We narrowed to 11,225 results for: kars;
-
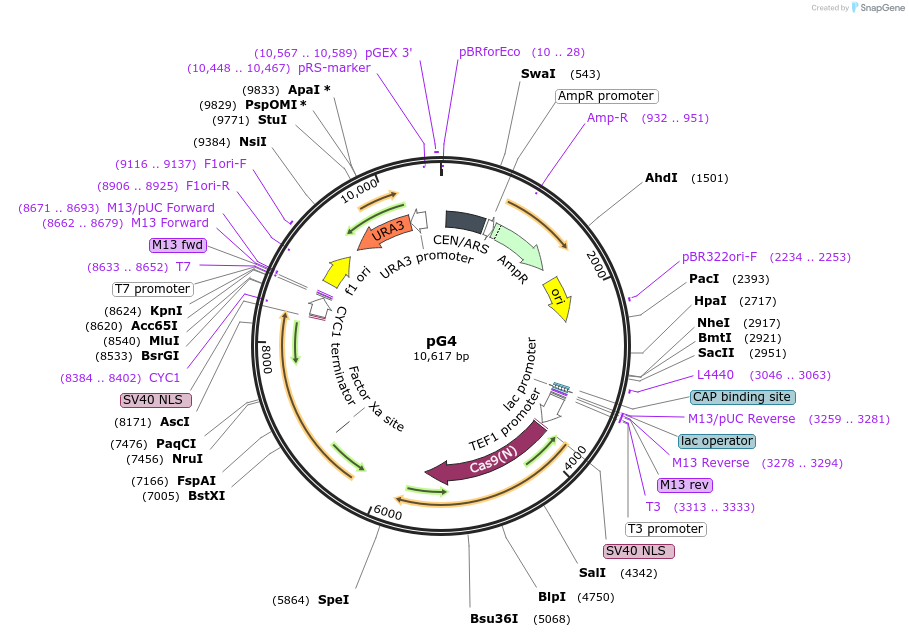
Plasmid#162605PurposeEdit Ade2 gene in yeastDepositorInsertTef1-Cas9 with RPL25 Intron
ExpressionYeastAvailable SinceJune 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
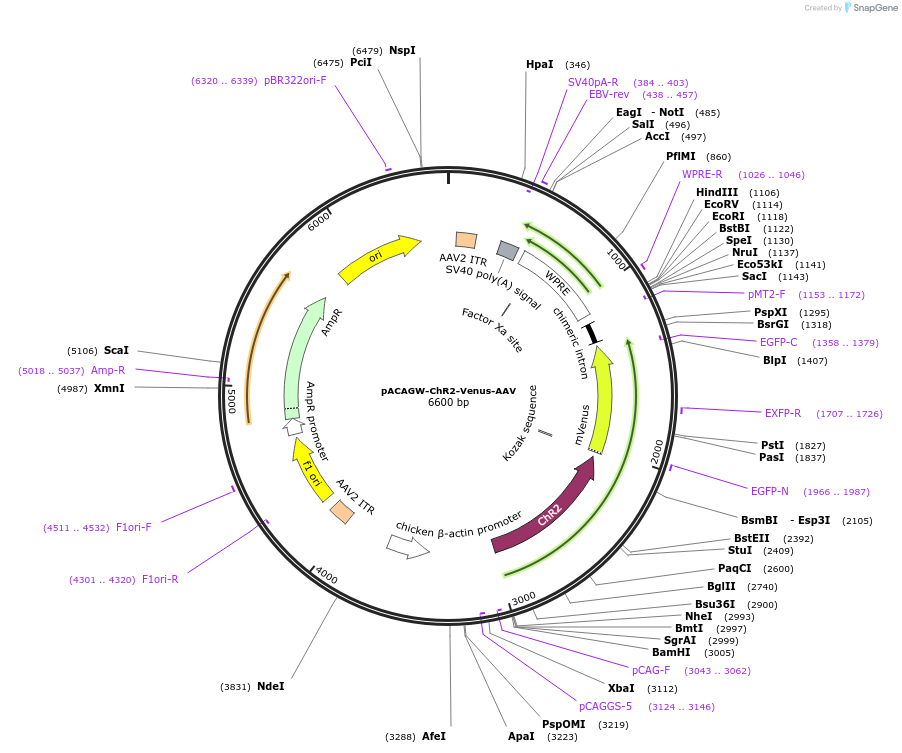
pACAGW-ChR2-Venus-AAV
Plasmid#20071PurposeAAV-mediated expression of ChR2-Venus for neuronal excitationDepositorHas ServiceAAV1 and AAV9InsertChannelrhodopsin-2 (CHLRE_02g085257v5 Chlamydomonas reinhardtii)
UseAAVTagsVenusExpressionMammalianPromoterCAGGAvailable SinceJan. 9, 2009AvailabilityAcademic Institutions and Nonprofits only -
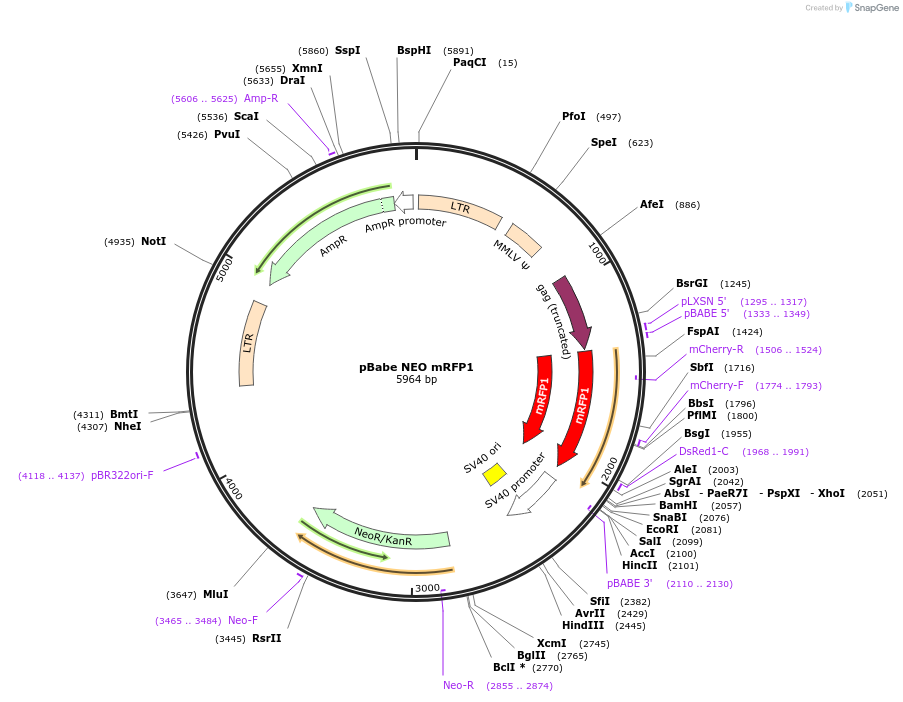
pBabe NEO mRFP1
Plasmid#98397PurposeRetroviral vector for constitutive mRFP1 expressionDepositorInsertmRFP1
UseRetroviralExpressionMammalianAvailable SinceSept. 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
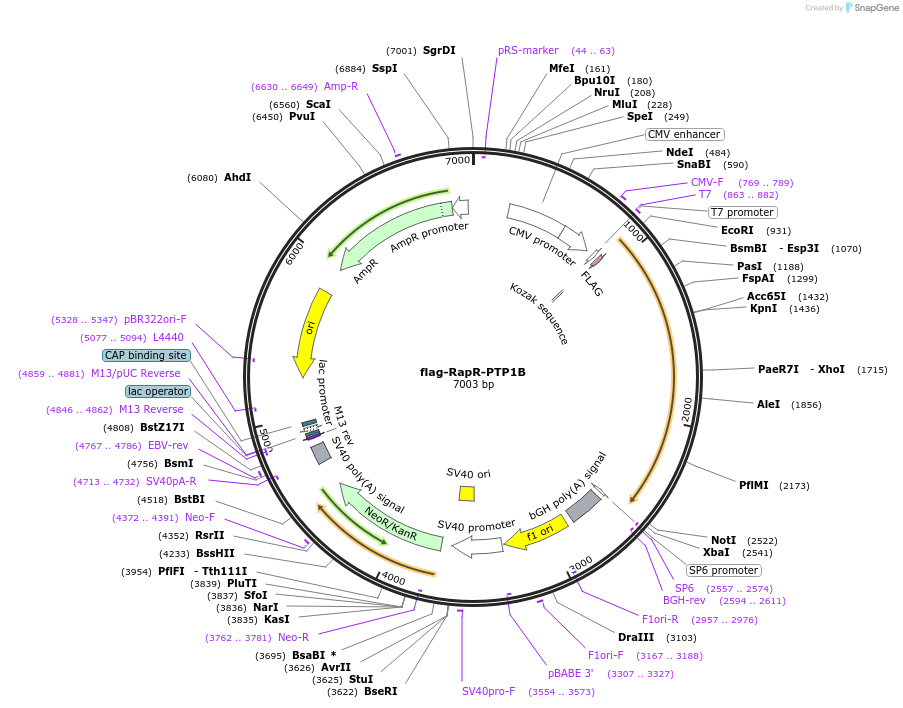
flag-RapR-PTP1B
Plasmid#188656PurposeRapamycin regulated PTP1B with flag tagDepositorInsertPTP1B
TagsflagExpressionMammalianPromoterCMVAvailable SinceAug. 19, 2022AvailabilityAcademic Institutions and Nonprofits only -
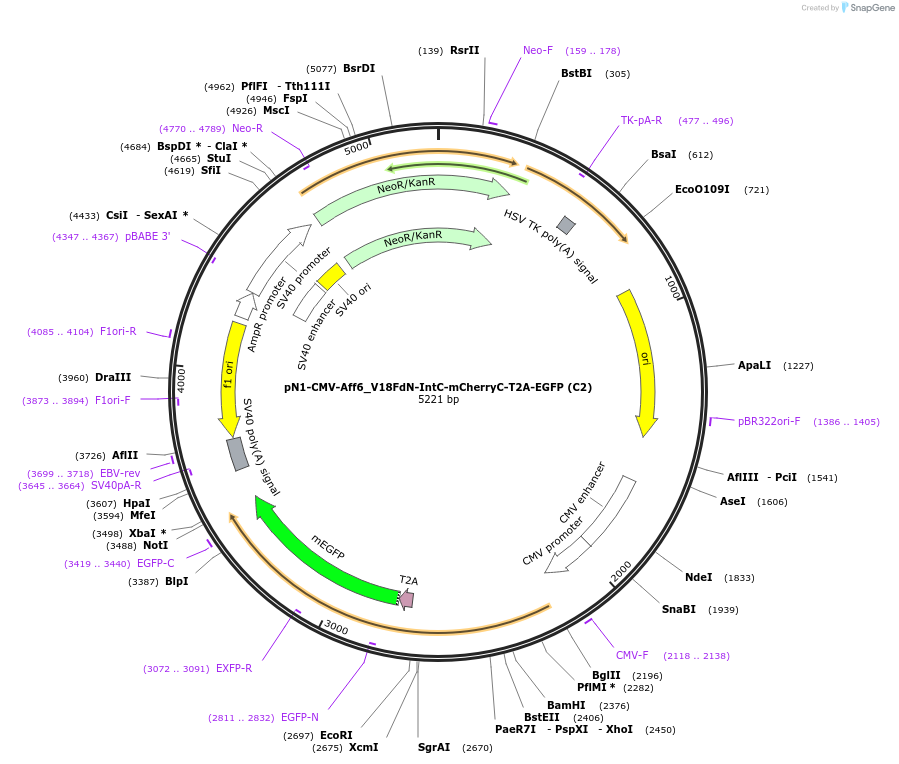
pN1-CMV-Aff6_V18FdN-IntC-mCherryC-T2A-EGFP (C2)
Plasmid#210725PurposeNIR optogenetic module for conditional protein splicing of split mCherry (C-part)DepositorInsertC2
ExpressionMammalianPromoterCMVAvailable SinceDec. 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
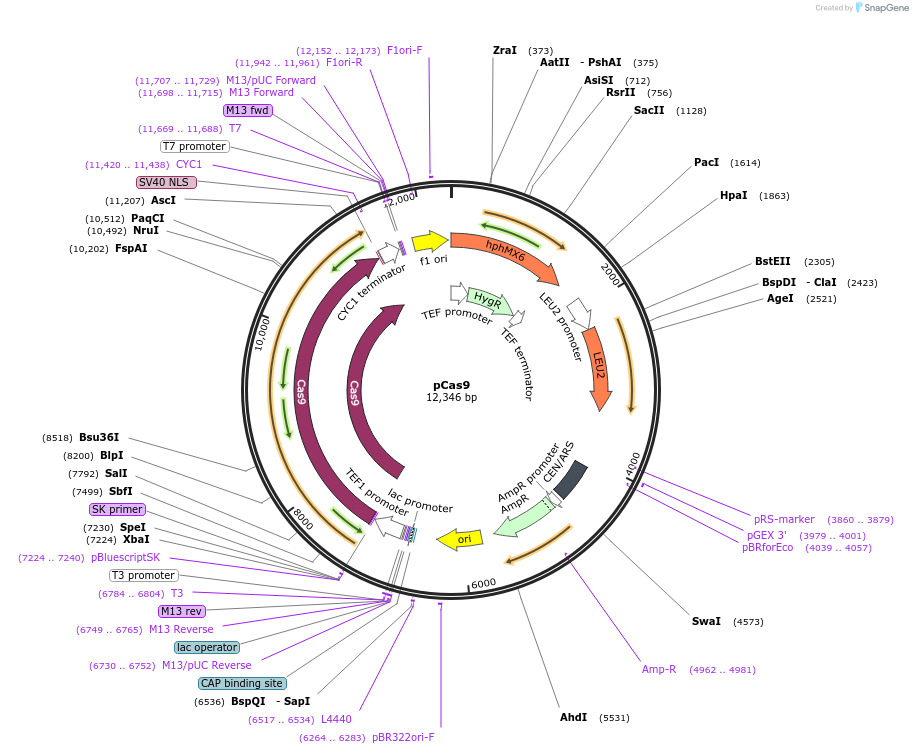
pCas9
Plasmid#162606PurposePre-express Cas9 in yeastDepositorInsertTef1-Cas9
ExpressionYeastAvailable SinceJune 11, 2021AvailabilityAcademic Institutions and Nonprofits only -
pUC118-FLIP-Puro
Plasmid#84538Purposea mock vector for generating FLIP targeting vectorsDepositorInsertFLIP - Puro
UseCre/Lox and Mouse TargetingAvailable SinceJan. 31, 2017AvailabilityAcademic Institutions and Nonprofits only -
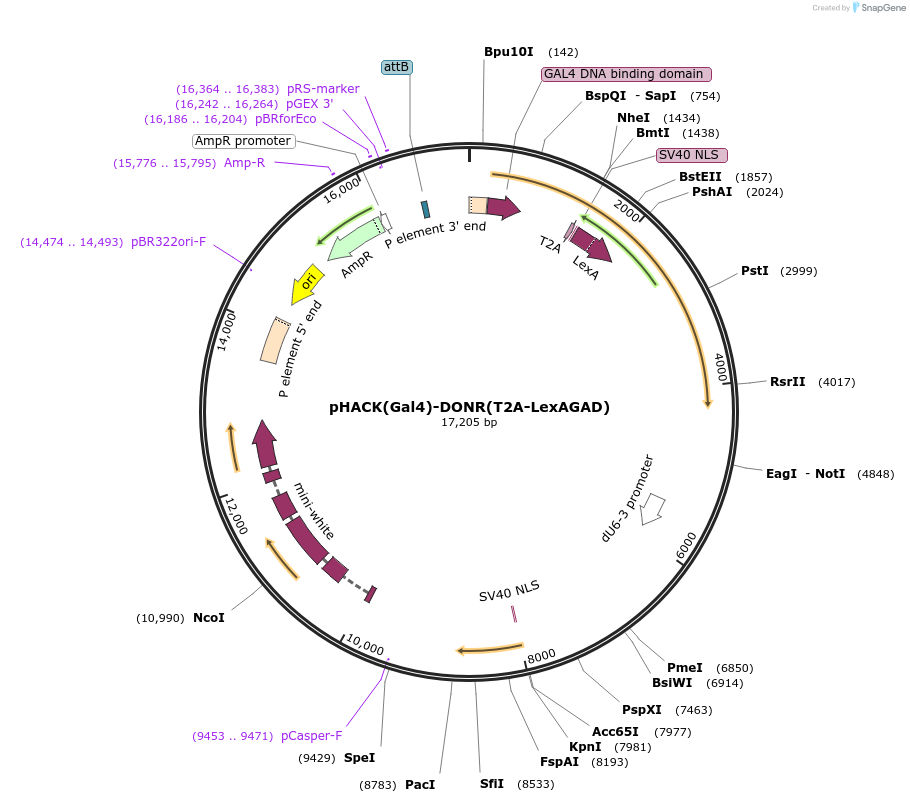
pHACK(Gal4)-DONR(T2A-LexAGAD)
Plasmid#194769PurposeTo insert T2A-LexAGAD into Gal4 coding sequence in Drosophila, converting Gal4 into a T2A-LexAGAD transgene.DepositorInsertT2A-LexAGAD
UseCRISPRExpressionInsectAvailable SinceJan. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
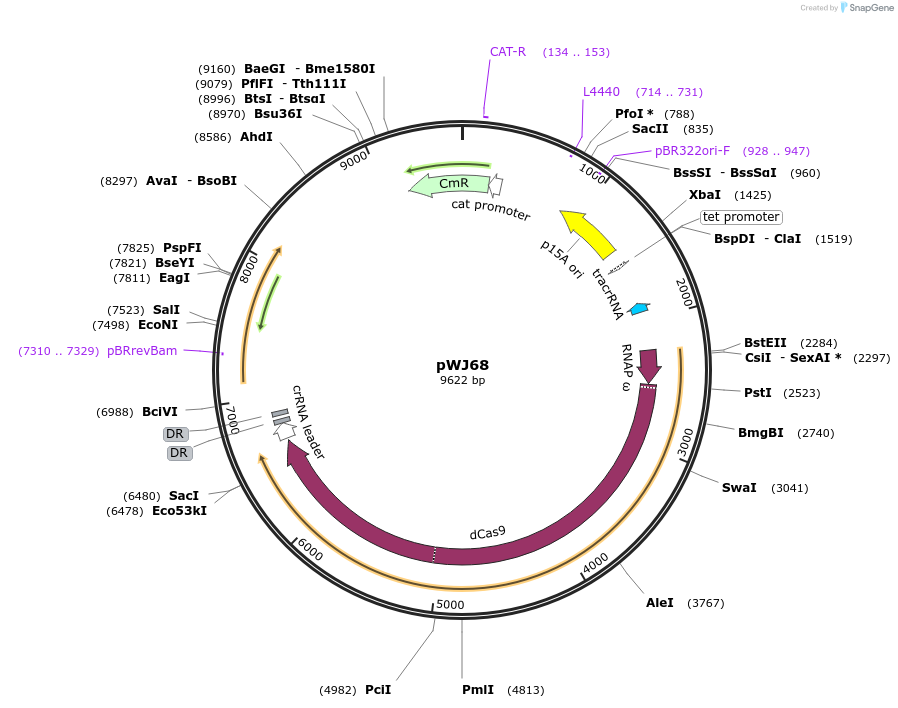
pWJ68
Plasmid#46571PurposeSame as pdCas9, but with the w-dCas9 fusion (w is fused at the N-terminal end of dCas9)DepositorInsertstracrRNA
w-dcas9
CRISPR array
UseCRISPRTagsrpoZExpressionBacterialMutationD10A, H840AAvailable SinceAug. 27, 2013AvailabilityAcademic Institutions and Nonprofits only -
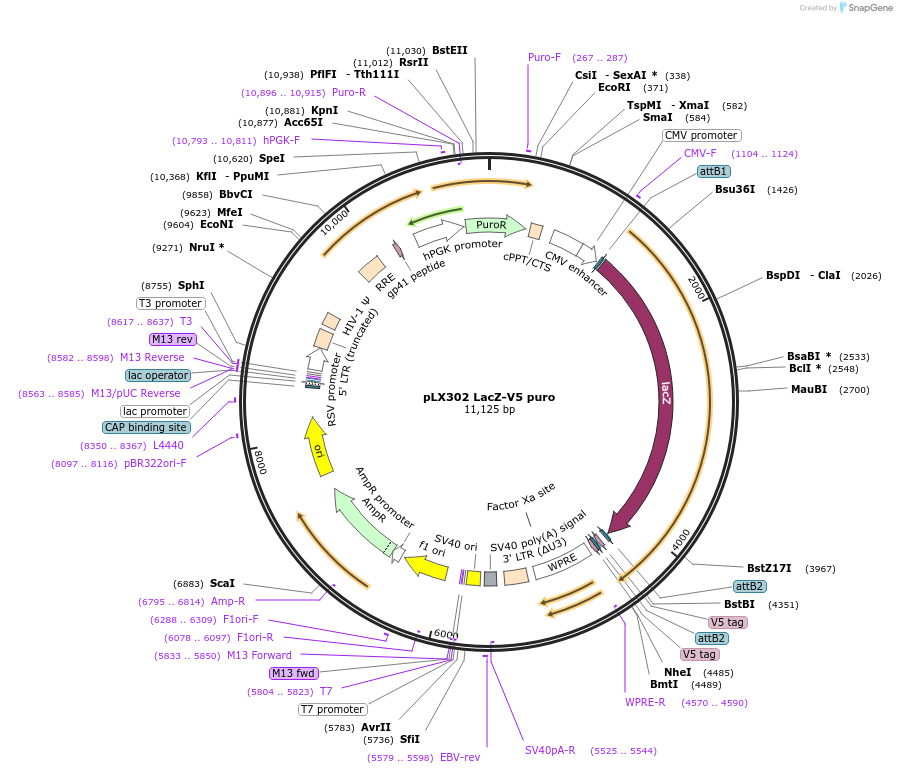
pLX302 LacZ-V5 puro
Plasmid#98579PurposeLentiviral vector for constitutive expression of human LacZ with C-terminal V5 tagDepositorInsertLacZ
UseLentiviralTags2x V5 tagExpressionMammalianAvailable SinceAug. 15, 2017AvailabilityAcademic Institutions and Nonprofits only -
mAzami Green-N1 (mAG)
Plasmid#54798PurposeLocalization: N1 Cloning Vector, Excitation: 492, Emission: 505DepositorTypeEmpty backboneTagsmAzami GreenExpressionMammalianAvailable SinceJune 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
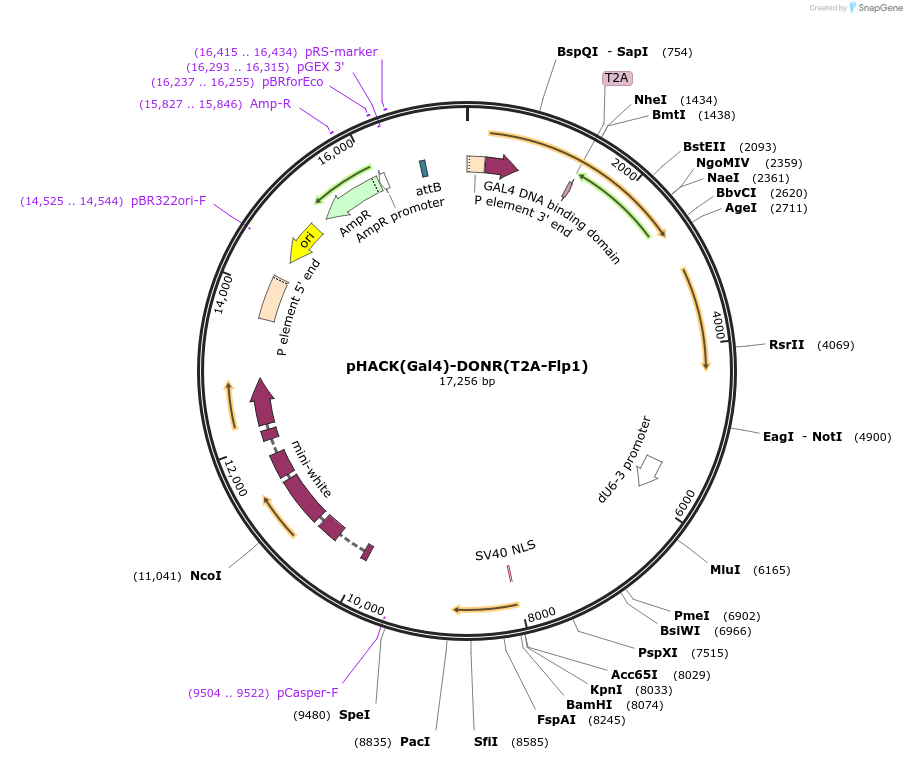
pHACK(Gal4)-DONR(T2A-Flp1)
Plasmid#194770PurposeTo insert T2A-Flp1 into Gal4 coding sequence in Drosophila, converting Gal4 into a T2A-Flp1 transgene.DepositorInsertT2A-Flp1
UseCRISPRExpressionInsectAvailable SinceJan. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
CrBlue-mcherry - pcDNA3.1
Plasmid#64204PurposeCrBlue fused to mcherry on the C terminusDepositorInsertCrBlue
TagsmcherryExpressionMammalianPromoterCMVAvailable SinceApril 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
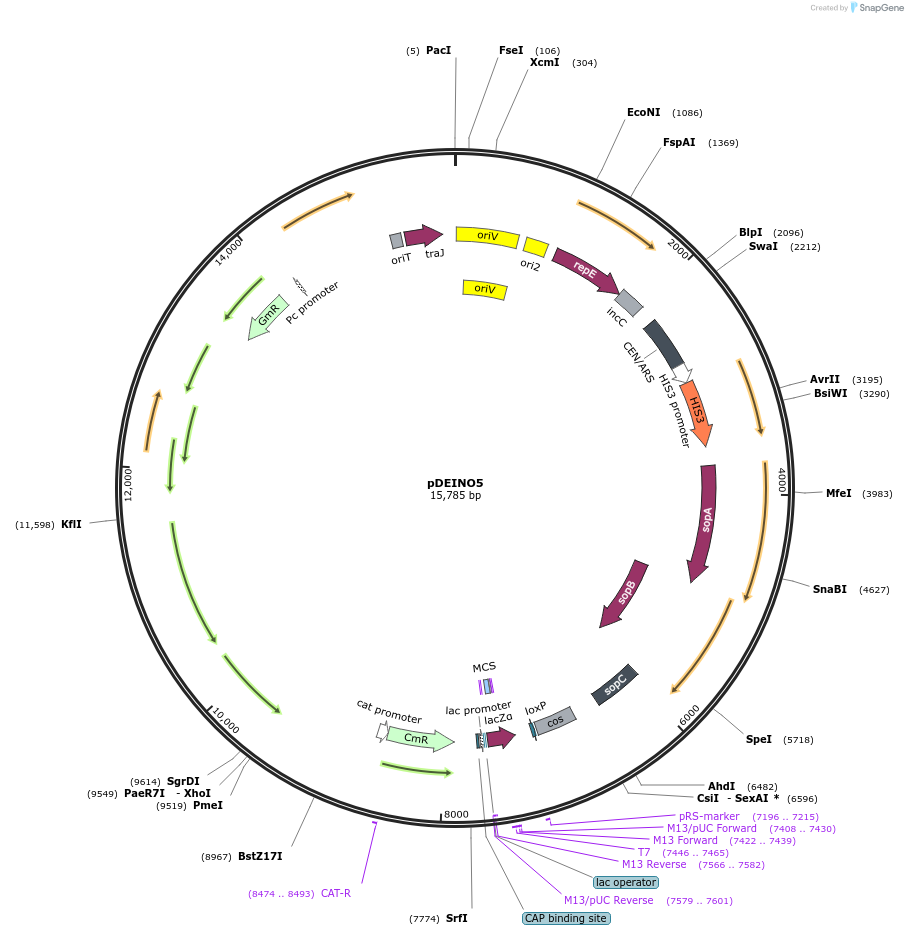
pDEINO5
Plasmid#179489PurposeMulti-host shuttle plasmid for E. coli, D. radiodurans, and S. cerevisiae; contains an oriT for transfer via conjugationDepositorInsertaacC1
UseSynthetic Biology; Bacterial and yeast cloningAvailable SinceMay 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
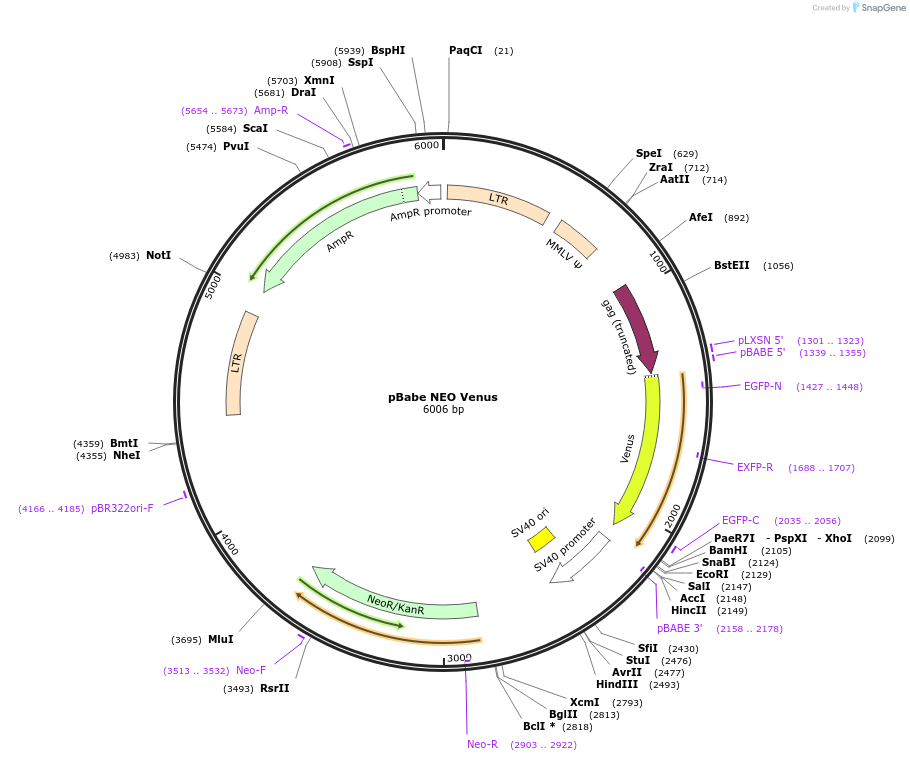
pBabe NEO Venus
Plasmid#98396PurposeRetroviral vector for constitutive Venus expressionDepositorInsertVenus
UseRetroviralExpressionMammalianAvailable SinceAug. 15, 2017AvailabilityAcademic Institutions and Nonprofits only -
mKO-pBAD
Plasmid#54735PurposeLocalization: Protein Expression Vector , Excitation: 548, Emission: 561DepositorTypeEmpty backboneTags6xHis and mKOExpressionBacterialAvailable SinceJuly 25, 2014AvailabilityAcademic Institutions and Nonprofits only -
pKS014
Plasmid#65467PurposepSC101 based plasmid where pl-tetO expression drives synthesis of construct expressing (N to C terminal) Ntag and beta-galactosidaseDepositorInsertNtag-beta galactosidase
UseSynthetic BiologyTagsFLAGExpressionBacterialPromoterpl-TetOAvailable SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
mAzami Green-C1 (mAG)
Plasmid#54797PurposeLocalization: C1 Cloning Vector, Excitation: 492, Emission: 505DepositorTypeEmpty backboneTagsmAzami GreenExpressionMammalianAvailable SinceJune 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
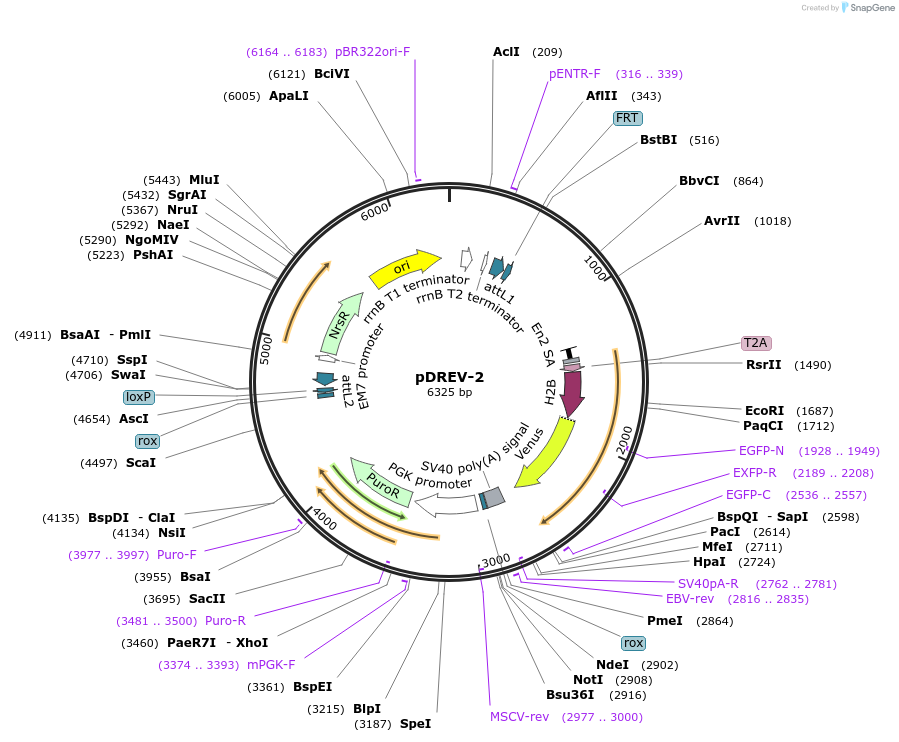
pDREV-2
Plasmid#26752DepositorAvailable SinceJan. 10, 2011AvailabilityAcademic Institutions and Nonprofits only -
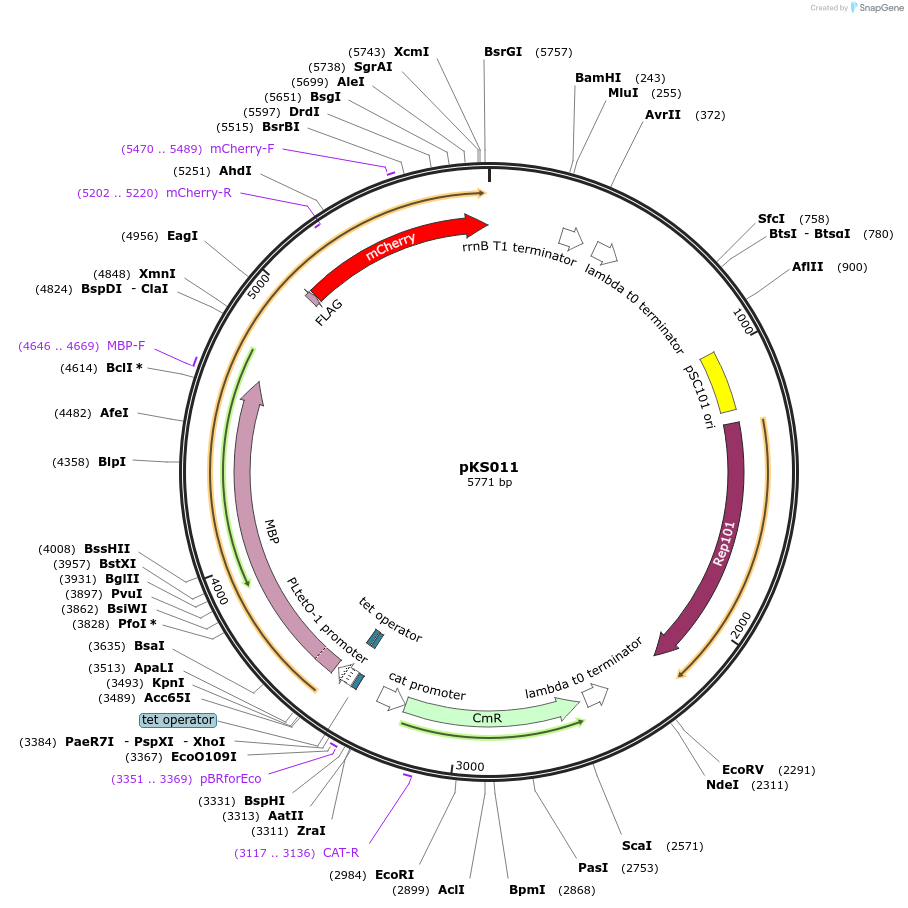
pKS011
Plasmid#65464PurposepSC101 based plasmid where pl-tetO expression drives synthesis of construct expressing (N to C terminal) MBP (Maltose Binding Protein), Ntag, and mCherryDepositorInsertMBP-Ntag-mCherry
TagsFLAG and MBPExpressionBacterialPromoterpl-TetOAvailable SinceMarch 1, 2016AvailabilityAcademic Institutions and Nonprofits only