We narrowed to 40,900 results for: Eras
-
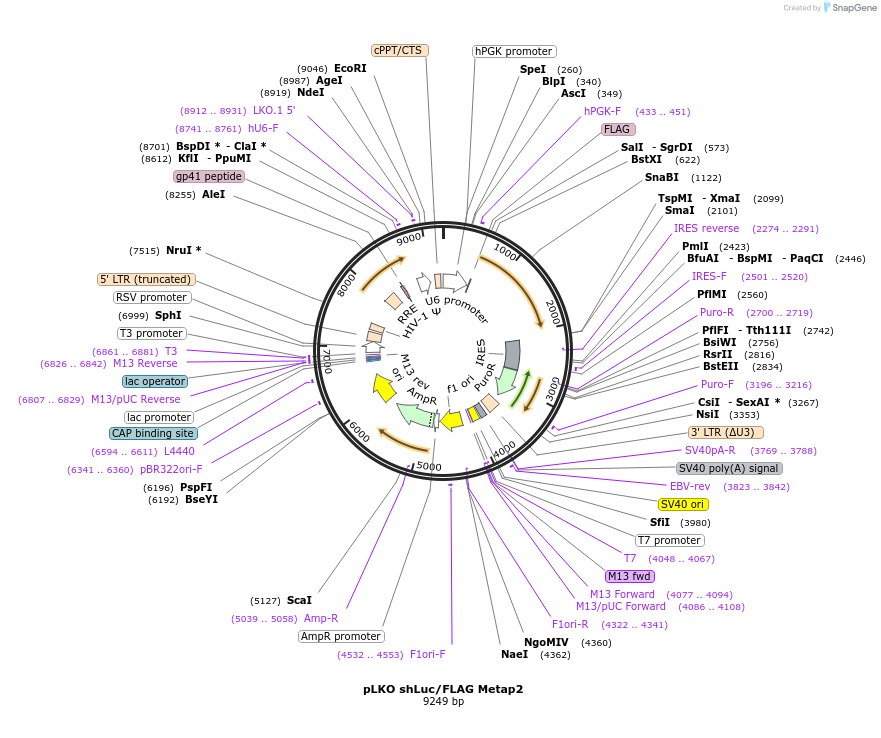
Plasmid#209316PurposeLentiviral construct expressing FLAG Metap2 and shRNA against Luciferase (control shRNA)DepositorAvailable SinceDec. 11, 2023AvailabilityAcademic Institutions and Nonprofits only
-
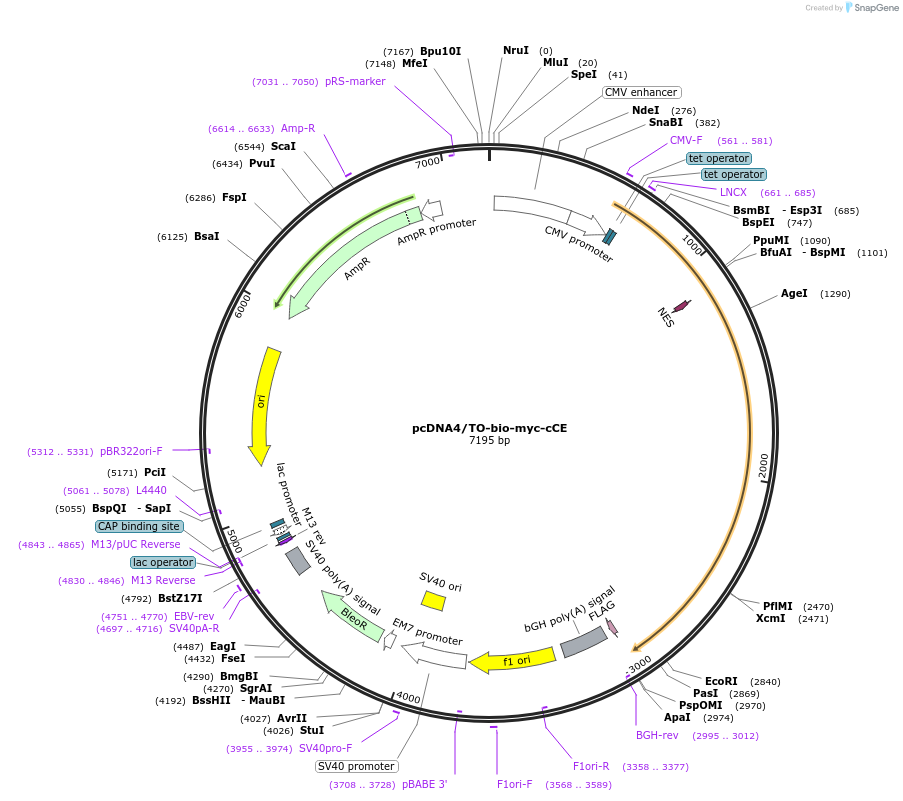
pcDNA4/TO-bio-myc-cCE
Plasmid#112711PurposeFor tetracycline-inducible expression of bio-myc-NES-mCE ΔNLS ("bio-myc-cCE") in mammalian cellsDepositorInsertRngtt (Rngtt Mouse)
TagsHIV Rev nuclear localization signal (NES), Stop c…ExpressionMammalianMutationSilent mutation at Arg 132 (CGT to CGC), deleted …PromoterCMV with Tet repressor binding sitesAvailable SinceSept. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
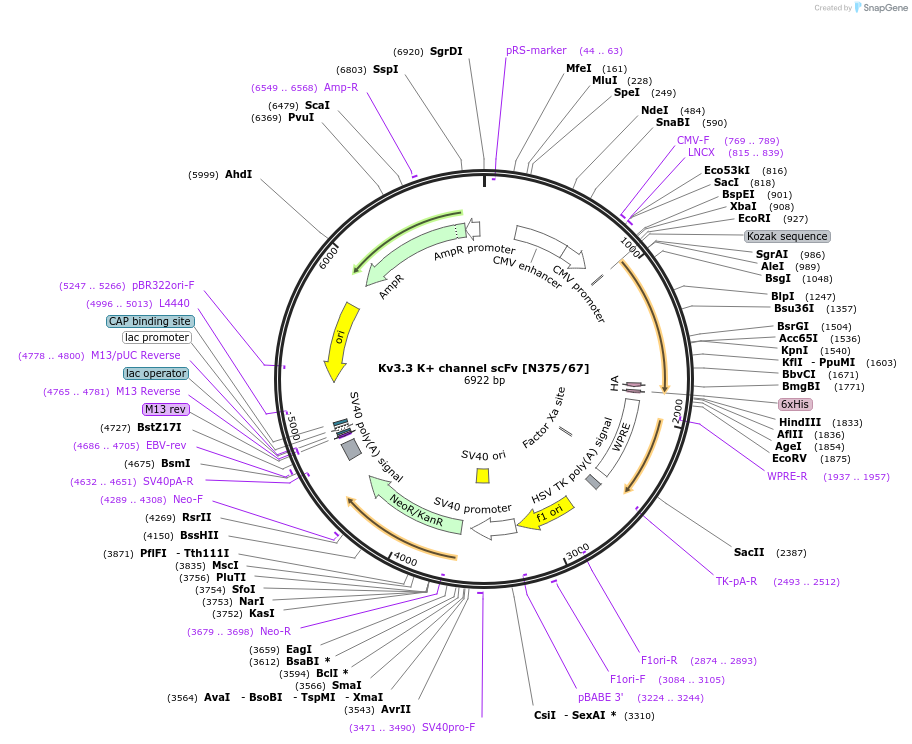
Kv3.3 K+ channel scFv [N375/67]
Plasmid#190549PurposeMammalian Expression of Kv3.3 K+ channel scFV. Derived from hybridoma N375/67.DepositorInsertKv3.3 K+ channel (Mus musculus) recombinant scFV (Kcnc3 Mouse)
TagsHA, Sortase, 6xHisExpressionMammalianPromoterCMVAvailable SinceNov. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
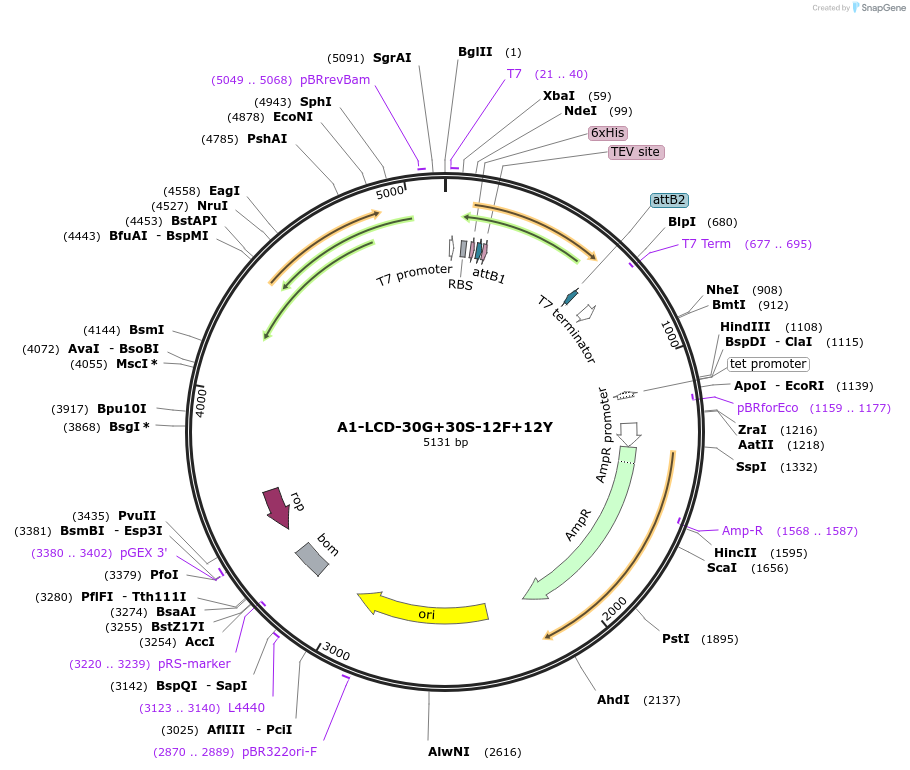
A1-LCD-30G+30S-12F+12Y
Plasmid#178687PurposeBacterial expression of N-terminally 6His tagged A1-LCD with thirty Gly residues replaced with Ser, twelve Phe residues replaced with Tyr (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD-30G+30S-12F+12Y (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationthirty Gly residues replaced with Ser, twelve Phe…Available SinceMarch 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
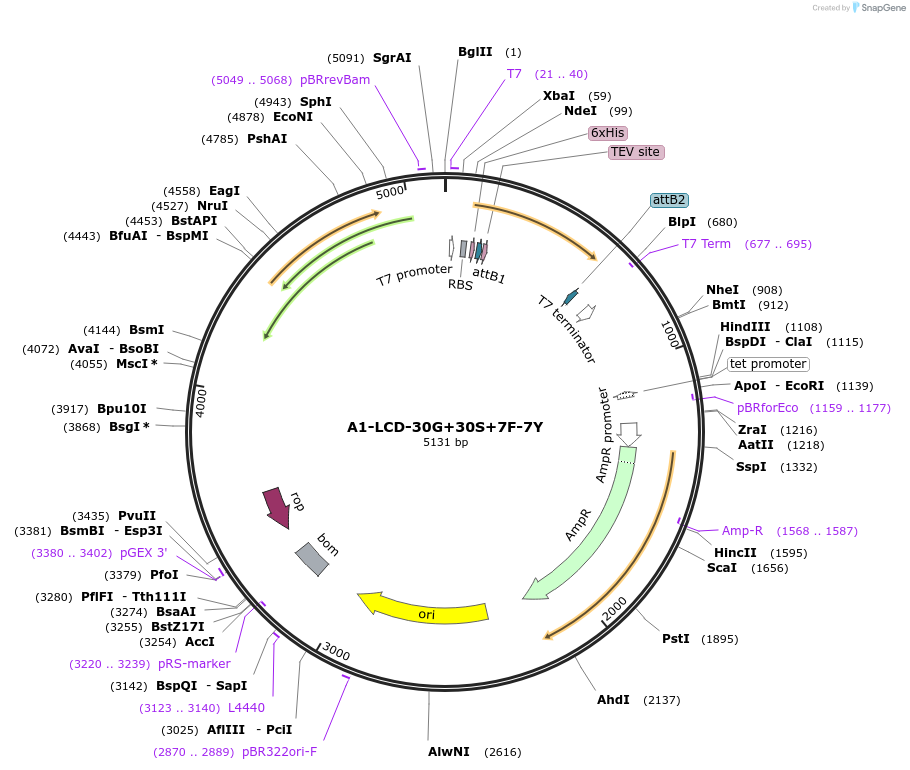
A1-LCD-30G+30S+7F-7Y
Plasmid#178686PurposeBacterial expression of N-terminally 6His tagged A1-LCD with thirty Gly residues replaced with Ser, seven Tyr residues replaced with Phe (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD-30G+30S+7F-7Y (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationthirty Gly residues replaced with Ser, seven Tyr …Available SinceMarch 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
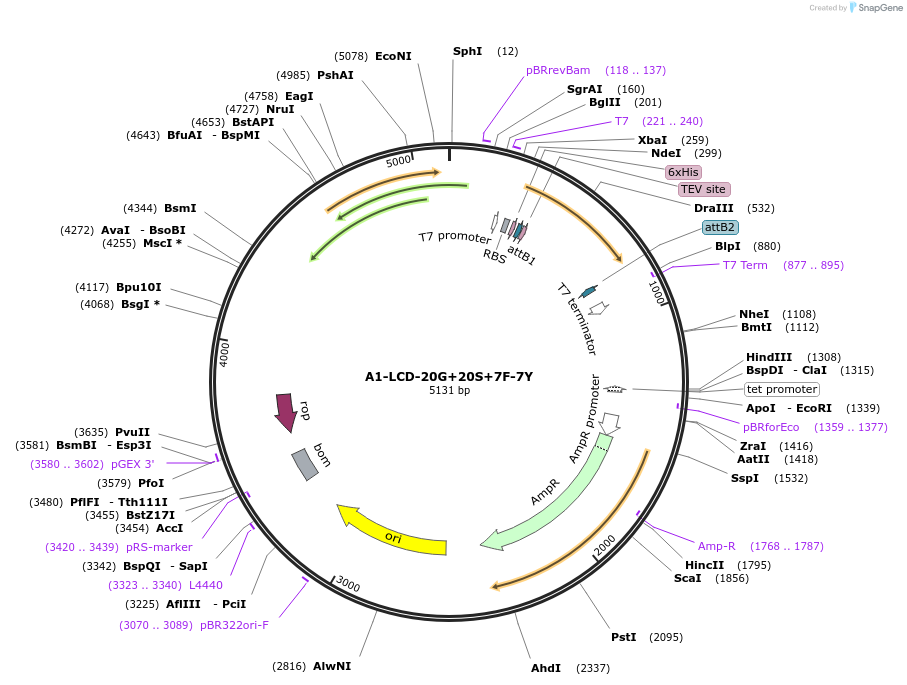
A1-LCD-20G+20S+7F-7Y
Plasmid#178688PurposeBacterial expression of N-terminally 6His tagged A1-LCD with twenty Gly residues replaced with Ser, seven Tyr residues replaced with Phe (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD-20G+20S+7F-7Y (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationtwenty Gly residues replaced with Ser, seven Tyr …Available SinceMarch 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
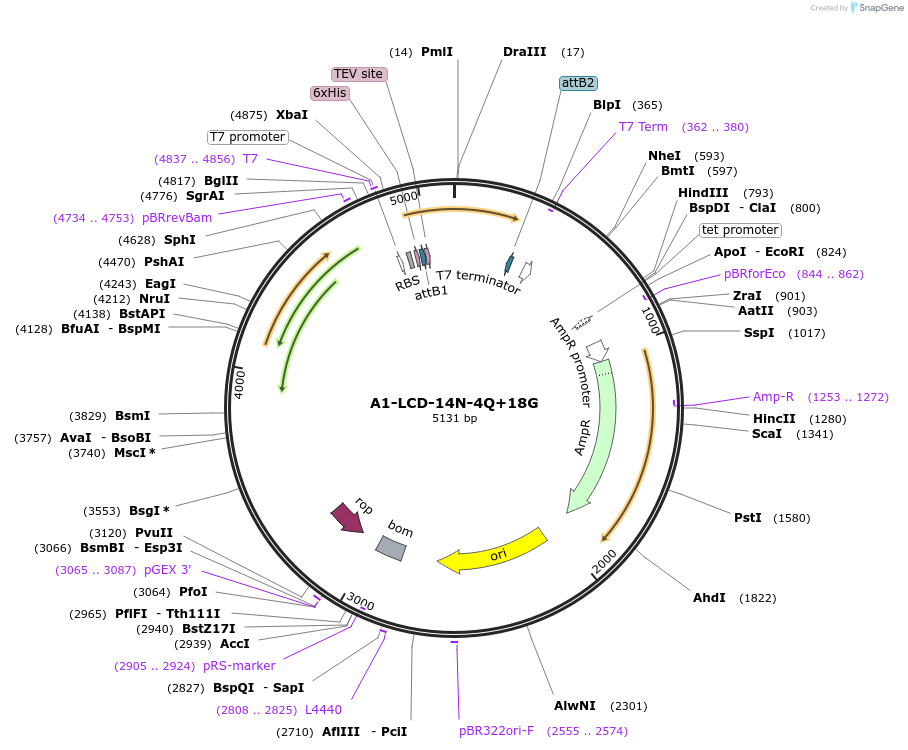
A1-LCD-14N-4Q+18G
Plasmid#178694PurposeBacterial expression of N-terminally 6His tagged A1-LCD with fourteen Gln residues removed, four Asn removed, replaced with eighteen Gly (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD-14N-4Q+18G (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationfourteen Gln residues removed, four Asn removed, …Available SinceMarch 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
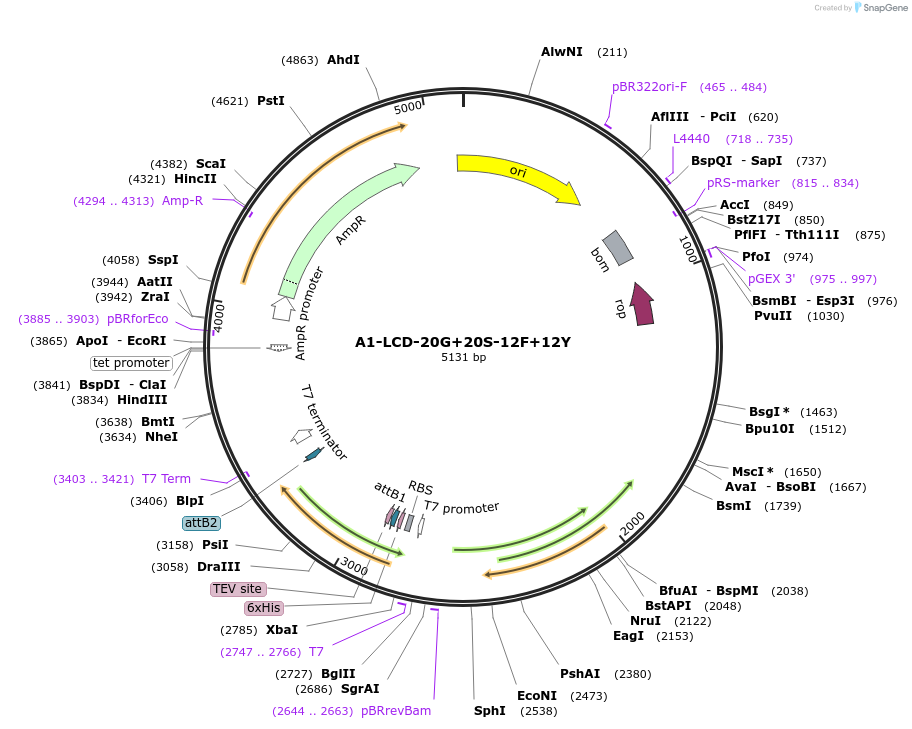
A1-LCD-20G+20S-12F+12Y
Plasmid#178689PurposeBacterial expression of N-terminally 6His tagged A1-LCD with twenty Gly residues replaced with Ser, twelve Phe residues replaced with Tyr (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD-20G+20S-12F+12Y (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationtwenty Gly residues replaced with Ser, twelve Phe…Available SinceMarch 9, 2022AvailabilityAcademic Institutions and Nonprofits only -
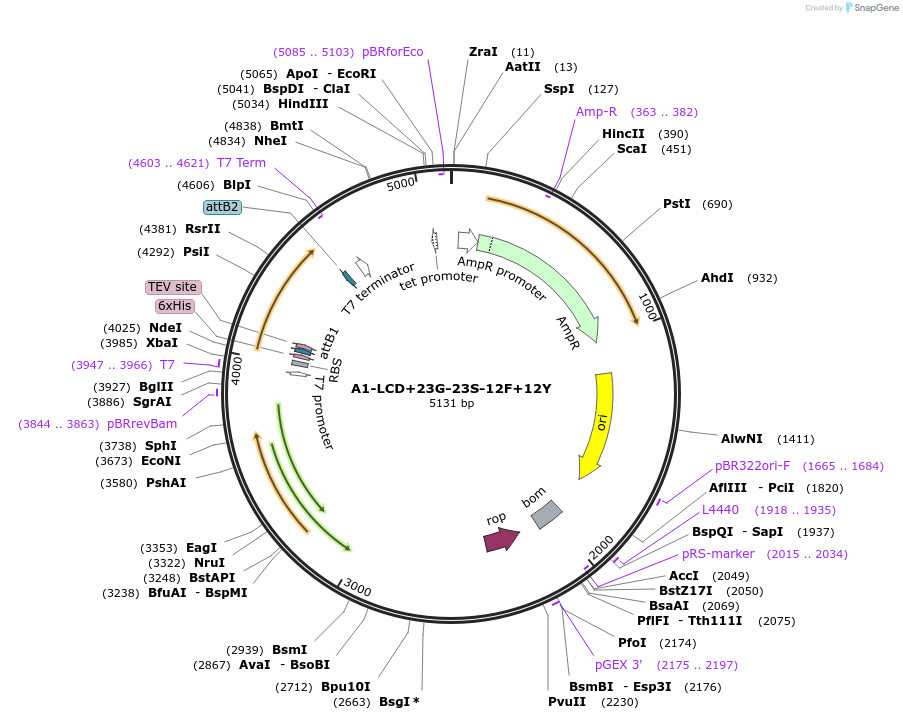
A1-LCD+23G-23S-12F+12Y
Plasmid#178691PurposeBacterial expression of N-terminally 6His tagged A1-LCD with twenty-three Ser residues replaced with Gly, twelve Phe residues replaced with Tyr (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD+23G-23S-12F+12Y (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationtwenty-three Ser residues replaced with Gly, twel…Available SinceMarch 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
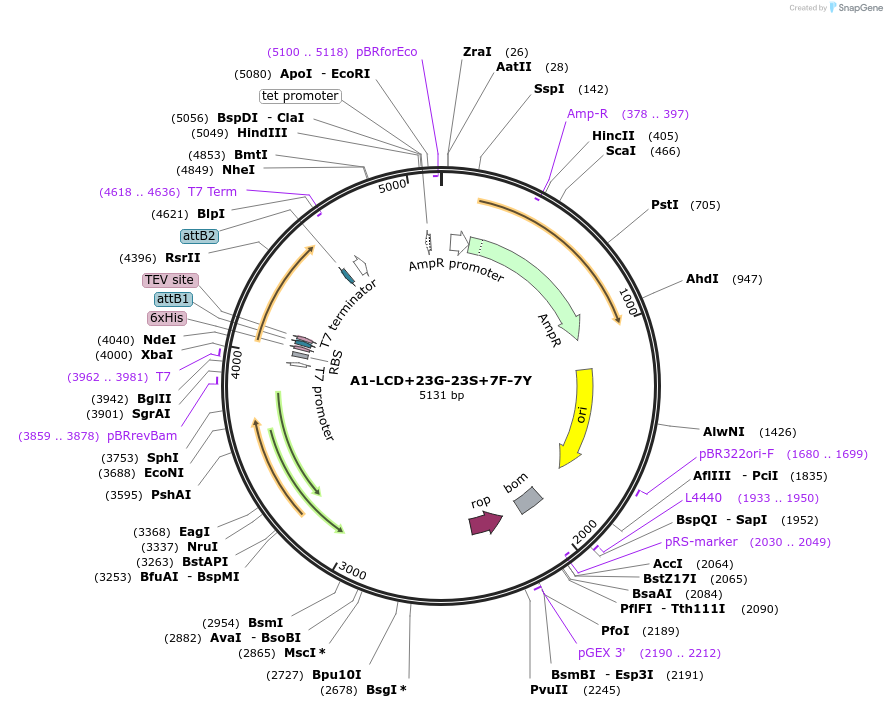
A1-LCD+23G-23S+7F-7Y
Plasmid#178690PurposeBacterial expression of N-terminally 6His tagged A1-LCD with twenty-three Ser residues replaced with Gly, seven Tyr replaced with Phe (LCD: low-complexity domain, aa 186-320)DepositorInsertA1-LCD+23G-23S+7F-7Y (HNRNPA1 Human)
Tags6xHisExpressionBacterialMutationtwenty-three Ser residues replaced with Gly, seve…Available SinceMarch 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
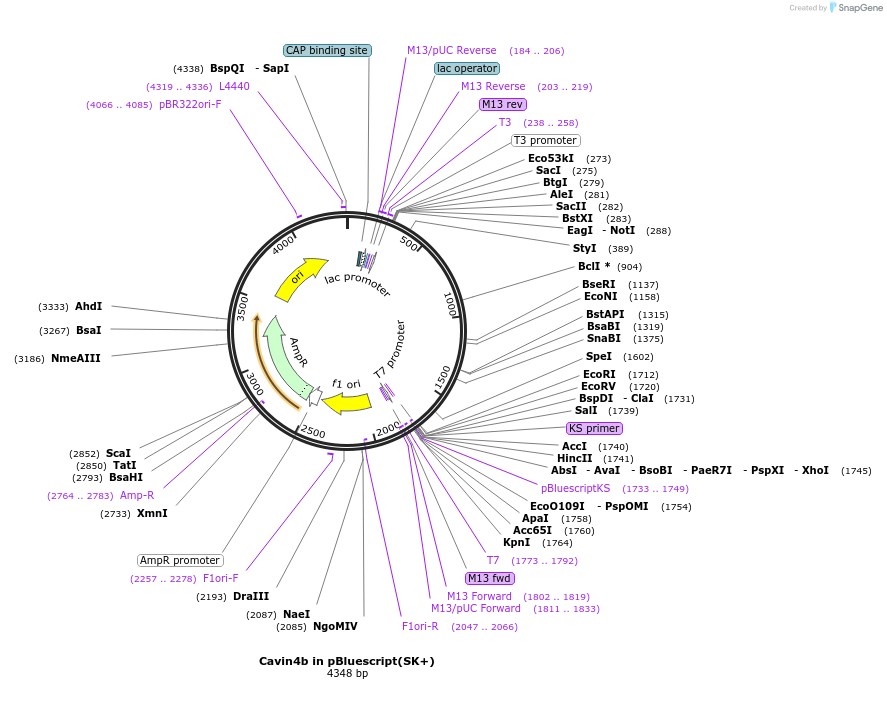
Cavin4b in pBluescript(SK+)
Plasmid#126566PurposeFor generating RNA probes for in situ hybridisation. Parton lab clone IRZDepositorInsertCavin4b (cavin4b Zebrafish)
UseGateway entry cloneAvailable SinceNov. 4, 2021AvailabilityAcademic Institutions and Nonprofits only -
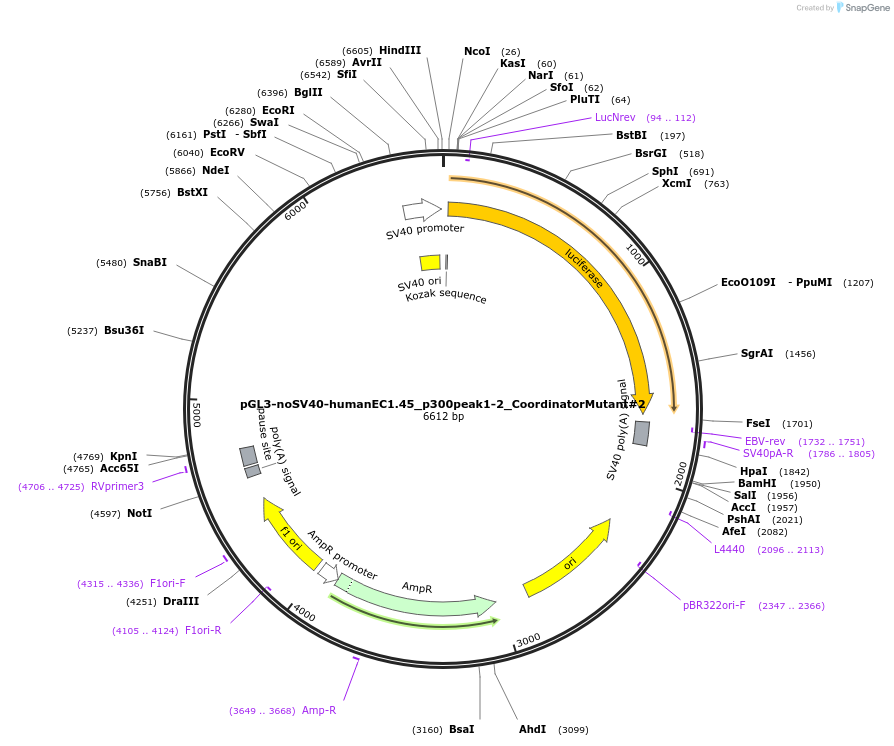
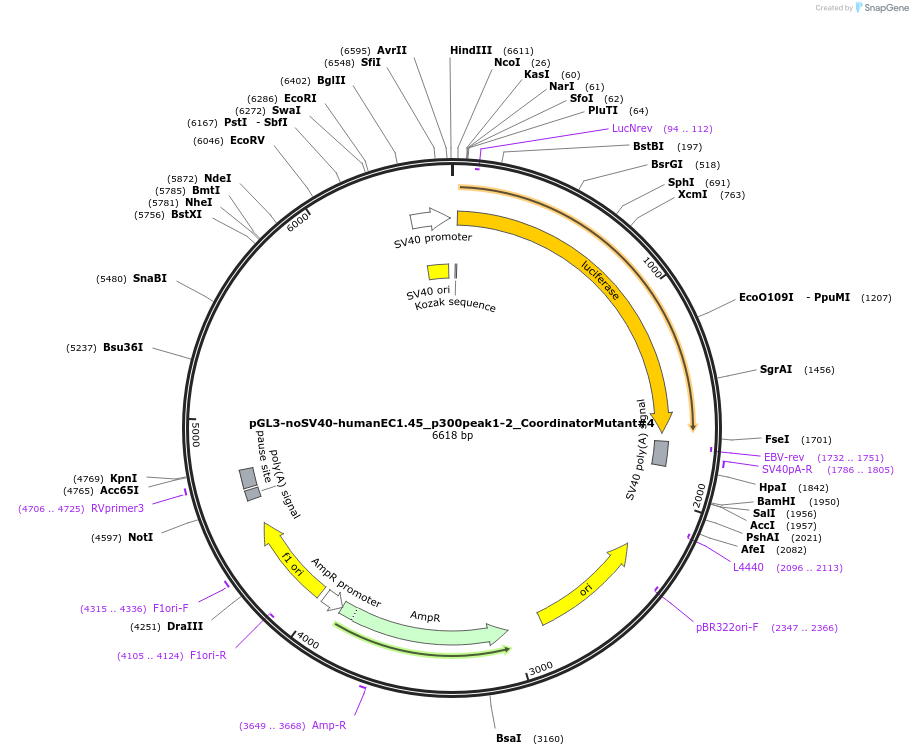
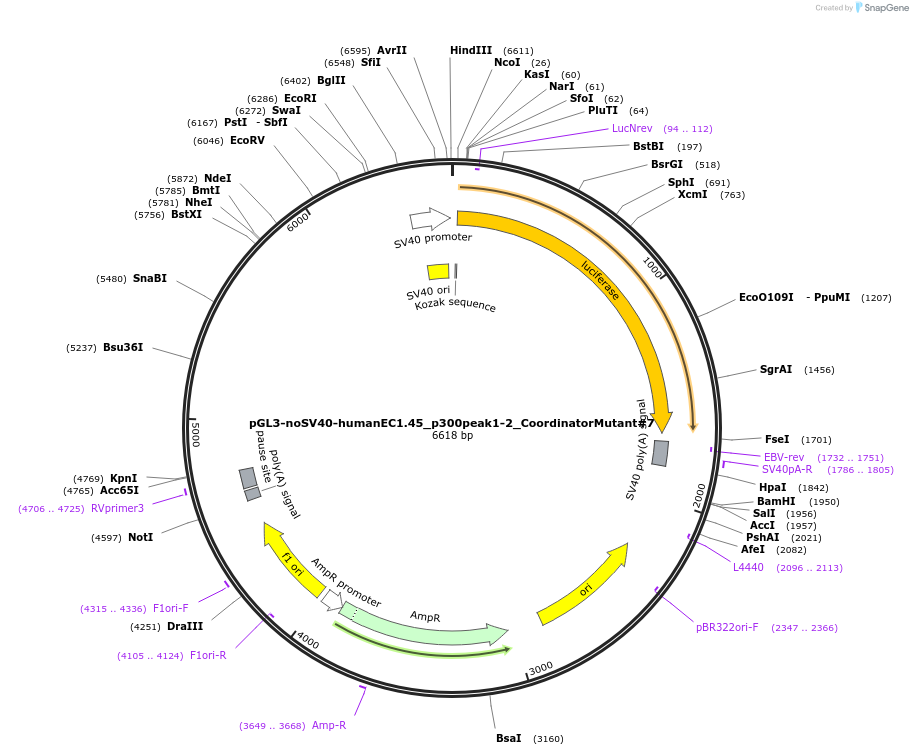
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#2
Plasmid#173986PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #2 mutated
UseLuciferaseMutationCoordinator motif mutation #2PromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
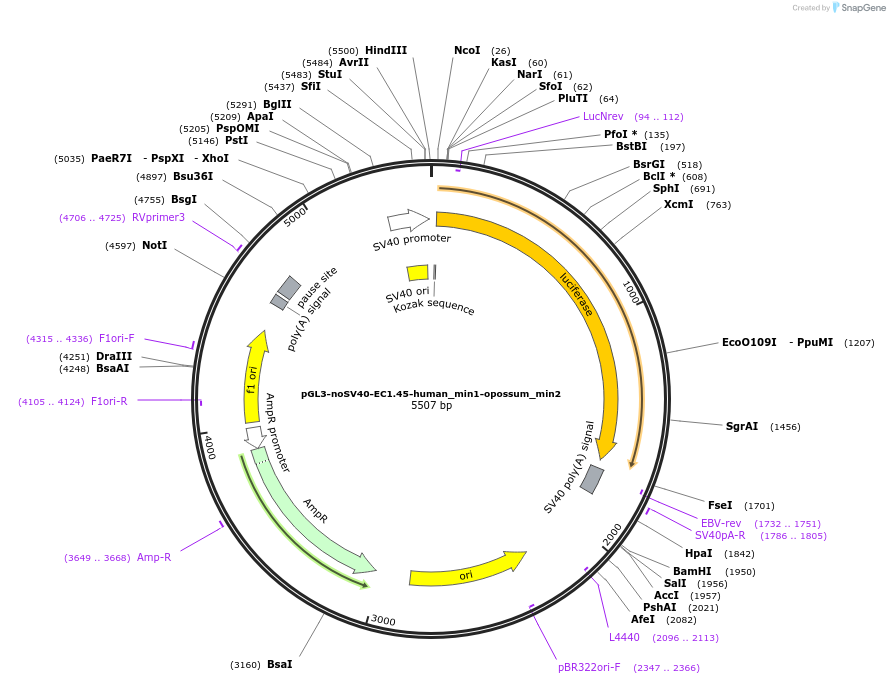
pGL3-noSV40-EC1.45-human_min1-opossum_min2
Plasmid#173966PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and opossum minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationSee Depositor Comments BelowPromoterSV40 promoterAvailable SinceSept. 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
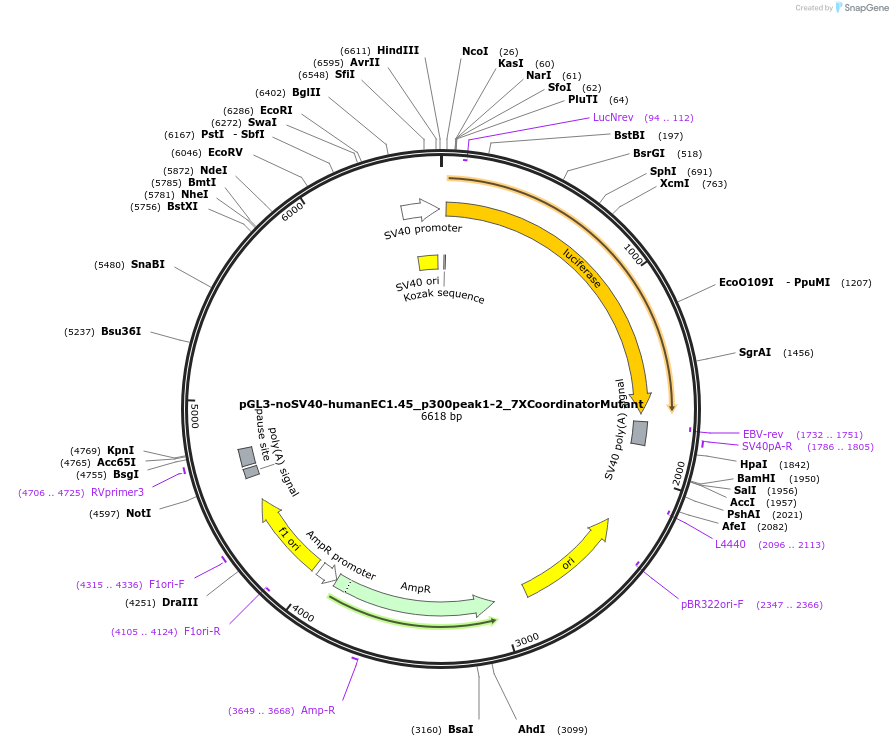
pGL3-noSV40-humanEC1.45_p300peak1-2_7XCoordinatorMutant
Plasmid#173994PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with all 7 Coordinator motifs mutated
UseLuciferaseMutation7X Coordinator motif mutation #1-2-3-4-5-6-7PromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
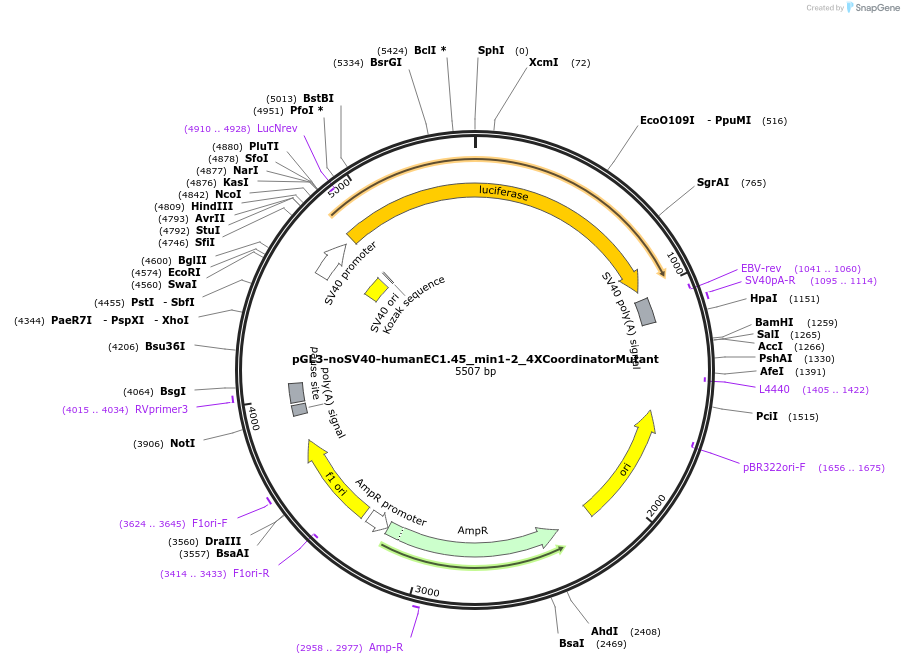
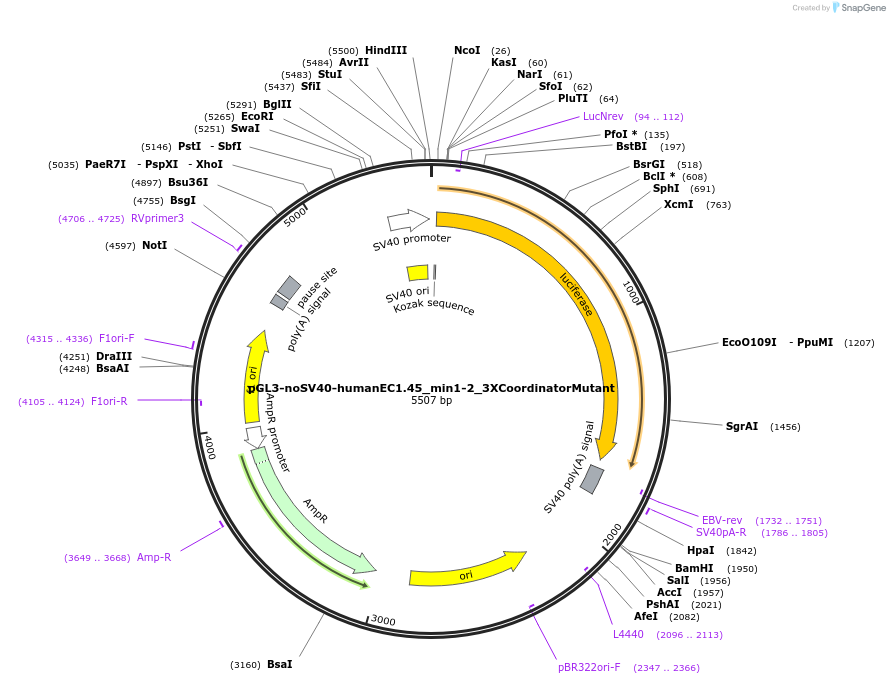
pGL3-noSV40-humanEC1.45_min1-2_4XCoordinatorMutant
Plasmid#173962PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer regions 1 and 2 (min1 and min2) from SOX9 enhancer cluster EC1.45 with 4 mutated Coordinator motifs
UseLuciferaseMutation4 mutated Coordinator motifsPromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
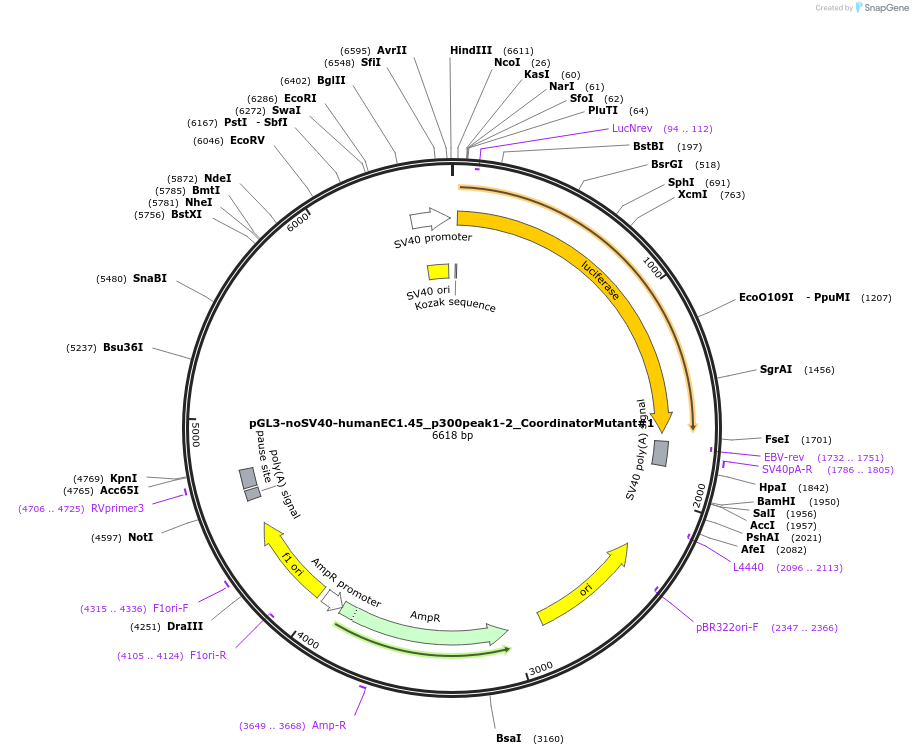
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#1
Plasmid#173985PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #1 mutated
UseLuciferaseMutationCoordinator motif mutation #1PromoterSV40 promoterAvailable SinceSept. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
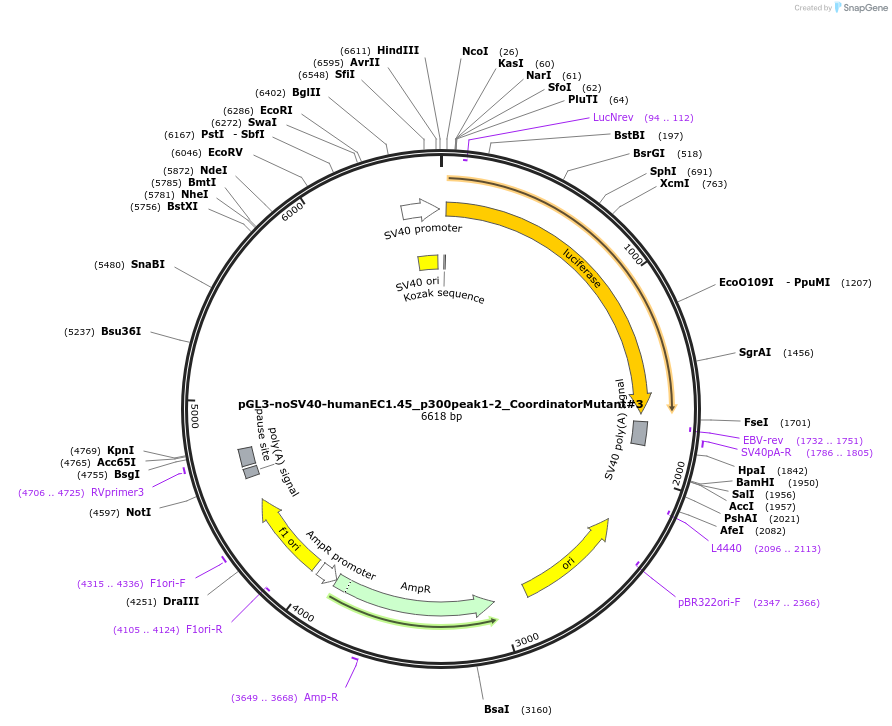
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#3
Plasmid#173987PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #3 mutated
UseLuciferaseMutationCoordinator motif mutation #3PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
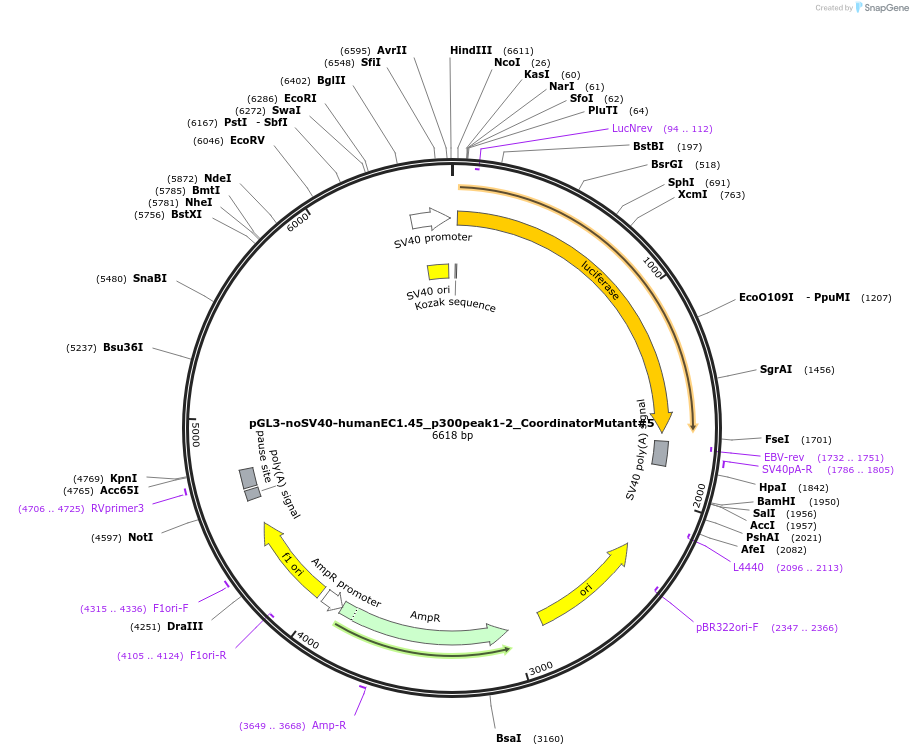
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#5
Plasmid#173989PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #5 mutated
UseLuciferaseMutationCoordinator motif mutation #5PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
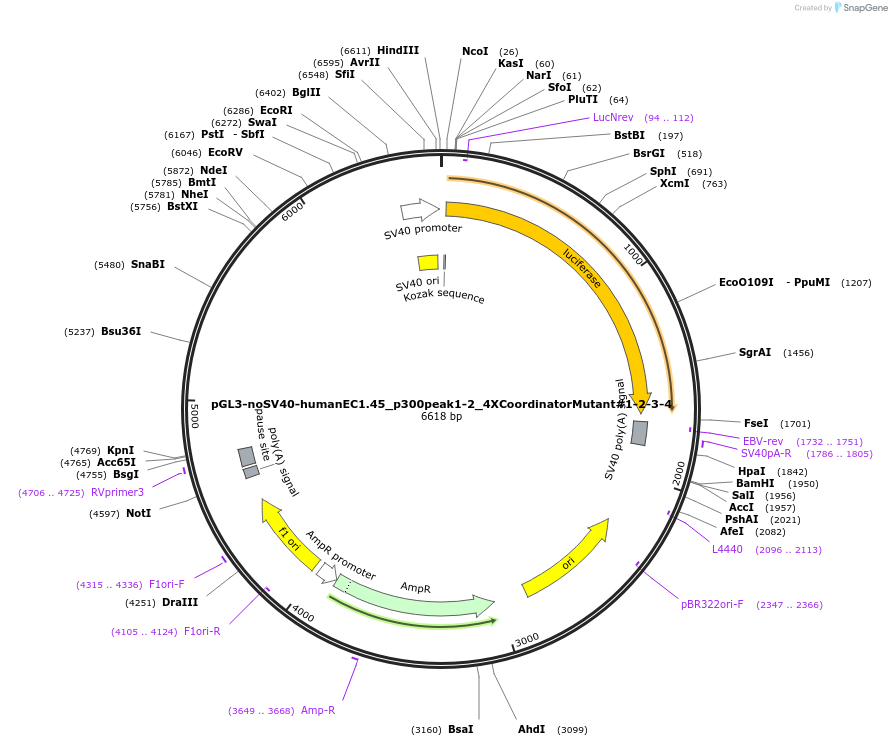
pGL3-noSV40-humanEC1.45_p300peak1-2_4XCoordinatorMutant#1-2-3-4
Plasmid#173992PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motifs #1, #2, #3 and #4 mutated
UseLuciferaseMutation4X Coordinator motif mutations #1-2-3-4PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
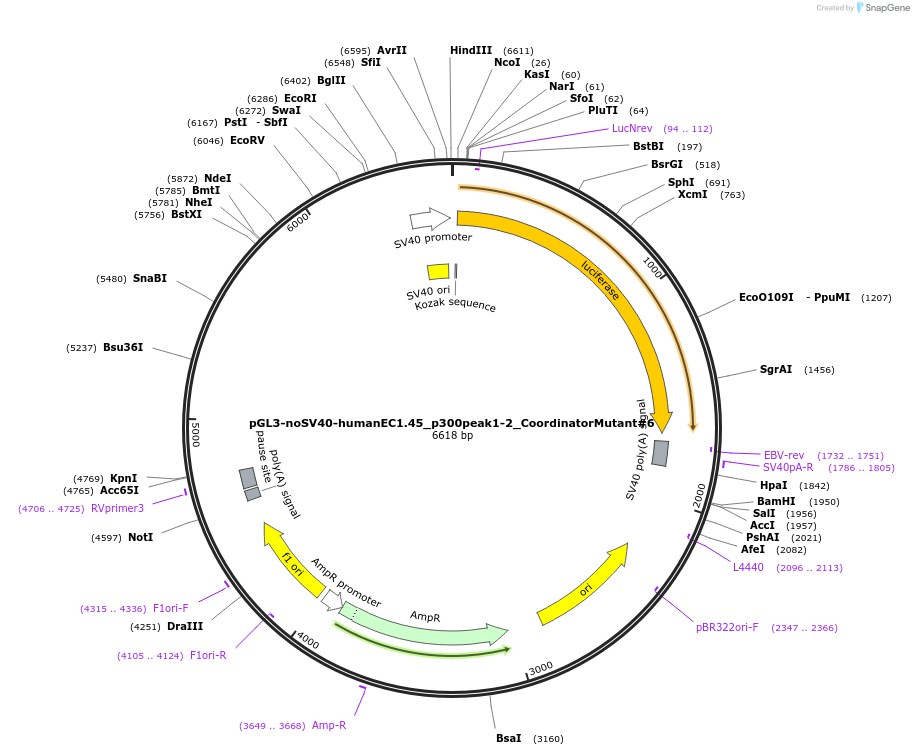
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#6
Plasmid#173990PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #6 mutated
UseLuciferaseMutationCoordinator motif mutation #6PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#4
Plasmid#173988PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #4 mutated
UseLuciferaseMutationCoordinator motif mutation #4PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
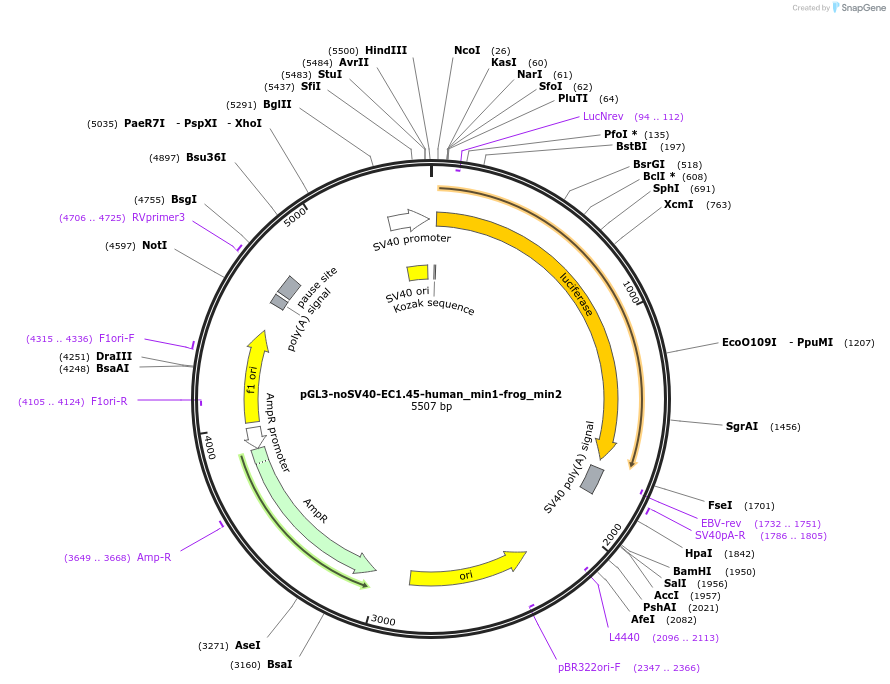
pGL3-noSV40-EC1.45-human_min1-frog_min2
Plasmid#173970PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and frog minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
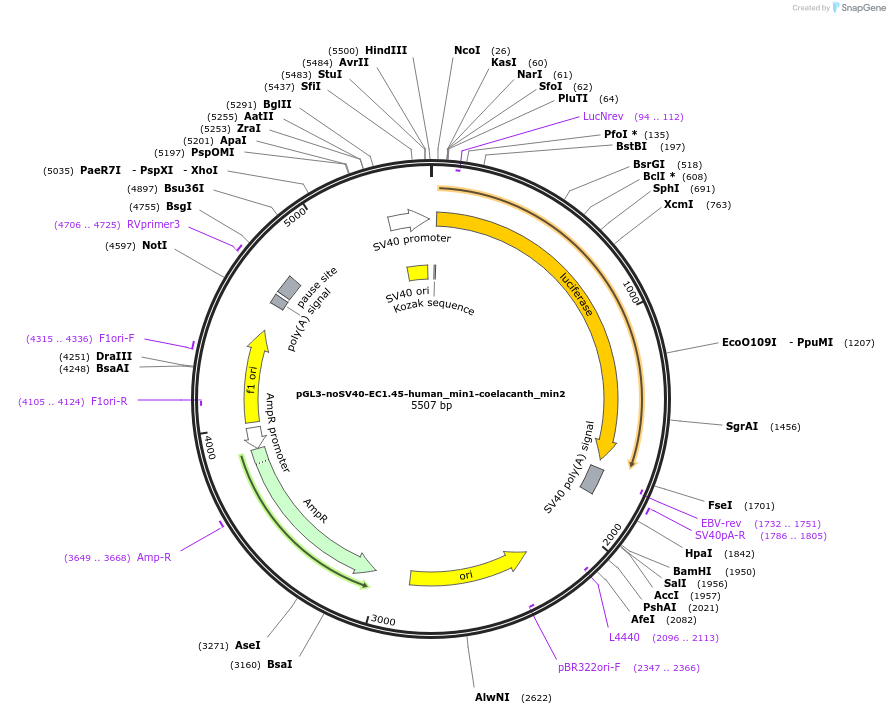
pGL3-noSV40-EC1.45-human_min1-coelacanth_min2
Plasmid#173971PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and coelacanth minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
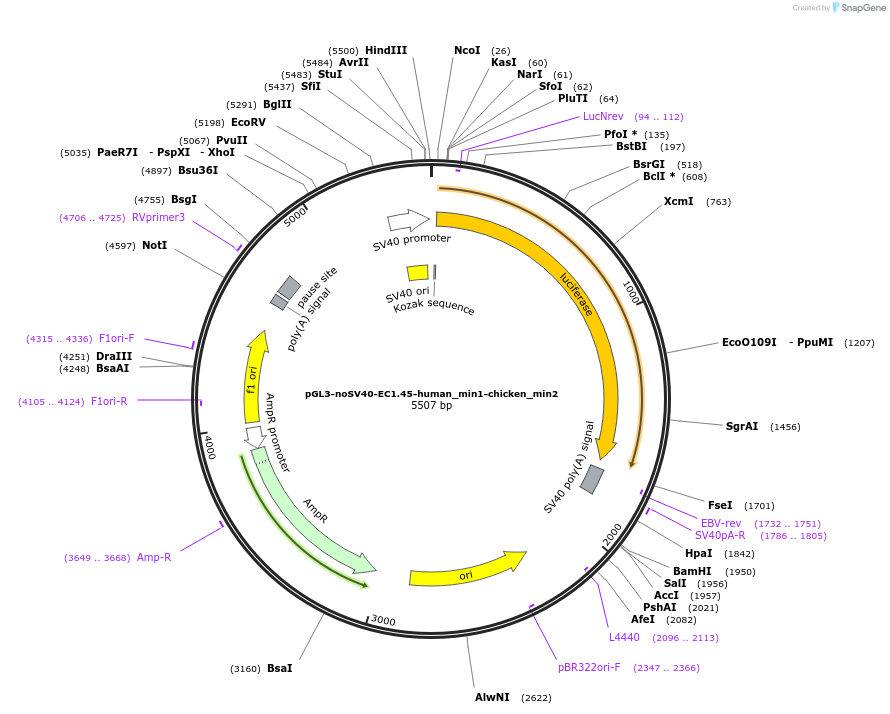
pGL3-noSV40-EC1.45-human_min1-chicken_min2
Plasmid#173968PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and chicken minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_p300peak1-2_CoordinatorMutant#7
Plasmid#173991PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motif #7 mutated
UseLuciferaseMutationCoordinator motif mutation #7PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
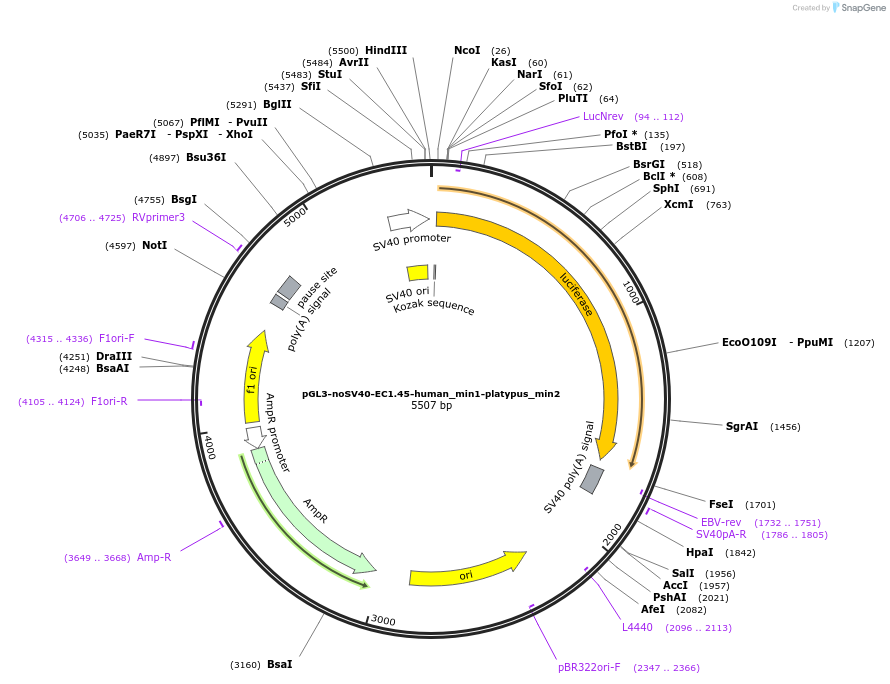
pGL3-noSV40-EC1.45-human_min1-platypus_min2
Plasmid#173967PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and platypus minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
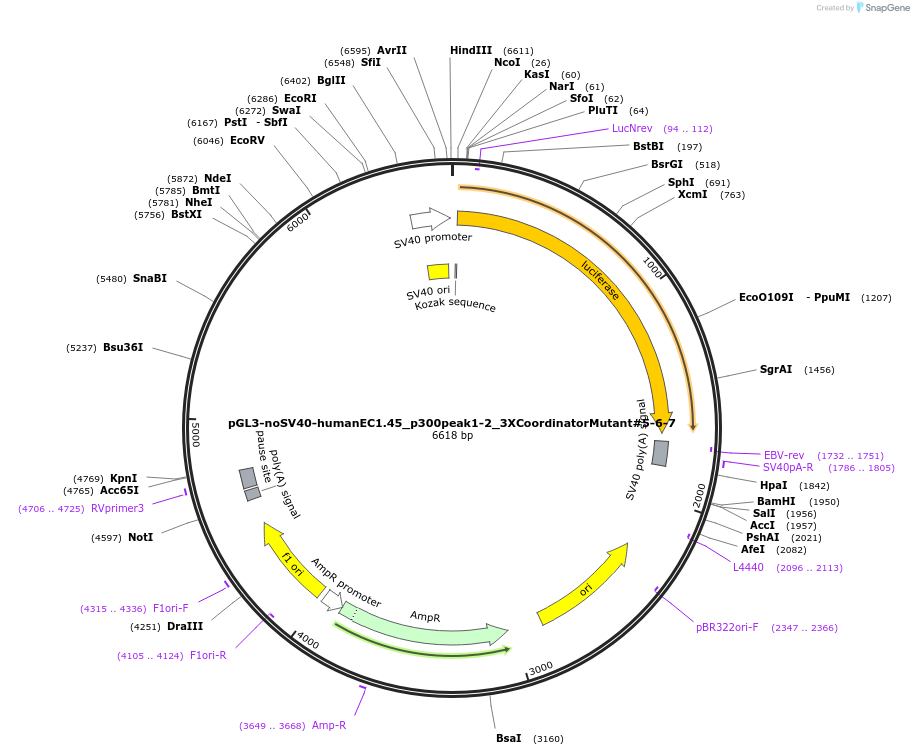
pGL3-noSV40-humanEC1.45_p300peak1-2_3XCoordinatorMutant#5-6-7
Plasmid#173993PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertp300 peak 1 and peak 2 from SOX9 enhancer cluster EC1.45 with Coordinator motifs #5, #6 and #7 mutated
UseLuciferaseMutation3X Coordinator motif mutation #5-6-7PromoterSV40 promoterAvailable SinceSept. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
pGL3-noSV40-humanEC1.45_min1-2_3XCoordinatorMutant
Plasmid#173964PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertMinimal enhancer regions 1 and 2 (min1 and min2) from SOX9 enhancer cluster EC1.45 with 3 mutated Coordinator motifs
UseLuciferaseMutation3 mutated Coordinator motifsPromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
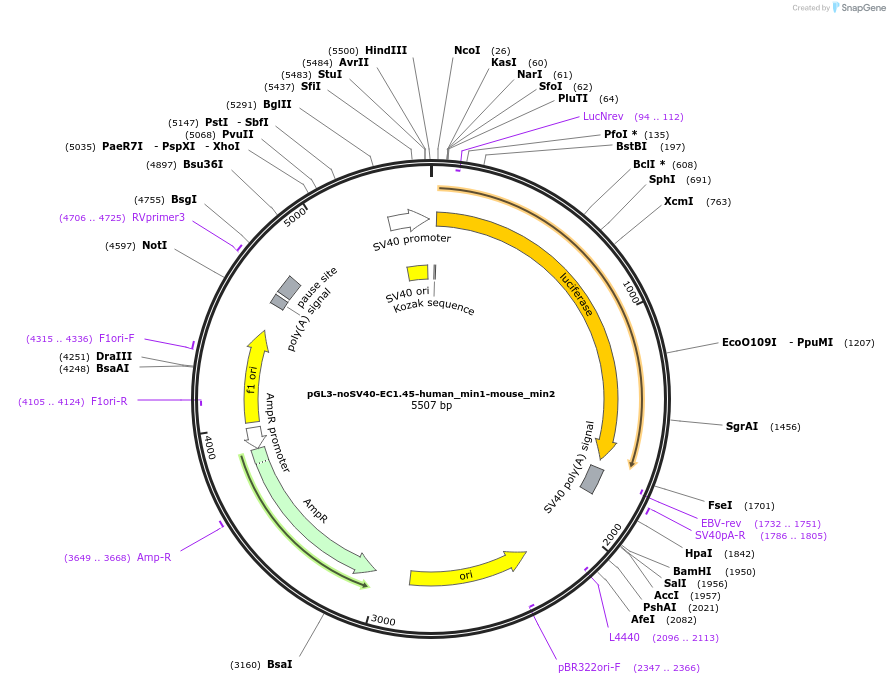
pGL3-noSV40-EC1.45-human_min1-mouse_min2
Plasmid#173965PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and mouse minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
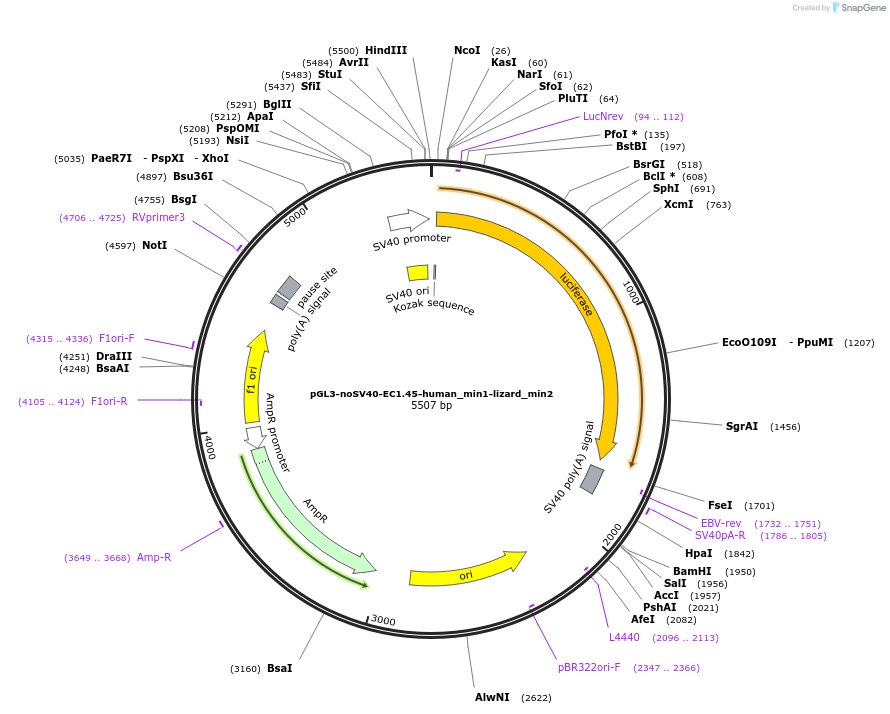
pGL3-noSV40-EC1.45-human_min1-lizard_min2
Plasmid#173969PurposeFirefly luciferase enhancer reporter plasmid.DepositorInsertHuman minimal enhancer region 1 (min1) and lizard minimal enhancer region 2 (min2) from SOX9 enhancer cluster EC1.45
UseLuciferaseMutationNonePromoterSV40 promoterAvailable SinceSept. 1, 2021AvailabilityAcademic Institutions and Nonprofits only