We narrowed to 30,195 results for: REP
-
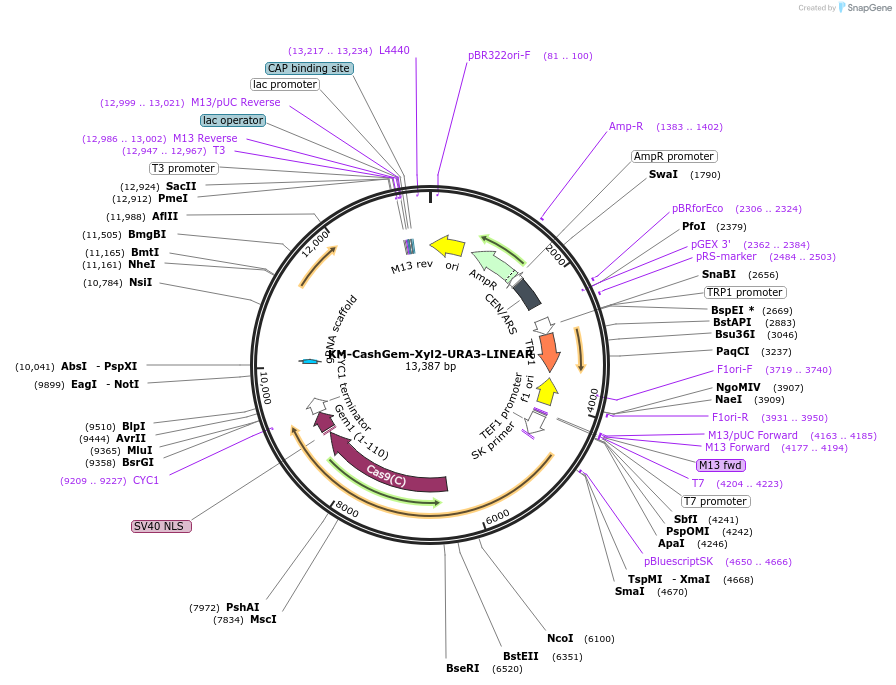
Plasmid#174838PurposepRS414 backbone containing LINEAR fragment targeting XYL2 in K. marxianusDepositorInsertCodon optimized Cas9:hGem
UseCRISPR and Synthetic BiologyTagshGeminin tagExpressionBacterial and YeastPromoterTEF1pAvailable SinceJan. 5, 2022AvailabilityAcademic Institutions and Nonprofits only -
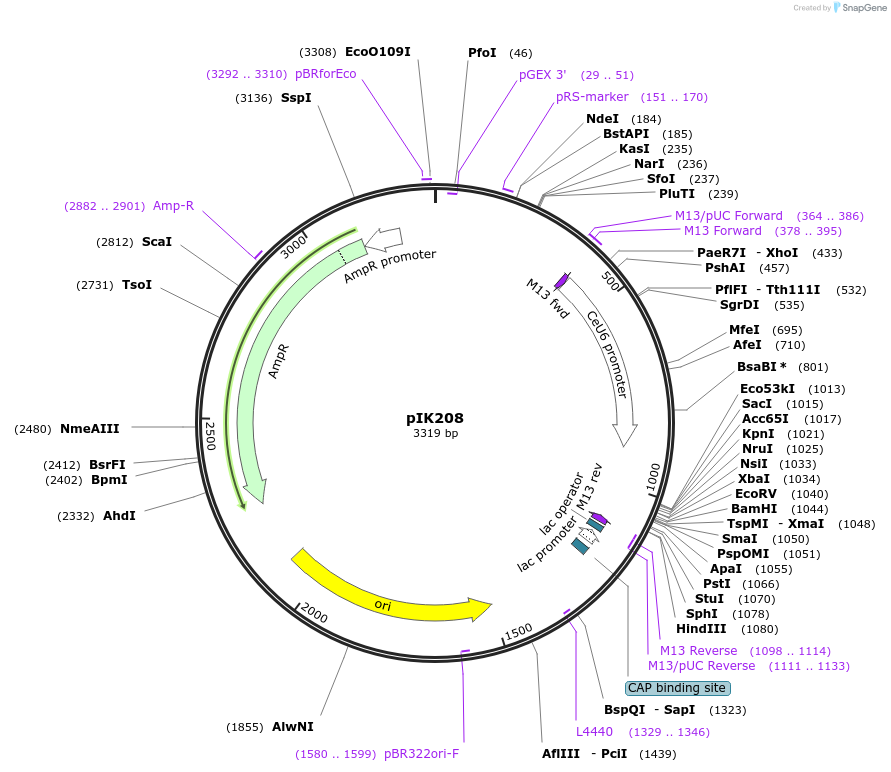
pIK208
Plasmid#65630PurposesgRNA targeting dpy-10 (Arribere et al., 2014) expressed from a C. elegans U6 promoter (Friedland et al., 2013), in a "flipped and extended" sgRNA backbone (Chen et al., 2013)"DepositorInsertU6 promoter::dpy-10 sgRNA with a "flipped and extended" sgRNA backbone
UseCRISPRExpressionWormAvailable SinceJune 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
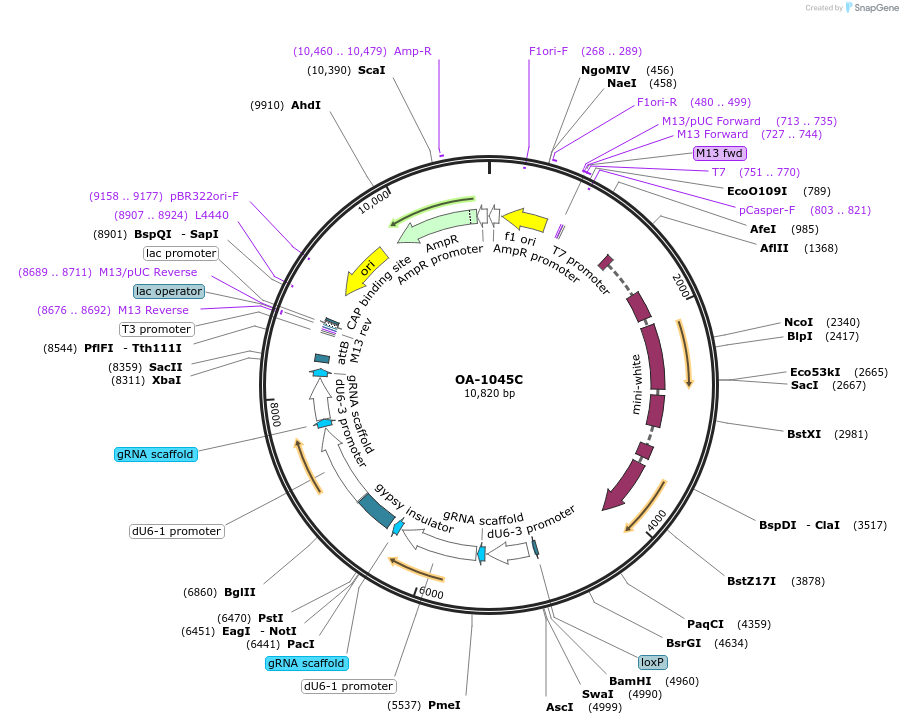
OA-1045C
Plasmid#125005PurposeExpresses gRNAs targeting hid, eve, and winglessDepositorAvailable SinceOct. 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
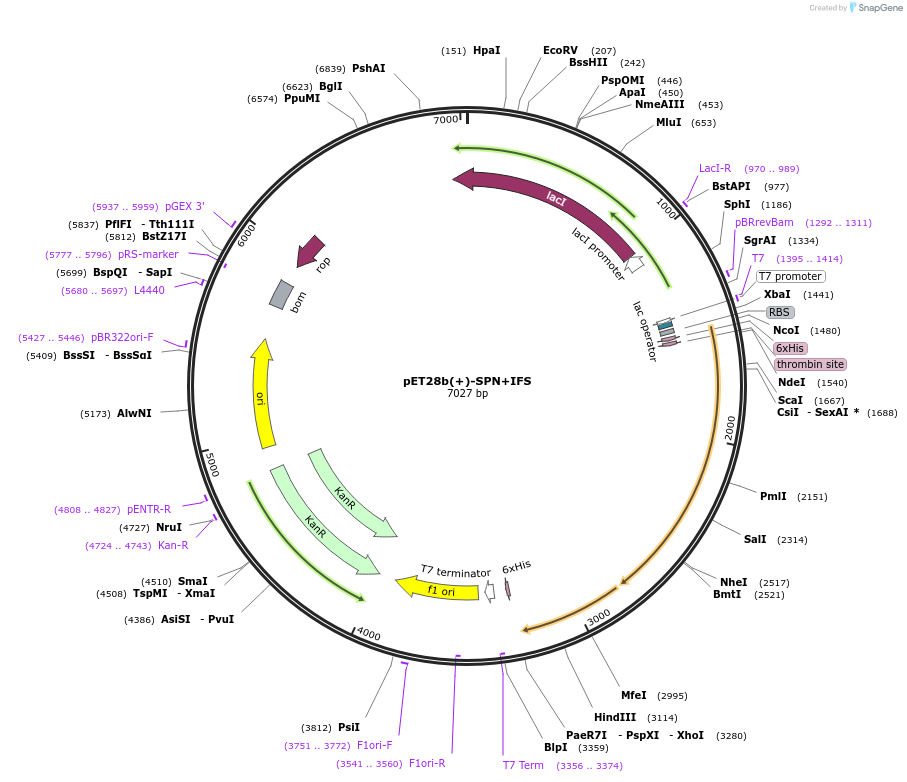
pET28b(+)-SPN+IFS
Plasmid#83372PurposeExpresses SPN and IFS complex in E. coli cellDepositorInsertsSPN
IFS
TagsHexahistidine tagExpressionBacterialPromoterT7Available SinceJan. 30, 2017AvailabilityAcademic Institutions and Nonprofits only -
pMALp2x-SCO5461(43-204)
Plasmid#42509DepositorInsertguanosine ADP-ribosyltransferase (SCO5461 Streptomyces coelicolor A3(2))
Tagsmaltose-binding proteinExpressionBacterialMutationdeleted amino acids 1-42 (nucleotide 1-126)PromotertacAvailable SinceApril 10, 2013AvailabilityAcademic Institutions and Nonprofits only -
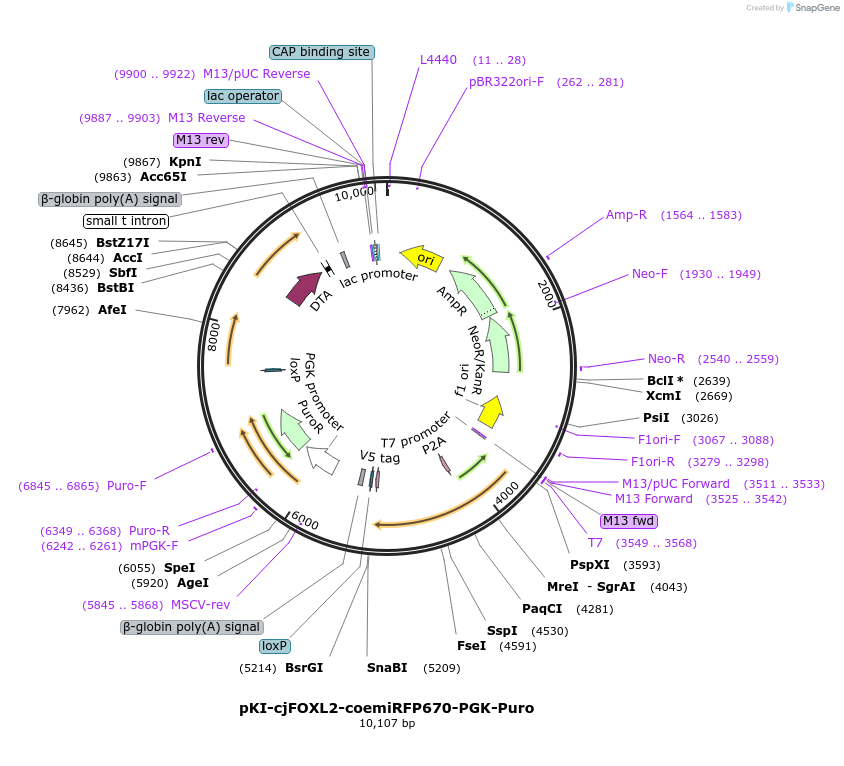
pKI-cjFOXL2-coemiRFP670-PGK-Puro
Plasmid#186168PurposeKnock-in gene targeting in marmoset cells at FOXL2 locusDepositorInsertemiRFP670
UseMarmoset targetingMutationhuman codon-optimizedAvailable SinceNov. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
-
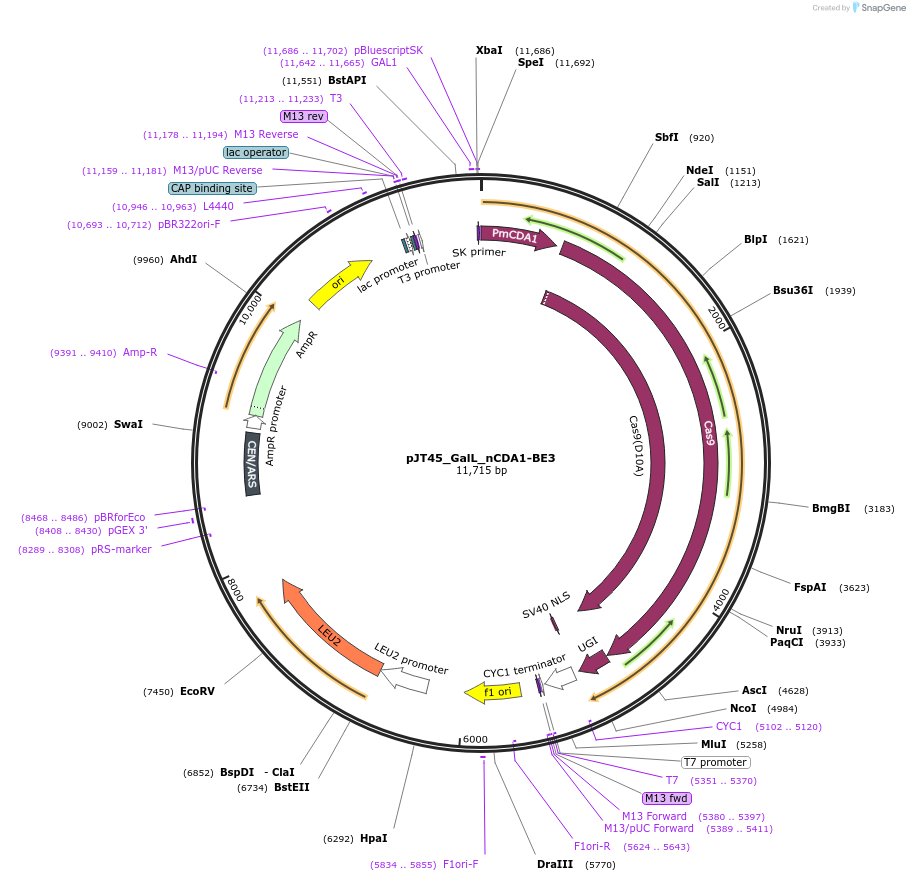
pJT45_GalL_nCDA1-BE3
Plasmid#145038PurposeExpresses nCDA1-BE3 in yeast cellsDepositorInsertnCDA1-BE3
UseCRISPRExpressionYeastMutationspCas9(D10A)PromoterGalLAvailable SinceSept. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
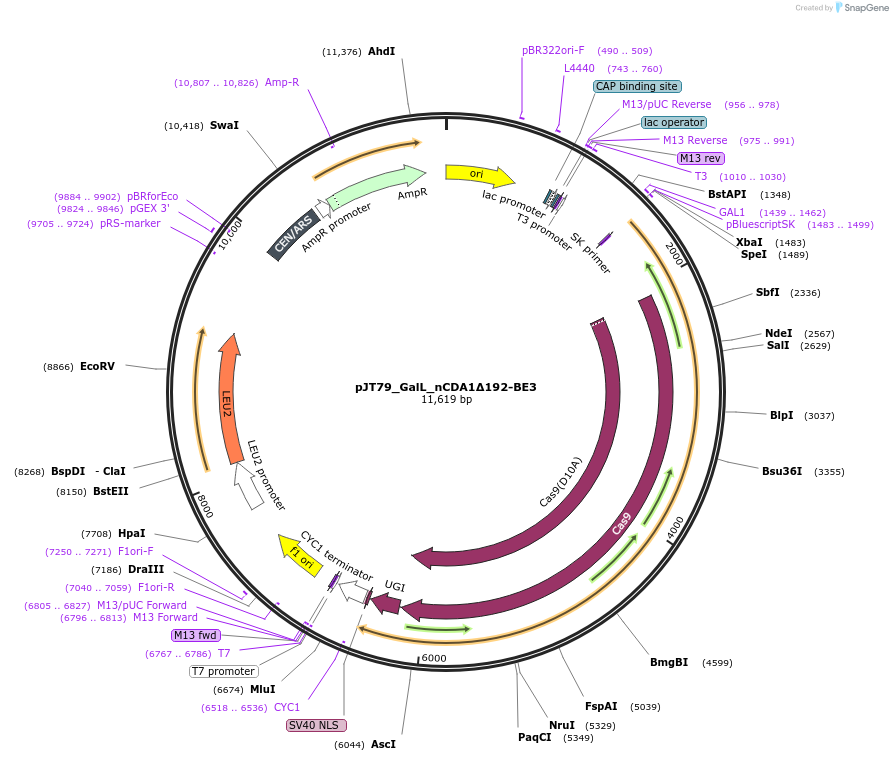
pJT79_GalL_nCDA1Δ192-BE3
Plasmid#145045PurposeExpresses nCDA1Δ192-BE3 in yeast cellsDepositorInsertnCDA1Δ192-BE3
UseCRISPRExpressionYeastMutationpmCDA1(1-192AA); spCas9(D10A)PromoterGalLAvailable SinceJuly 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
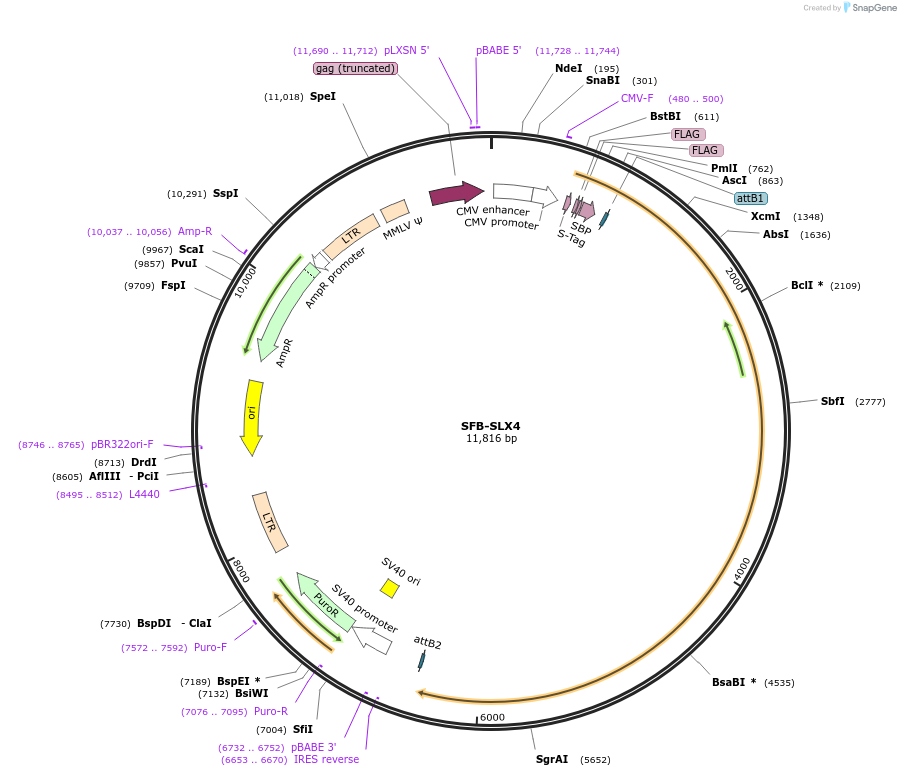
SFB-SLX4
Plasmid#247854PurposeMammalian expression construct for N-terminal S protein-Flag-Streptavidin binding peptide (SFB)-tagged SLX4DepositorInsertSLX4 (SLX4 Human)
TagsS protein-Flag-Streptavidin binding peptide (SFB)ExpressionMammalianAvailable SinceDec. 9, 2025AvailabilityAcademic Institutions and Nonprofits only -
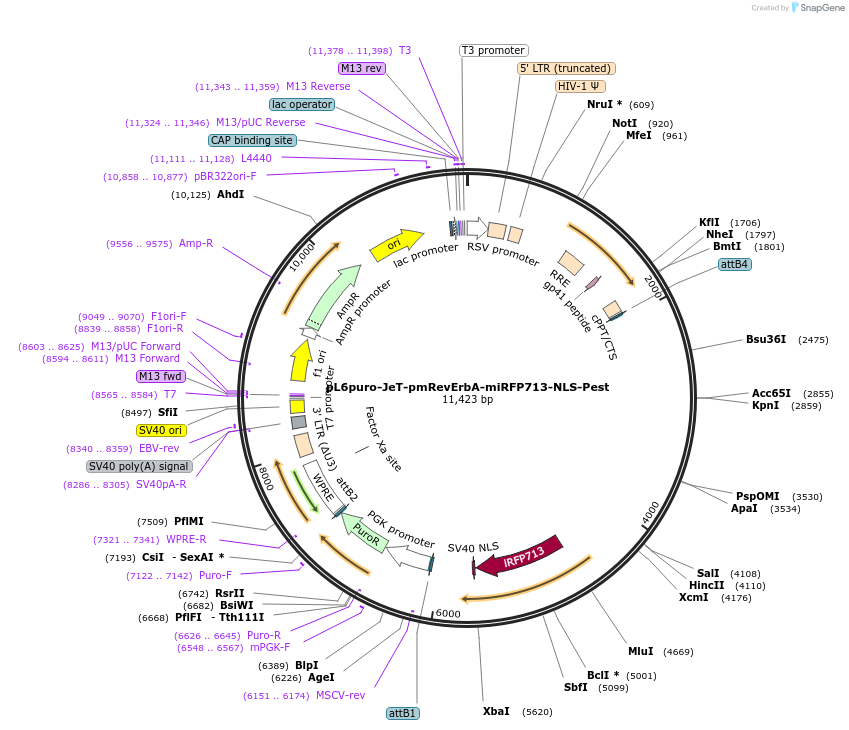
pL6puro-JeT-pmRevErbA-miRFP713-NLS-Pest
Plasmid#240110PurposeA lentiviral (infrared) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by synthtic JeT promoter.DepositorInsertmurine Nr1d1 promoter+miRFP713-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded JeT PromoterAvailable SinceSept. 8, 2025AvailabilityAcademic Institutions and Nonprofits only -
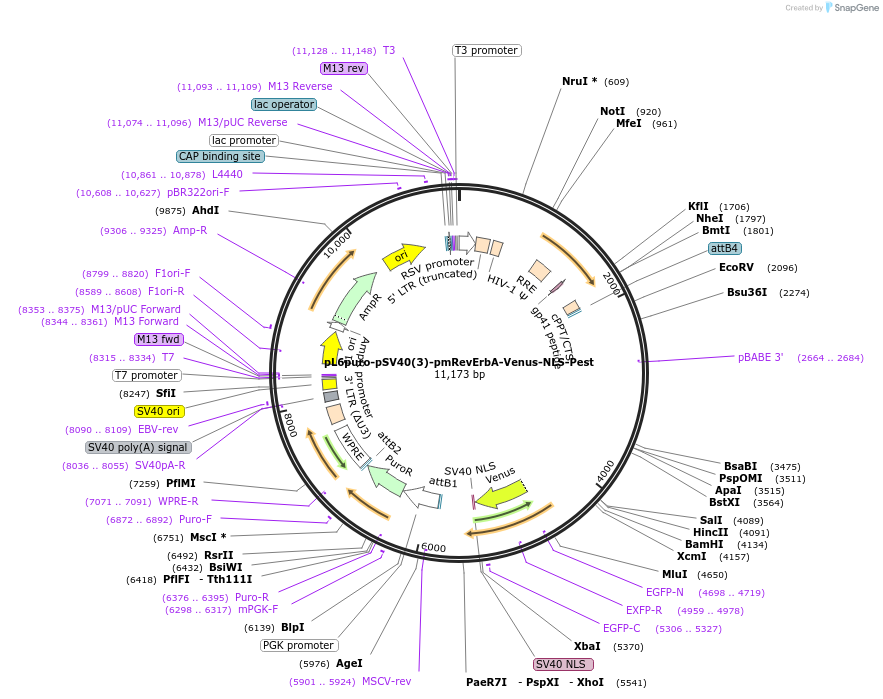
pL6puro-pSV40(3)-pmRevErbA-Venus-NLS-Pest
Plasmid#240114PurposeA lentiviral (yellow) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter fragment.DepositorInsertmurine Nr1d1 promoter+Venus-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-EnhancerAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
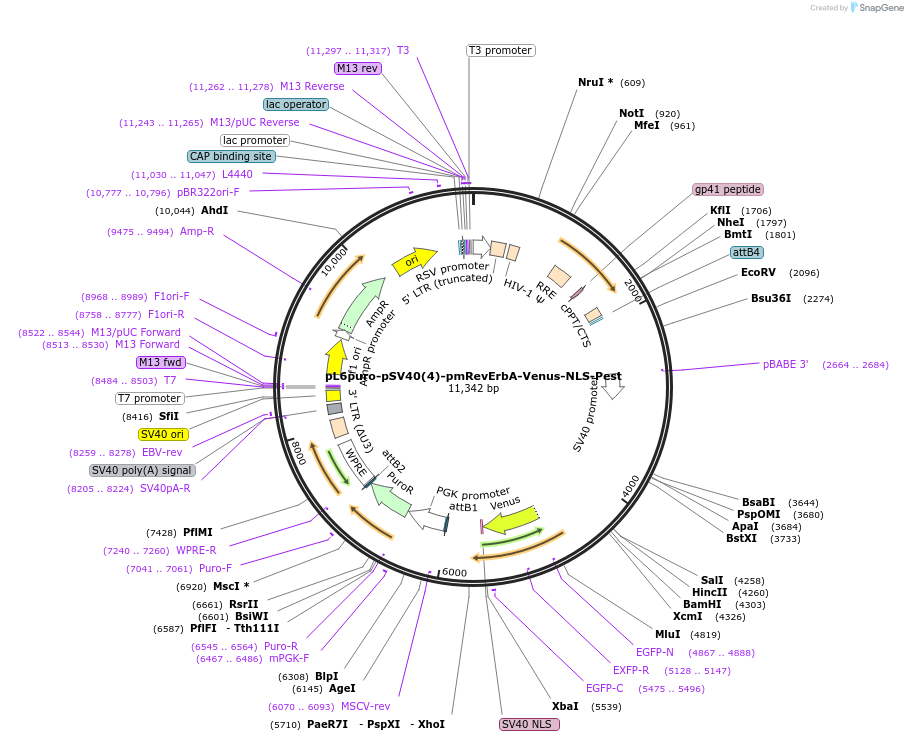
pL6puro-pSV40(4)-pmRevErbA-Venus-NLS-Pest
Plasmid#240115PurposeA lentiviral (yellow) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter.DepositorInsertmurine Nr1d1 promoter+Venus-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-Enhancer+OriAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
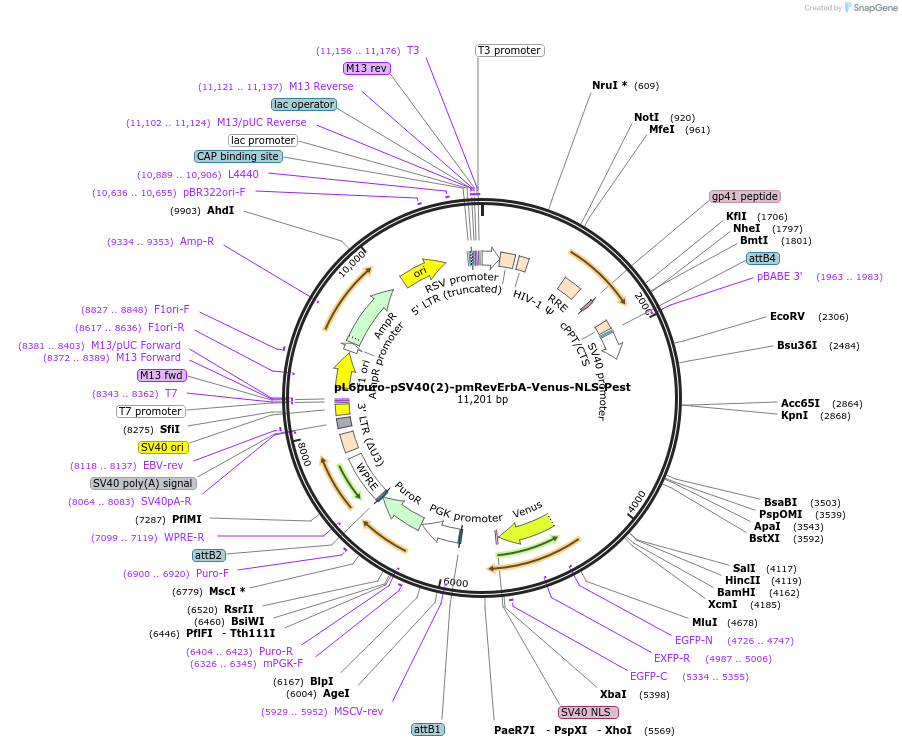
pL6puro-pSV40(2)-pmRevErbA-Venus-NLS-Pest
Plasmid#240113PurposeA lentiviral (yellow) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter.DepositorInsertmurine Nr1d1 promoter+Venus-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-Enhancer+OriAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
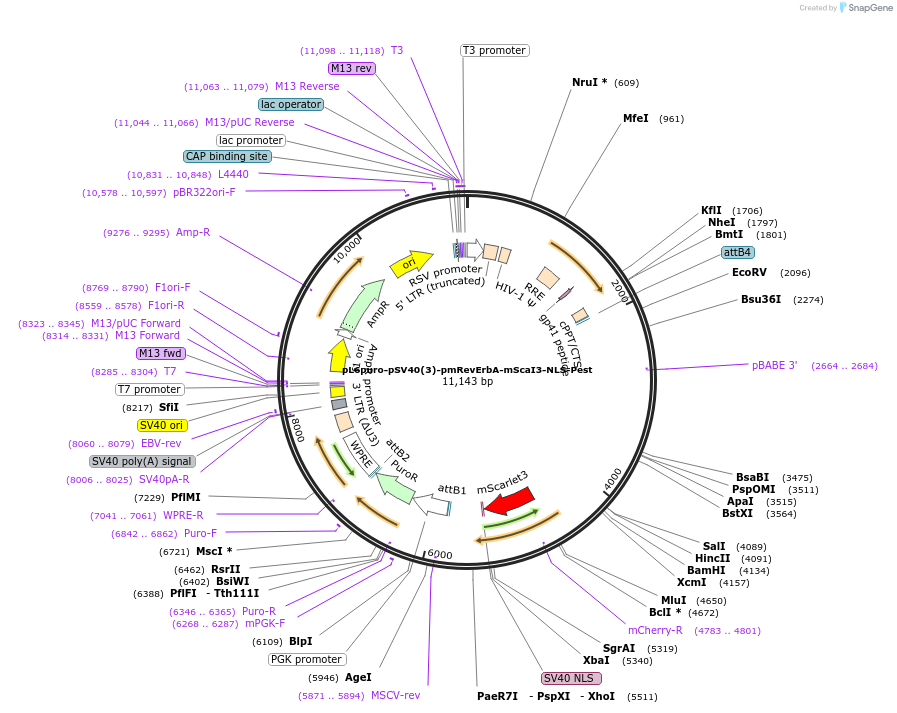
pL6puro-pSV40(3)-pmRevErbA-mScaI3-NLS-Pest
Plasmid#240117PurposeA lentiviral (red) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by SV40 promoter fragment.DepositorInsertmurine Nr1d1 promoter+mScarlet-I3-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded SV40-EnhancerAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
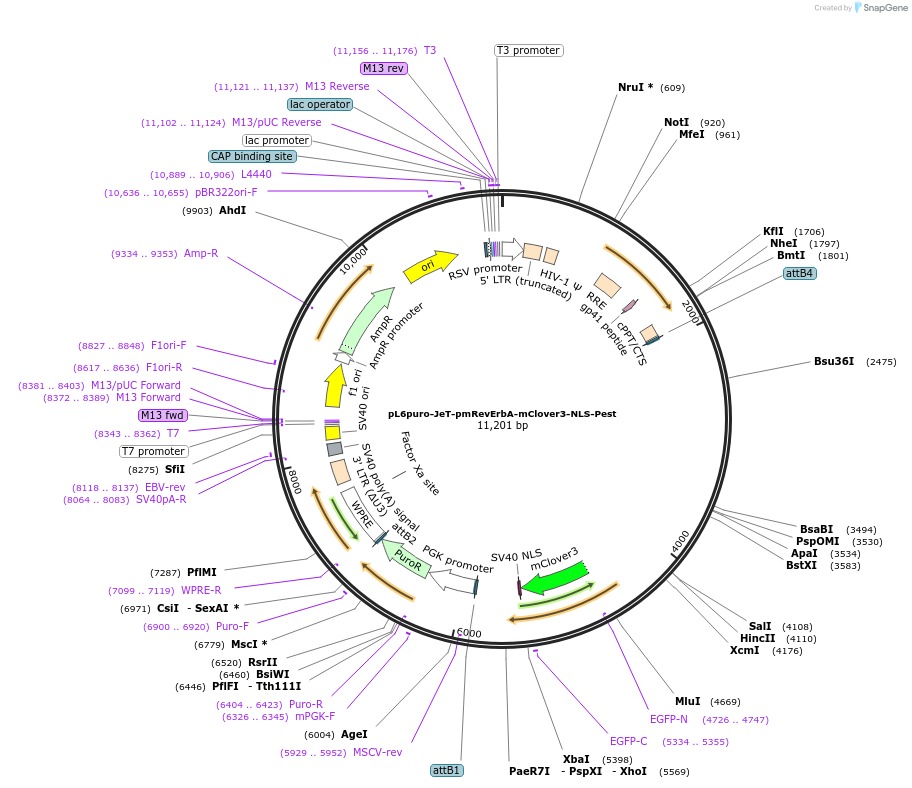
pL6puro-JeT-pmRevErbA-mClover3-NLS-Pest
Plasmid#240109PurposeA lentiviral (yellow-green) fluorescent reporter of circadian rhythm, based on the murine Nr1d1-gene (REVERBa protein), expression enhanced by synthtic JeT promoter.DepositorInsertmurine Nr1d1 promoter+mClover3-NLS-Pest-Reporter (Nr1d1 Mouse)
UseLentiviralMutationadded JeT PromoterAvailable SinceAug. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
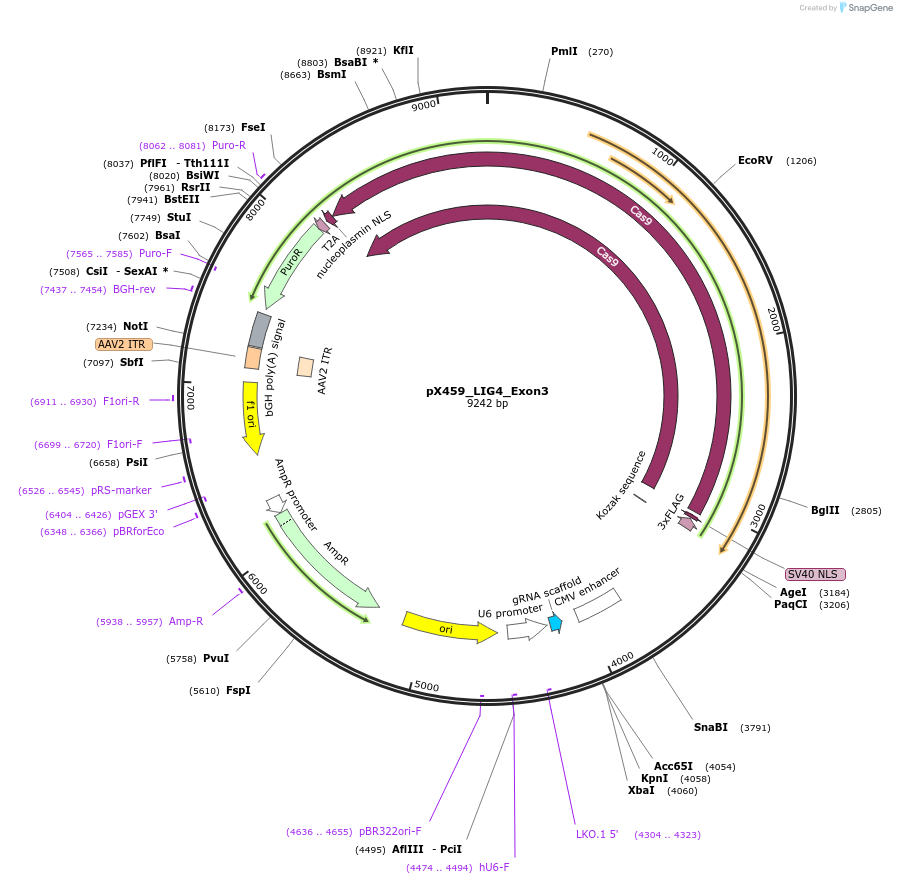
pX459_LIG4_Exon3
Plasmid#225363PurposepX459 plasmid encoding the sgRNA protospacer sequence “5’-GCATAATGTCACTACAGATC-3’ to target the LIG4 gene.DepositorAvailable SinceJuly 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
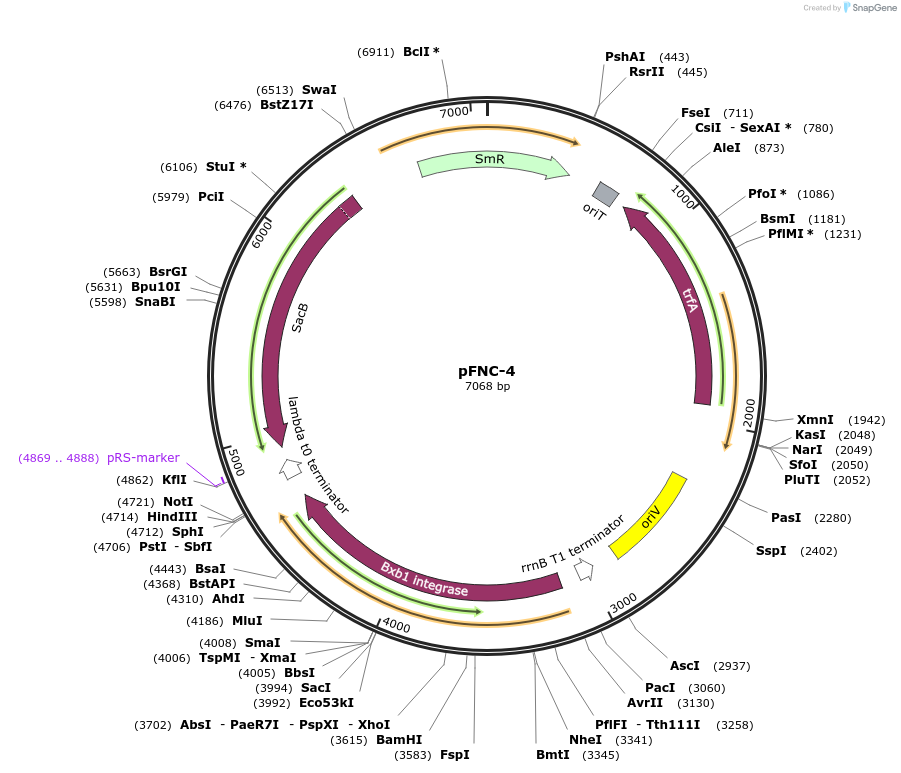
pFNC-4
Plasmid#227668PurposePlasmid encoding the BxbI phage integrase. It can be used to remove the antibiotic resistance cassette integrated with pCIFR. SmR, easily curable via sucrose counterselection.DepositorInsertSmR
ExpressionBacterialAvailable SinceJan. 6, 2025AvailabilityAcademic Institutions and Nonprofits only -
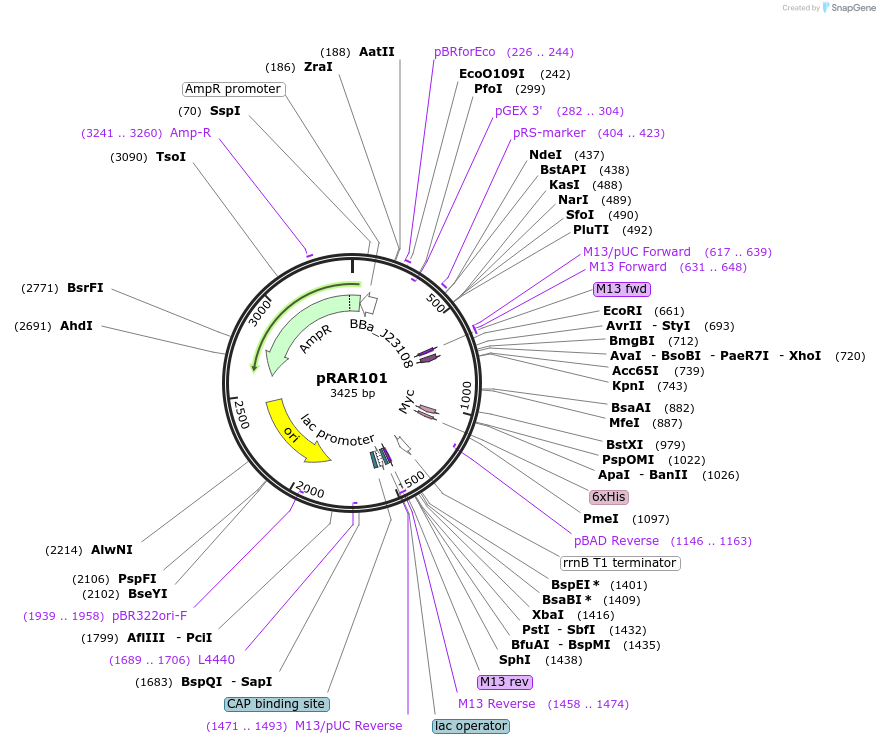
pRAR101
Plasmid#185046PurposeJ23108(spel), pHP14 stability hairpin, strong RBS, L31p coding region with amino acids 2-8 deleted, trrnBDepositorInsertL31p
ExpressionBacterialMutationAmino acids 2-8 deleted of L31p coding regionAvailable SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
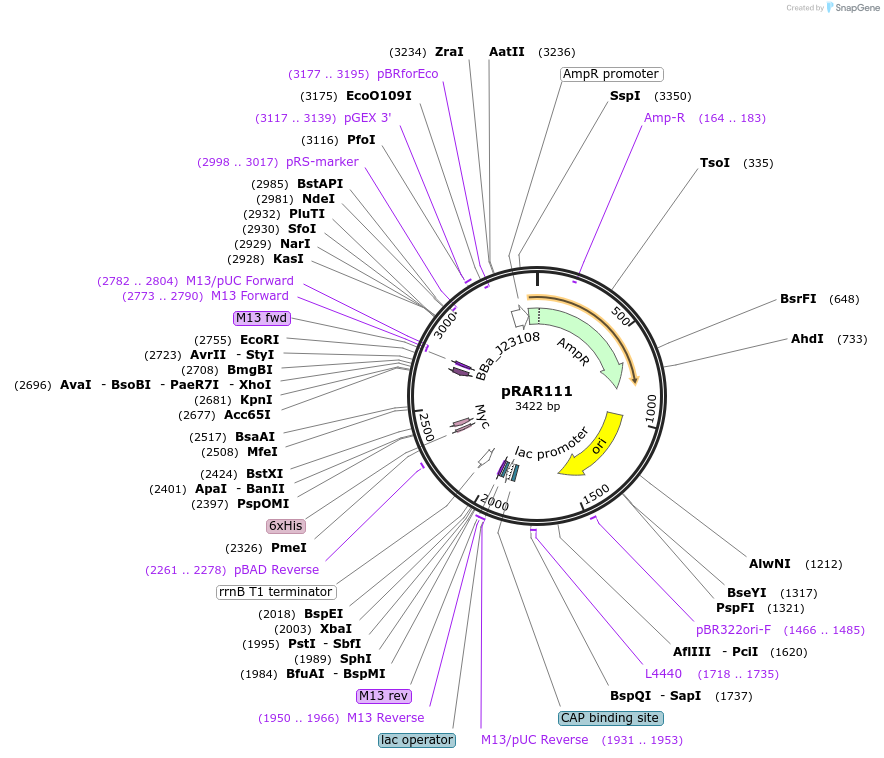
pRAR111
Plasmid#185047PurposeJ23108(spel), pHP14 stability hairpin, strong RBS, L31p coding region with last 8 amino acids deleted, trrnBDepositorInsertL31p
ExpressionBacterialMutationLast 8 amino acids deleted of L31p coding regionAvailable SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only