We narrowed to 1,720 results for: plasmids pcas
-
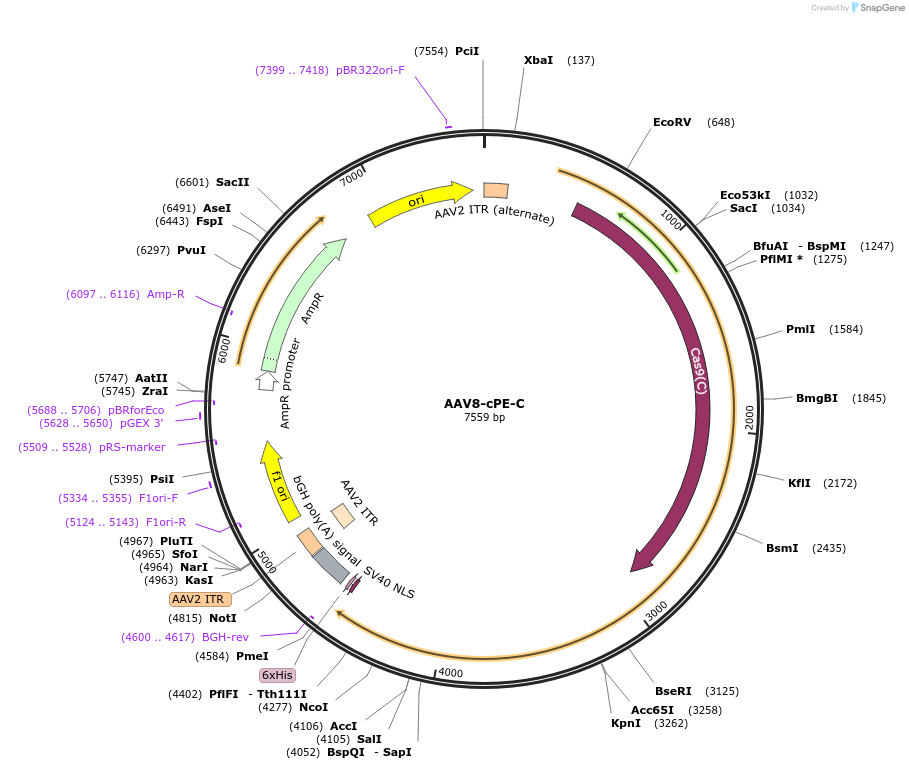
Plasmid#183539PurposeDeliver a split compact Prime Editor in vivoDepositorInsertC-terminal fragment of SpCas9 nickase (H840A), Reverse Transcriptase
UseAAVExpressionMammalianPromoterU1AAvailable SinceJune 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
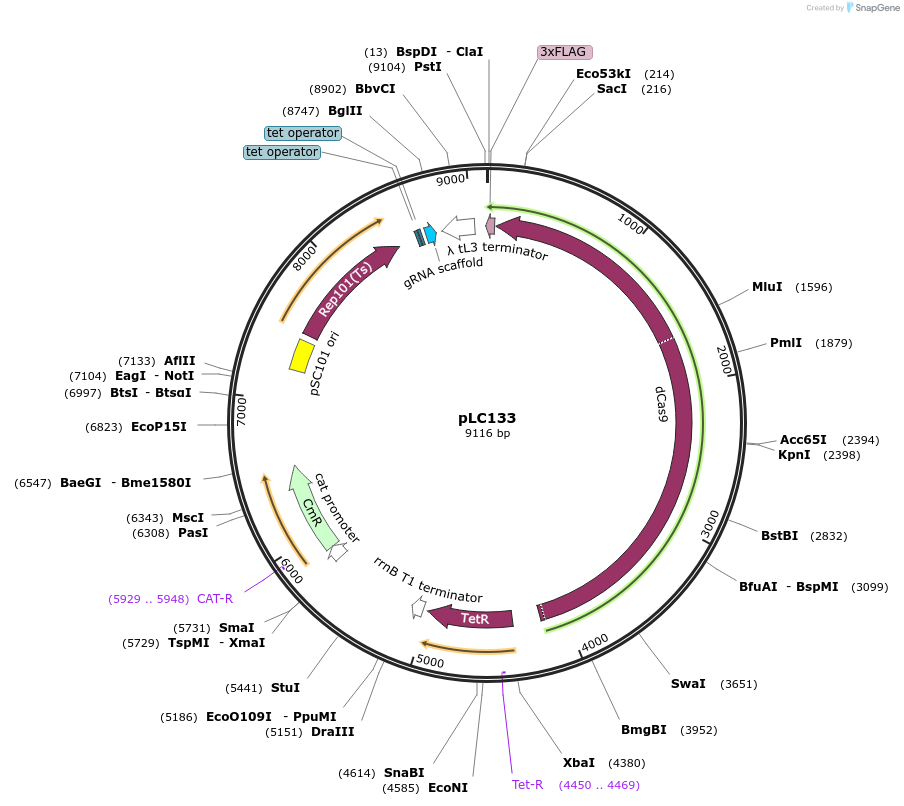
pLC133
Plasmid#198639PurposeExpression plasmid for dCas9 in E. coli used in Chip-seq experiments. dCas9 is tagged with a C-terminal 3x FLAG tag. A guide RNA can be cloned using BsaI restriction sites.DepositorInsertdCas9
Tags3x-FLAGExpressionBacterialPromoterpTetAvailable SinceApril 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
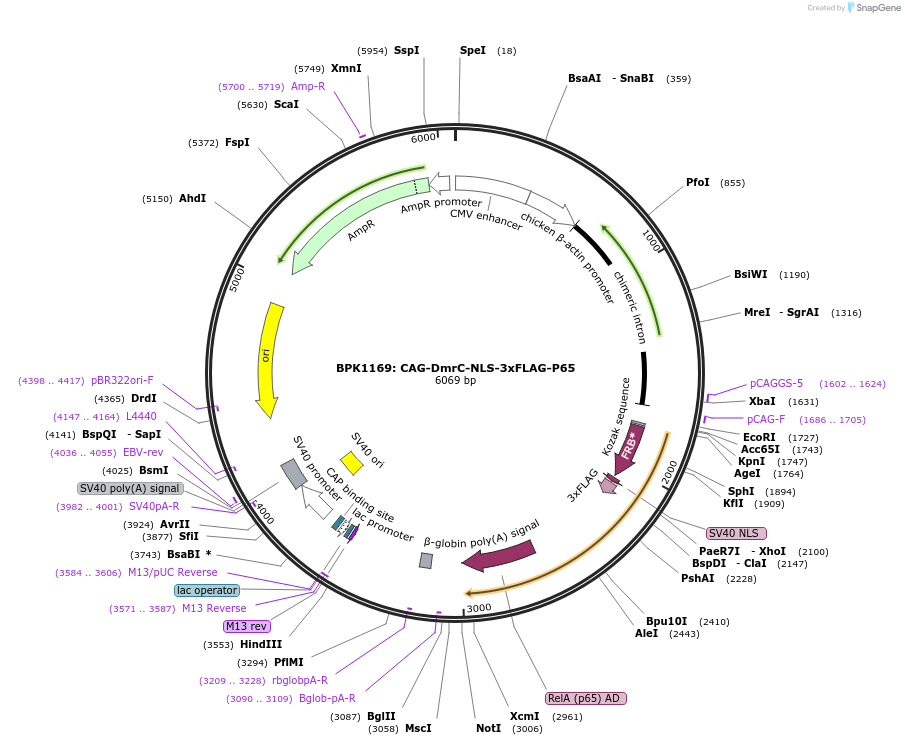
BPK1169: CAG-DmrC-NLS-3xFLAG-P65
Plasmid#104564PurposeMammalian expression vector for DmrC fused to p65 activation domainDepositorInsertDmrC-p65
Tags3x FLAG and NLSExpressionMammalianPromoterCAGAvailable SinceJan. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
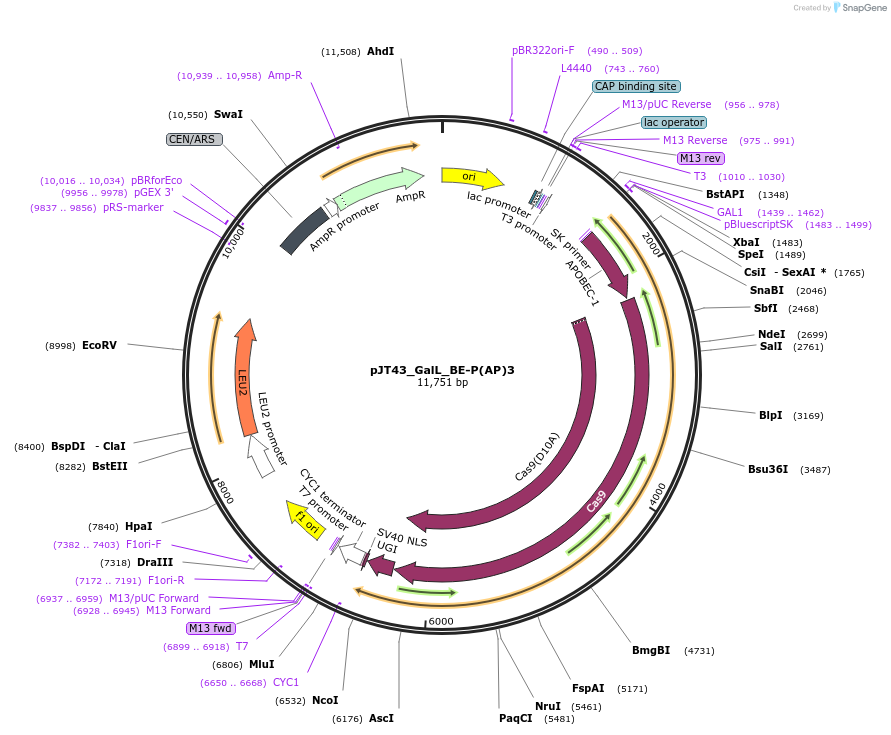
pJT43_GalL_BE-P(AP)3
Plasmid#145036PurposeExpresses BE-P(AP)3 in yeast cellsDepositorInsertBE-(AP)3
UseCRISPRExpressionYeastMutationspCas9(D10A)PromoterGalLAvailable SinceJuly 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
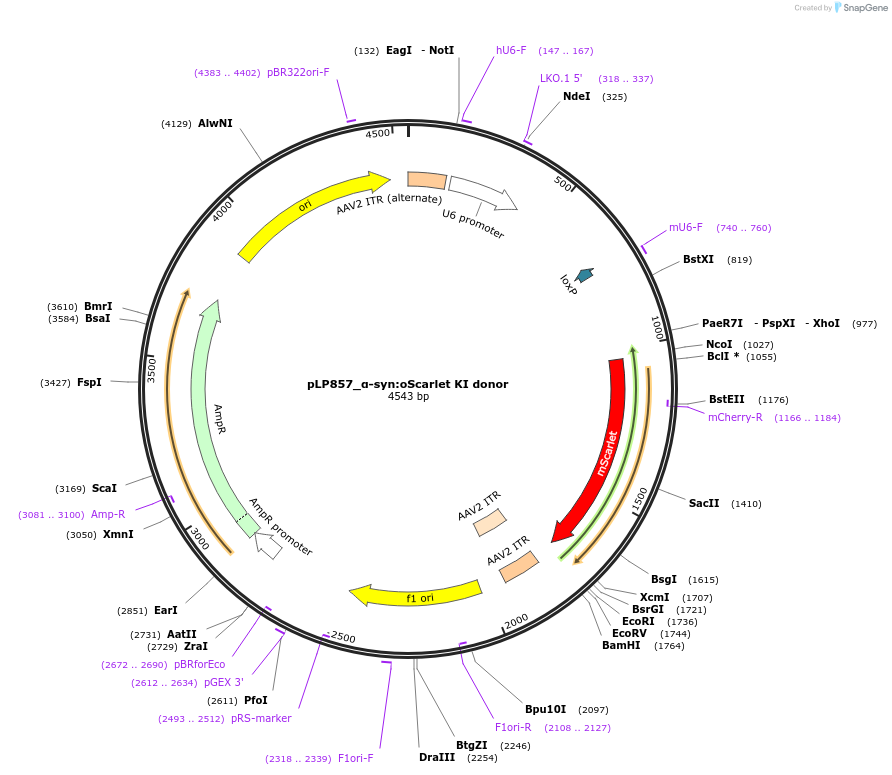
pLP857_α-syn:oScarlet KI donor
Plasmid#239403PurposeAAV vector for SpCas9-mediated CRISPR KI of oScarlet at the C-terminus of mouse α-synDepositorInsertoScarlet
UseAAV and CRISPRTagsoScarletExpressionMammalianAvailable SinceSept. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
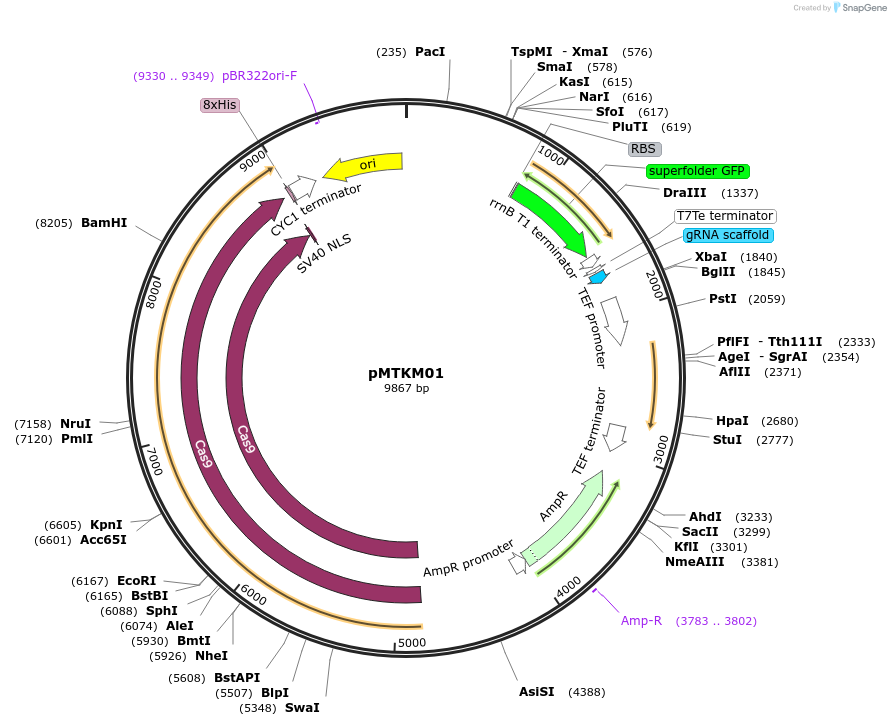
pMTKM01
Plasmid#233473PurposeSpCAS9-Tyr-[gRNA: BsaI GFP dropout] with pan(OPT)ARS replication origin and Nat selection.DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
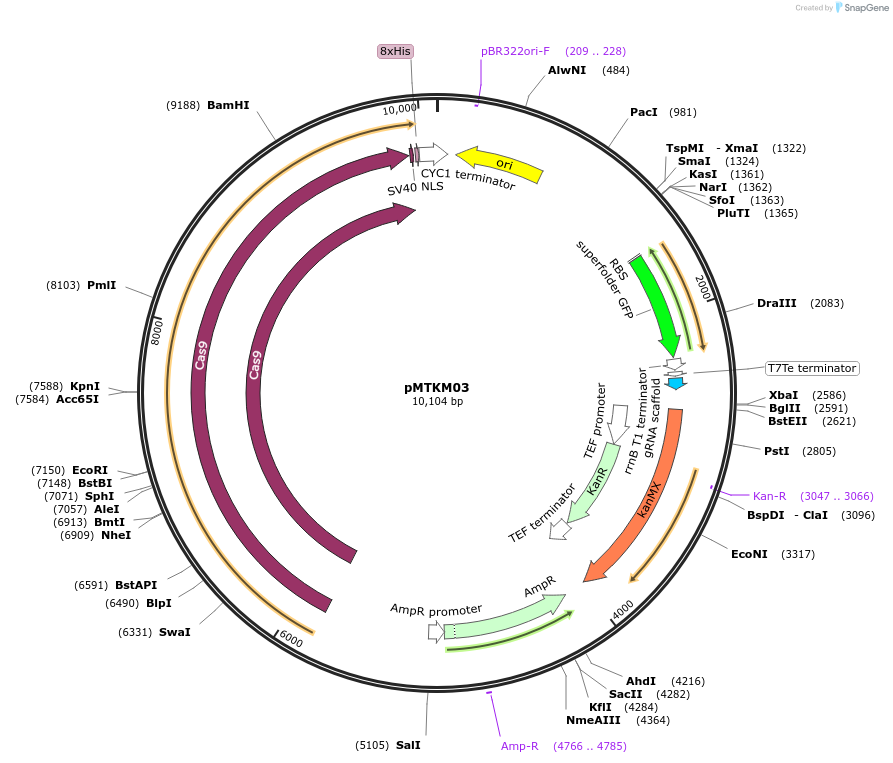
pMTKM03
Plasmid#233475PurposeSpCAS9-Tyr-[gRNA: BsaI GFP dropout] with pan(OPT)ARS replication origin and G418 selection.DepositorTypeEmpty backboneUseCRISPR and Synthetic BiologyExpressionYeastAvailable SinceJuly 2, 2025AvailabilityAcademic Institutions and Nonprofits only -
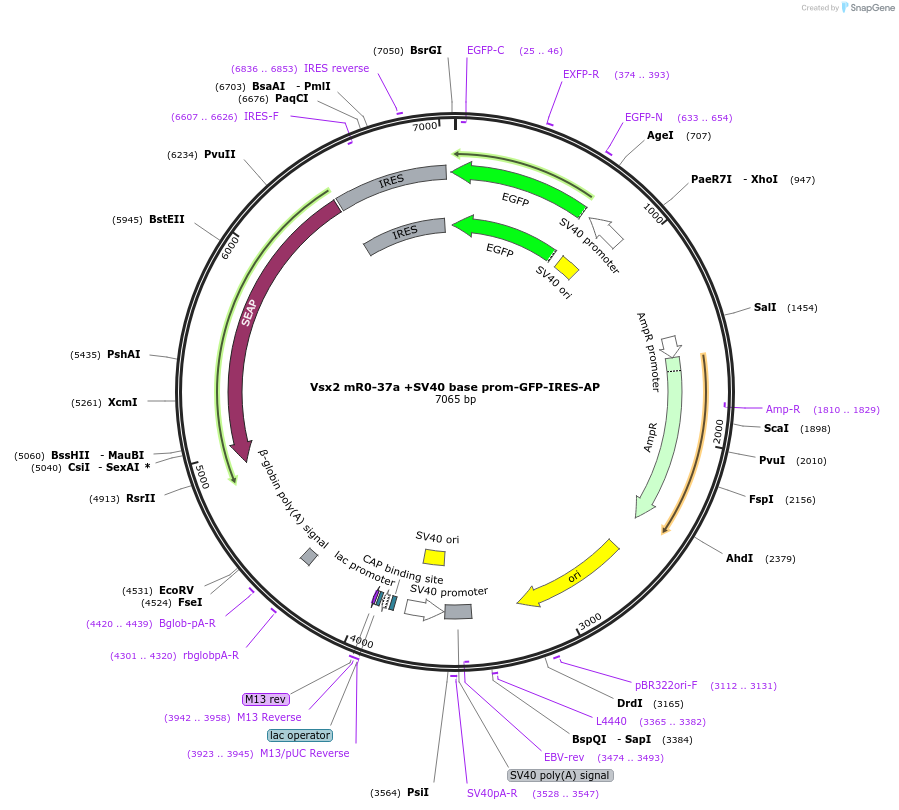
Vsx2 mR0-37a +SV40 base prom-GFP-IRES-AP
Plasmid#225889PurposeEnhancer reporterDepositorInsertmR0-37a
TagsEGFPExpressionMammalianAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
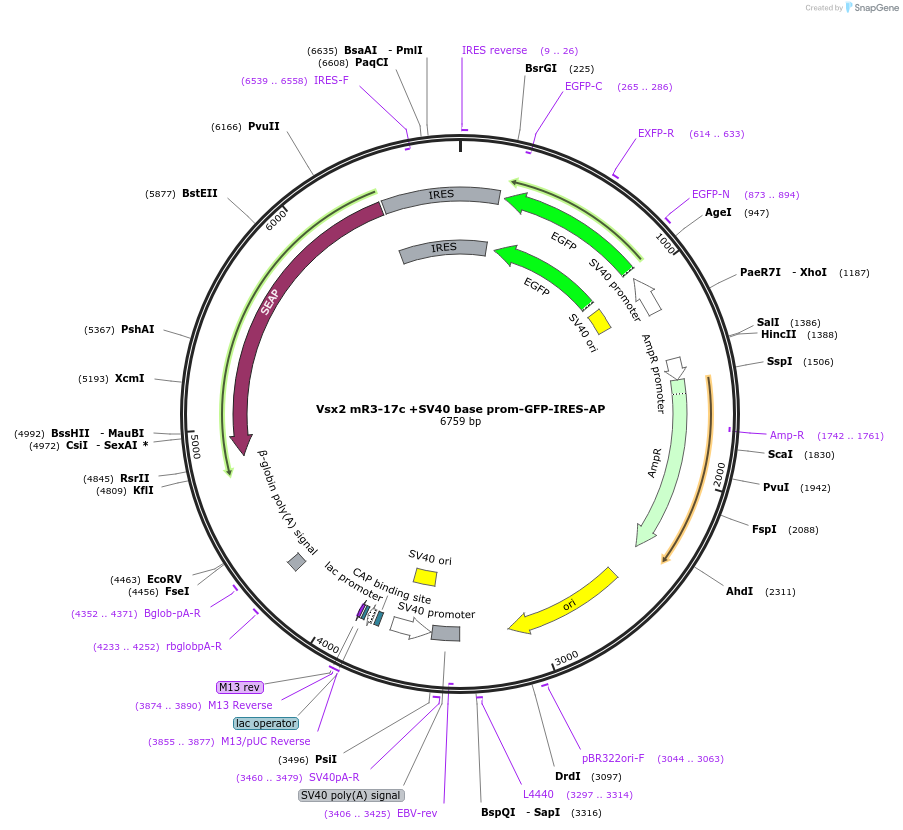
Vsx2 mR3-17c +SV40 base prom-GFP-IRES-AP
Plasmid#225890PurposeEnhancer reporterDepositorInsertmR3-17c
TagsEGFPExpressionMammalianAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
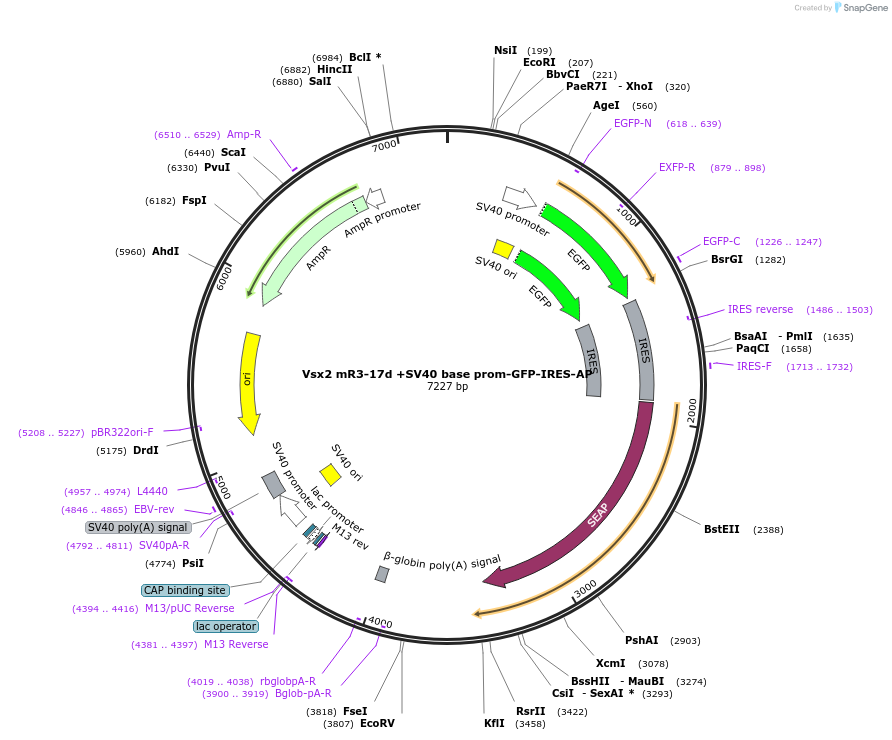
Vsx2 mR3-17d +SV40 base prom-GFP-IRES-AP
Plasmid#225891PurposeEnhancer reporterDepositorInsertmR3-17d
TagsEGFPExpressionMammalianAvailable SinceApril 18, 2025AvailabilityAcademic Institutions and Nonprofits only -
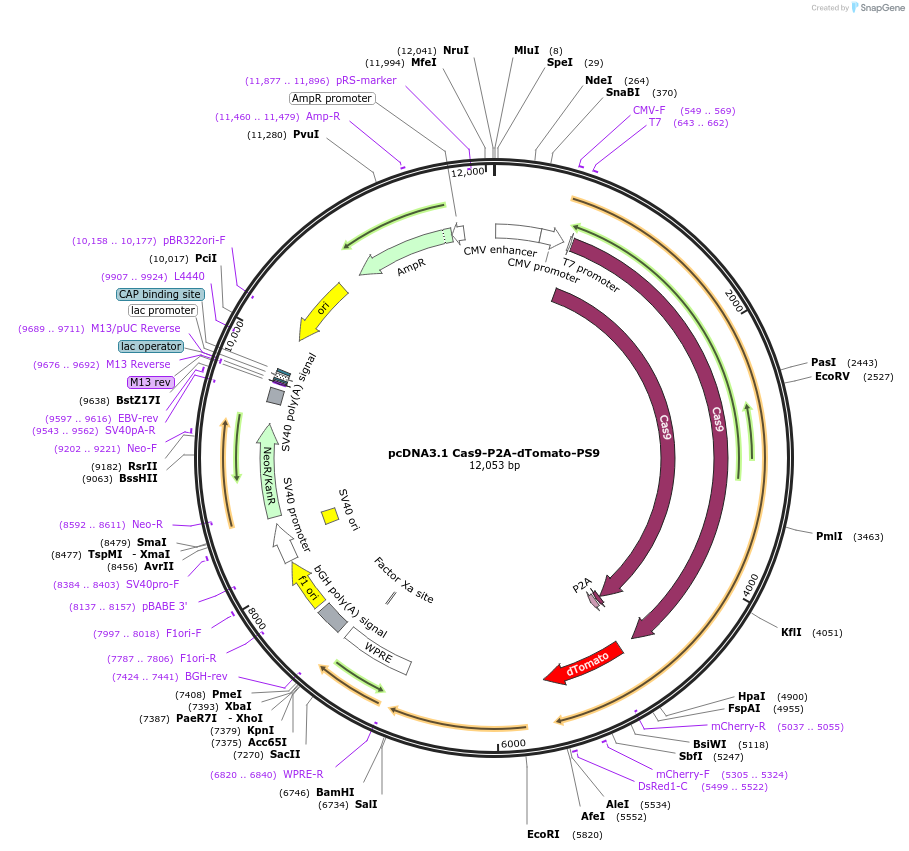
pcDNA3.1 Cas9-P2A-dTomato-PS9
Plasmid#231911PurposeFor packaging spCas9, dTomato mRNA into SARS-CoV-2 virus-like particles (VLPs) with a 'PS9' packaging signalDepositorInsertCas9-P2A-dTomato
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceMarch 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
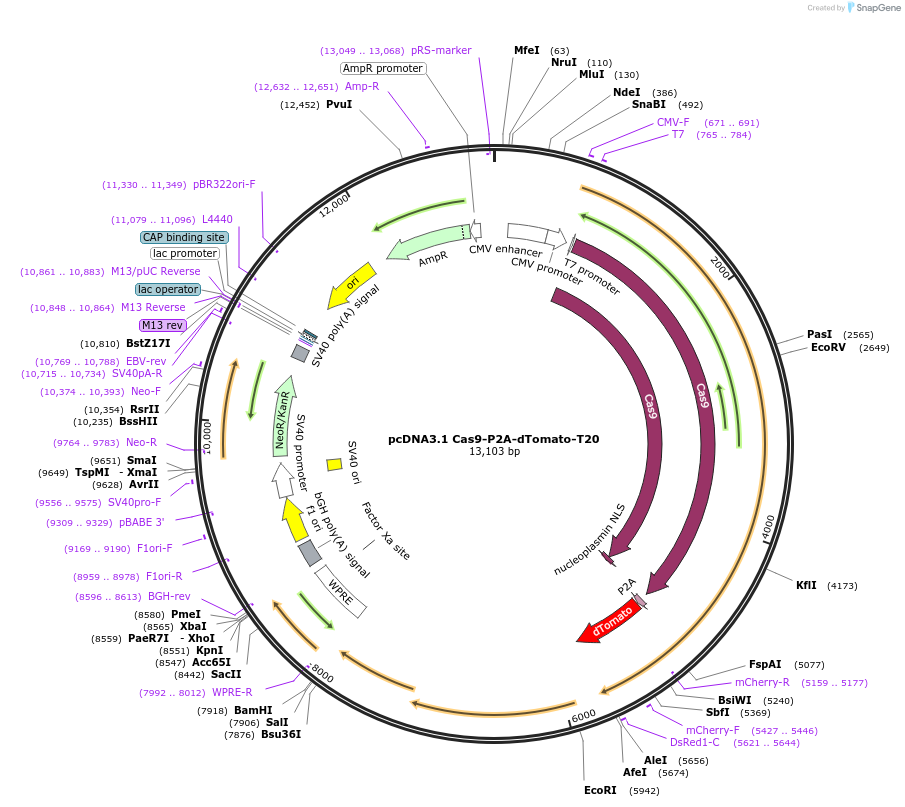
pcDNA3.1 Cas9-P2A-dTomato-T20
Plasmid#231912PurposeFor packaging spCas9, dTomato mRNA into SARS-CoV-2 virus-like particles (VLPs) with a 'T20' packaging signalDepositorInsertCas9-P2A-dTomato
UseCRISPRExpressionMammalianPromoterCMVAvailable SinceMarch 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
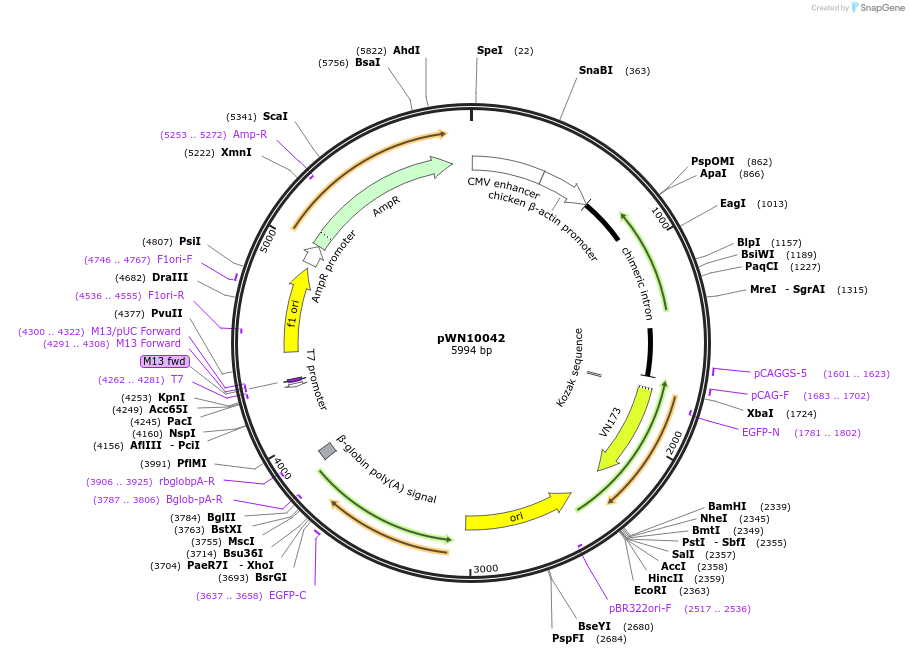
pWN10042
Plasmid#89052PurposeGF-ori-FP plasmid for GFxFP assayDepositorInsertsEGFP part 1
EGFP part 2
UseCRISPRExpressionMammalianPromoterCAG and noneAvailable SinceApril 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
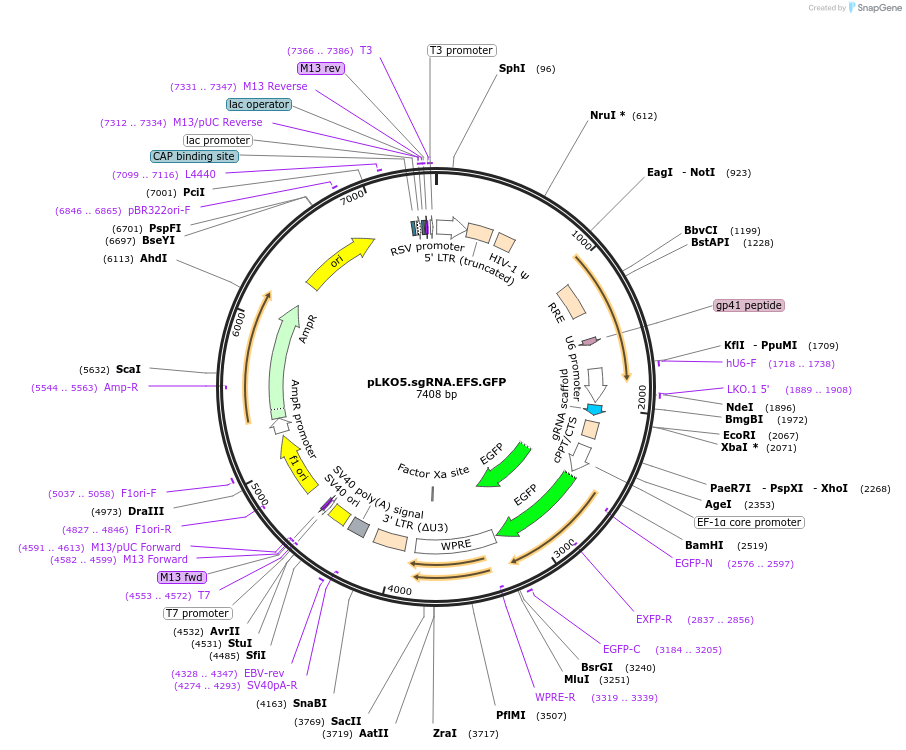
pLKO5.sgRNA.EFS.GFP
Plasmid#57822Purpose3rd generation lentiviral Vector for Sp sgRNA delivery without SpCas9, GFP, EFS Promoter drivenDepositorInsertssgRNA
EFS
eGFP
UseCRISPR and LentiviralAvailable SinceAug. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
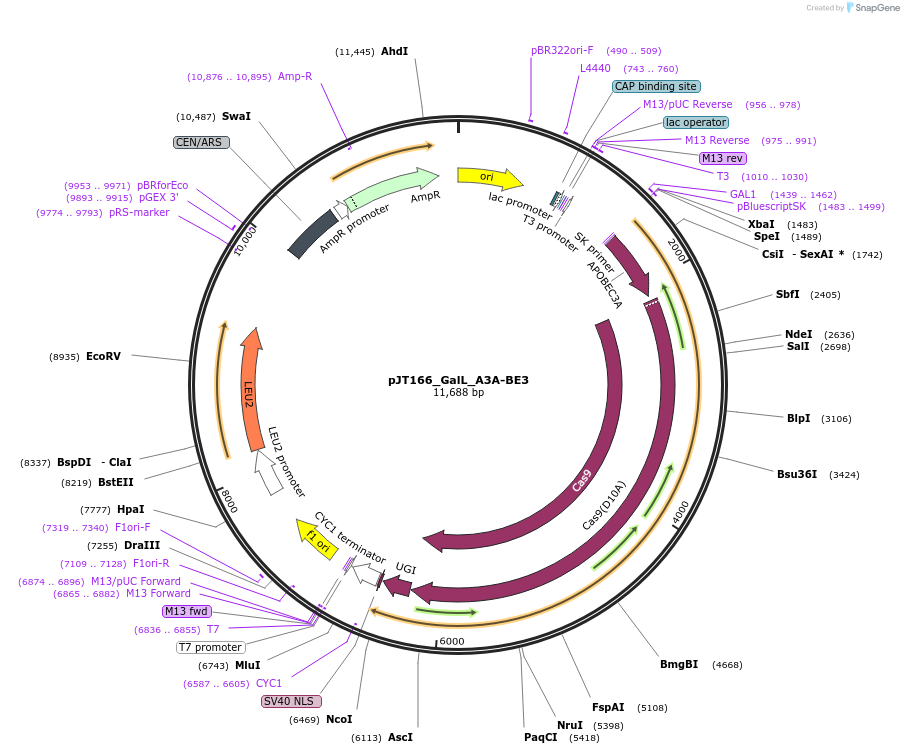
pJT166_GalL_A3A-BE3
Plasmid#145116PurposeExpressing base editor A3A-BE3 in yeast cellsDepositorInsertA3A-BE3
UseCRISPRExpressionYeastMutationspCas9(D10A)PromoterGalLAvailable SinceJuly 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
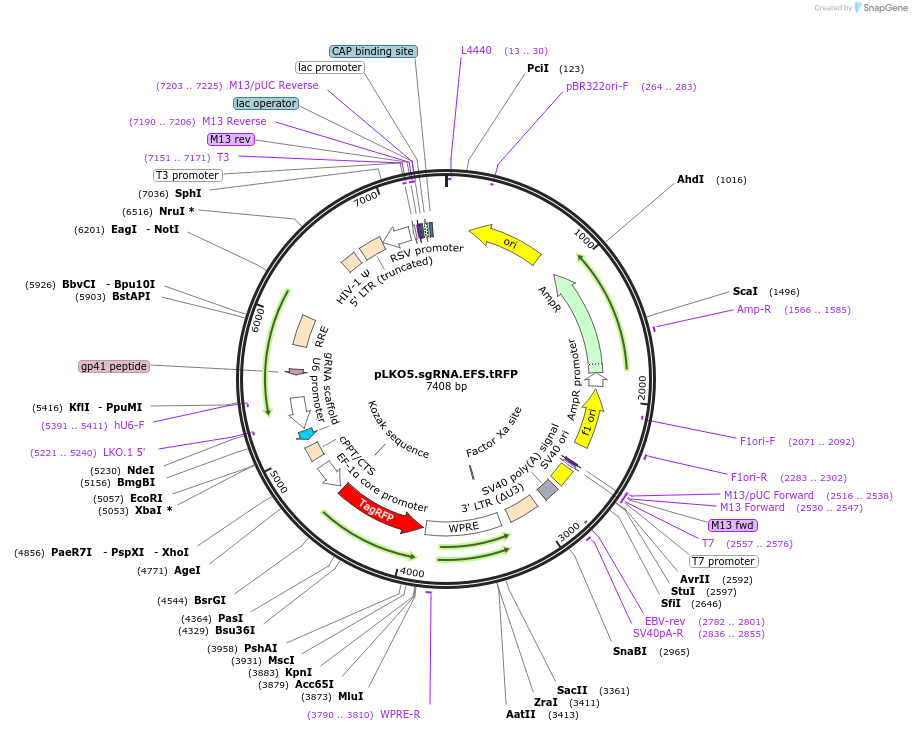
pLKO5.sgRNA.EFS.tRFP
Plasmid#57823Purpose3rd generation Lentiviral Vector for Sp sgRNA delivery without SpCas9, tagRFP, EFS Promoter drivenDepositorInsertssgRNA
EFS
tagRFP
UseCRISPR and LentiviralAvailable SinceAug. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
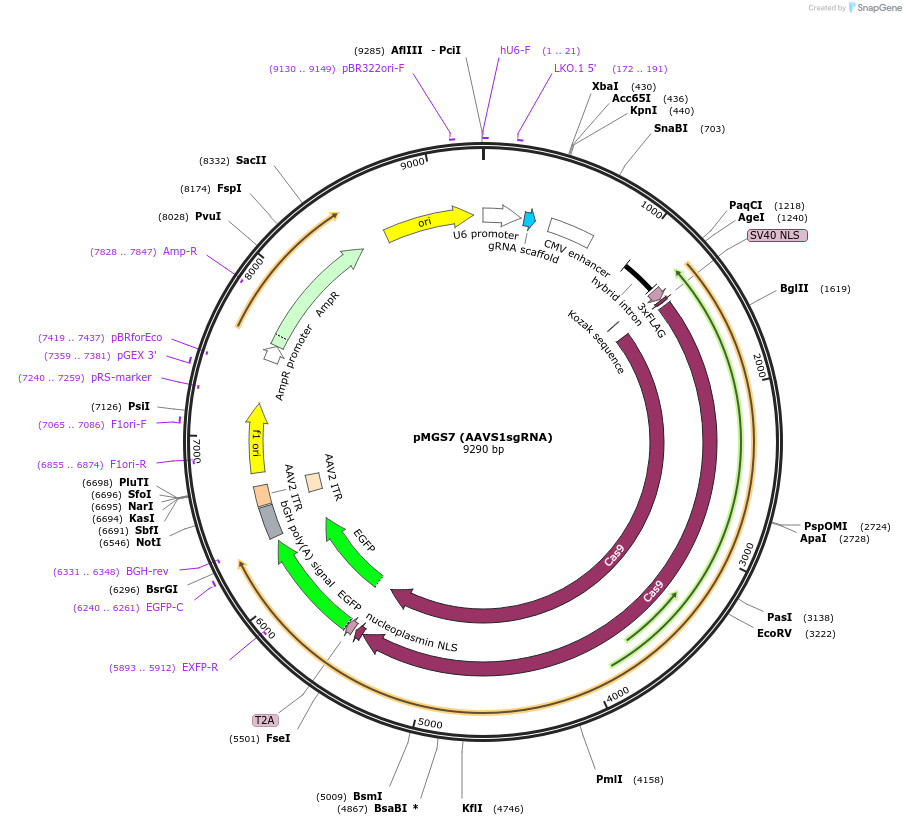
pMGS7 (AAVS1sgRNA)
Plasmid#126582PurposeA CRISPR/Cas9 construct to target the AAVS1 locus in humansDepositorInsertAAVS1 sgRNA
ExpressionMammalianAvailable SinceJune 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
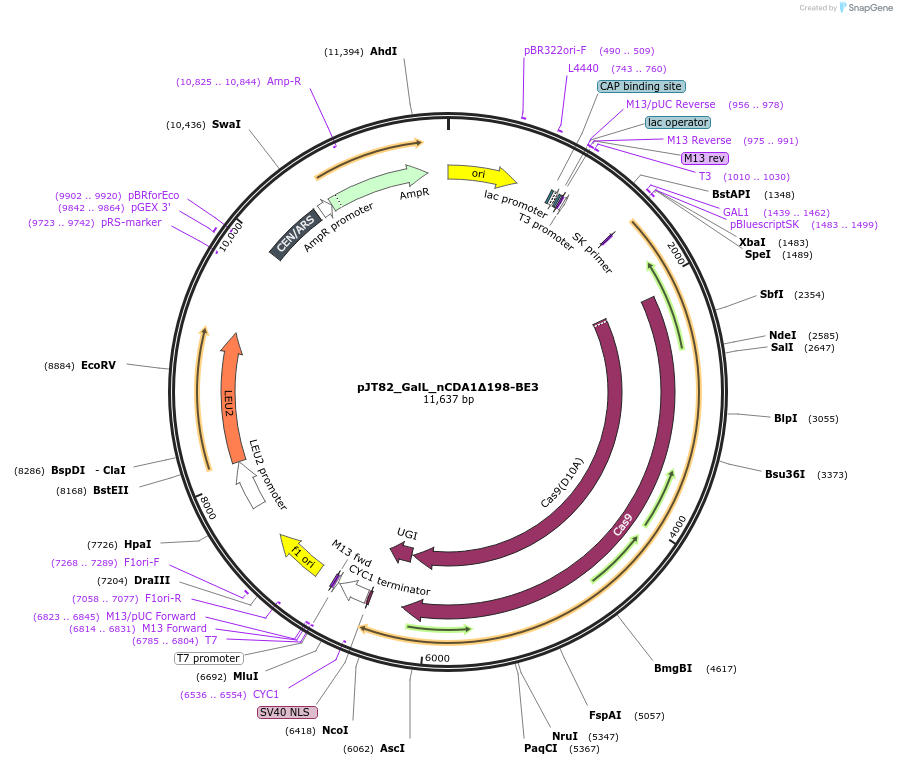
pJT82_GalL_nCDA1Δ198-BE3
Plasmid#145046PurposeExpresses nCDA1Δ198-BE3 in yeast cellsDepositorInsertnCDA1Δ198-BE3
UseCRISPRExpressionYeastMutationpmCDA1(1-198AA); spCas9(D10A)PromoterGalLAvailable SinceJuly 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
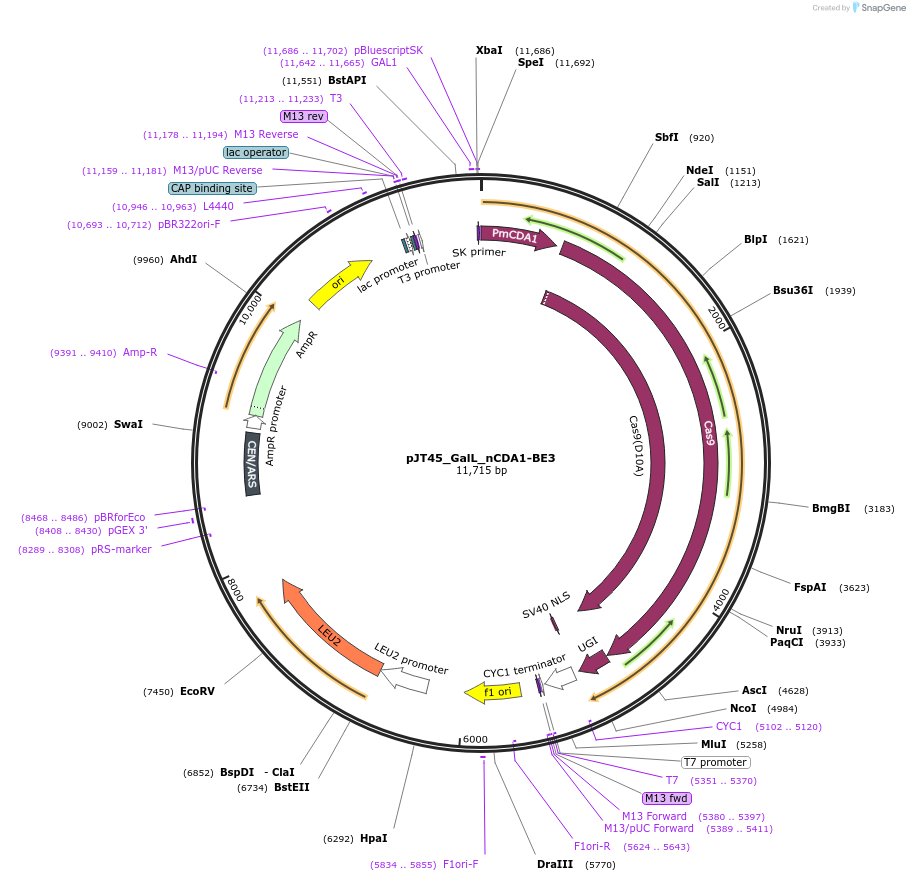
pJT45_GalL_nCDA1-BE3
Plasmid#145038PurposeExpresses nCDA1-BE3 in yeast cellsDepositorInsertnCDA1-BE3
UseCRISPRExpressionYeastMutationspCas9(D10A)PromoterGalLAvailable SinceSept. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
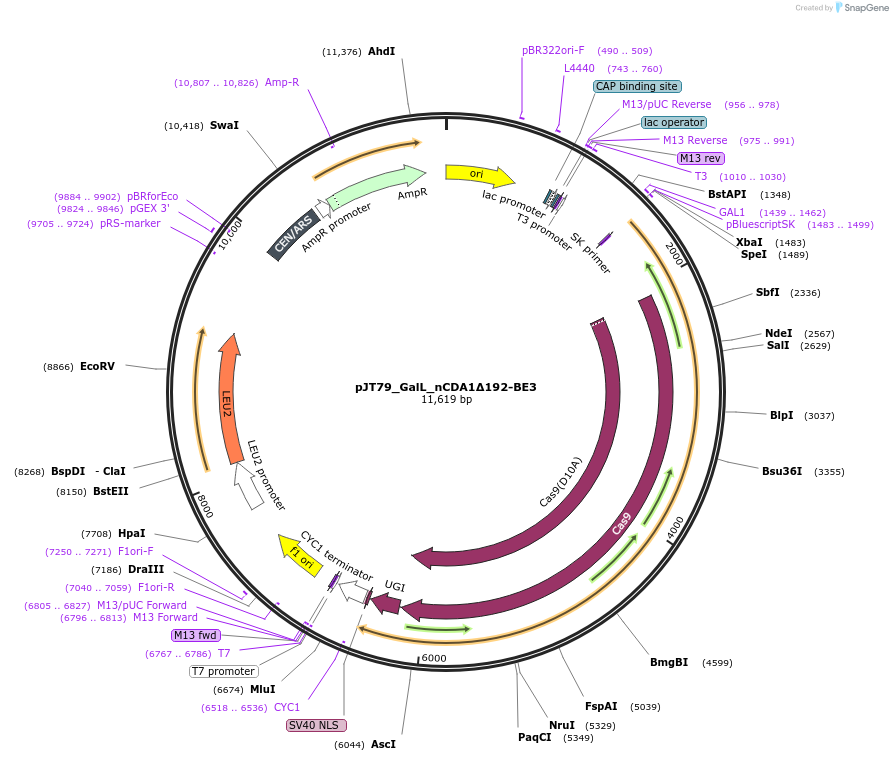
pJT79_GalL_nCDA1Δ192-BE3
Plasmid#145045PurposeExpresses nCDA1Δ192-BE3 in yeast cellsDepositorInsertnCDA1Δ192-BE3
UseCRISPRExpressionYeastMutationpmCDA1(1-192AA); spCas9(D10A)PromoterGalLAvailable SinceJuly 10, 2020AvailabilityAcademic Institutions and Nonprofits only