We narrowed to 28,253 results for: Tat
-
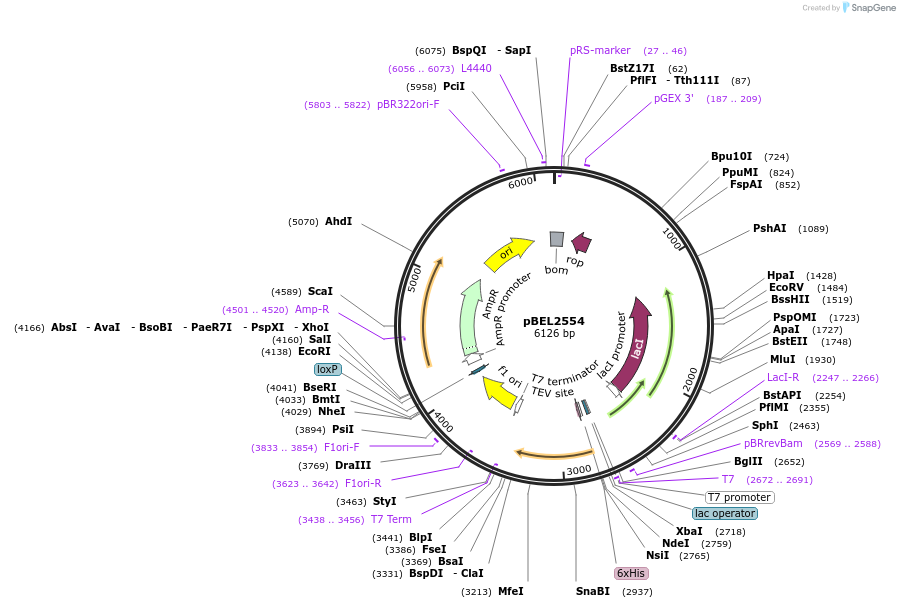
Plasmid#195672PurposeExpression/purification of 6xHis-TEV-MlaC(E169R)DepositorInsertMlaC
ExpressionBacterialMutation6xHis-TEV-MlaC(E169R)Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
pBEL1160
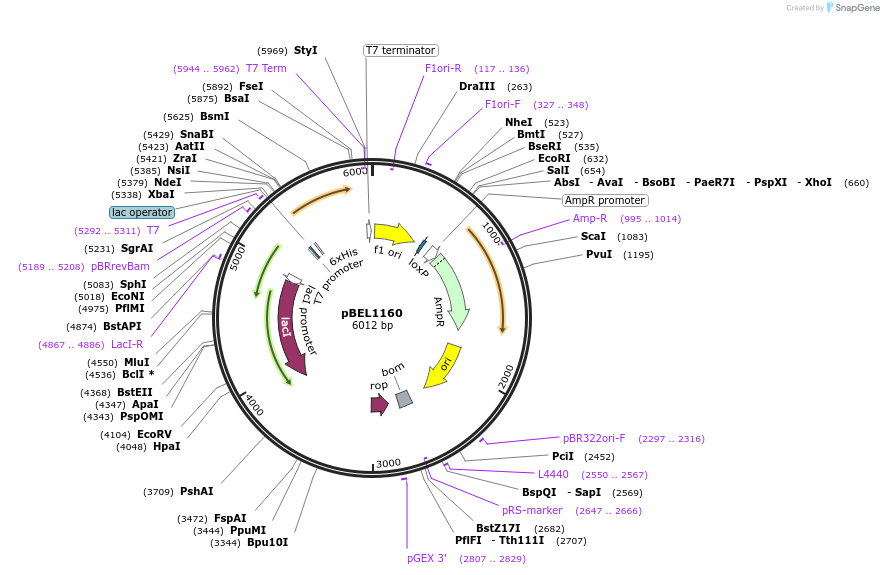
Plasmid#196025PurposeExpression/purification of 6xHis-TEV-MlaDDepositorInsertMlaD
ExpressionBacterialAvailable SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
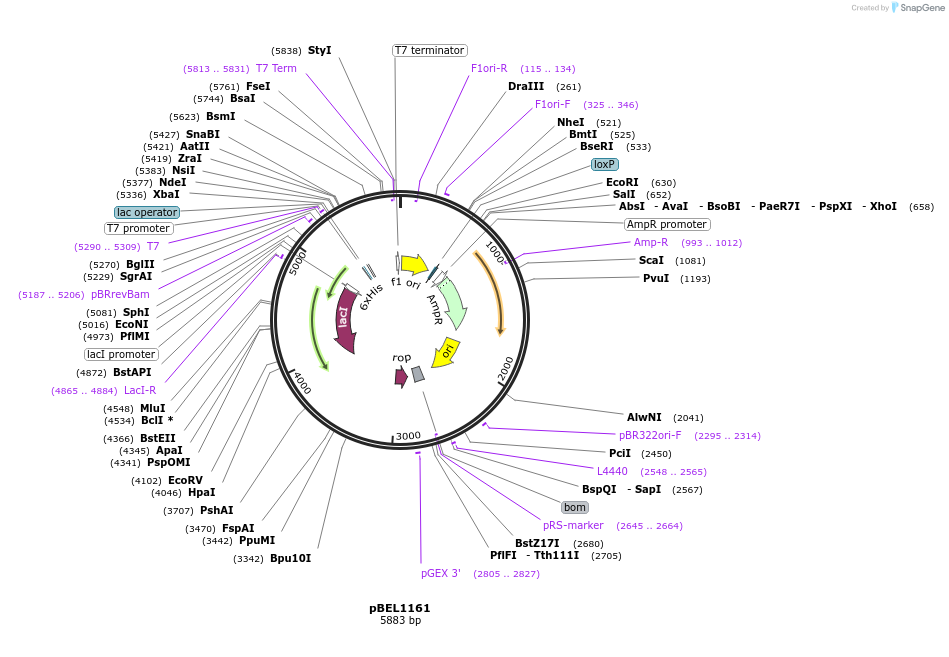
pBEL1161
Plasmid#196026PurposeExpression/purification of 6xHis-TEV-MlaDdelta141-183DepositorInsertMlaD
ExpressionBacterialMutation6xHis-TEV-MlaDdelta141-183Available SinceMay 2, 2023AvailabilityAcademic Institutions and Nonprofits only -
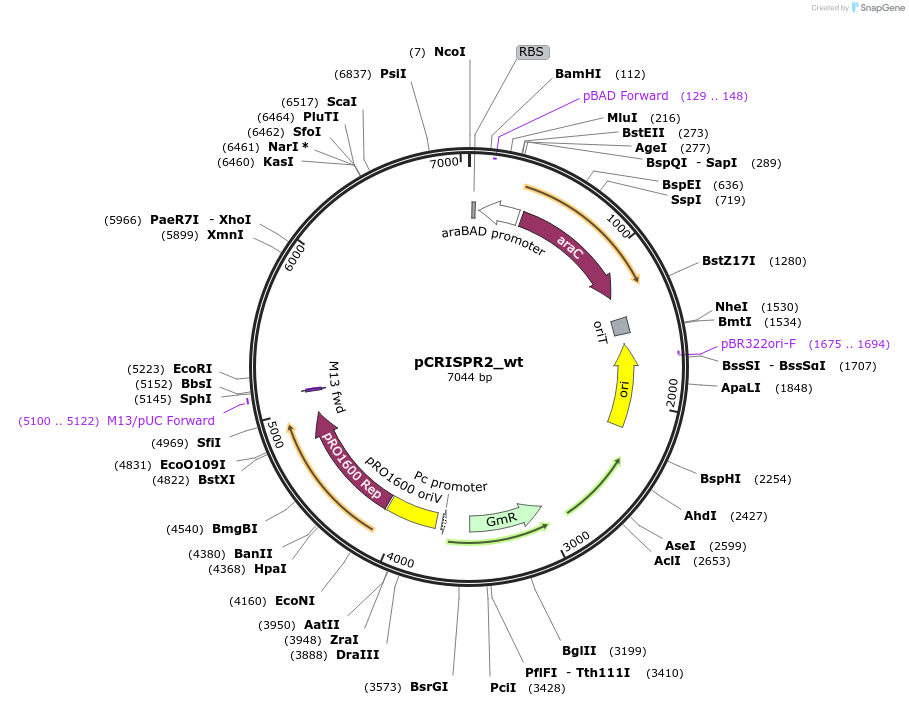
pCRISPR2_wt
Plasmid#149386PurposeP. aeruginosa PA14 CRISPR2 locusDepositorInsertPseudomonas aeruginosa PA14 CRISPR2 locus
ExpressionBacterialPromoterpBADAvailable SinceAug. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
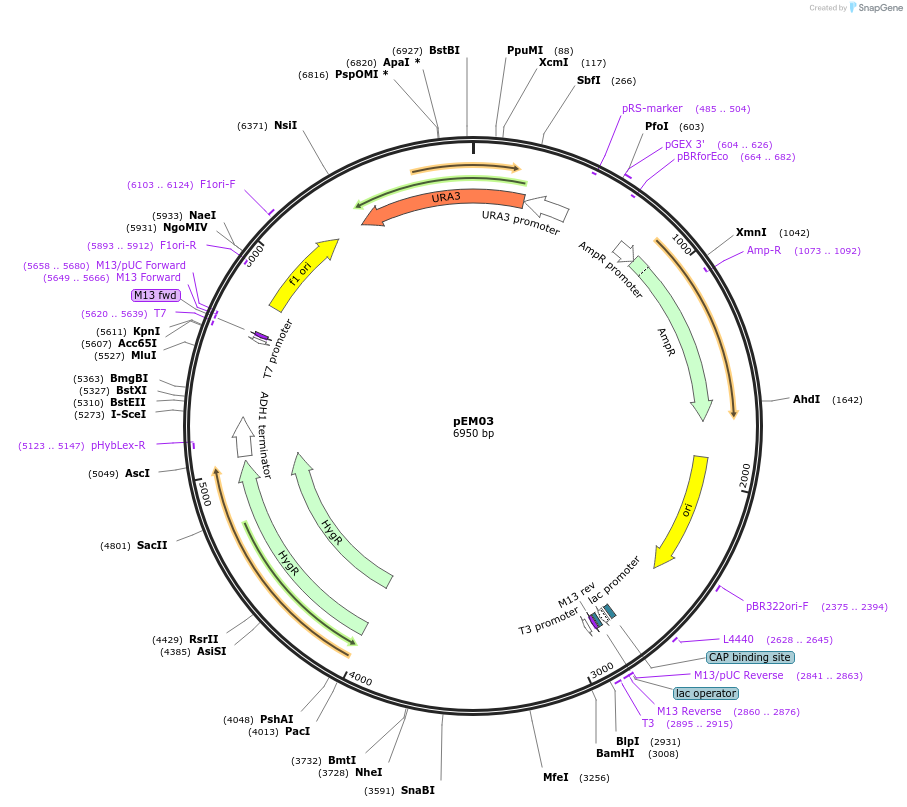
pEM03
Plasmid#135417PurposeExpression of Hygromycin resistance from Spizellomyces Hsp70 promoterDepositorInsertSpunHsp70pr
ExpressionBacterial and YeastPromoterHsp70prAvailable SinceMay 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
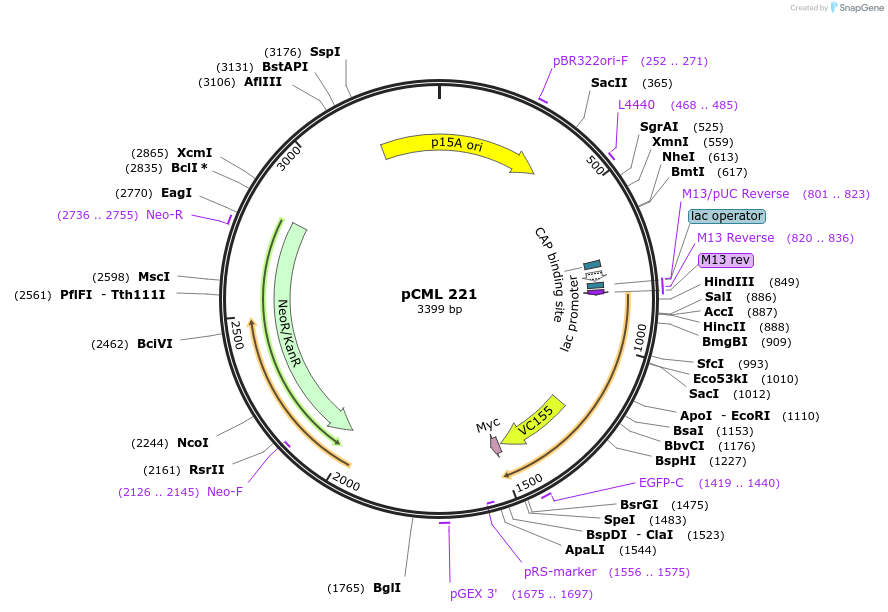
pCML 221
Plasmid#98587PurposepMS2-CYFP. Expresses MS2 protein-CYFP fusion in E. coli for TriFC assay (kanR).DepositorInsertMS2
PromoterpLacOAvailable SinceSept. 27, 2017AvailabilityAcademic Institutions and Nonprofits only -
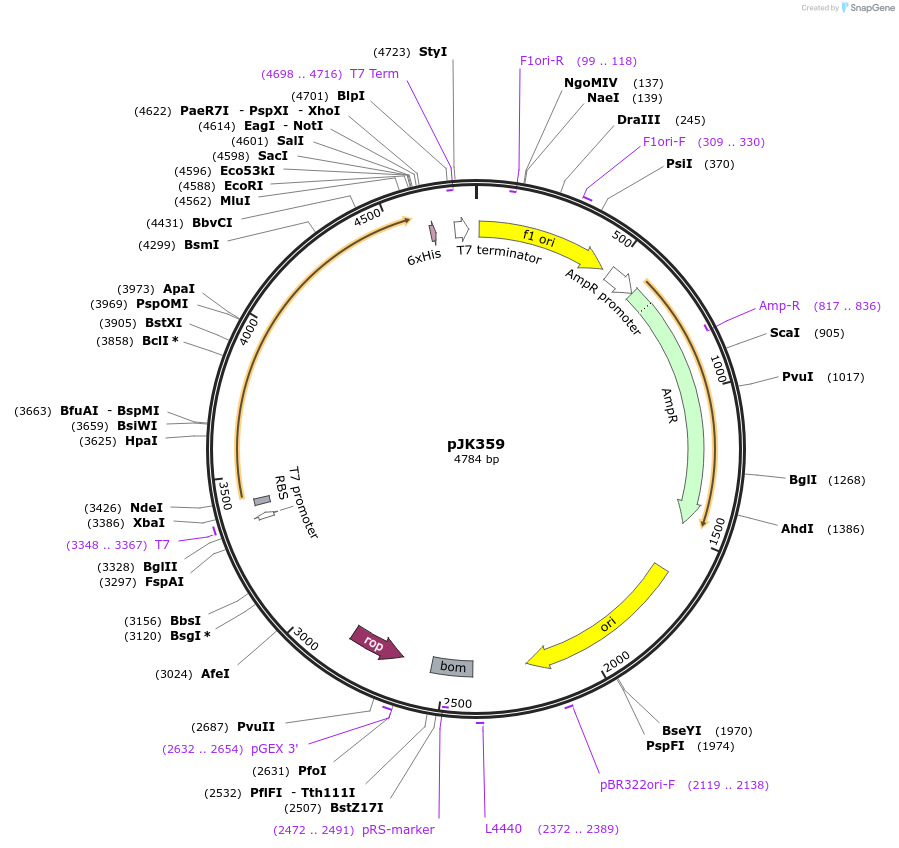
pJK359
Plasmid#71570PurposeProduces Acetobacter aceti 1023 putative acyl-CoA:carboxylate CoA-transferase (UctA)DepositorInsertputative CoA-transferase
ExpressionBacterialPromoterT7Available SinceFeb. 26, 2016AvailabilityAcademic Institutions and Nonprofits only -
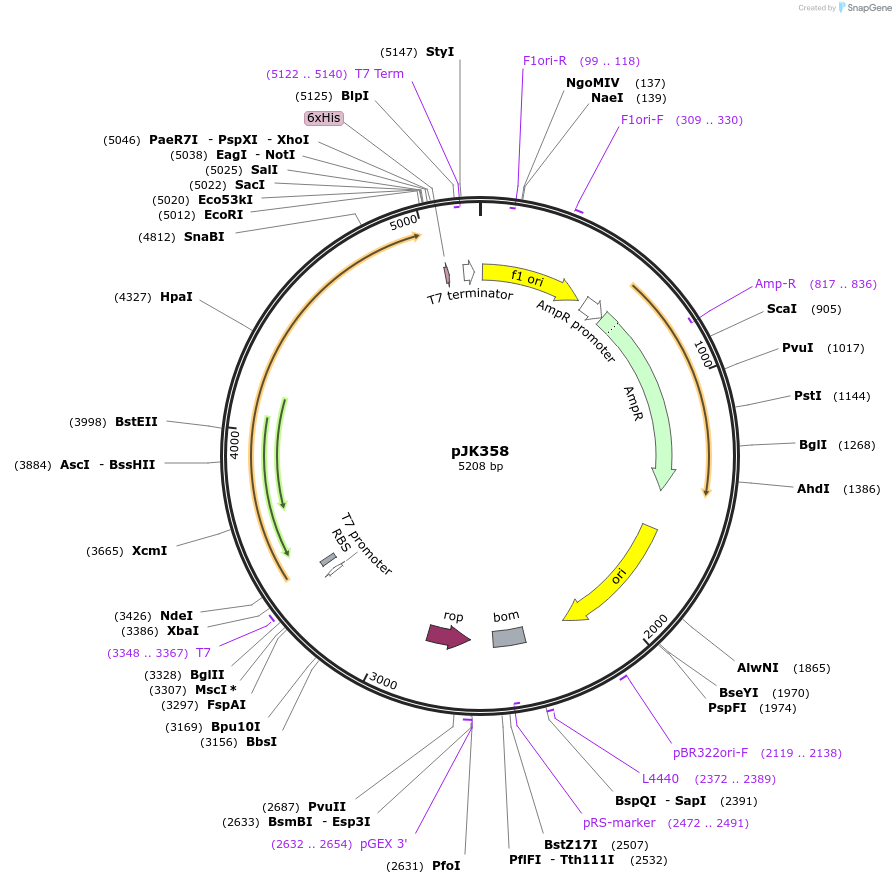
pJK358
Plasmid#71569PurposeProduces Acetobacter aceti 1023 putative acyl-CoA:carboxylate CoA-transferase (UctD)DepositorInsertputative CoA-transferase
ExpressionBacterialPromoterT7Available SinceJan. 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
pSL7 SMR1791
Plasmid#28030DepositorAvailable SinceApril 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
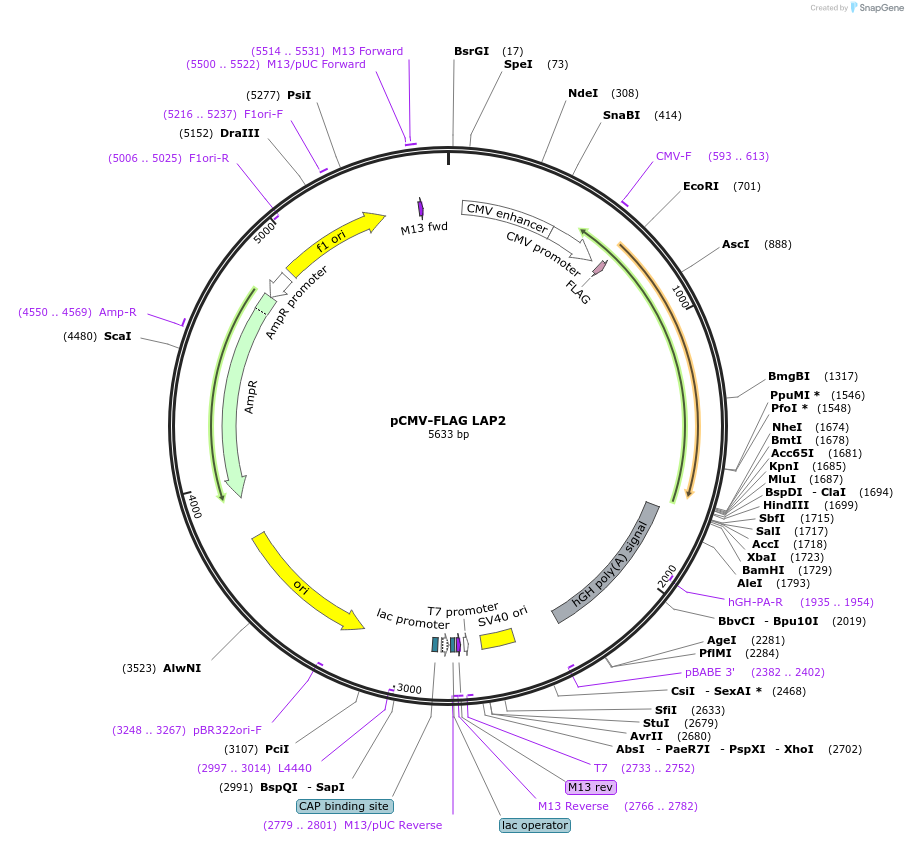
pCMV-FLAG LAP2
Plasmid#15738DepositorAvailable SinceSept. 7, 2007AvailabilityAcademic Institutions and Nonprofits only -
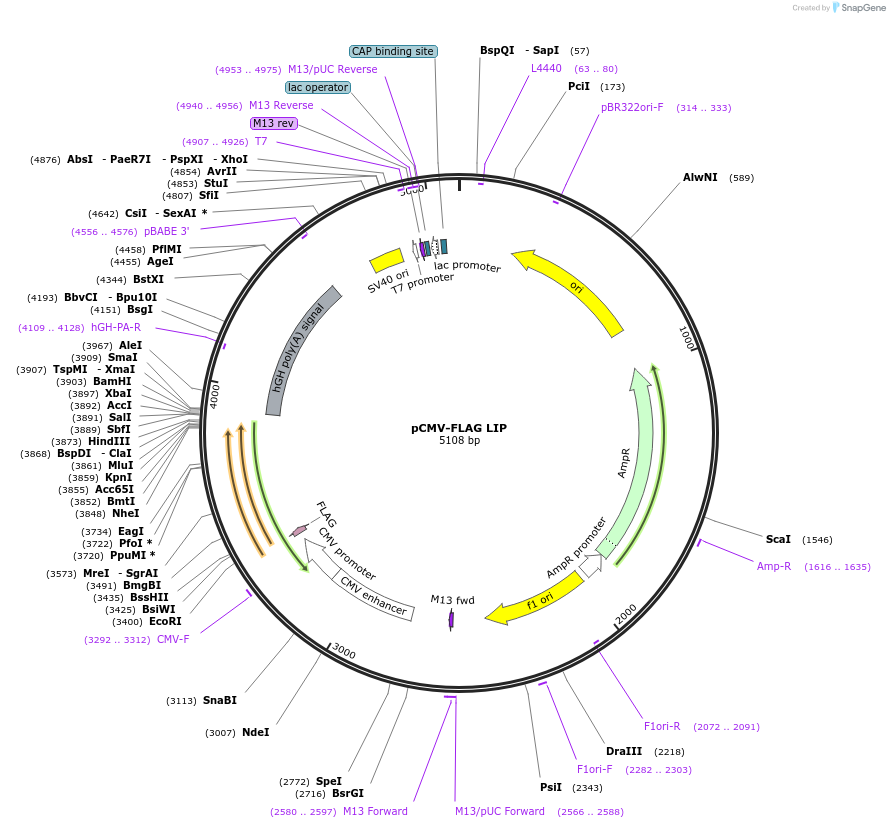
pCMV-FLAG LIP
Plasmid#15737DepositorAvailable SinceSept. 7, 2007AvailabilityAcademic Institutions and Nonprofits only -
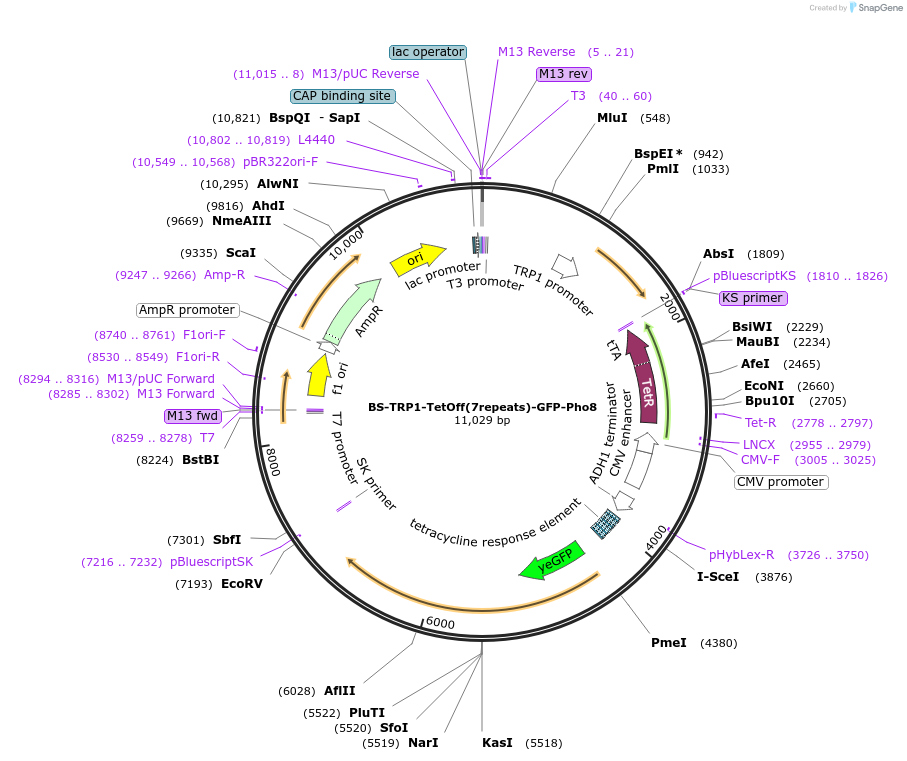
BS-TRP1-TetOff(7repeats)-GFP-Pho8
Plasmid#207011PurposeTet-Off controlled GFP-Pho8 with TRP1 selection. Tetracycline-controlled transactivator (tTA) present on same plasmid.DepositorInsertPho8
TagsGFPExpressionYeastAvailable SinceJune 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
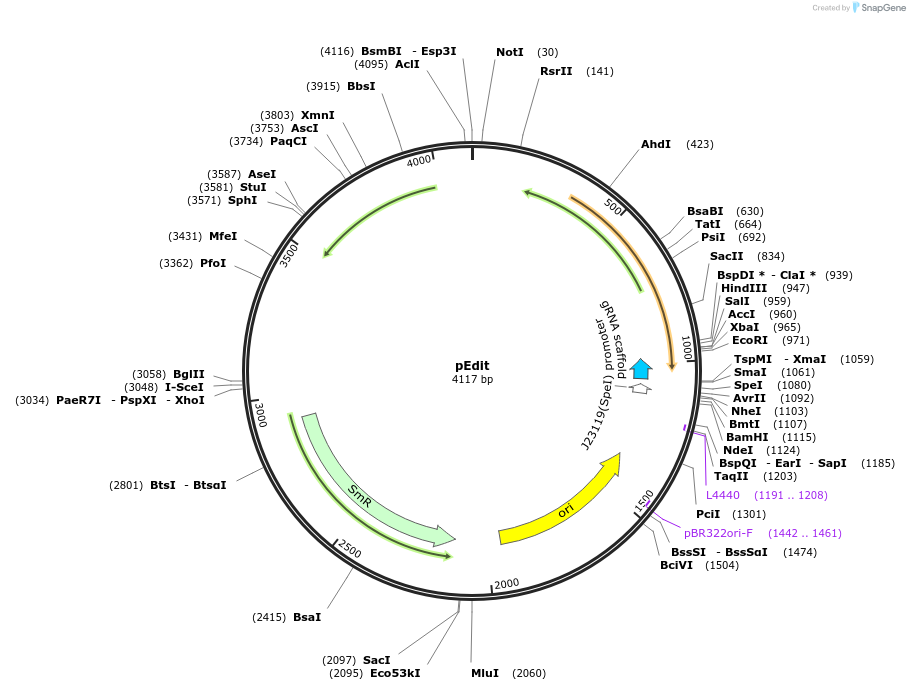
pEdit
Plasmid#232355PurposeThe pEdit series plasmids are derived from the pTarget series plasmids by inserting homologous recombination arms targeting the gene of interest, which enables gene knockout or gene insertion at the tDepositorInsertsgRNA
UseCRISPRAvailable SinceNov. 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
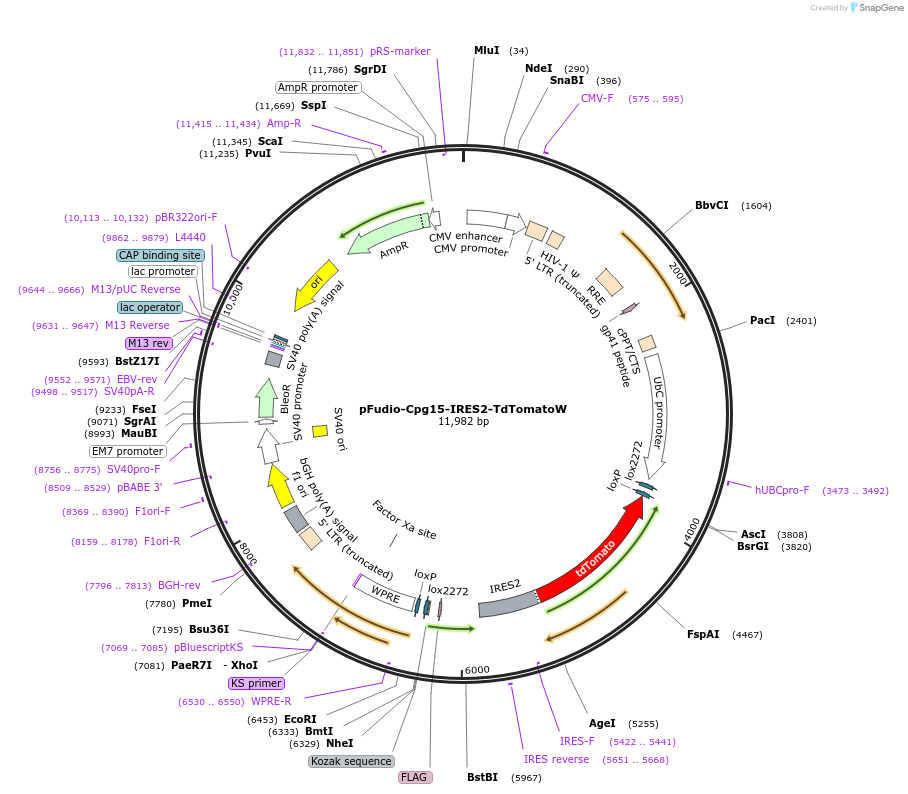
pFudio-Cpg15-IRES2-TdTomatoW
Plasmid#134309PurposeExpresses cpg15 and TdTomato upon coexpression with Cre recombinaseDepositorInsertsUseCre/Lox and LentiviralExpressionMammalianPromoterUbCAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
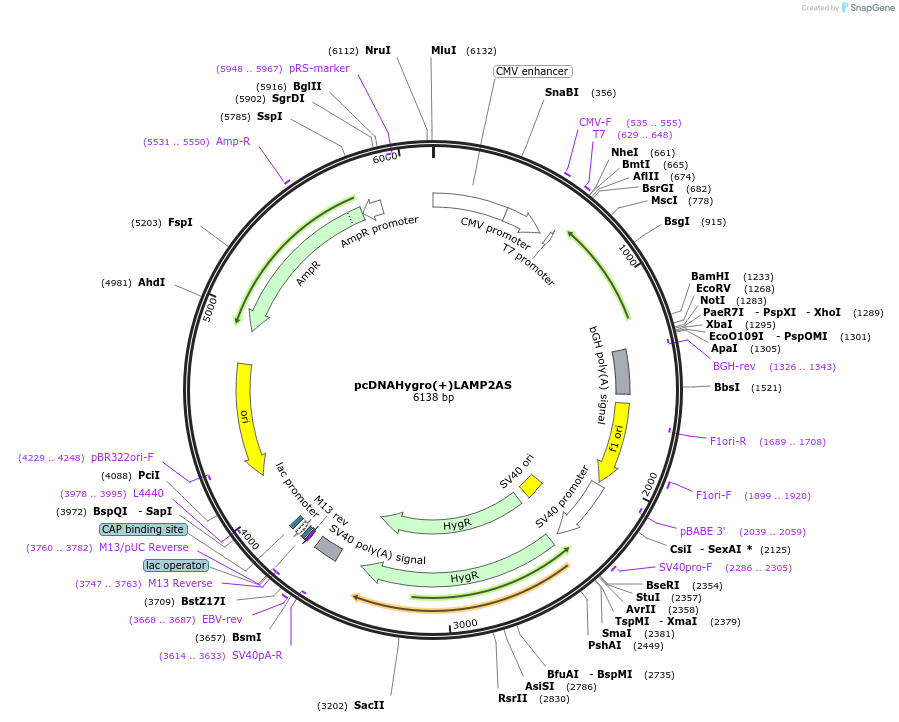
pcDNAHygro(+)LAMP2AS
Plasmid#86146PurposeAnti sense for LAMP2DepositorInsertLAMP2a (LAMP2 Human)
ExpressionMammalianMutationcontains first 525 bases of LAMP cDNAPromoterCMVAvailable SinceFeb. 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
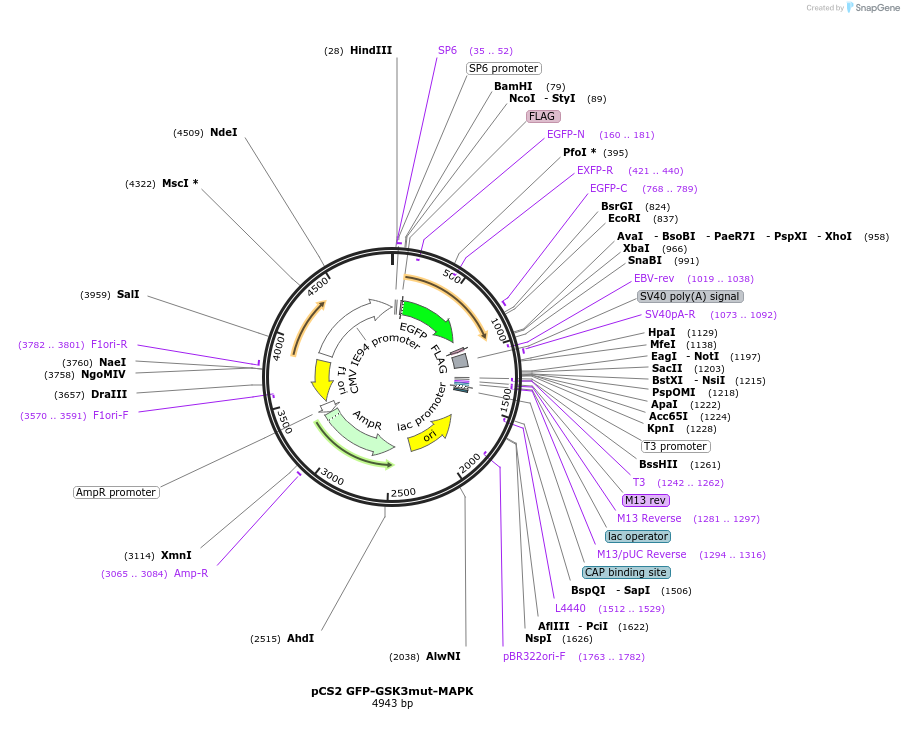
pCS2 GFP-GSK3mut-MAPK
Plasmid#29690PurposeGFP reporter protein fused to three mutated GSK3 phosphorylation sitesDepositorInsertGFP glycogen synthase kinase 3 recognition site mutated primed by MAPK
TagsFlag and Flag-EGFPExpressionMammalianMutationrecognition sites only, and E3 ligase site, GSK3 …PromoterSP6Available SinceJan. 6, 2012AvailabilityAcademic Institutions and Nonprofits only -
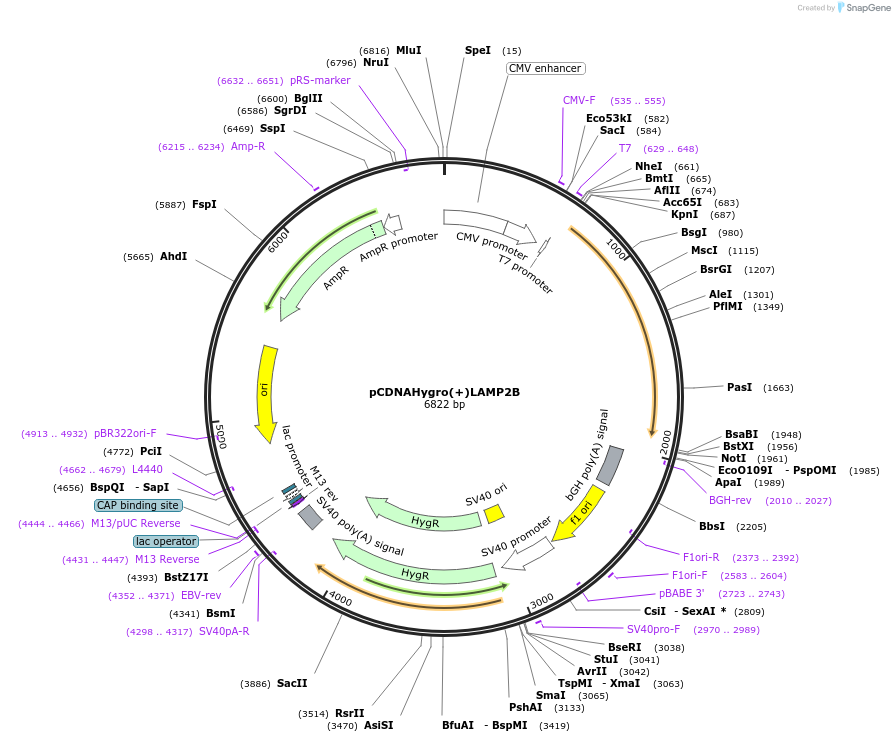
pCDNAHygro(+)LAMP2B
Plasmid#86029PurposeExpresses LAMP2B in Mammalian cellsDepositorAvailable SinceFeb. 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
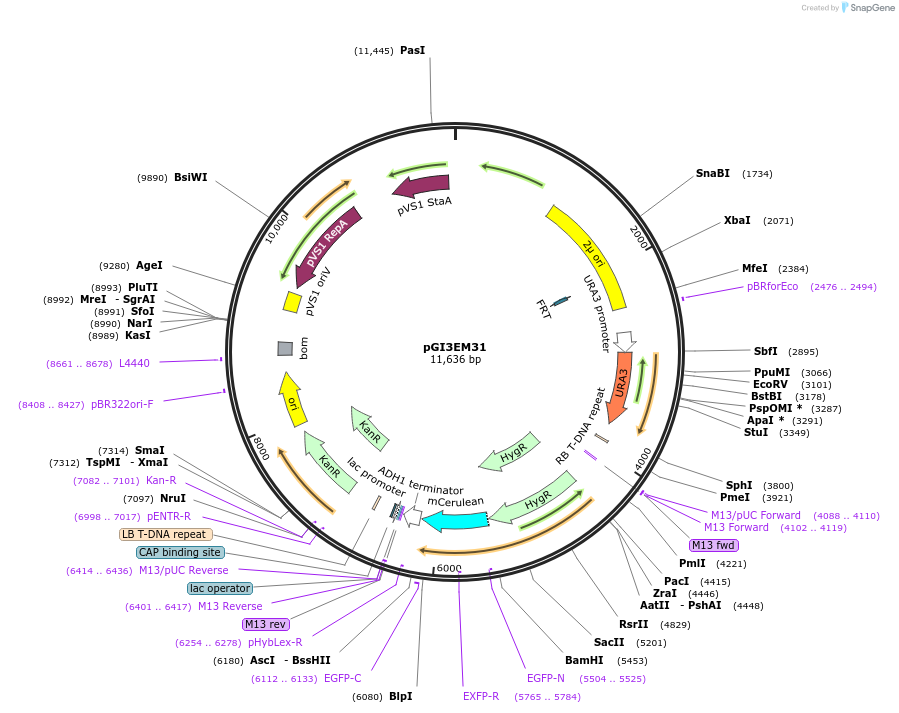
pGI3EM31
Plasmid#135491PurposeAgrobacterium T-DNA with Spun H2Bpr driving hph-mCerulean3DepositorInserthph SpunH2A/Bpr hph-mCerulean3
TagsmCerulean3ExpressionBacterial and YeastPromoterSpunH2A/Bpr (divergent)Available SinceMay 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
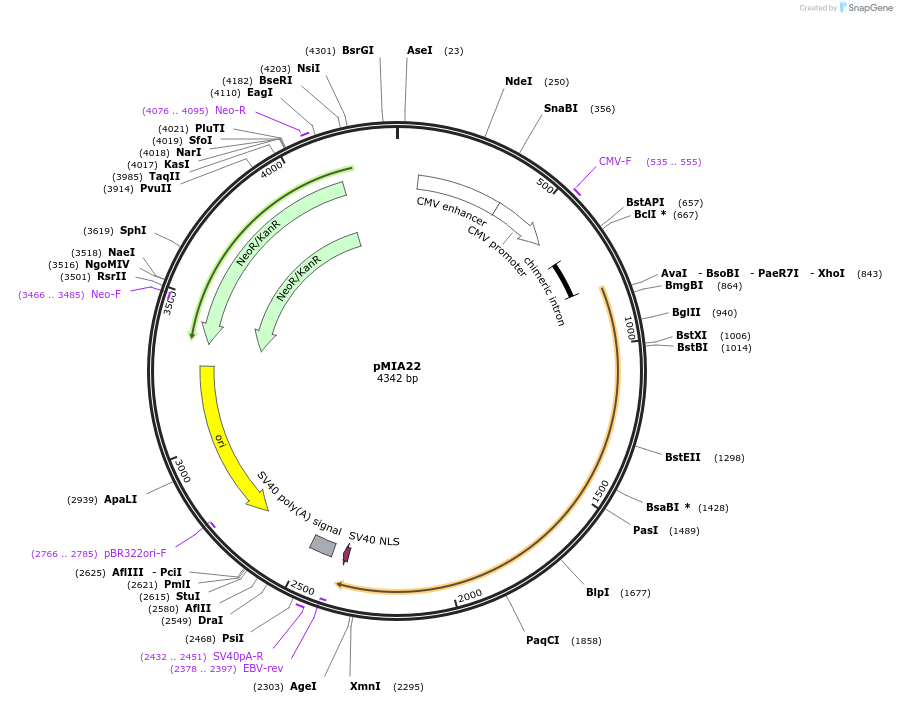
pMIA22
Plasmid#214448PurposeExpresses BxbI-BP-NLSDepositorInsertBxbI
TagsBP-NLSExpressionMammalianPromoterCMVAvailable SinceApril 29, 2024AvailabilityAcademic Institutions and Nonprofits only -
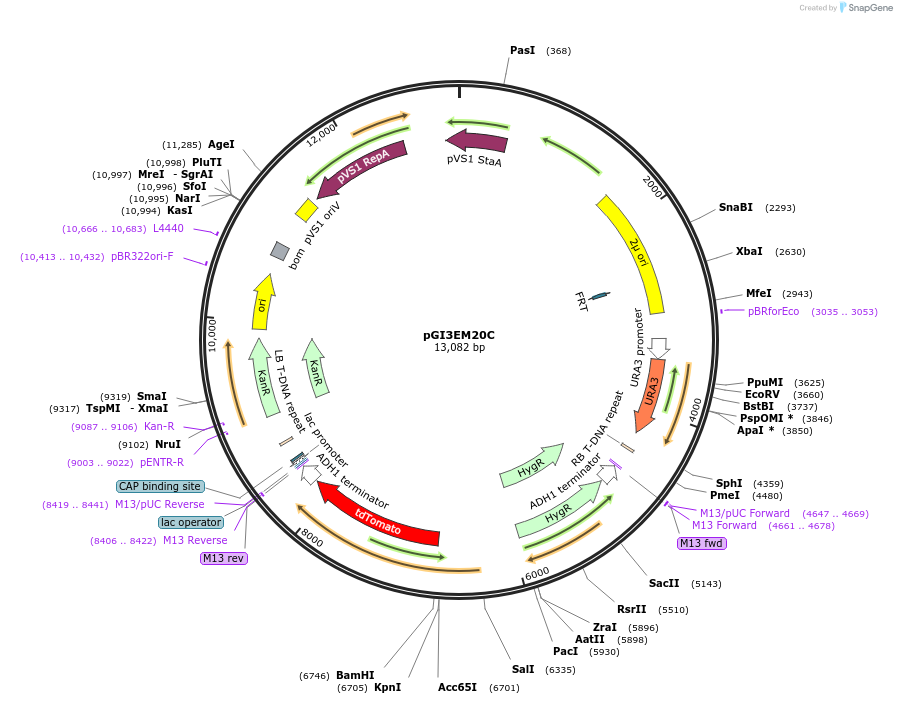
pGI3EM20C
Plasmid#135487PurposeAgrobacterium T-DNA with divergent Spun H2A/Bpr driving H2B-tdTomato and hphDepositorInserthph SpunH2A/Bpr H2B-tdTomato
TagstdTomatoExpressionBacterial and YeastPromoterSpunH2A/Bpr (divergent)Available SinceMay 19, 2020AvailabilityAcademic Institutions and Nonprofits only