We narrowed to 4,187 results for: LYC;
-
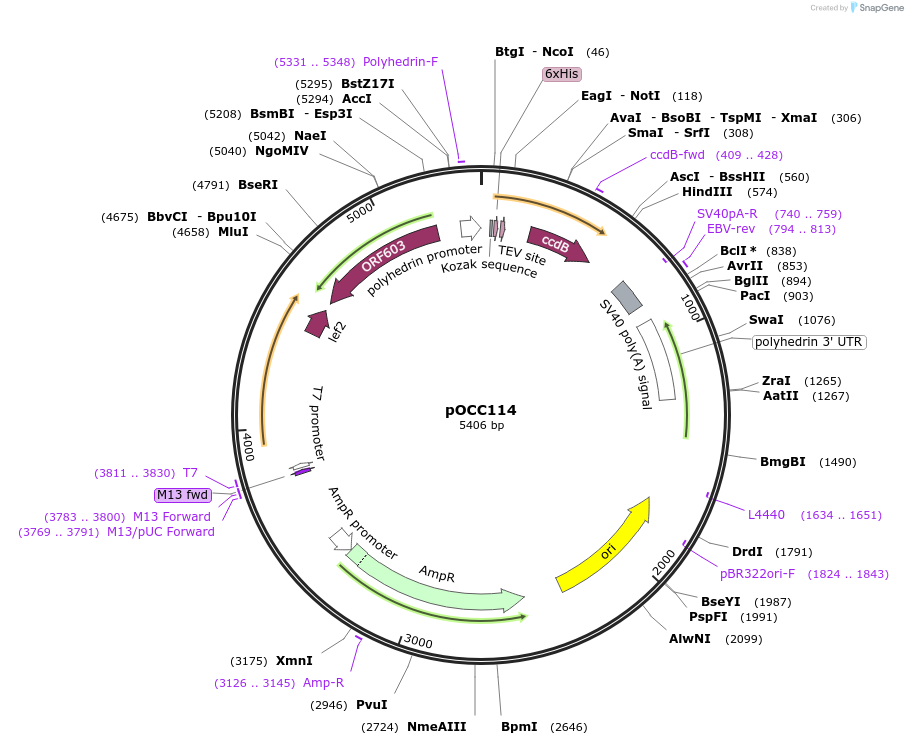
Plasmid#118904Purposeshuttle vector for baculovirus production, using FlashBac bacmid; for recombinant protein with N-terminal sortase siteDepositorInsertNcoI-HIS6-TEV-GG-NotI-ccdB-AscI-stop-HindIII cassette
TagsHIS6, cleavable with TEV protease, two glycinesExpressionInsectPromoterpolHAvailable SinceJan. 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
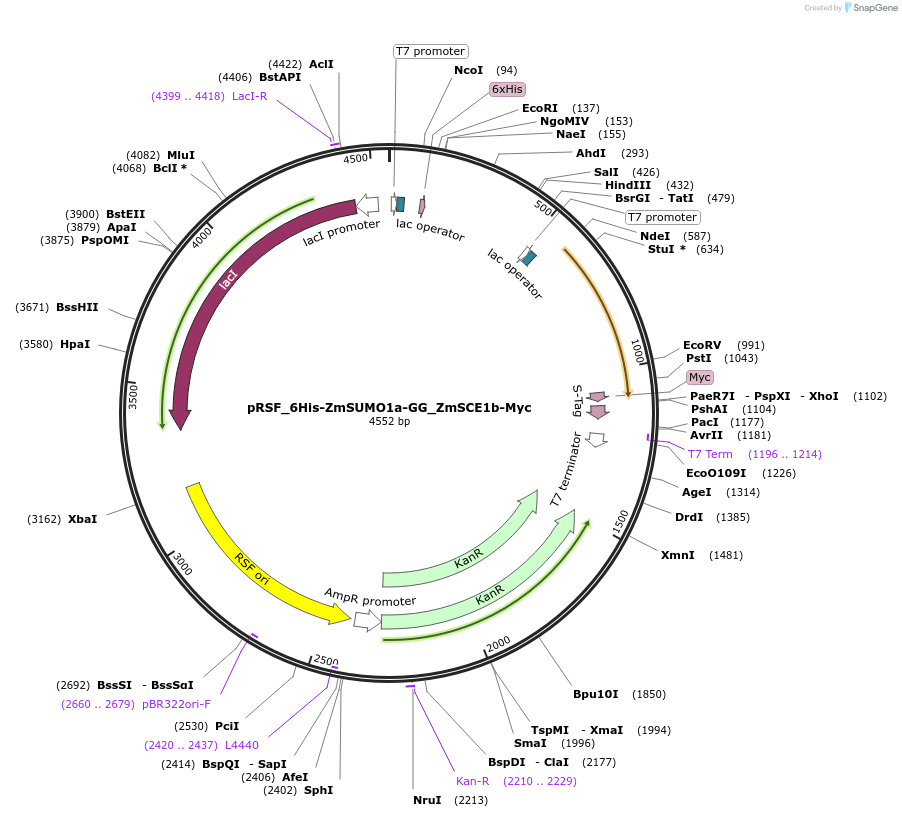
pRSF_6His-ZmSUMO1a-GG_ZmSCE1b-Myc
Plasmid#91932PurposeBacterial expression of maize SUMO1a and SCE1bDepositorInserts6His-ZmSUMO1a-GG
ZmSCE1b-Myc
Tags6His and MycExpressionBacterialMutationTruncation of ZmSUMO1a after the diGlycine motifAvailable SinceJune 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
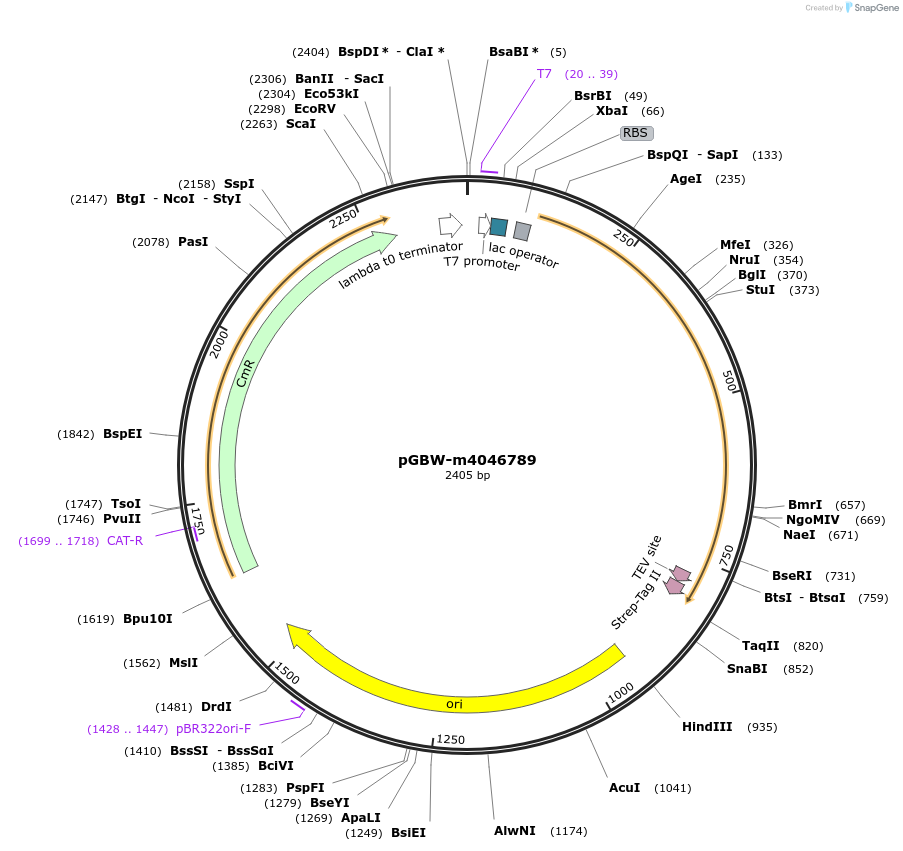
pGBW-m4046789
Plasmid#145660PurposeBacterial Expression plasmid for SARS-CoV-2 M (membrane)DepositorInsertSARS-CoV-2 M (membrane) (M Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;StrepIIExpressionBacterialMutationEscherichia coli recode 1PromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
pV2A-T
Plasmid#153022PurposeSingle-plasmid multi-gene expression system suitable for fungi. The obtained polycistronic gene is expressed under the control of a TetON inducible promoterDepositorTypeEmpty backboneExpressionBacterial and YeastAvailable SinceOct. 8, 2020AvailabilityAcademic Institutions and Nonprofits only -
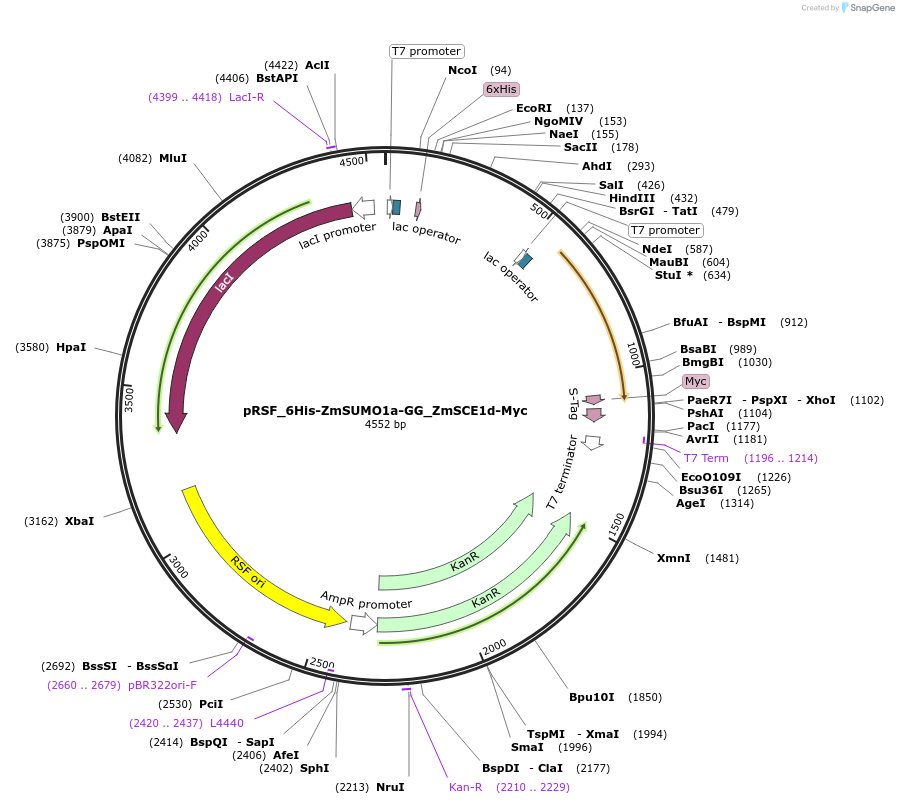
pRSF_6His-ZmSUMO1a-GG_ZmSCE1d-Myc
Plasmid#91934PurposeBacterial expression of maize SUMO1a and SCE1dDepositorInserts6His-ZmSUMO1a-GG
ZmSCE1d-Myc
Tags6His and MycExpressionBacterialMutationTruncation of ZmSUMO1a after the diGlycine motifAvailable SinceJune 28, 2017AvailabilityAcademic Institutions and Nonprofits only -
pGBW-m4046763
Plasmid#145606PurposeBacterial Expression plasmid for SARS-CoV-2 M (membrane)DepositorInsertSARS-CoV-2 M (membrane) (M Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
TagsCleavable TEV;FLAG_2ExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceApril 29, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
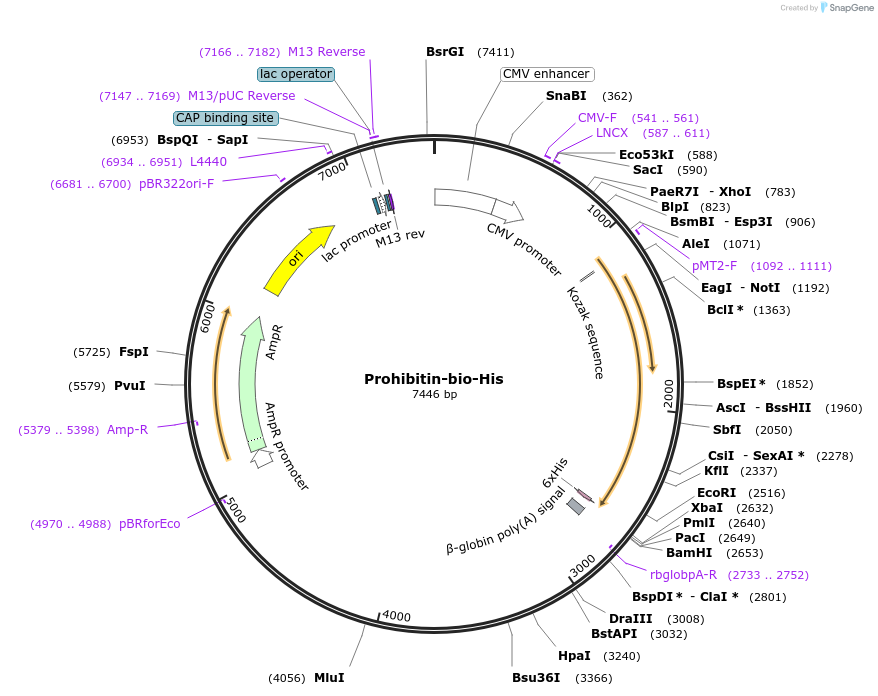
Prohibitin-bio-His
Plasmid#50800PurposeExpresses enzymatically monobiotinylated full length ProhibitinDepositorInsertProhibitin
TagsratCD4d3+4,biotinylation sequence, 6xHisExpressionMammalianMutationall potential N-linked glycosylation sites (N-X-S…PromoterCMVAvailable SinceFeb. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
3155 pGEM-4Z Bcl-2 cDNA G145A
Plasmid#8751DepositorAvailable SinceNov. 14, 2005AvailabilityAcademic Institutions and Nonprofits only -
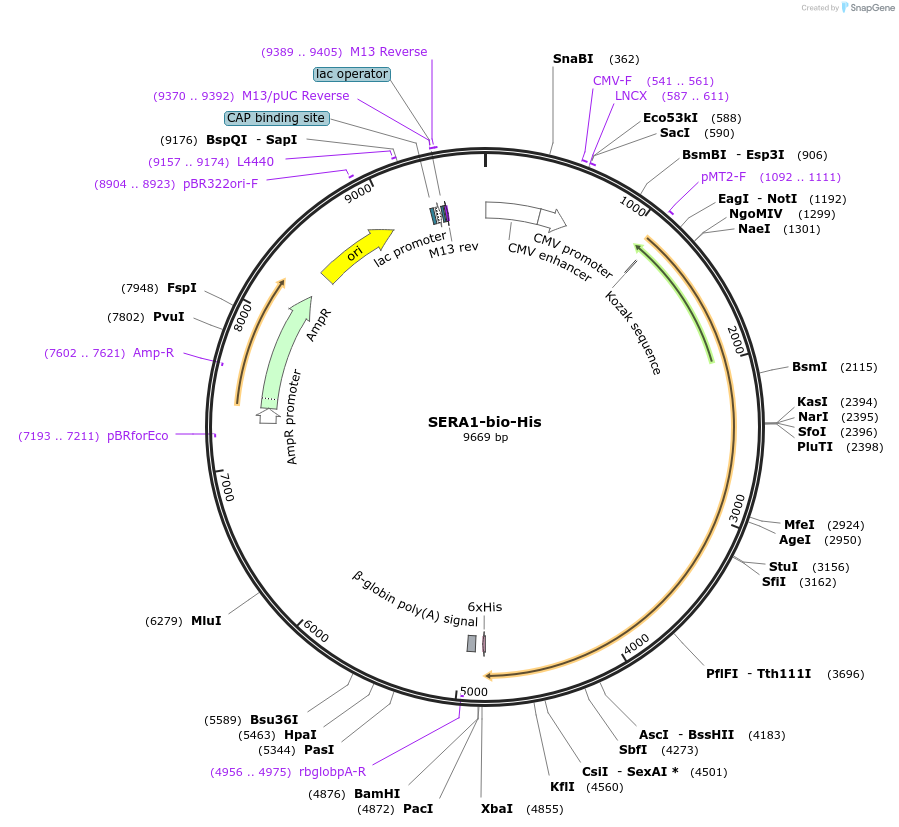
SERA1-bio-His
Plasmid#50812PurposeExpresses enzymatically monobiotinylated full length SERA1DepositorInsertSERA1
TagsratCD4d3+4,biotinylation sequence, 6xHisExpressionMammalianMutationall potential N-linked glycosylation sites (N-X-S…PromoterCMVAvailable SinceFeb. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
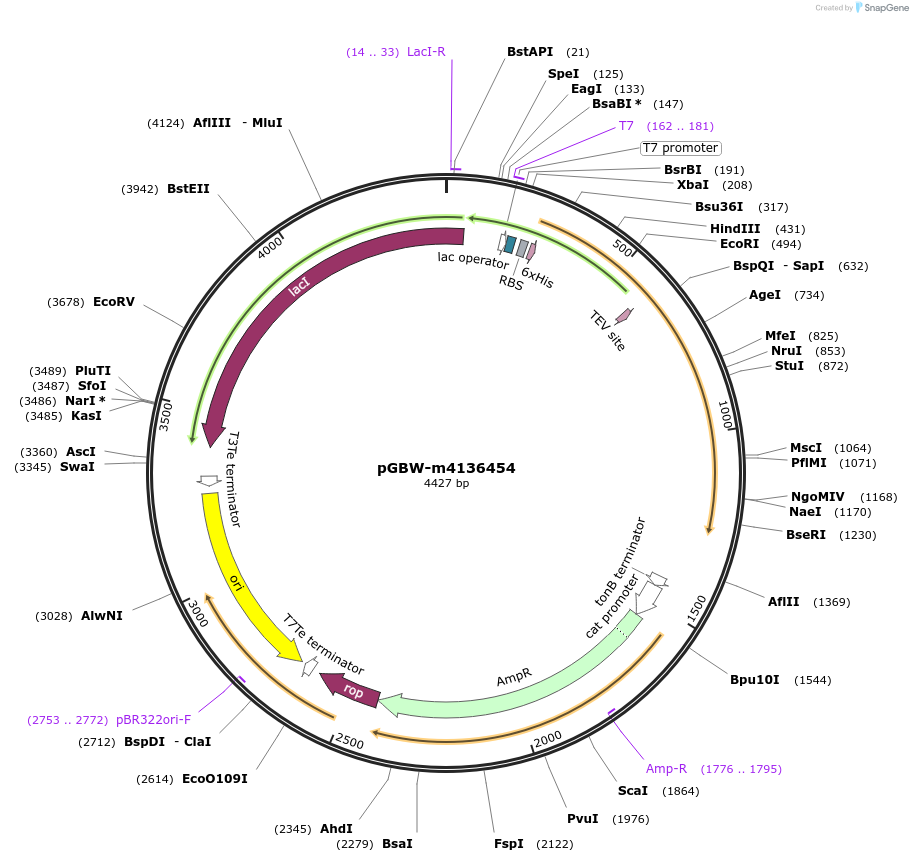
pGBW-m4136454
Plasmid#149529PurposeBacterial expression plasmid for SARS-CoV-2 M (membrane)DepositorInsertSARS-CoV-2 M (membrane) (M Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
Tags6xHIS;Linker_GGSG;SUMO tag;Linker_GGSG;Cleavable …ExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceMay 13, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
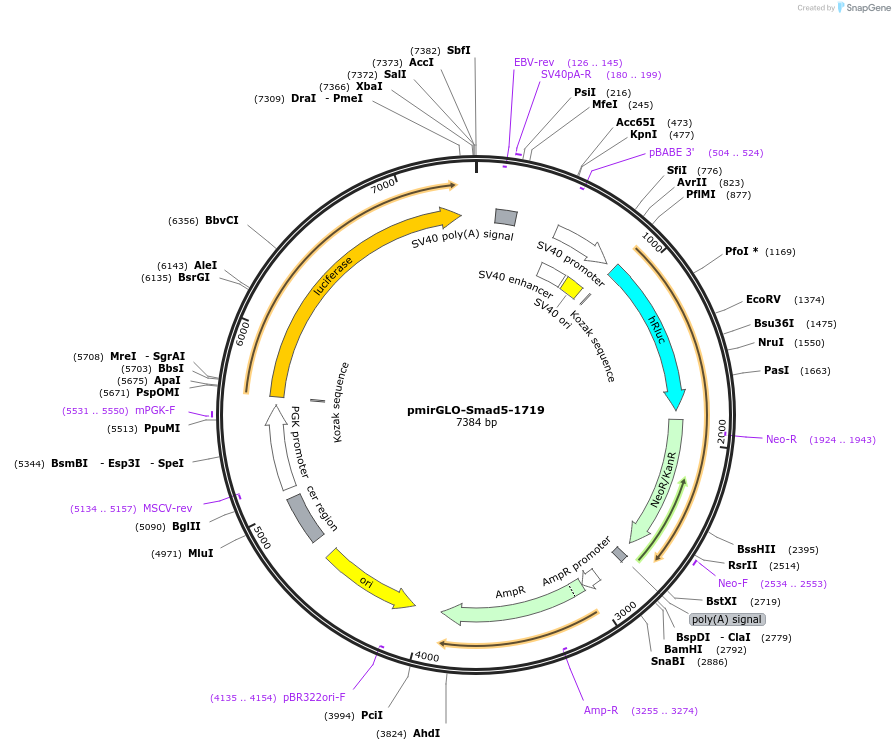
pmirGLO-Smad5-1719
Plasmid#49382PurposeInsert is a fragment of the Smad5 3'-UTR that contains miR-17 and miR-19 binding sites. To be used for 3'UTR assay.DepositorInsertSmad5 (Smad5 Human, Mouse)
UseLuciferasePromoterHuman phosphoglycerate kinase (PGK) promoterAvailable SinceOct. 17, 2016AvailabilityAcademic Institutions and Nonprofits only -
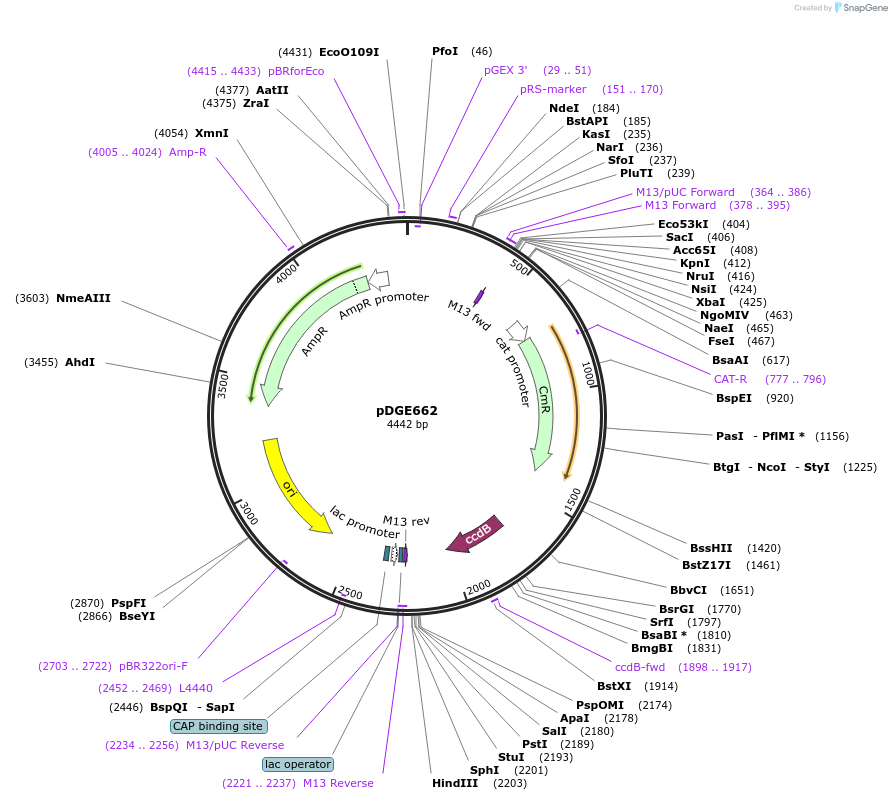
pDGE662
Plasmid#153260PurposeM6 shuttle vector containing the Solanum lycopersicum U6 promoter for preparation of single guide RNA transcriptional units by cloning of hybridized oligonucleotides.DepositorTypeEmpty backboneUseCRISPRAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
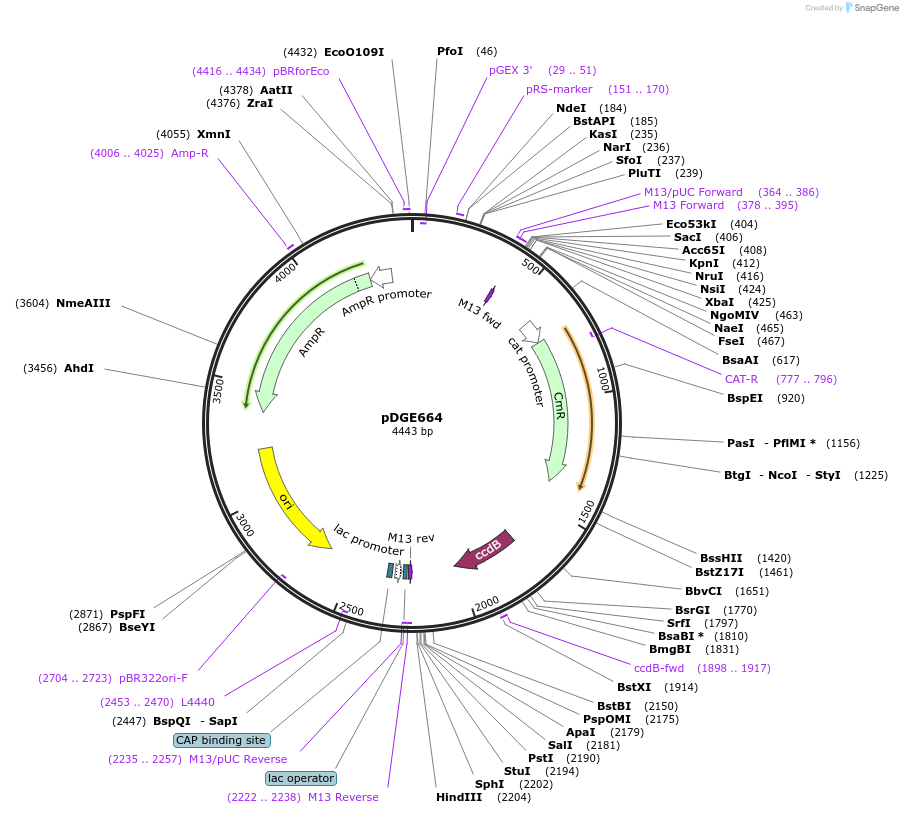
pDGE664
Plasmid#153262PurposeM7 shuttle vector containing the Solanum lycopersicum U6 promoter for preparation of single guide RNA transcriptional units by cloning of hybridized oligonucleotides.DepositorTypeEmpty backboneUseCRISPRAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
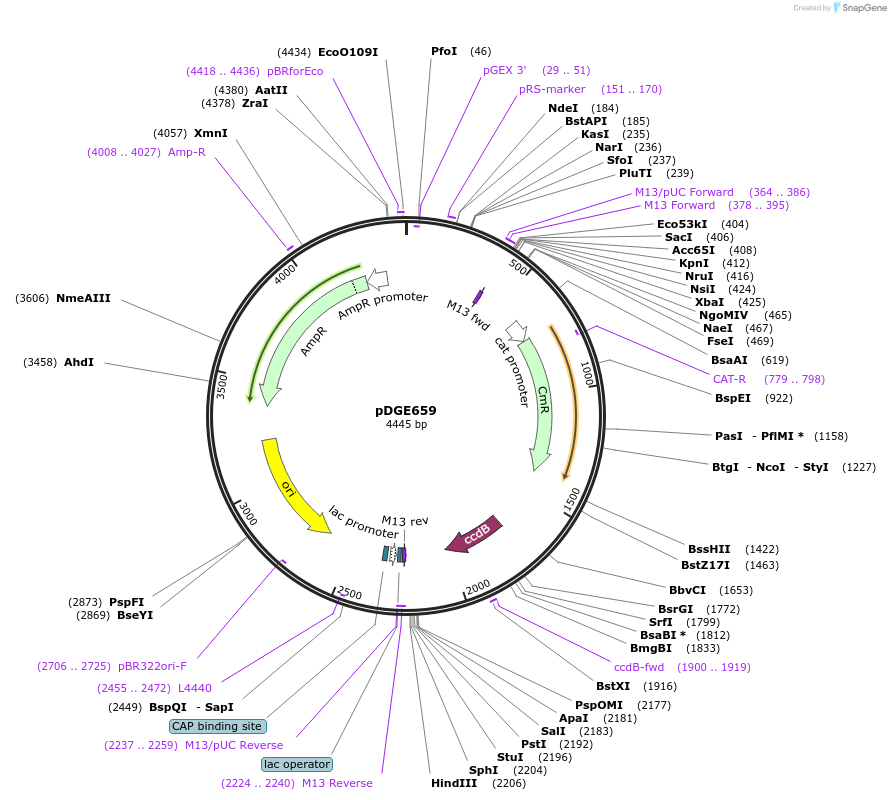
pDGE659
Plasmid#153257PurposeM4 shuttle vector containing the Solanum lycopersicum U6 promoter for preparation of single guide RNA transcriptional units by cloning of hybridized oligonucleotides.DepositorTypeEmpty backboneUseCRISPRAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
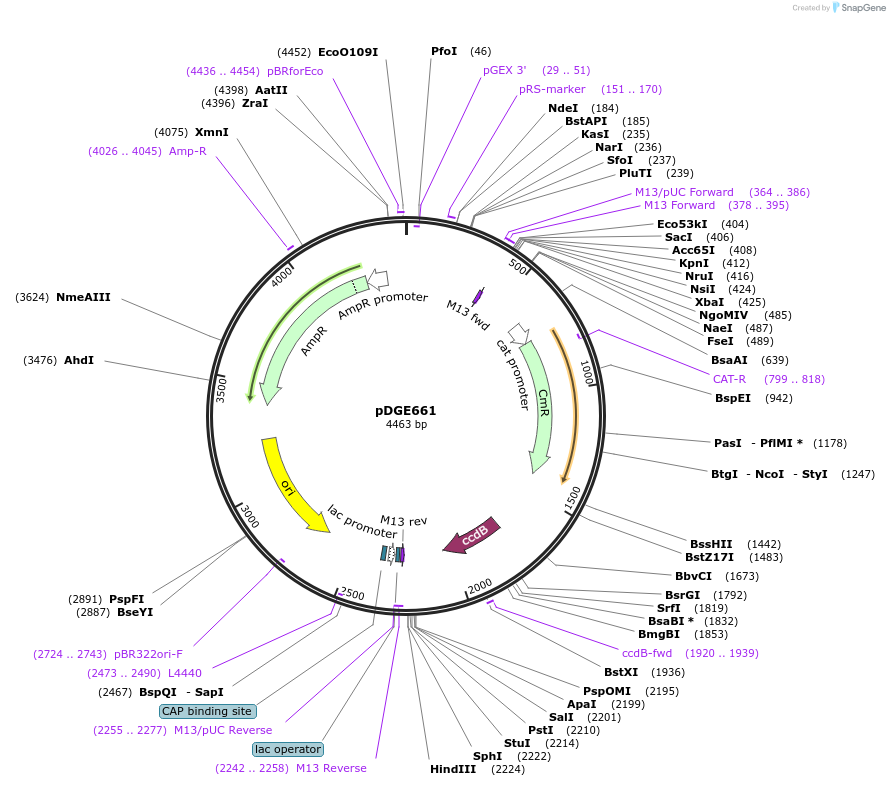
pDGE661
Plasmid#153259PurposeM5 shuttle vector containing the Solanum lycopersicum U6 promoter for preparation of single guide RNA transcriptional units by cloning of hybridized oligonucleotides.DepositorTypeEmpty backboneUseCRISPRAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
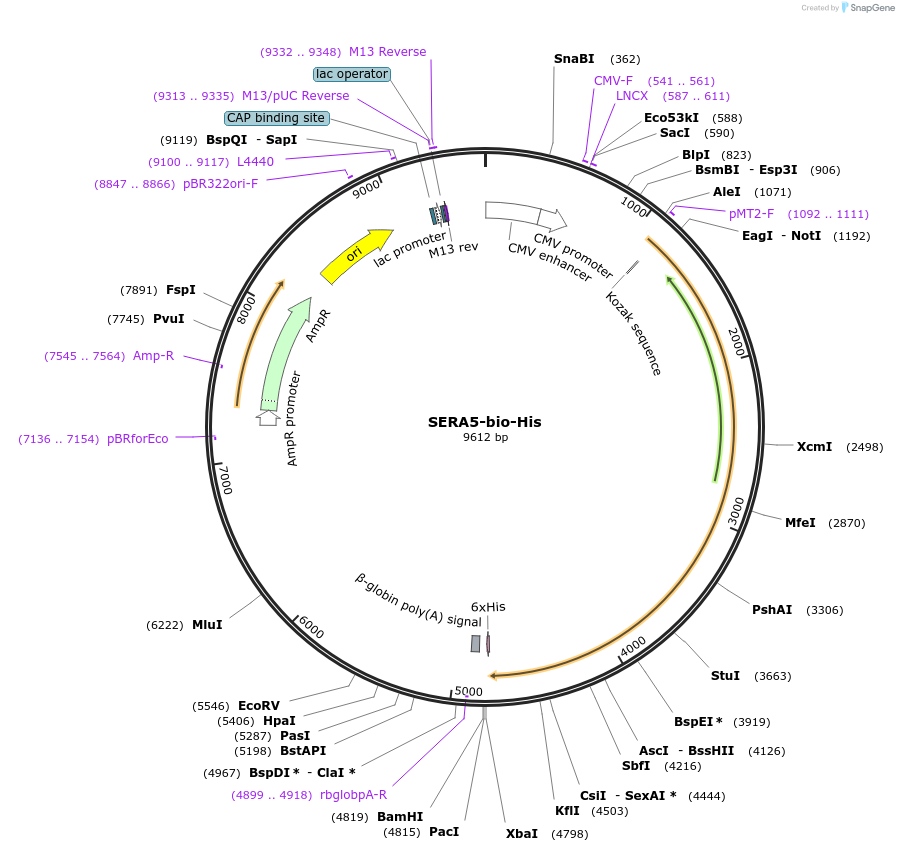
SERA5-bio-His
Plasmid#50817PurposeExpresses enzymatically monobiotinylated full length SERA5DepositorInsertSERA5
TagsratCD4d3+4,biotinylation sequence, 6xHisExpressionMammalianMutationall potential N-linked glycosylation sites (N-X-S…PromoterCMVAvailable SinceFeb. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
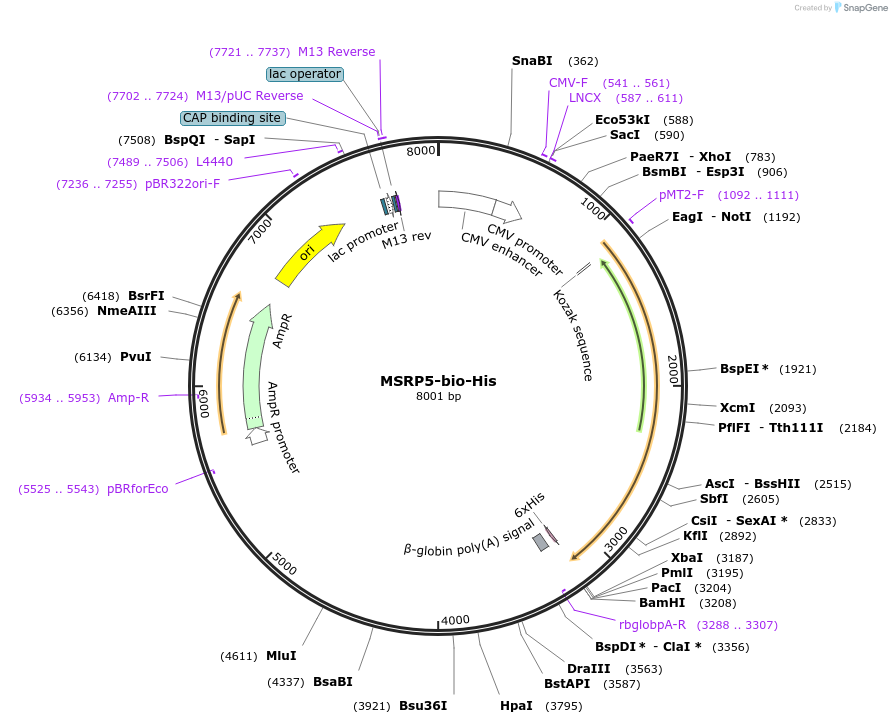
MSRP5-bio-His
Plasmid#50805PurposeExpresses enzymatically monobiotinylated full length MSRP5DepositorInsertMSRP5
TagsratCD4d3+4,biotinylation sequence, 6xHisExpressionMammalianMutationall potential N-linked glycosylation sites (N-X-S…Available SinceFeb. 13, 2014AvailabilityAcademic Institutions and Nonprofits only -
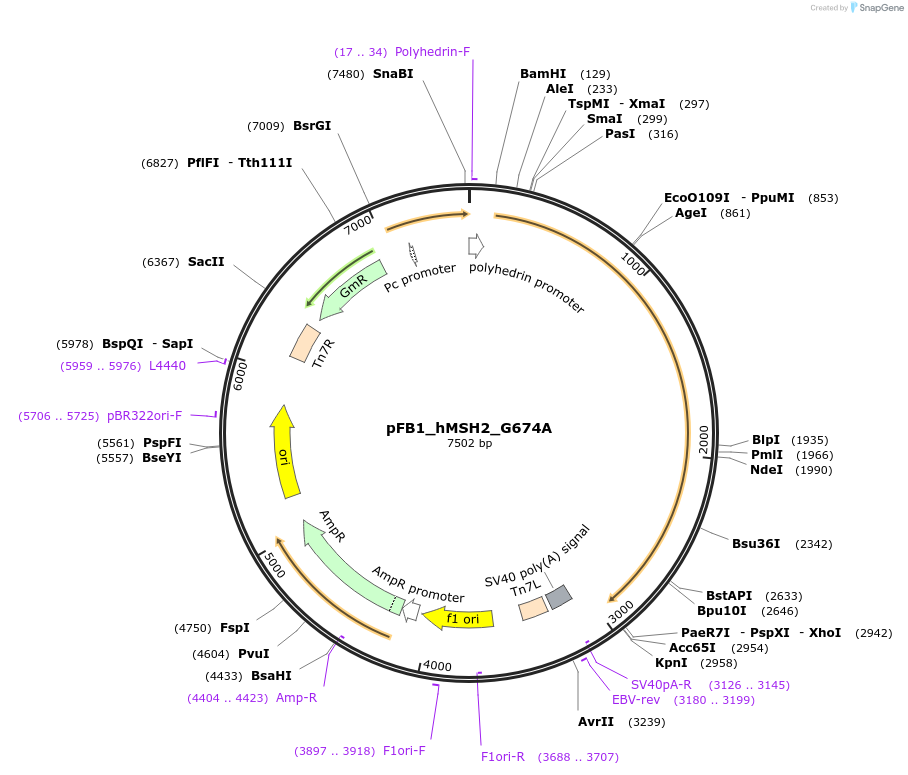
pFB1_hMSH2_G674A
Plasmid#134874PurposeExpression human MSH2 G674 mutant protein using baculovirusDepositorInserthuman MSH2_G674A (MSH2 Human)
ExpressionInsectMutationChanged Glycine to Alanine at aa#674Available SinceJan. 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
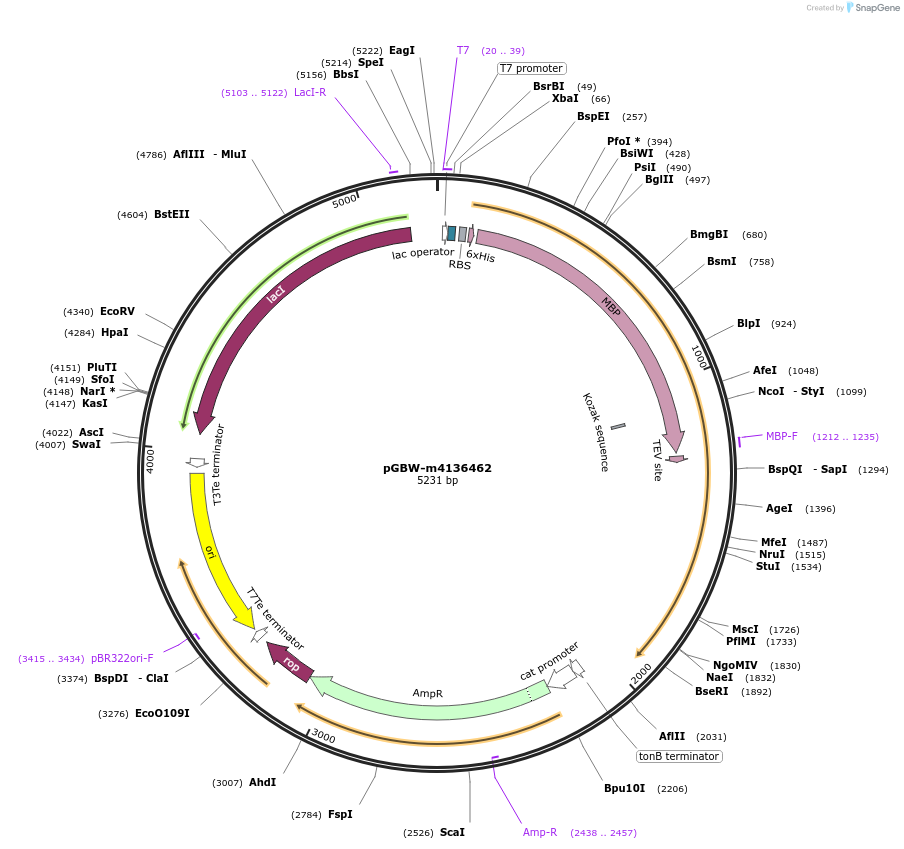
pGBW-m4136462
Plasmid#149533PurposeBacterial expression plasmid for SARS-CoV-2 M (membrane)DepositorInsertSARS-CoV-2 M (membrane) (M Escherichia coli str. K-12 substr. MG1655; Severe acute respiratory syndrome coronavirus 2)
Tags6xHIS;Linker_GGSG;MBP;Linker_GGSG;Cleavable TEVExpressionBacterialMutationwtPromoterPromoter | pT7 ; Operator | lacO ; RBS | T7_rbs ;…Available SinceMay 14, 2020AvailabilityIndustry, Academic Institutions, and Nonprofits -
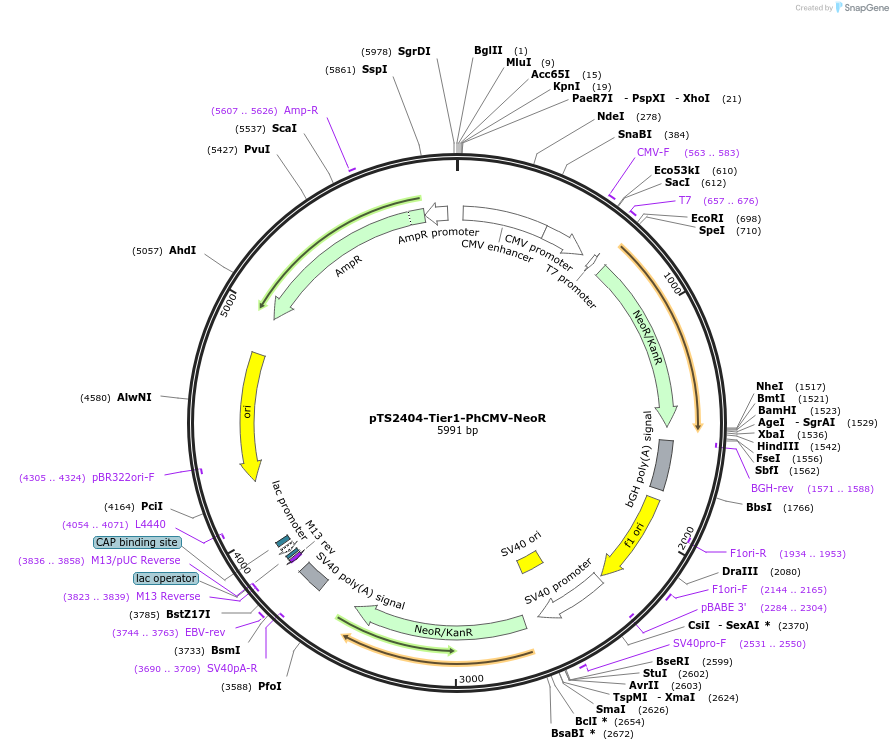
pTS2404-Tier1-PhCMV-NeoR
Plasmid#169496PurposeTier-1 vector encoding PhCMV-driven neomycin resistance gene NeoR (PhCMV-NeoR-pA).DepositorInsertPCMV-driven aminoglycoside 3'-phosphotransferase, APH 3' II
ExpressionMammalianPromoterPhCMVAvailable SinceJune 16, 2022AvailabilityAcademic Institutions and Nonprofits only