We narrowed to 75,518 results for: LAS
-
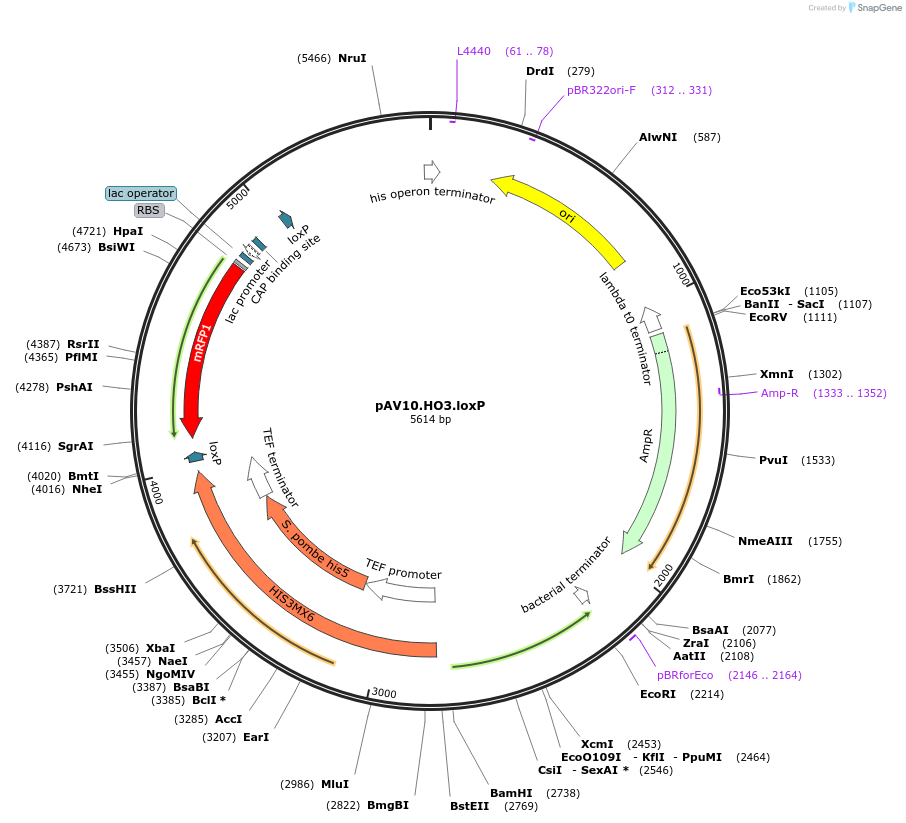
Plasmid#63205PurposepCACS backbone (Amp), SpHIS5, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into HO locus (NotI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only
-
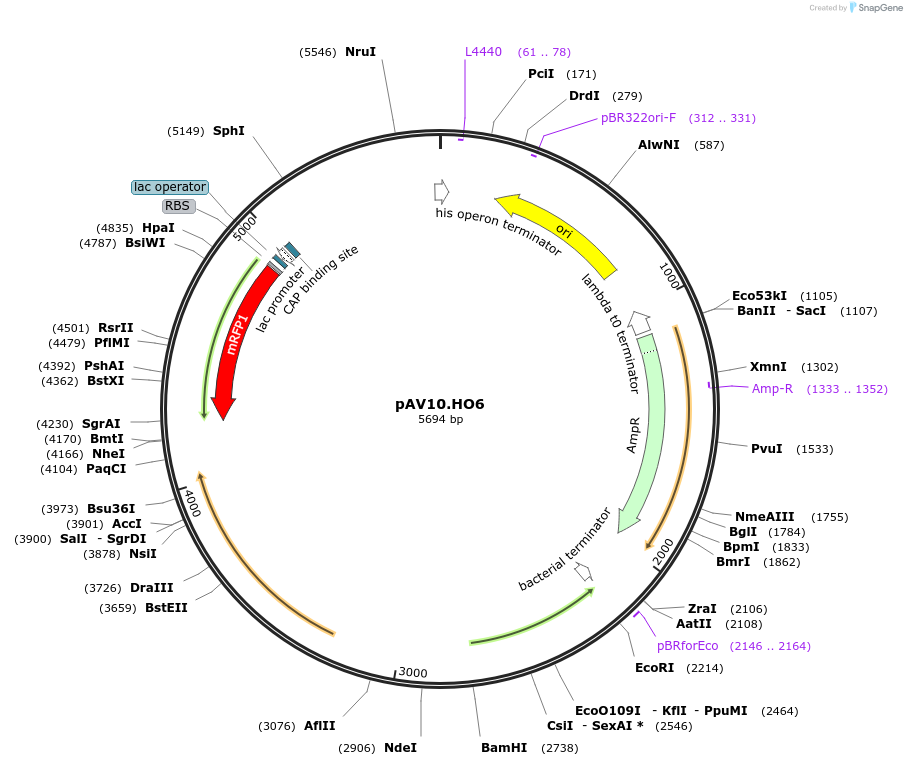
pAV10.HO6
Plasmid#63211PurposepCACS backbone (Amp), KlURA3, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. Integration into HO locus (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
delta guaB
Bacterial Strain#113653PurposeBase strain that the pDCAF plasmids (Addgene #113646-113652) must be in.DepositorBacterial ResistanceKanamycinAvailable SinceNov. 18, 2019AvailabilityIndustry, Academic Institutions, and Nonprofits -
UQ4802
Bacterial Strain#37160DepositorBacterial ResistanceAmpicillinSpeciesBurkholderia xenovoransAvailable SinceJune 21, 2012AvailabilityAcademic Institutions and Nonprofits only -
Strain HK022 JQ929581
Bacterial Strain#38204DepositorBacterial ResistanceKanamycinSpeciesSyntheticAvailable SinceJuly 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
Strain Phi80 JQ929581
Bacterial Strain#38202DepositorBacterial ResistanceKanamycinSpeciesSyntheticAvailable SinceJuly 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
UQ4884
Bacterial Strain#37161DepositorBacterial ResistanceAmpicillinSpeciesBurkholderia xenovoransAvailable SinceJune 21, 2012AvailabilityAcademic Institutions and Nonprofits only -
Strain Phi80 JQ929582
Bacterial Strain#38203DepositorBacterial ResistanceKanamycinSpeciesSyntheticAvailable SinceJuly 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
UQ4861
Bacterial Strain#37163DepositorBacterial ResistanceAmpicillinSpeciesBurkholderia xenovoransAvailable SinceJuly 16, 2012AvailabilityAcademic Institutions and Nonprofits only -
LX-miR
Pooled Library#112200PurposeTargets 1,594 primary human miRNA genes. The library contains 4-5 gRNAs per gene and 1,000 non-targeting control gRNAs for a total of 8,382 gRNAs.DepositorExpressionMammalianSpeciesHomo sapiensUseLentiviralAvailable SinceSept. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
Human Whole Genome sgRNA iBAR Library
Pooled Library#140633PurposeEnables genome-wide CRISPR screening at high MOI through use of barcoded gRNA plasmids.DepositorUseCRISPR and LentiviralAvailable SinceApril 21, 2020AvailabilityAcademic Institutions and Nonprofits only -
TB204
Bacterial Strain#230033PurposeFluorescently labelled (monomeric sfGFP) E. coliDepositorBacterial ResistanceNoneAvailable SinceFeb. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
BL21∆serB
Bacterial Strain#34929DepositorBacterial ResistanceNoneAvailable SinceFeb. 24, 2012AvailabilityAcademic Institutions and Nonprofits only -
TB201
Bacterial Strain#230031PurposeFluorescently labelled (YFP) E. coliDepositorBacterial ResistanceNoneAvailable SinceFeb. 3, 2025AvailabilityAcademic Institutions and Nonprofits only -
Top10∆serB
Bacterial Strain#34928DepositorBacterial ResistanceNoneAvailable SinceFeb. 24, 2012AvailabilityAcademic Institutions and Nonprofits only -
HL 3325 (=HL 770 + rhyB knockout_KanR)
Bacterial Strain#30065DepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceAug. 20, 2012AvailabilityAcademic Institutions and Nonprofits only -
Z956
Bacterial Strain#200838PurposeHost strain for plasmids pYG205 and pYG215 or derivatives of the latterDepositorBacterial ResistanceNoneSpeciesEscherichia coli str. k-12 substr. mg1655Available SinceJuly 25, 2023AvailabilityAcademic Institutions and Nonprofits only -
p11-5’_10xN_PAM-site3 (RTW572)
Pooled Library#160134PurposePlasmid library harboring a randomized 10nt PAM 5’ of target site 3. Can be used to profile CRISPR-Cas PAM requirements of enzymes that recognize 5’ PAMs (e.g. Cas12a)DepositorSpeciesSyntheticUseCRISPRAvailable SinceFeb. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
p11-3’_8xN_PAM-site1 (RTW554)
Pooled Library#160132PurposePlasmid library harboring a randomized 8nt PAM 3’ of target site 1. Can be used to profile CRISPR-Cas PAM requirements of enzymes that recognize 3’ PAMs (e.g. Cas9)DepositorSpeciesSyntheticUseCRISPRAvailable SinceFeb. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
p11-5’_10xN_PAM-site4 (RTW574)
Pooled Library#160135PurposePlasmid library harboring a randomized 10nt PAM 5’ of target site 4. Can be used to profile CRISPR-Cas PAM requirements of enzymes that recognize 5’ PAMs (e.g. Cas12a)DepositorSpeciesSyntheticUseCRISPRAvailable SinceFeb. 8, 2021AvailabilityAcademic Institutions and Nonprofits only