Safe Haven 1 locus, contains ptxD marker for selection on media containing phosphite as a sole phosphorus sourceDepositorTypeEmpty backboneExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
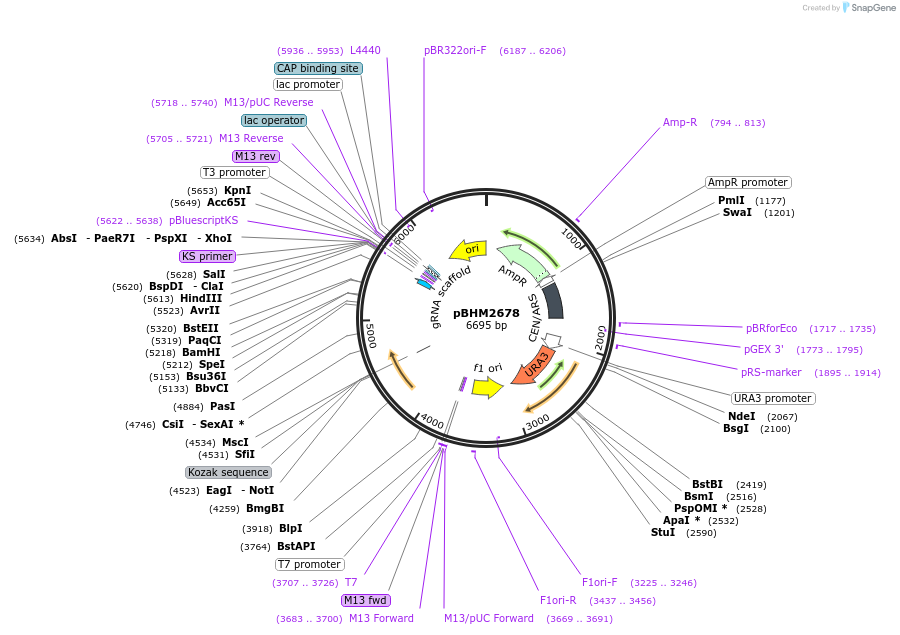
pBHM2678
Plasmid#241723PurposeConstruction of fused marker-sgRNA inserts for Cryptococcus neoformans genome editing, contains BLE marker for selection on phleomycinDepositorTypeEmpty backboneUseCRISPRExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
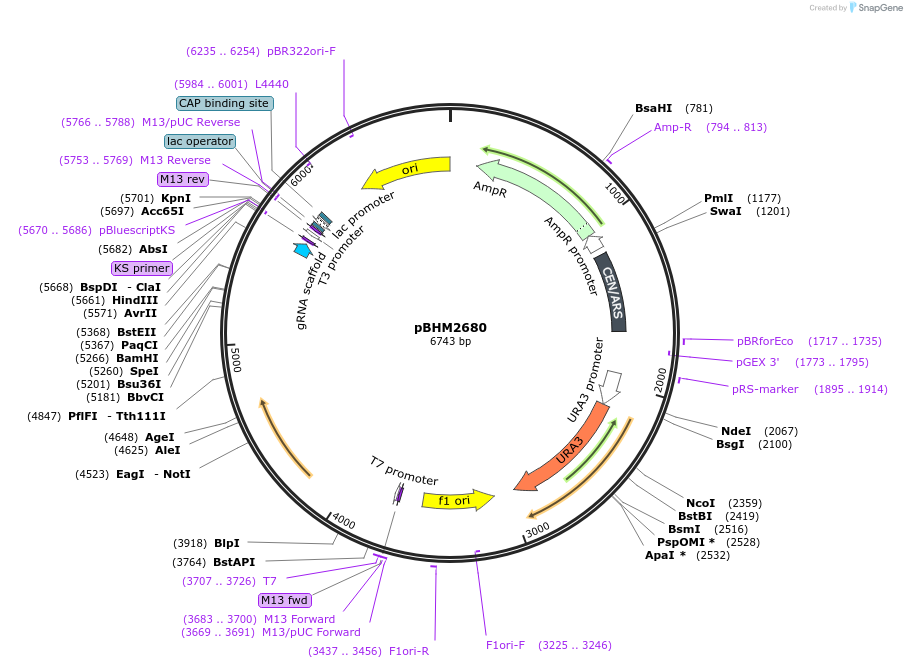
pBHM2680
Plasmid#241725PurposeConstruction of fused marker-sgRNA inserts for Cryptococcus neoformans genome editing, contains BSR marker for selection on blasticidin SDepositorTypeEmpty backboneUseCRISPRExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
pBHM2674
Plasmid#241727PurposeIntegration of DNA into Cryptococcus neoformans Safe Haven 1 locus, contains BSR marker for selection on blasticidin SDepositorTypeEmpty backboneExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
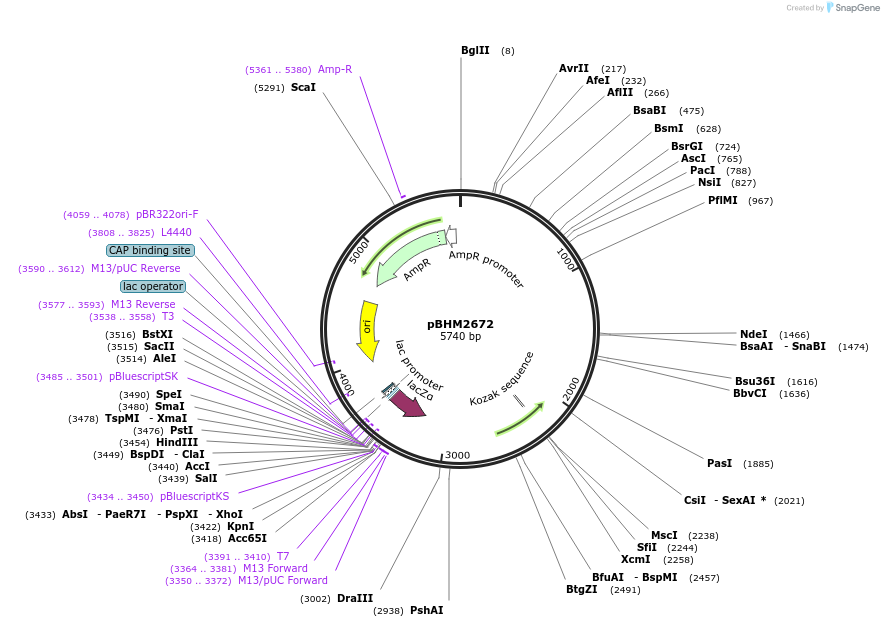
pBHM2672
Plasmid#241719PurposeIntegration of DNA into Cryptococcus neoformans Safe Haven 1 locus, contains BLE marker for selection on phleomycinDepositorTypeEmpty backboneExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
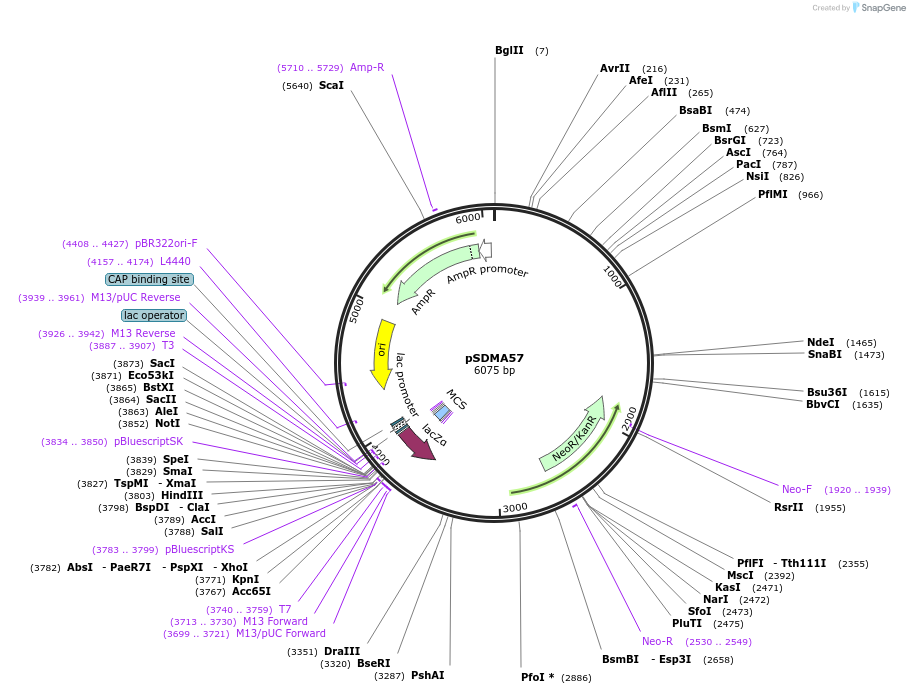
pSDMA57
Plasmid#67941PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site, a small gene-free region. Contains NEO resistance marker.DepositorTypeEmpty backboneUseCryptococcal complementation vectorPromoterACT1 for Cryptococcal markerAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
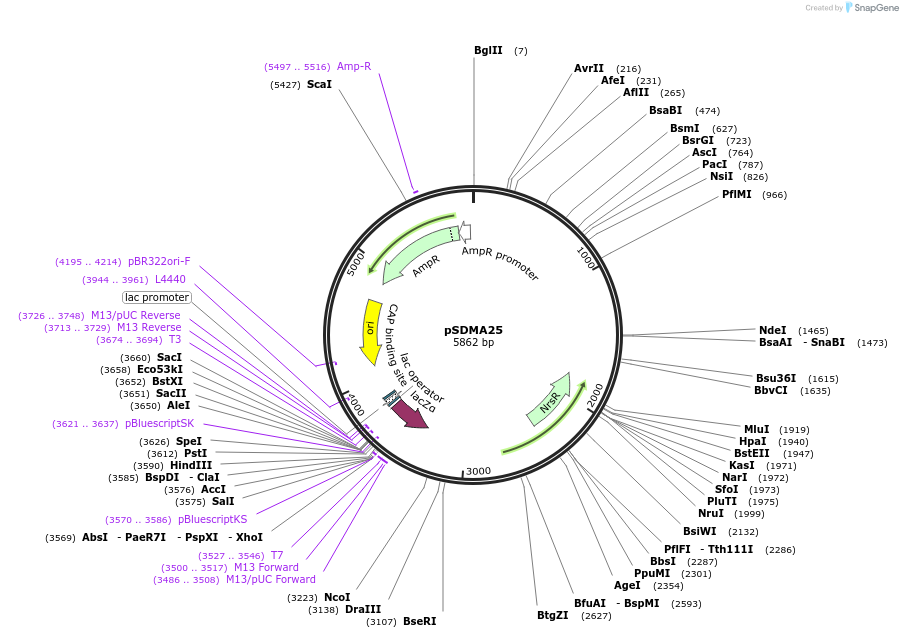
pSDMA25
Plasmid#67940PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site, a small gene-free region. Contains nourseothricin resistance marker (NAT)DepositorTypeEmpty backboneUseCryptococcal complementation vectorPromoterACT1 for Cryptococcal markerAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
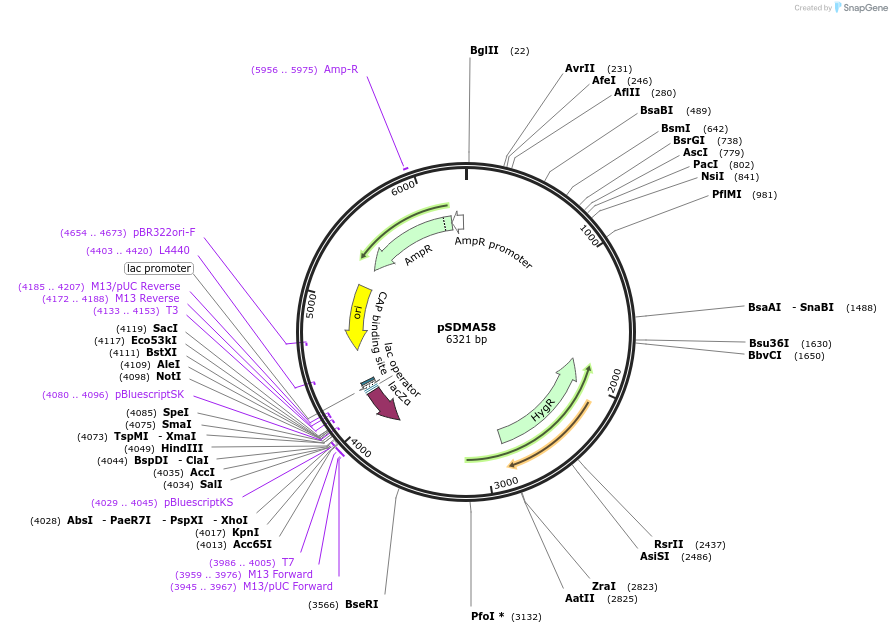
pSDMA58
Plasmid#67942PurposeComplementation in Cryptococcus neoformans. Targets DNA constructs to a Safe Haven Site, a small gene-free region. Contains HYG resistance marker.DepositorTypeEmpty backboneUseCryptococcal complementation vectorPromoterACT1 for Cryptococcal markerAvailable SinceOct. 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
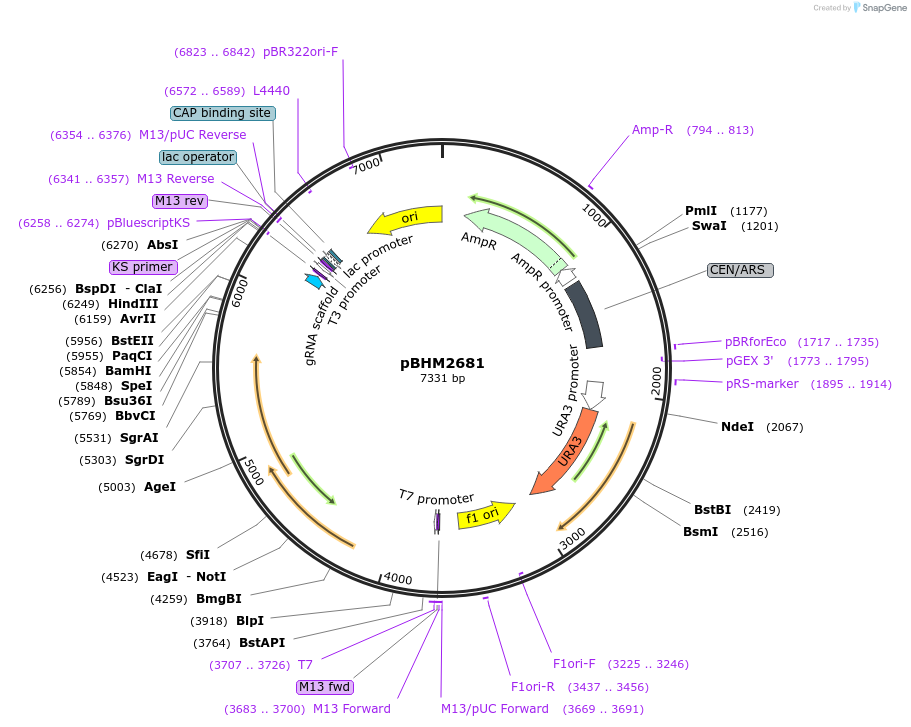
pBHM2681
Plasmid#241726PurposeConstruction of fused marker-sgRNA inserts for Cryptococcus neoformans genome editing, contains ptxD marker for selection on media containing phosphite as a sole phosphorus sourceDepositorTypeEmpty backboneUseCRISPRExpressionYeastAvailable SinceSept. 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
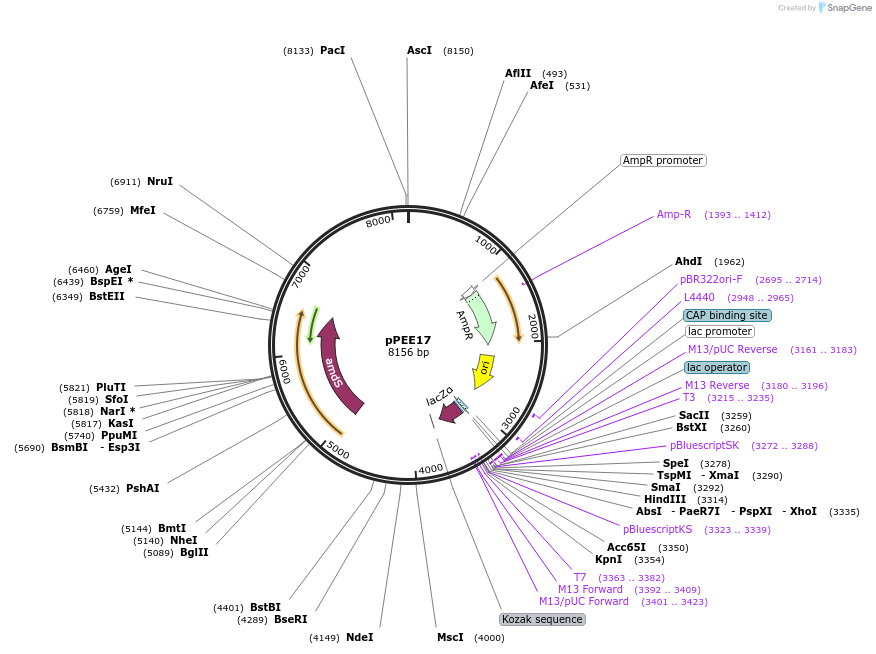
pPEE17
Plasmid#128361PurposeDetecting gene essentiality in Cryptococcus neoformans. Targets DNA constructs to the Safe Haven Site 1, a small gene-free region. Contains amdS2 counterselectable marker. This version of the Safe Haven vector includes direct repeats, enabling the formation of popouts on fluoroacetamide.DepositorInsertAcetamidase (amdS)
UseUnspecifiedPromoterTEF1Available SinceJan. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
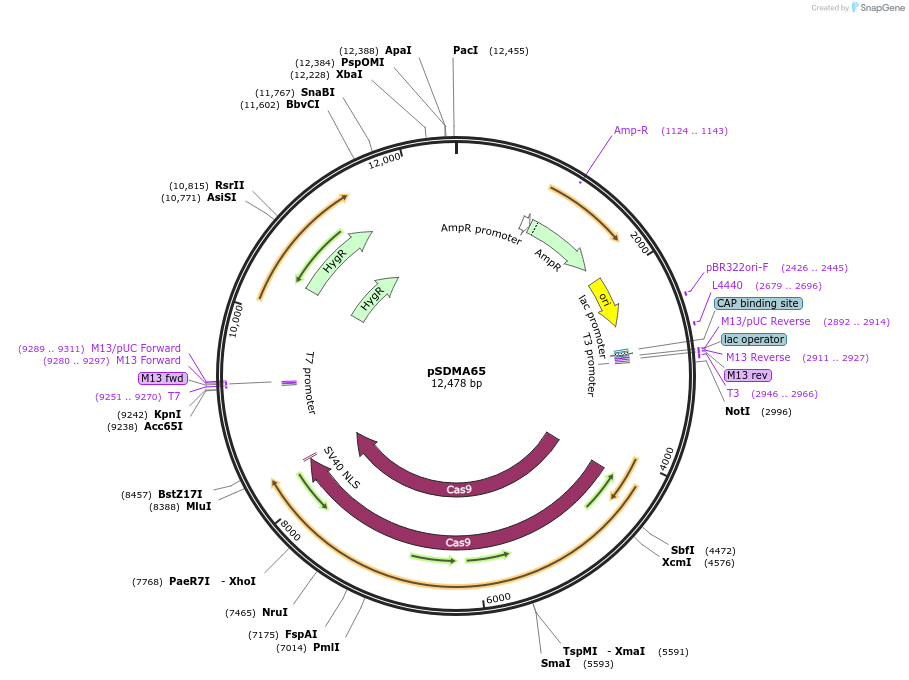
pSDMA65
Plasmid#128354PurposeCodon optimised Streptococcus pyogenes CAS9 ORF regulated by the Cryptococcus neoformans TEF1 promoter and terminator.DepositorTypeEmpty backboneUseCRISPRPromoterACT1Available SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
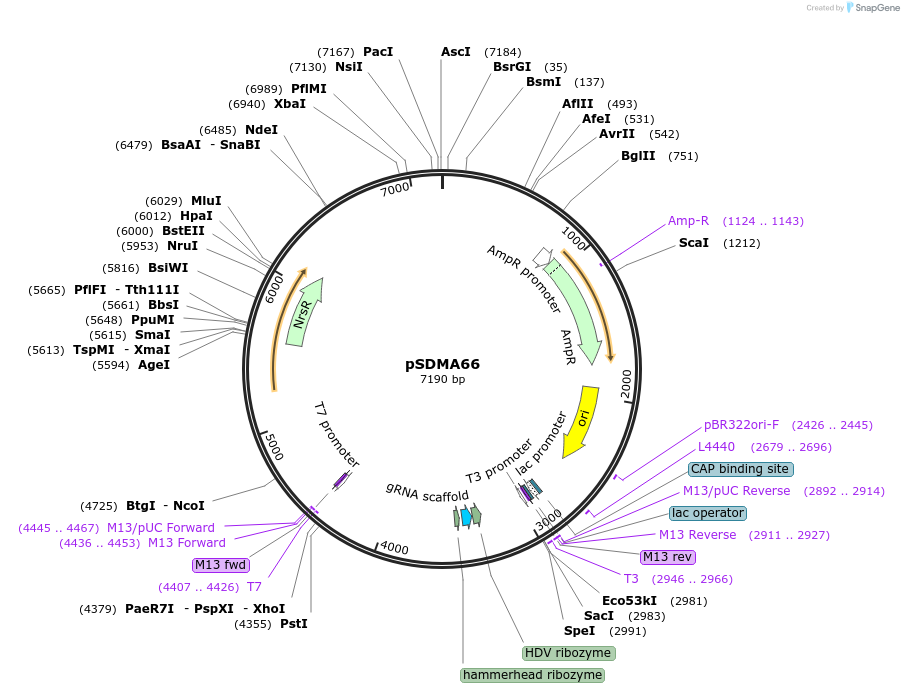
pSDMA66
Plasmid#128355PurposegRNA ribozyme construct with Cryptococcus neoformans ACT1 promter and TRP1 terminator. Following ribozyme cleavage, a gRNA targeting ADE2 is liberated.DepositorInsertNoureothricin (NAT)
UseUnspecifiedPromoterACT1Available SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
pSDMA67
Plasmid#128356PurposegRNA ribozyme construct with Cryptococcus neoformans ACT1 promter and TRP1 terminator. Following ribozyme cleavage, a gRNA targeting ADE2 is liberated.DepositorInsertNoureothricin (NAT)
UseUnspecifiedPromoterACT1Available SinceAug. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
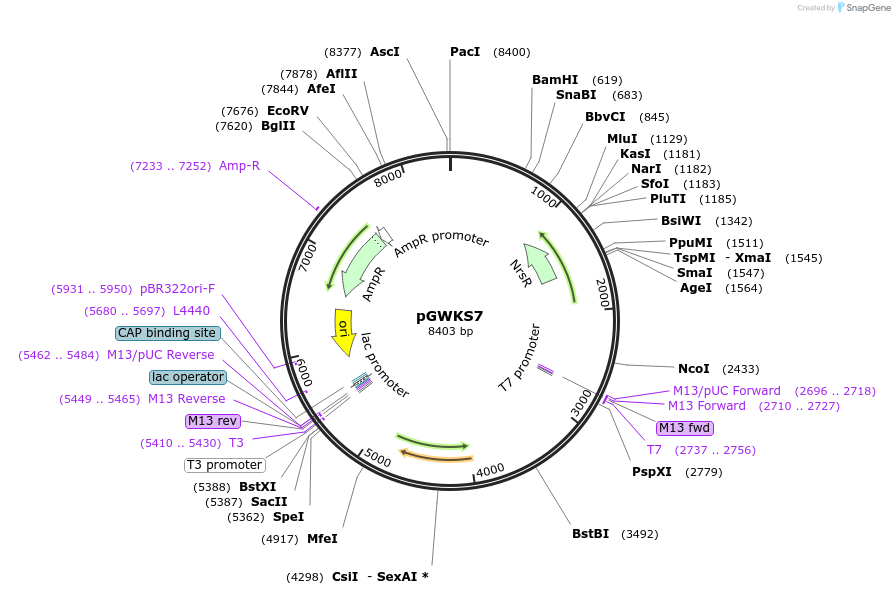
pGWKS7
Plasmid#139414PurposeFluoroescent marker for use in Cryptococcus neoformans. Contains a codon optimised mRuby3 marker.DepositorInsertmRuby3
ExpressionYeastPromoterTEF1Available SinceMay 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
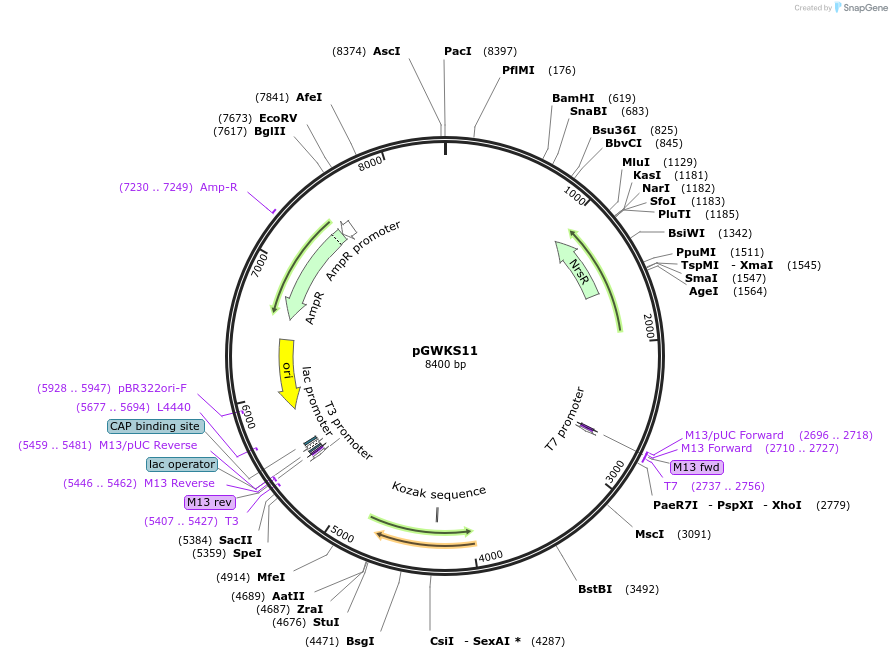
pGWKS11
Plasmid#139418PurposeFluoroescent marker for use in Cryptococcus neoformans. Contains a codon optimised mNeonGreen marker.DepositorInsertmNeonGreen
ExpressionYeastPromoterTEF1Available SinceMay 4, 2020AvailabilityAcademic Institutions and Nonprofits only -
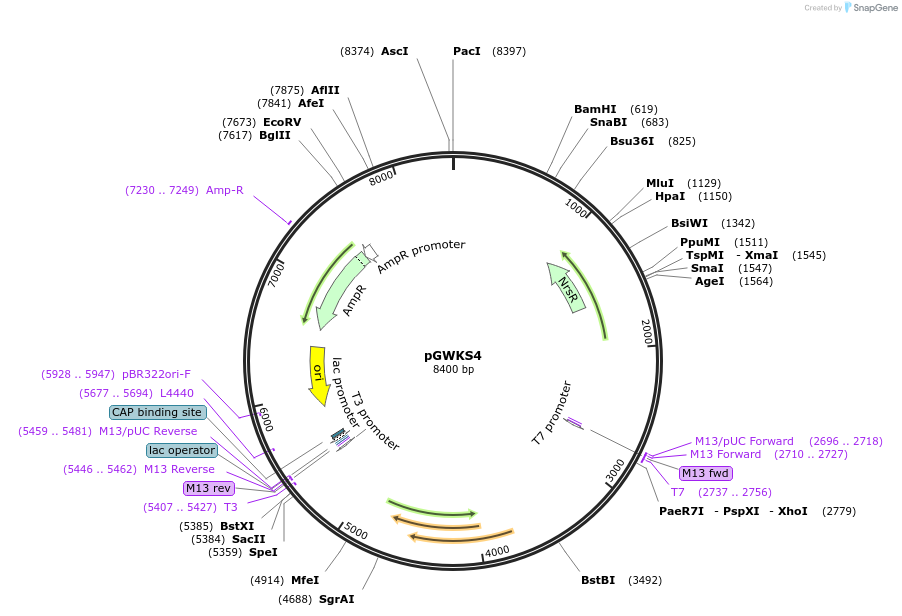
pGWKS4
Plasmid#139411PurposeFluoroescent marker for use in Cryptococcus neoformans. Contains a codon optimised mCherry marker.DepositorInsertmCherry
ExpressionYeastPromoterTEF1Available SinceMay 4, 2020AvailabilityAcademic Institutions and Nonprofits only