We narrowed to 421 results for: pet-28
-
Bacterial Strain#30030DepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceAug. 20, 2012AvailabilityAcademic Institutions and Nonprofits only
-
HL 849 (=HL 716 + micC knockout_KanR)
Bacterial Strain#30009DepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceJuly 5, 2012AvailabilityAcademic Institutions and Nonprofits only -
HL 713
Bacterial Strain#52931PurposeMG 1655 + lacIq_KanR inserted into the chromosome at the intS siteDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
HL 751
Bacterial Strain#52932PurposeMG 1655 + lacIq inserted into the chromosome at the intS site + Δhfq_KanRDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
HL 3221
Bacterial Strain#52936PurposeMG 1655 + lacIq inserted into the chromosome at the intS site + ΔoxyS_KanRDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
HL 3387
Bacterial Strain#52937PurposeMG 1655 + lacIq inserted into the chromosome at the intS site + Δhfq + ΔoxyS_KanRDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
HL 852
Bacterial Strain#52933PurposeMG 1655 + lacIq inserted into the chromosome at the intS site + ΔdsrA_KanRDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
HL 1120
Bacterial Strain#52934PurposeMG 1655 + lacIq inserted into the chromosome at the intS site + ΔmicC + Δhfq_KanRDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
HL 1179
Bacterial Strain#52935PurposeMG 1655 + lacIq inserted into the chromosome at the intS site + ΔdsrA + Δhfq_KanRDepositorBacterial ResistanceKanamycinSpeciesEscherichia coliAvailable SinceSept. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
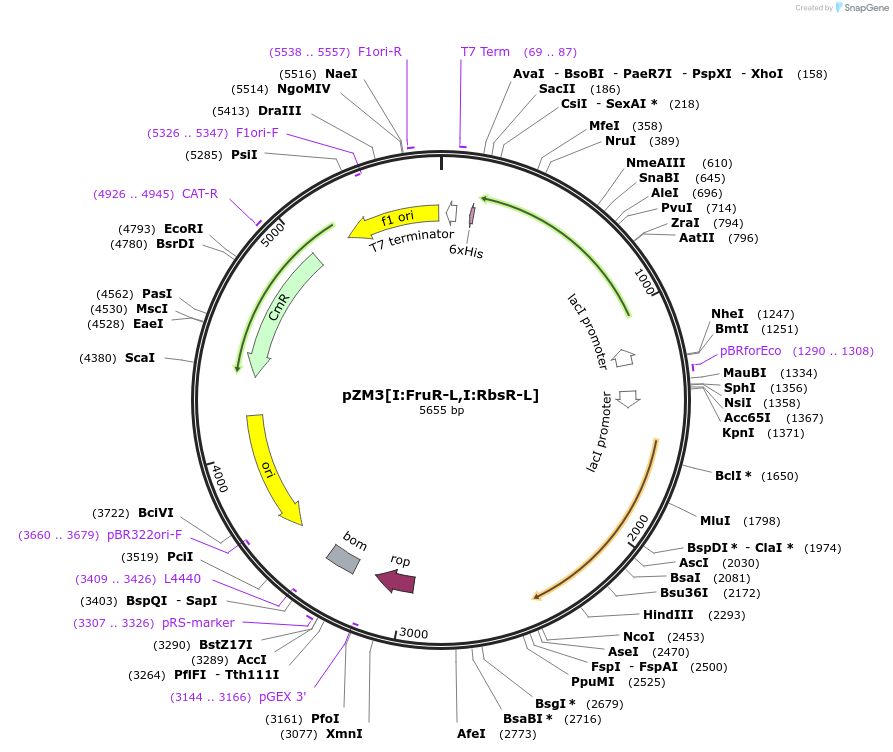
pZM3[I:FruR-L,I:RbsR-L]
Plasmid#60777PurposeContains PI driving expression FruR-L, and PI driving expression of RbsR-L.DepositorInsertsFruR-L
RbsR-L
MutationLacI/GalR repressor chimera. LacI DBD with FruR L…Available SinceJuly 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
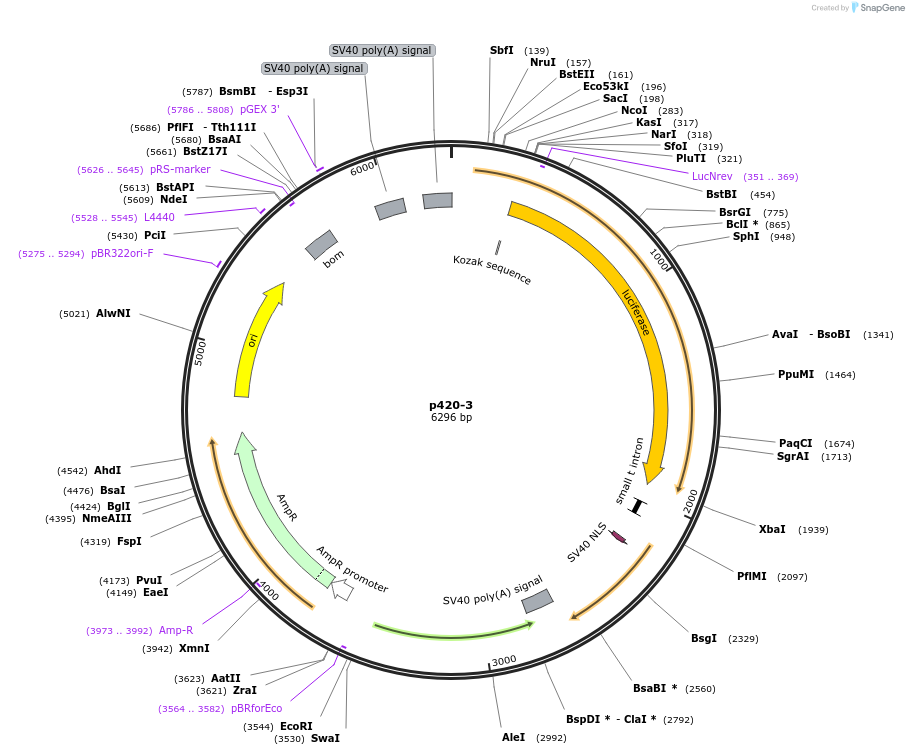
p420-3
Plasmid#71257Purposeluciferase reporter with 3 copies of the GM420 NFAT-AP-1 site in front of hGM-CSF -55 to +28 minimal promoterDepositorInsert3 copies of the GM420 NFAT-AP-1 site (CSF2 Human)
UseLuciferaseTagsluciferaseExpressionMammalianPromoterGM-CSF minimalAvailable SinceDec. 4, 2015AvailabilityAcademic Institutions and Nonprofits only -
GFP-CLCb (EED/QQN)
Plasmid#47422DepositorInsertCLCb EED/QQN (CLTB Human)
TagsEGFPExpressionMammalianMutationEED changed to QQN, in aa20-41 regionPromoterCMVAvailable SinceJuly 29, 2014AvailabilityAcademic Institutions and Nonprofits only -
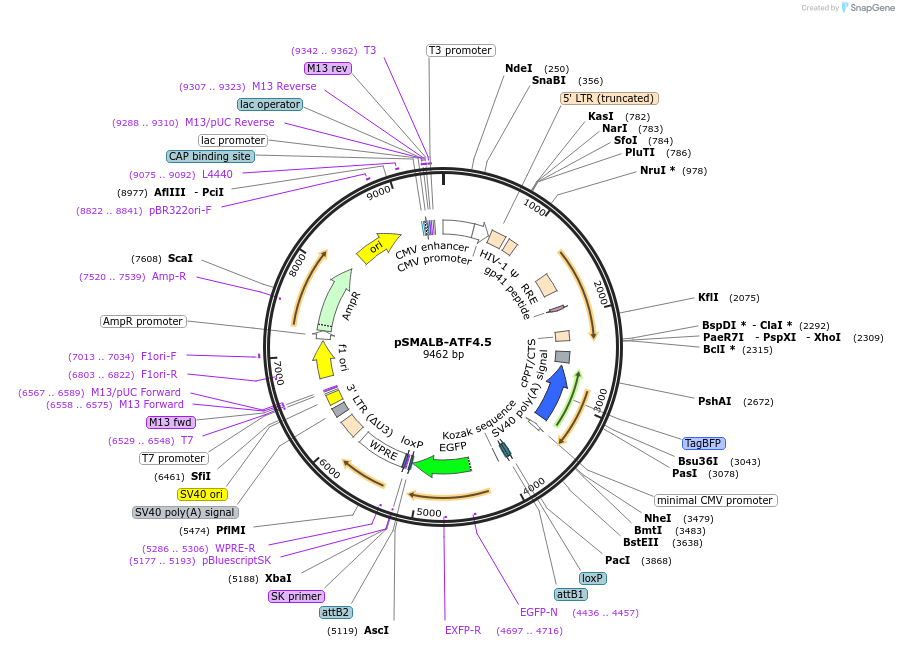
pSMALB-ATF4.5
Plasmid#155032PurposeFluorescent reporter for ATF4 translationDepositorAvailable SinceAug. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
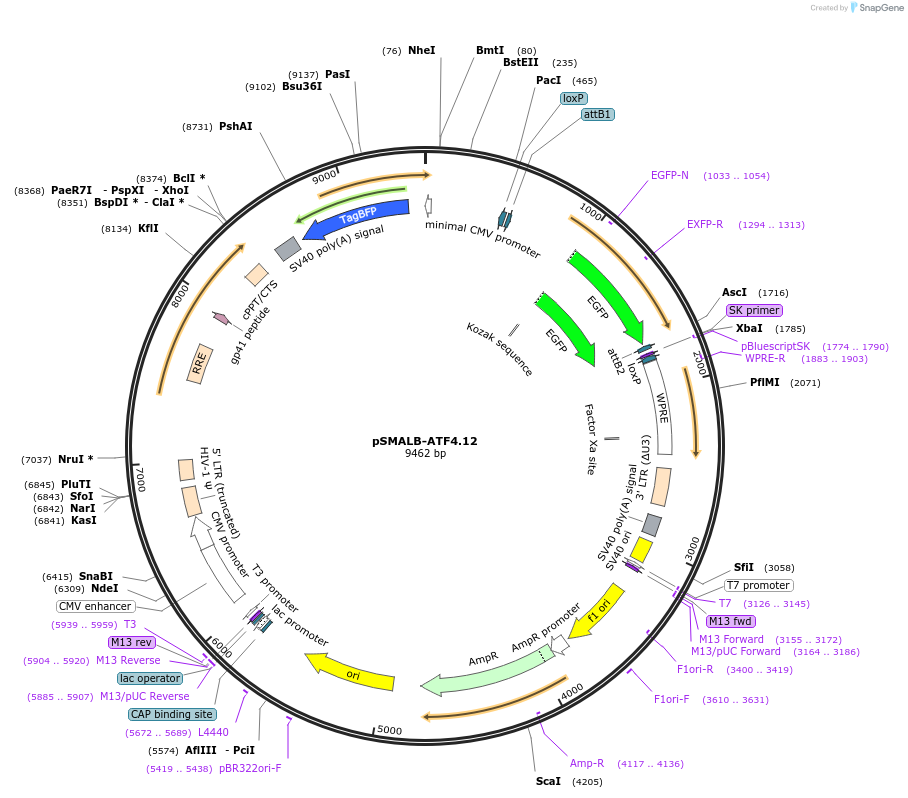
pSMALB-ATF4.12
Plasmid#155033PurposeFluorescent reporter for ATF4 translation, Negative ControlDepositorInsertsUseLentiviralExpressionMammalianMutationuORF1 start codon mutatedAvailable SinceSept. 15, 2020AvailabilityAcademic Institutions and Nonprofits only -
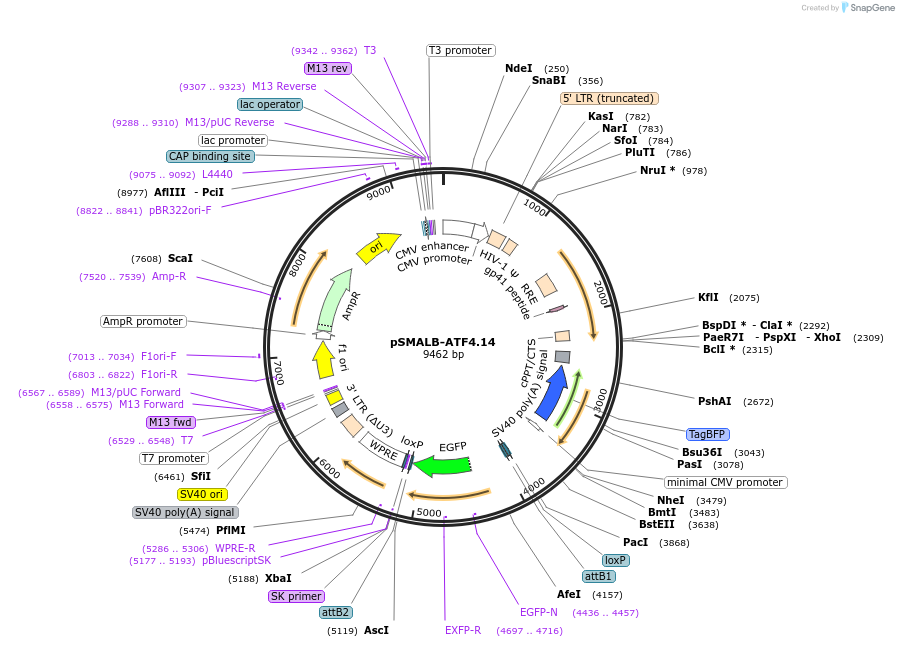
pSMALB-ATF4.14
Plasmid#155034PurposeFluorescent reporter for ATF4 translation, Positive ControlDepositorInsertsUseLentiviralExpressionMammalianMutationuORF1 and uORF2 start codons mutatedAvailable SinceAug. 18, 2020AvailabilityAcademic Institutions and Nonprofits only -
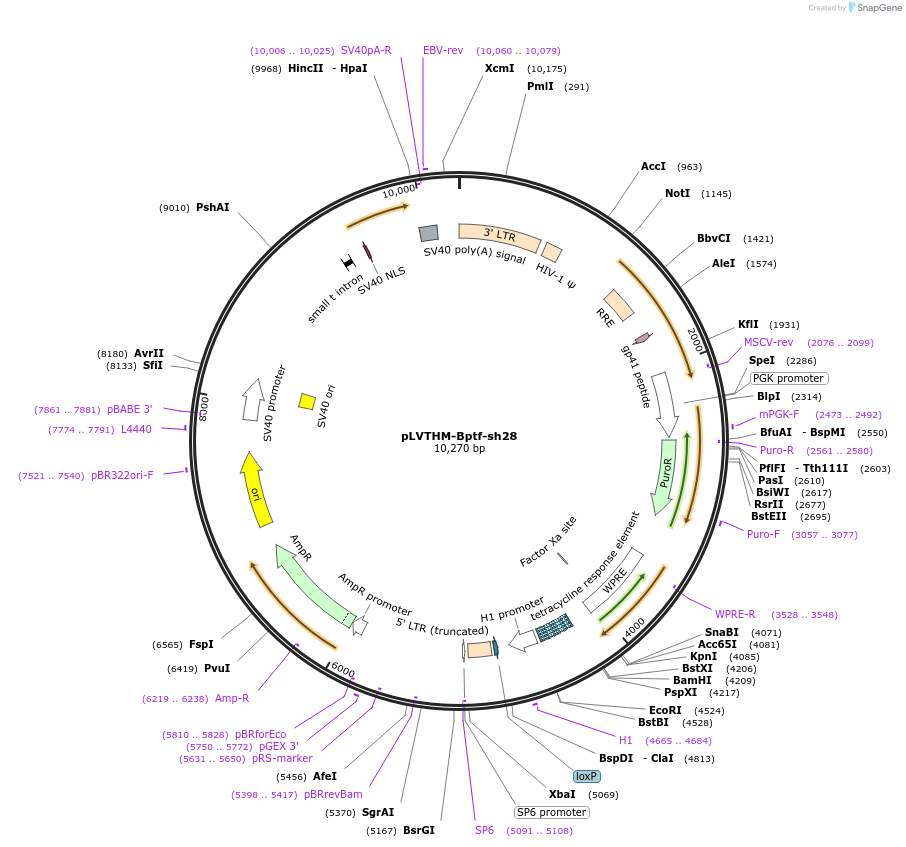
pLVTHM-Bptf-sh28
Plasmid#83278PurposeshRNA targeting BPTFDepositorAvailable SinceOct. 25, 2016AvailabilityIndustry, Academic Institutions, and Nonprofits -
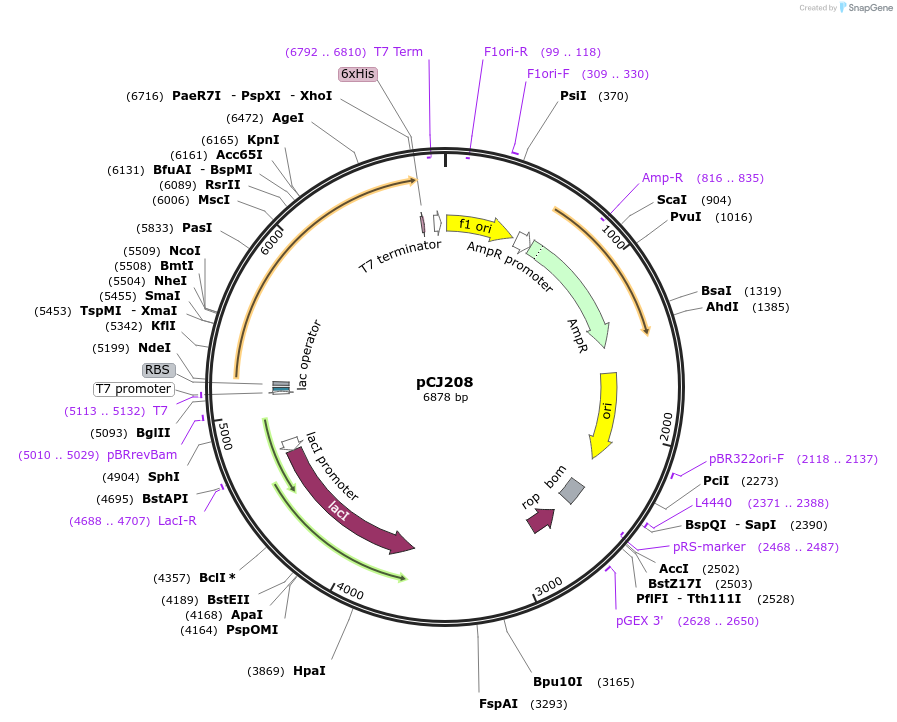
pCJ208
Plasmid#162681PurposeLidded variant of PETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1) removing a seven-residue loop of PETase (Trp185:Phe191, PETase numbering) and replacing it with Gly251:Thr472 from MHETase; codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertPETase from Ideonella sakaiensis 201-F6 (Genbank GAP38373.1) incorporating the MHETase lid
TagsHisExpressionBacterialMutationReplacement of W185-F191 with G251-T472 from MHET…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
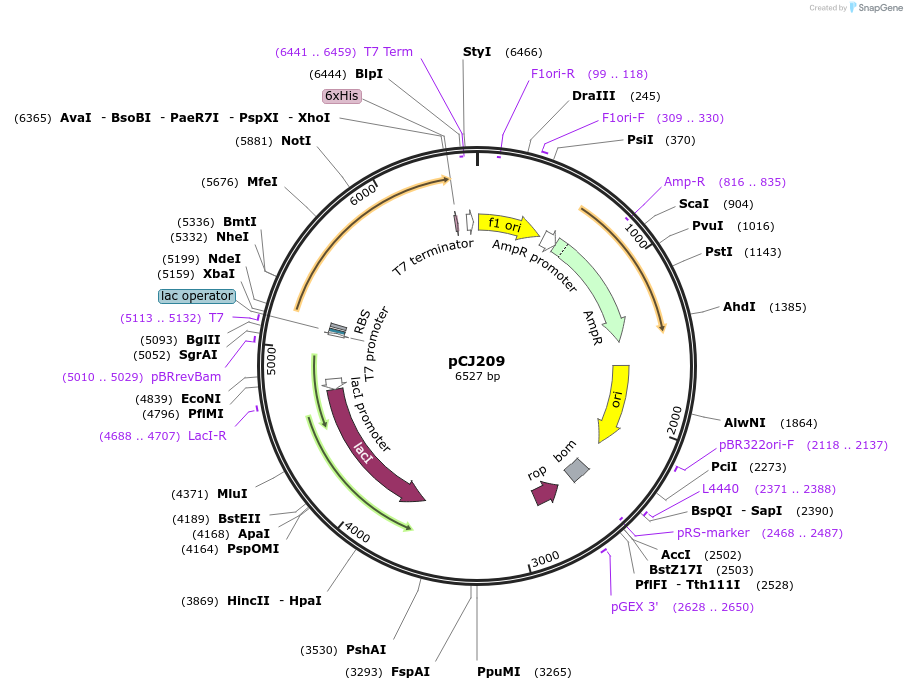
pCJ209
Plasmid#162682PurposeLidless variant of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) replacing Gly251:Thr472 of MHETase with the 7-residue loop (Trp185:Phe191, PETase numbering) of PETase , codon optimized for expression in E. coli K12, with C-terminal His tag.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) with the lid removed
TagsHisExpressionBacterialMutationReplacement of G251-T472 with W185-F191 from PETa…PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
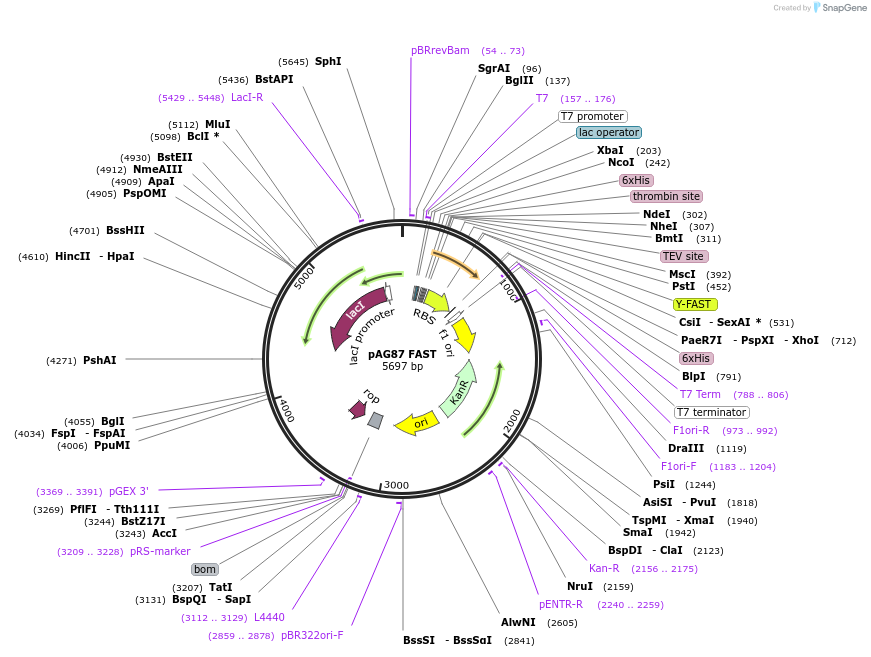
pAG87 FAST
Plasmid#130719PurposeExpresses His-tagged FAST (also called YFAST) in bacteriaDepositorInsertFAST
TagsHis-tagExpressionBacterialPromoterT7Available SinceSept. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
Gucy1b2-IRES-nlsCre-IRES-taulacZ-FNF TV
Plasmid#105196Purposetargeting vector to generate a Gucy1b2-IRES-nlsCre-IRES-taulacZ mouse strain.DepositorInsertGucy1b2-IRES-nlsCre-IRES-taulacZ-FNF (Gucy1b2 Mouse)
UseMouse TargetingAvailable SinceApril 4, 2022AvailabilityAcademic Institutions and Nonprofits only