We narrowed to 821 results for: mcherry reporter
-
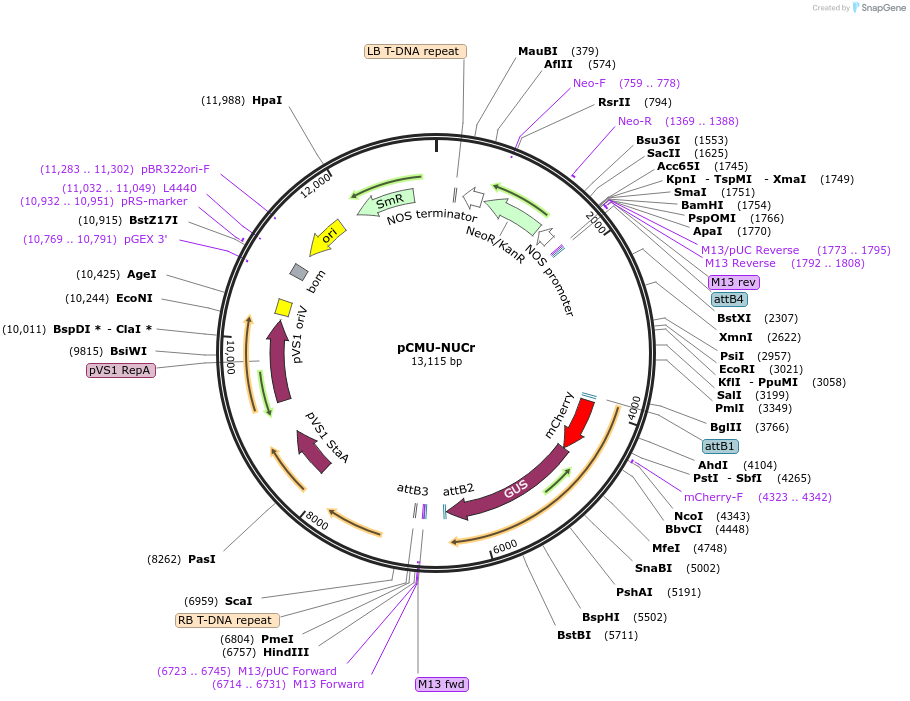
Plasmid#61168PurposeFluorescent reporter for nucleus NLS-mCherry-GUS expressed under AtUBQ10 promoterDepositorInsertNLS-mCherry-GUS
ExpressionPlantPromoterAtUBQ10Available SinceFeb. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
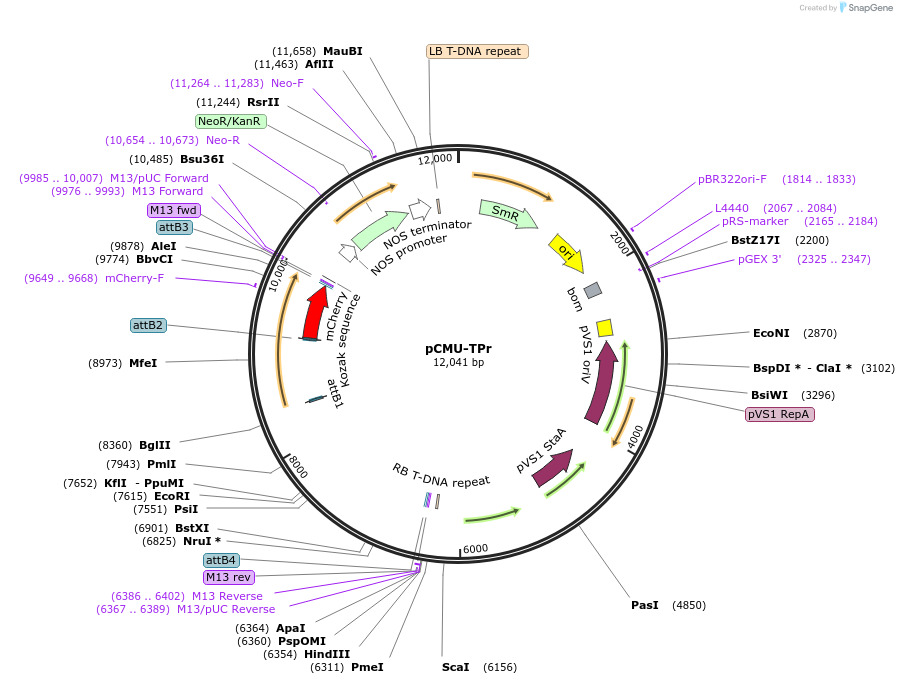
pCMU-TPr
Plasmid#61189PurposeFluorescent reporter for tonoplast AtTIP1-1-mCherry expressed under AtUBQ10 promoterDepositorInsertAtTIP1-1
TagsmCherryExpressionPlantPromoterAtUBQ10Available SinceFeb. 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
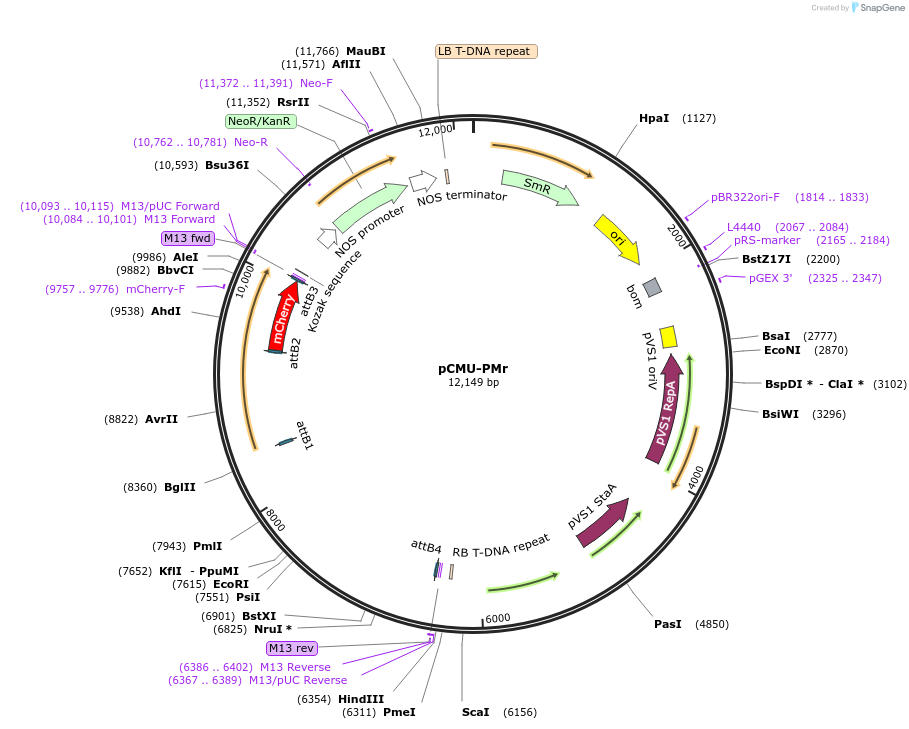
pCMU-PMr
Plasmid#61180PurposeFluorescent reporter for plasma membrane AtPIP2a-mCherry expressed under AtUBQ10 promoterDepositorInsertAtPIP2a
TagsmCherryExpressionPlantPromoterAtUBQ10Available SinceApril 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
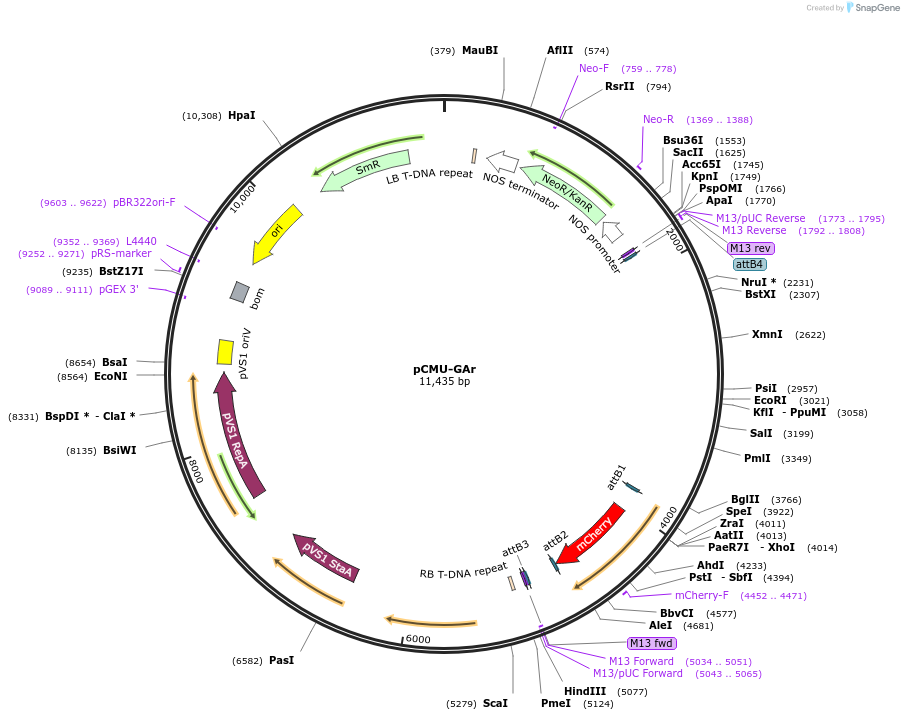
pCMU-GAr
Plasmid#61172PurposeFluorescent reporter for Golgi apparatus GmMAN49-mCherry expressed under AtUBQ10 promoterDepositorInsertGmMAN49-mCherry
ExpressionPlantPromoterAtUBQ10Available SinceApril 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
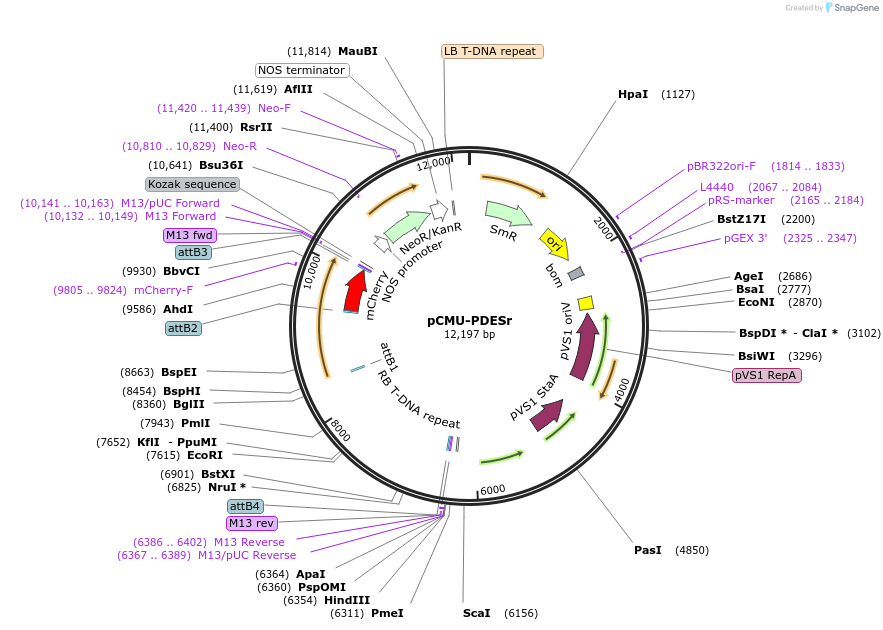
pCMU-PDESr
Plasmid#61206PurposeFluorescent reporter for plasmodesmata AtPDLP1-mCherry expressed under AtUBQ10 promoterDepositorInsertAtPDLP1
TagsmCherryExpressionPlantPromoterAtUBQ10Available SinceFeb. 23, 2015AvailabilityAcademic Institutions and Nonprofits only -
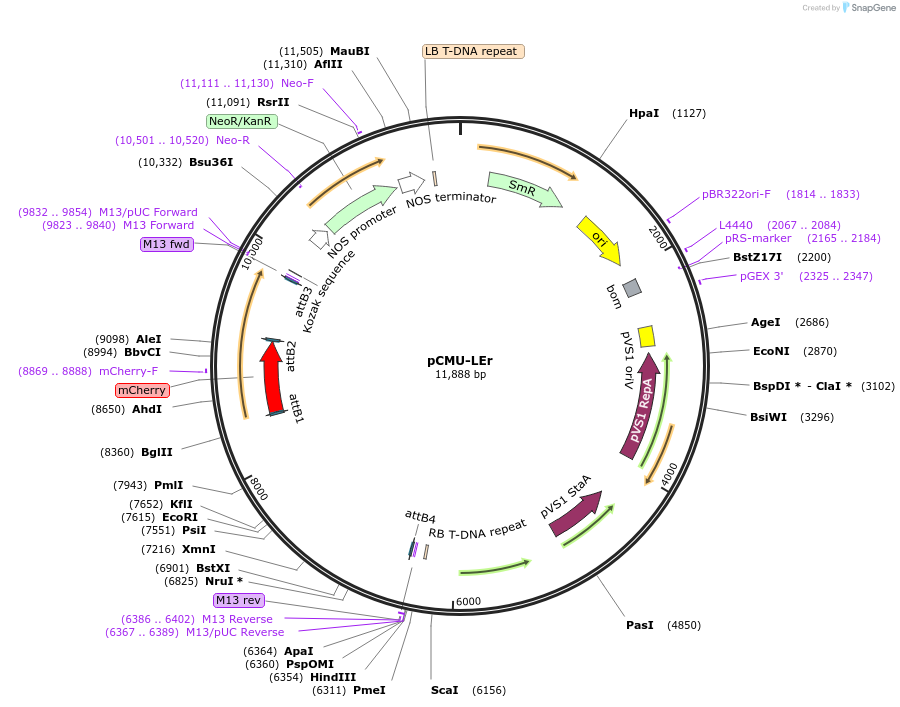
pCMU-LEr
Plasmid#61185PurposeFluorescent reporter for late endosome mCherry-MtRAB5A2 expressed under AtUBQ10 promoterDepositorInsertMtRAB5A2
TagsmCherryExpressionPlantPromoterAtUBQ10Available SinceFeb. 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
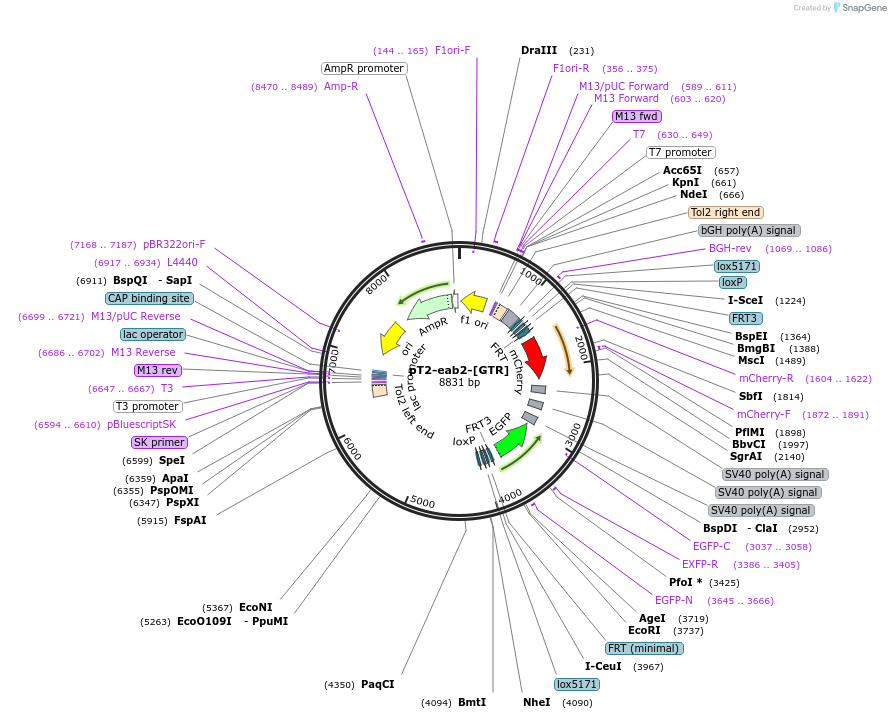
pT2-eab2-[GTR]
Plasmid#46144DepositorInsertEGFP/mCherry
UseCre/Lox; Tol2 zebrafish transgenic expressionAvailable SinceJuly 9, 2013AvailabilityAcademic Institutions and Nonprofits only -
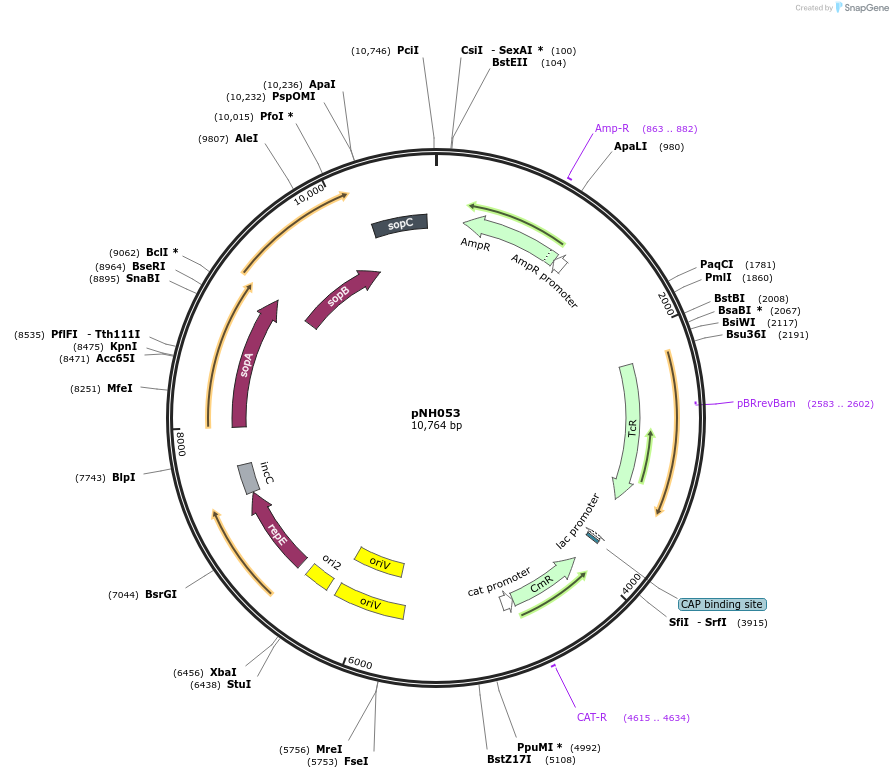
pNH053
Plasmid#42159DepositorInsertmCherry-RT-mCherry
Available SinceMarch 14, 2013AvailabilityAcademic Institutions and Nonprofits only -
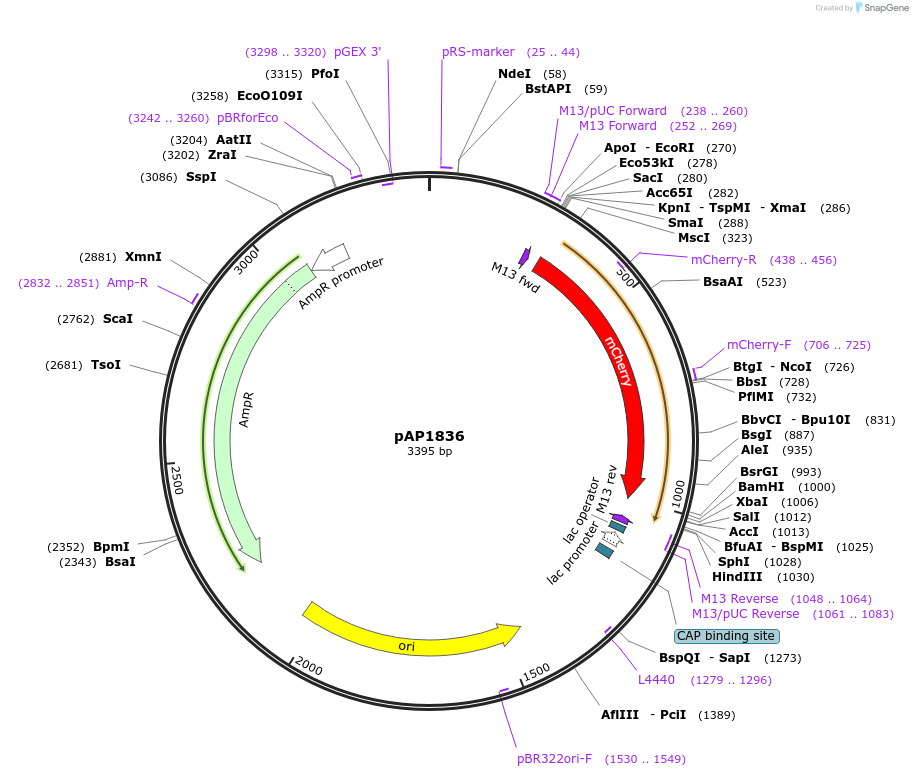
pAP1836
Plasmid#137087PurposePlasmid for PCR-generated donor DNA for rapid tagging of proteins with mCherryDepositorInsertmCherry
UsePcr template for donor dnaExpressionBacterialAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
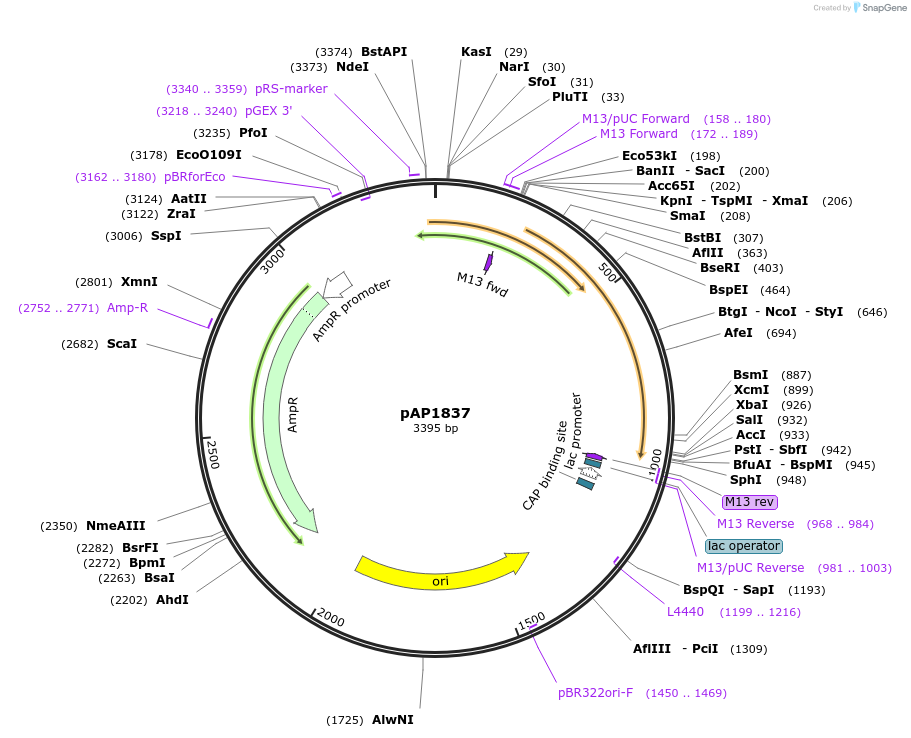
pAP1837
Plasmid#137086PurposePlasmid for PCR-generated donor DNA for rapid tagging of proteins with mCherryDepositorInsertmCherry
UsePcr template for donor dnaExpressionBacterialAvailable SinceMarch 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
FD1
Bacterial Strain#125258PurposeDerivative of MG1655 used for the screening of clones able to survive the bad seed toxicity of dCas9, while still efficiently silencing the expression of mcherryDepositorBacterial ResistanceNoneAvailable SinceAug. 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
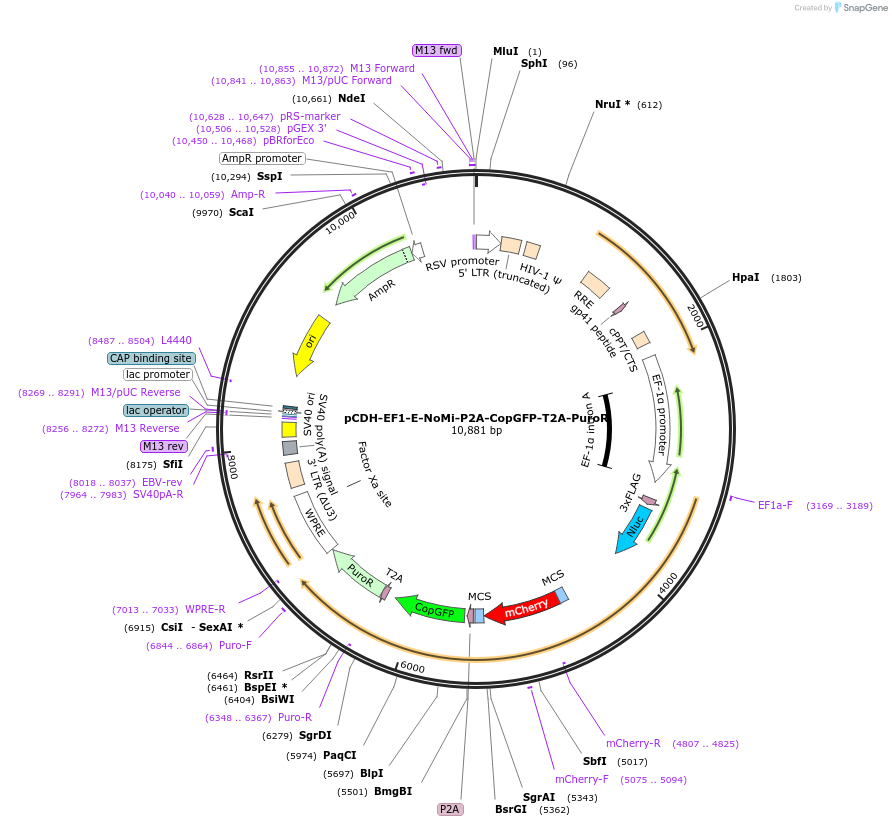
pCDH-EF1-E-NoMi-P2A-CopGFP-T2A-PuroR
Plasmid#207805PurposeE-NoMi is a re-engineered tetraspanin scaffold tagged with bioluminescent and fluorescent reporter proteins under an EF-1α promoter.DepositorInsertE-NoMi
UseLentiviralTags3xFlag tag, copGFP, mCherry, and nanoluciferaseExpressionMammalianPromoterEF-1alphaAvailable SinceJan. 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
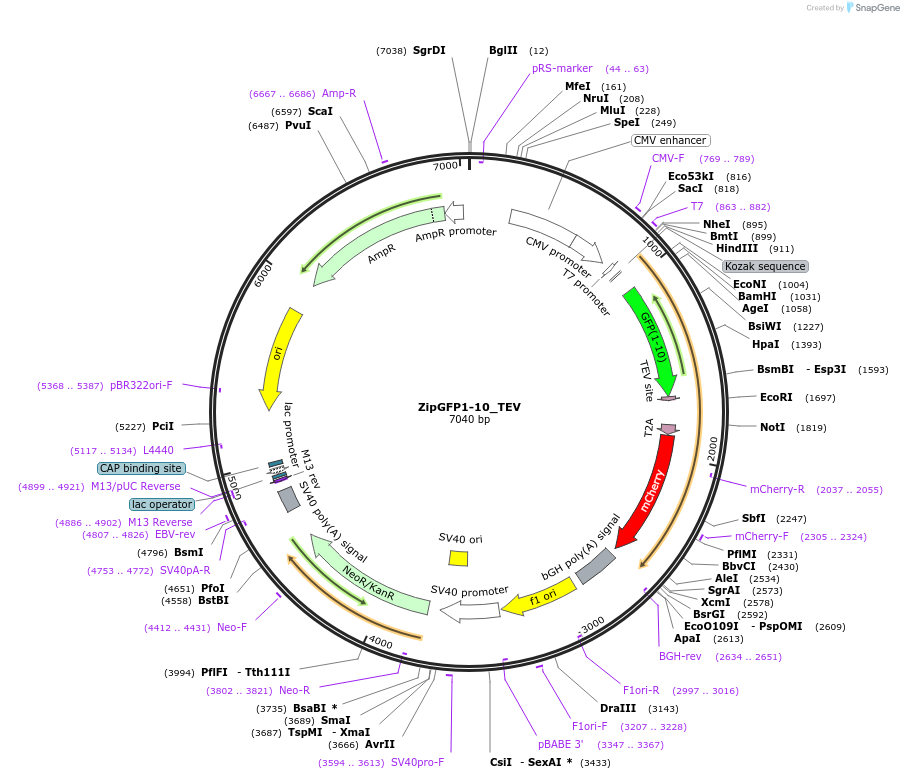
ZipGFP1-10_TEV
Plasmid#81242PurposeZipped GFP1-10 with TEV cleavage sequence T2A mCherryDepositorInsertzipped GFP11 with TEV cleavage sequence T2A mCherry
ExpressionMammalianAvailable SinceNov. 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
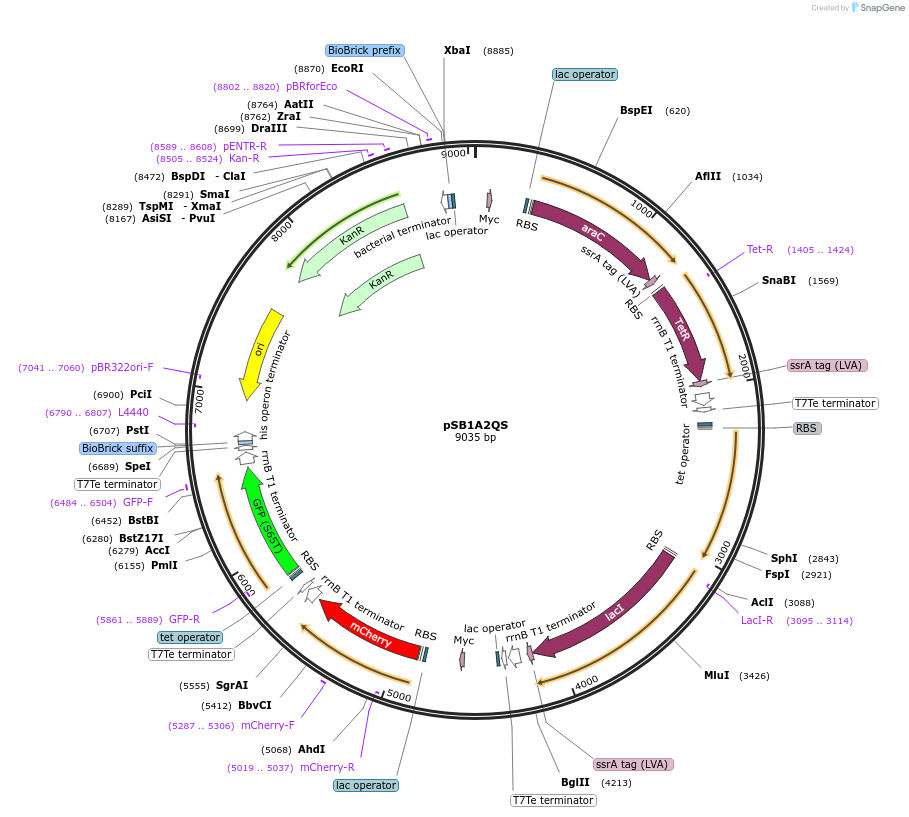
pSB1A2QS
Plasmid#138268PurposeMINPA Gene circuit expression in E. coli. GFP serves as a reporter for Plux/tet promoter activity. mCherry serves as reporter for Para/lac promoter activityDepositorInsertsAraC-TetR operon
LuxR-LacI operon
mCherry
GFP
UseSynthetic BiologyExpressionBacterialPromoterPara/lac and Plux/tetAvailable SinceApril 14, 2020AvailabilityAcademic Institutions and Nonprofits only -
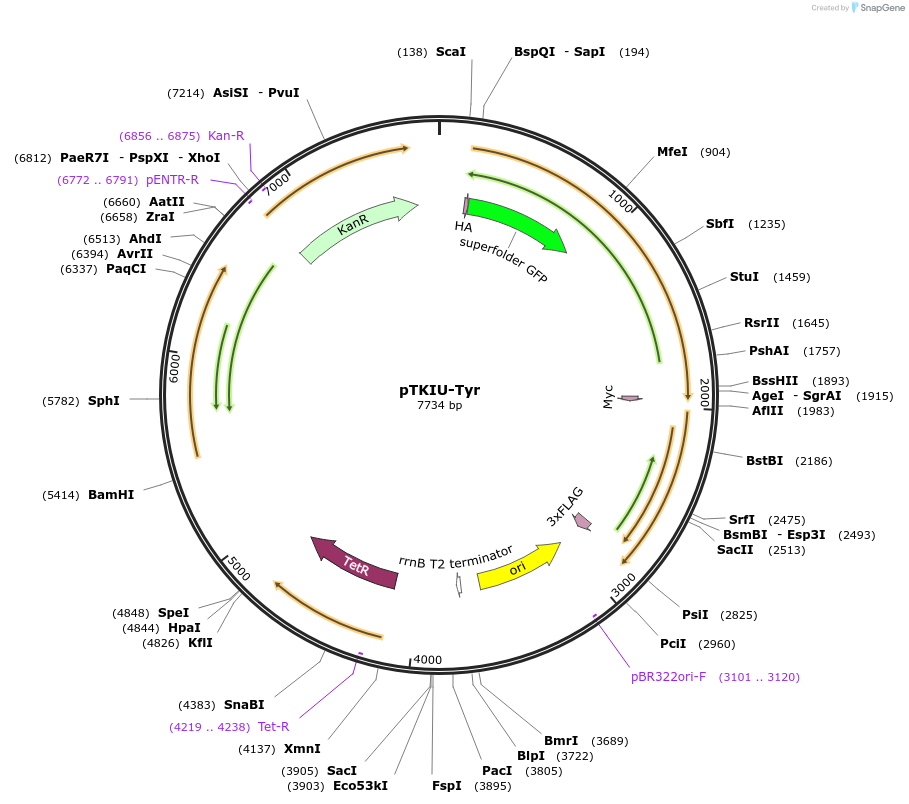
pTKIU-Tyr
Plasmid#247649PurposeExpresses GFP-SUMO-Tyr-mCherry N-degron reporter driven by aTc responsive promoter, and constitutively expresses Ulp1, from KanR integrating mycobacterial plasmidDepositorInsertsGFP-SUMO-Tyr-mCherry N-degron proteolysis dual-fluorescence reporter
Ulp1 SUMO protease
UseMycobacterial integratingTags3xFLAG tag, HA tag, and Myc tagExpressionBacterialPromoterPTet and PTet (co-operonic with GFP-SUMO-Ser-mChe…Available SinceDec. 9, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
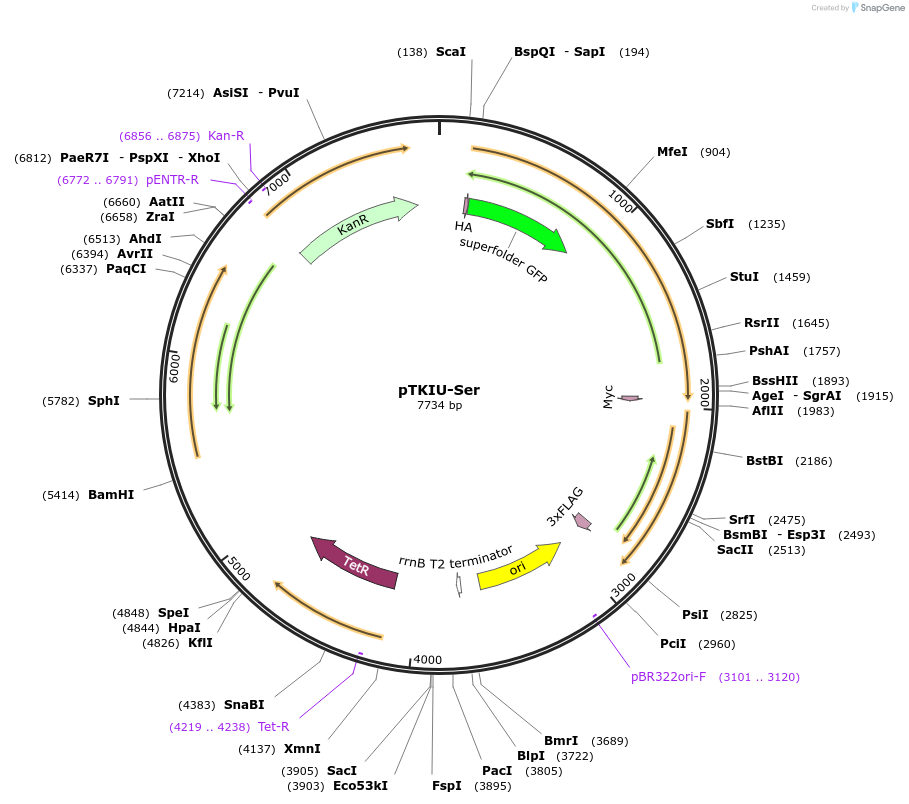
pTKIU-Ser
Plasmid#247650PurposeExpresses GFP-SUMO-Ser-mCherry N-degron reporter driven by aTc responsive promoter, and constitutively expresses Ulp1, from KanR integrating mycobacterial plasmidDepositorInsertsGFP-SUMO-Ser-mCherry N-degron proteolysis dual-fluorescence reporter
Ulp1 SUMO protease
UseMycobacterial integratingTags3xFLAG tag, HA tag, and Myc tagExpressionBacterialPromoterPTet and PTet (co-operonic with GFP-SUMO-Ser-mChe…Available SinceDec. 9, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
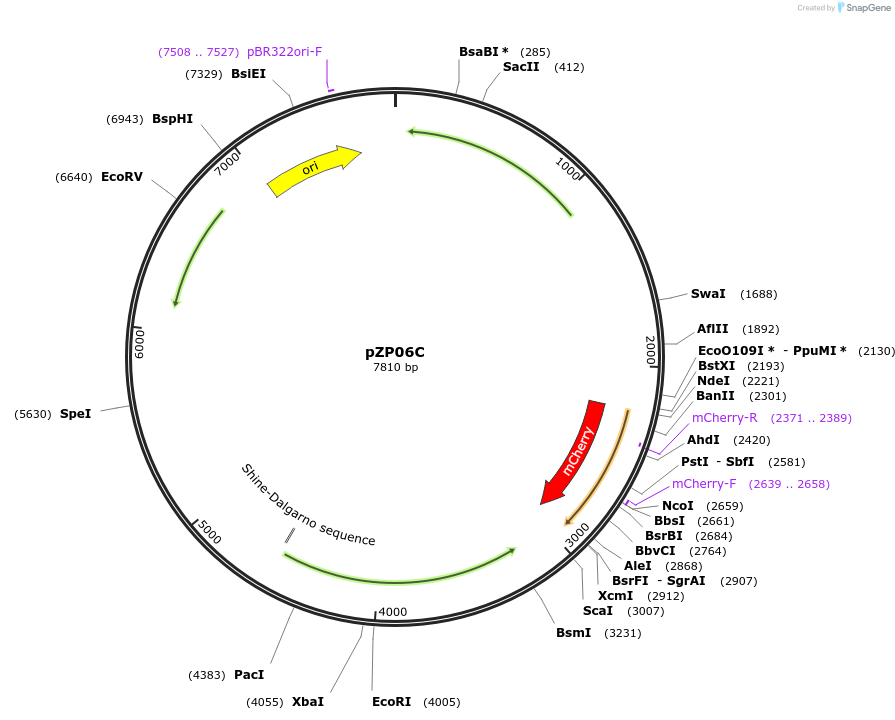
pZP06C
Plasmid#232322PurposepZP06C was constructed by adding the mCherry reporter gene driven by the rpsJ promoter into pZP06B (containing PrpsJ-sacB) to facilitate the screening of positive clones through red fluorescence.DepositorInsertmCherry
ExpressionBacterialPromoterrpsJAvailable SinceMarch 12, 2025AvailabilityAcademic Institutions and Nonprofits only -
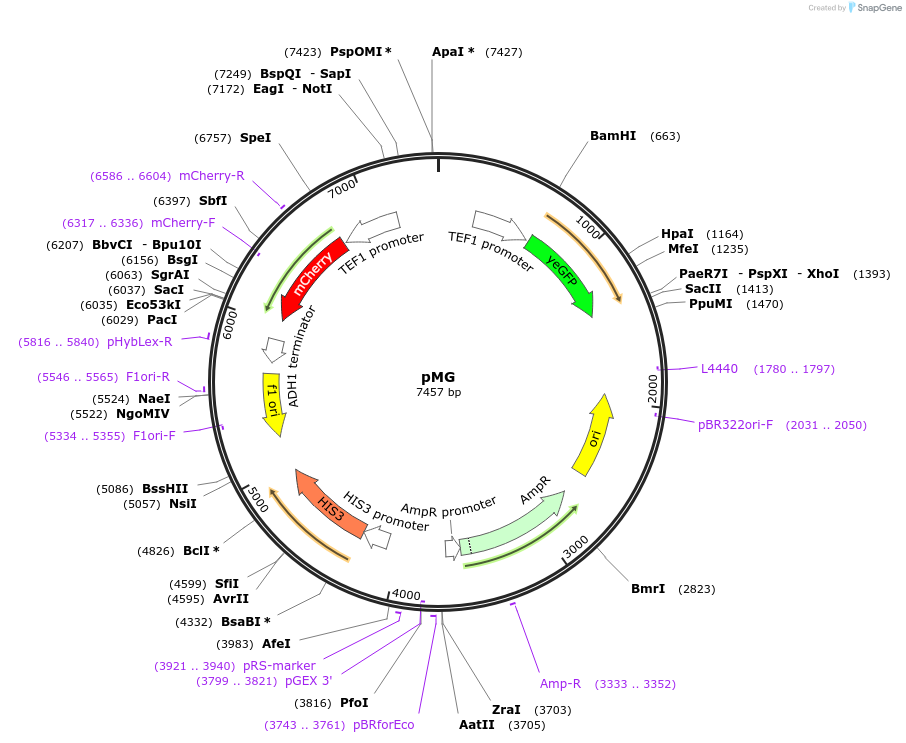
pMG
Plasmid#138264PurposeYeast intergration vector for integration of dual reporter system at the URA3 locus. Dual reporter system contains constitutive mCherry and constitutive eGFP flanked by iCas9 sites.DepositorInsertseGFP flanked by iCas9 sites
mCherry
UseCRISPRExpressionYeastPromoterTef1Available SinceMarch 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
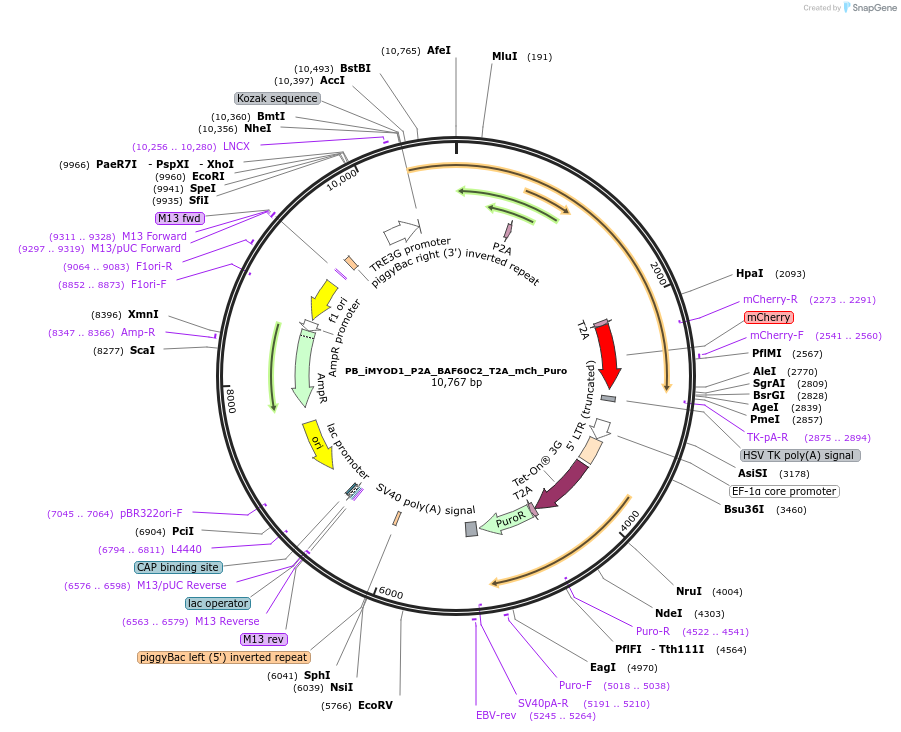
PB_iMYOD1_P2A_BAF60C2_T2A_mCh_Puro
Plasmid#168807PurposePiggyBac transposon construct with doxycycline inducible MYOD1, BAF60C + mCherry expressionDepositorInsertMYOD1 (MYOD1 Synthetic, Human)
UseSynthetic Biology; PiggybacExpressionMammalianPromoterTREAvailable SinceAug. 31, 2021AvailabilityAcademic Institutions and Nonprofits only