We narrowed to 2,626 results for: pyogenes Cas9
-
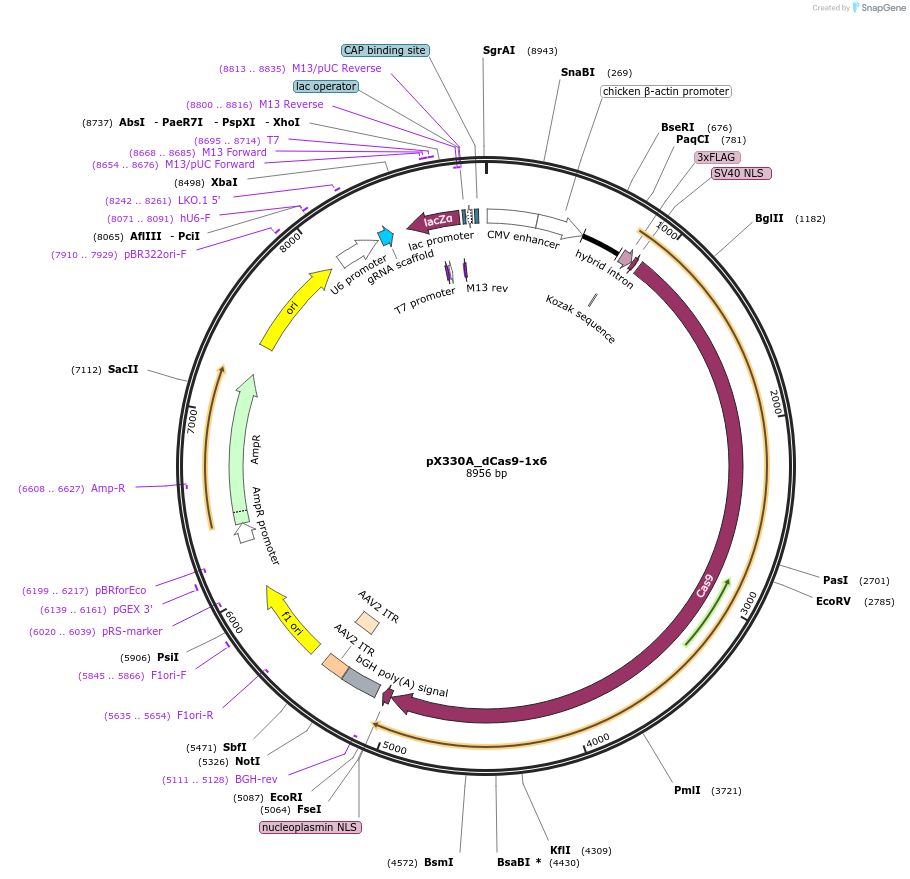
Plasmid#63600PurposeExpresses dCas9 and gRNADepositorInserthumanized S. pyogenes dCas9
UseCRISPRTags3xFLAGExpressionMammalianMutationD1361YPromoterCBhAvailable SinceJune 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
pX330A_dCas9-1x5
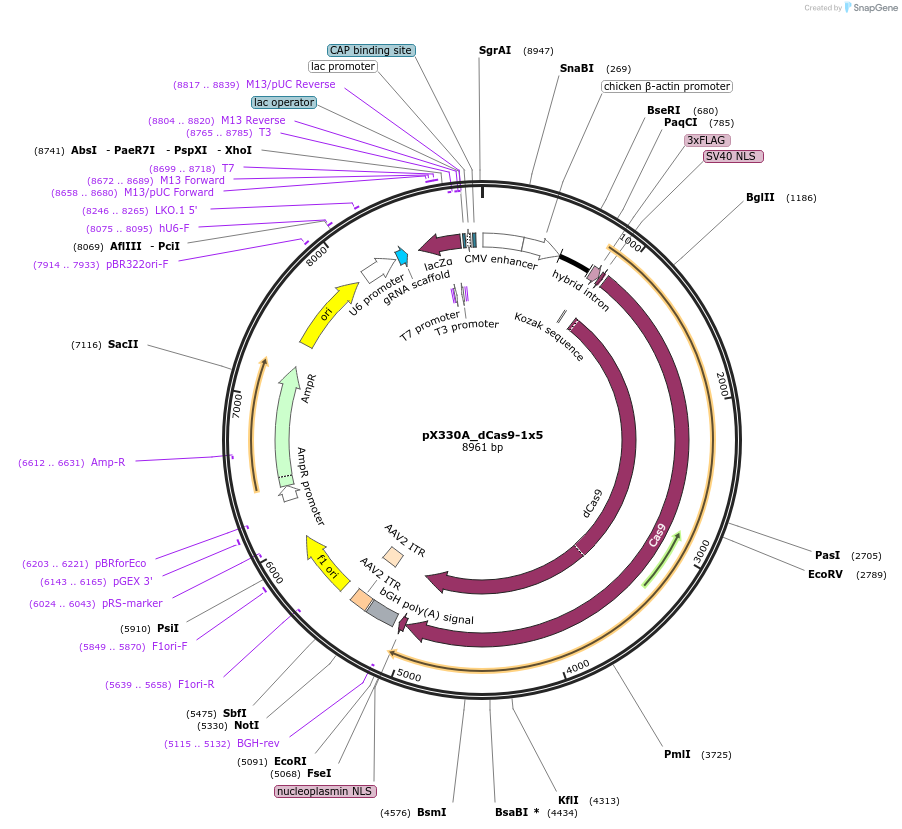
Plasmid#63599PurposeExpresses dCas9 and gRNADepositorInserthumanized S. pyogenes dCas9
UseCRISPRTags3xFLAGExpressionMammalianPromoterCBhAvailable SinceJune 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
pX330A_dCas9-1x3
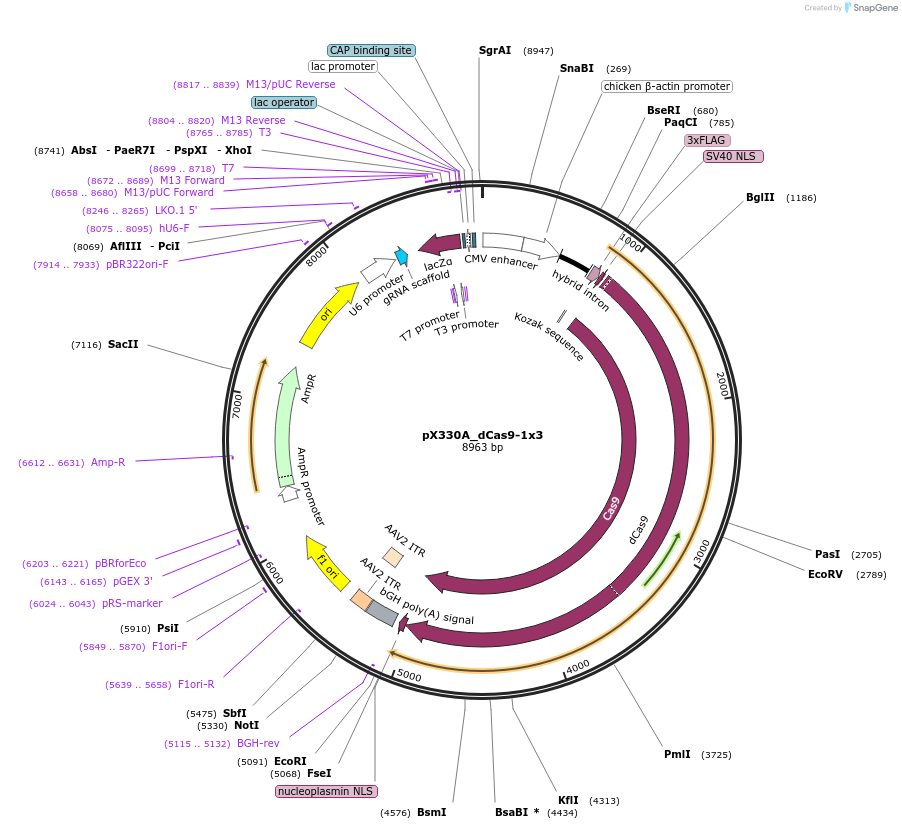
Plasmid#63597PurposeExpresses dCas9 and gRNADepositorInserthumanized S. pyogenes dCas9
UseCRISPRTags3xFLAGExpressionMammalianPromoterCBhAvailable SinceJune 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAN-PTet-dCas9
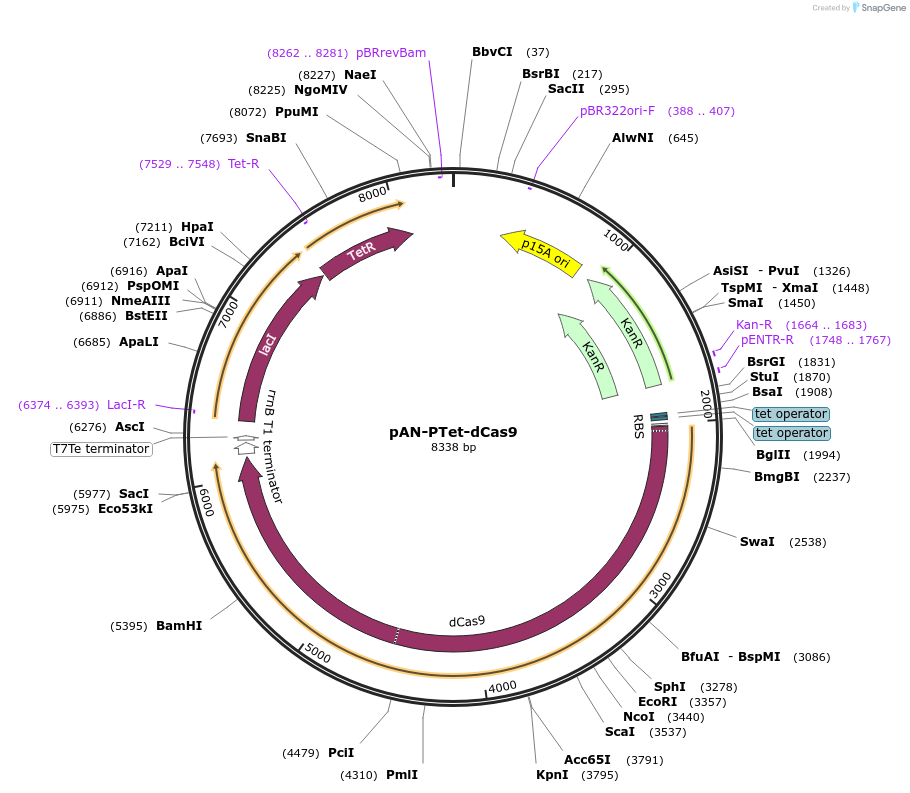
Plasmid#62244Purposecontrols the expression of S. pyogenes dCas9 from a Tc‐inducible PTet promoter.DepositorInsertdCas9
UseCRISPRExpressionBacterialMutationcatalytically inactive, D10A, H840APromoterPtetAvailable SinceJuly 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
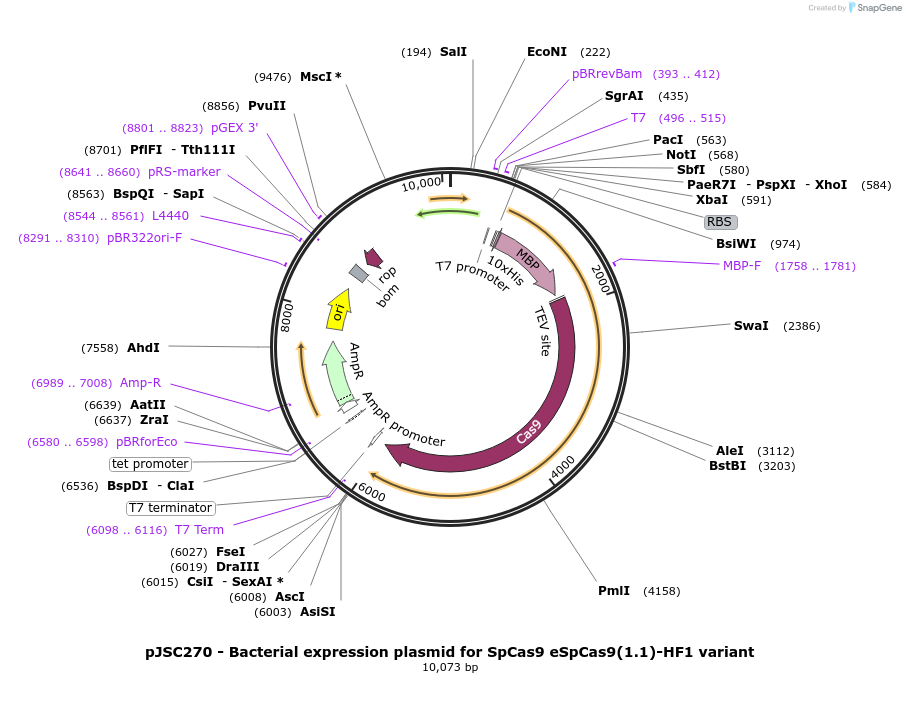
pJSC270 - Bacterial expression plasmid for SpCas9 eSpCas9(1.1)-HF1 variant
Plasmid#101217PurposeBacterial expression plasmid for SpCas9 eSpCas9(1.1)-HF1 variantDepositorInsertSpCas9 variant K848A/K1003A/R1060A/N497A/R661A/Q695A/Q926A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationK848A, K1003A, R1060A, N497A, R661A, Q695A and Q9…PromoterT7Available SinceOct. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
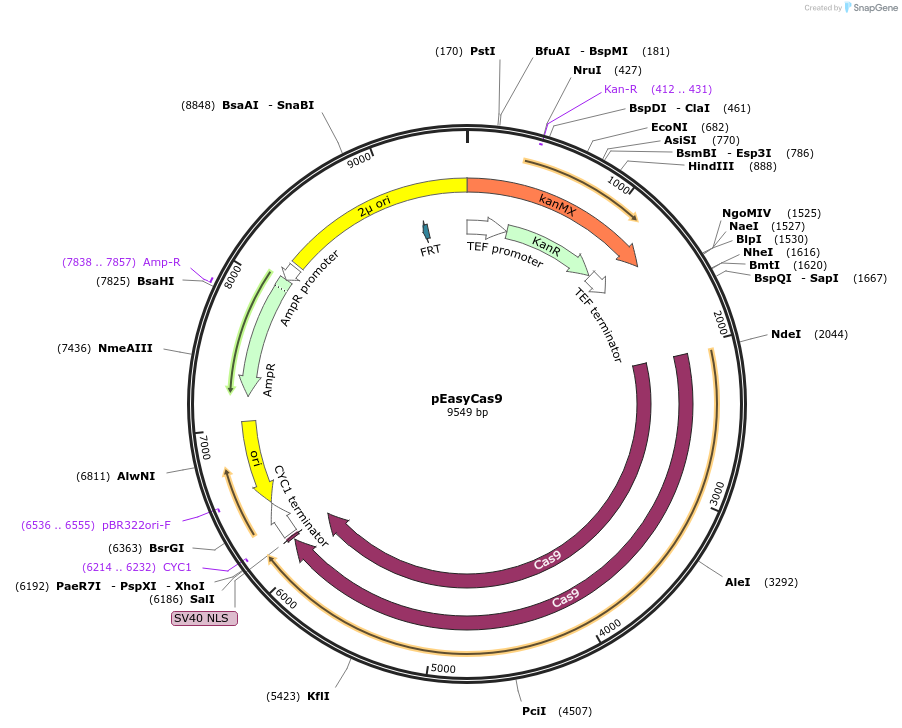
pEasyCas9
Plasmid#184909PurposeExpresses the Streptococcus pyogenes Cas9 proteinDepositorInsertCas9
UseCRISPRExpressionBacterial and YeastAvailable SinceJan. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
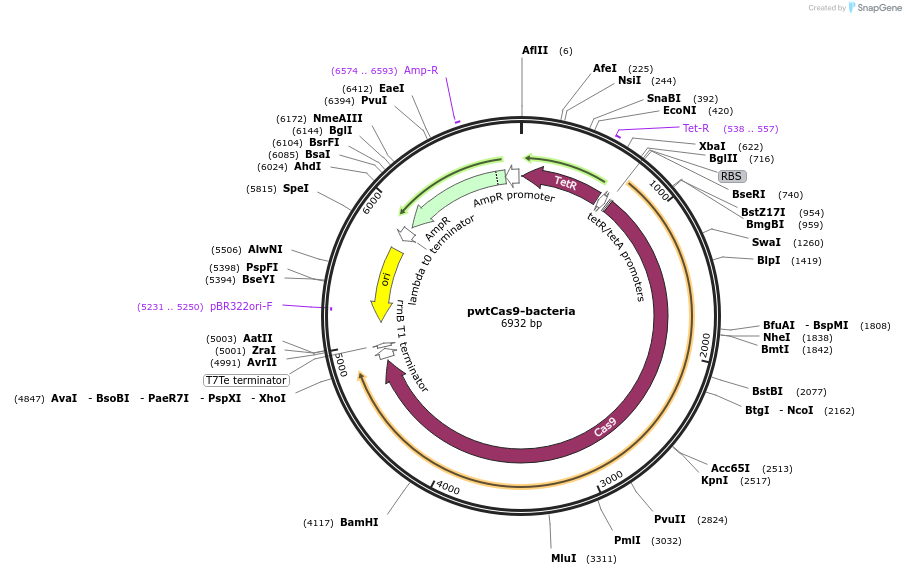
pwtCas9-bacteria
Plasmid#44250PurposeaTc-inducible expression of wild-type Cas9 (S .pyogenes) for bacterial gene knockdownDepositorInsertwild-type Cas9
UseCRISPRExpressionBacterialPromoterpLtetO-1Available SinceApril 11, 2013AvailabilityAcademic Institutions and Nonprofits only -
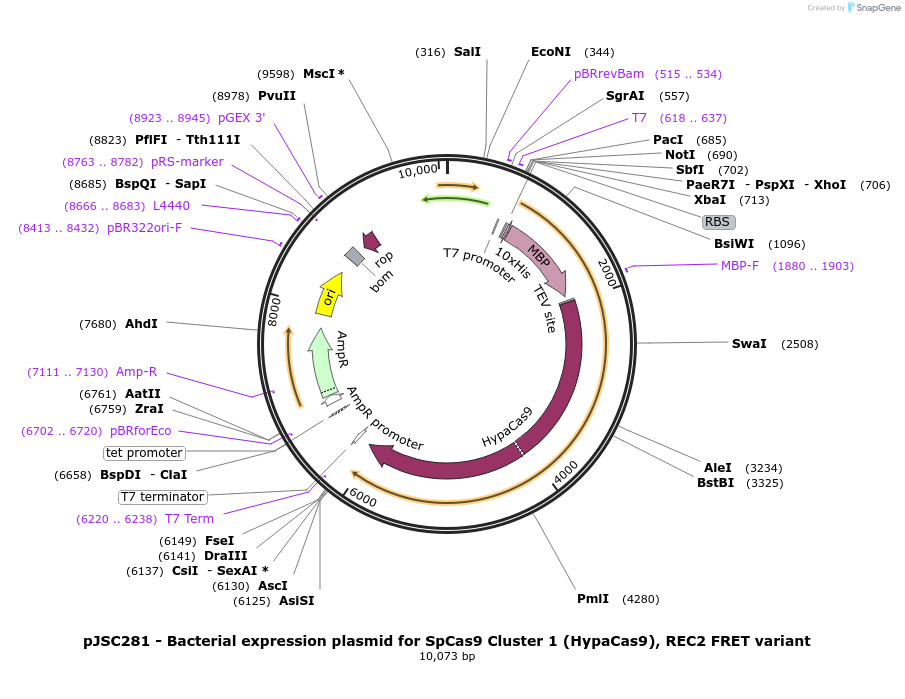
pJSC281 - Bacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC2 FRET variant
Plasmid#101230PurposeBacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC2 FRET variantDepositorInsertSpCas9 variant C80S/C574S/E60C/D273C/N692A/M694A/Q695A/H698A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, E60C, D273C, N692A, M694A, Q695A and…PromoterT7Available SinceNov. 7, 2017AvailabilityAcademic Institutions and Nonprofits only -
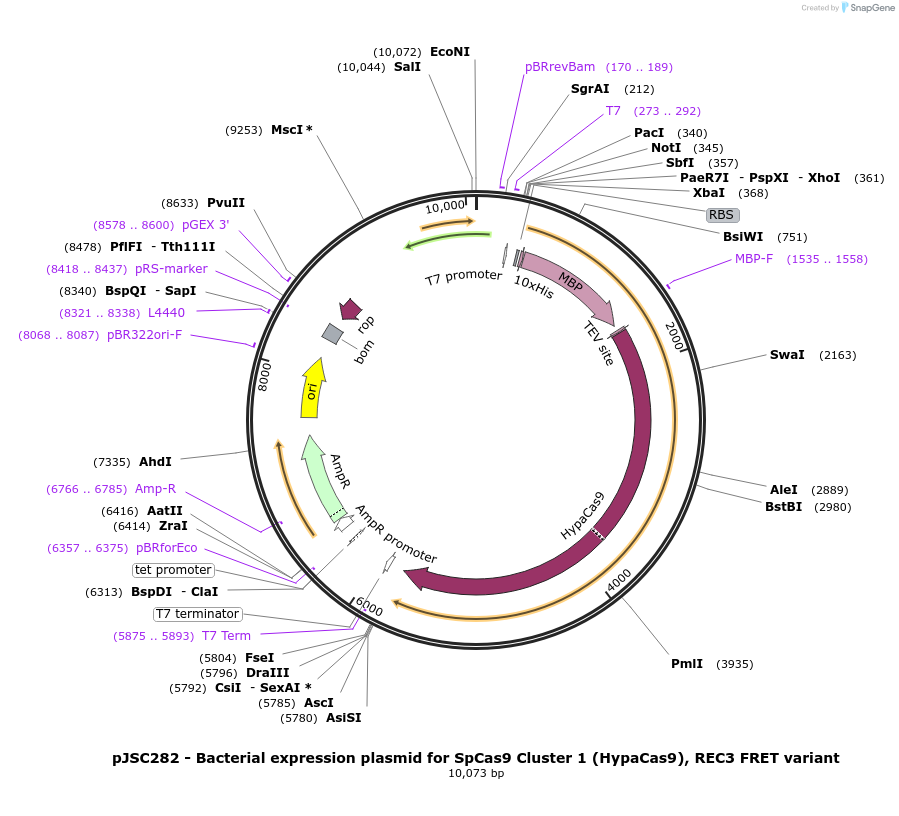
pJSC282 - Bacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC3 FRET variant
Plasmid#101231PurposeBacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), REC3 FRET variantDepositorInsertSpCas9 variant C80S/C574S/S701C/S960C/N692A/M694A/Q695A/H698A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S701C, S960C, N692A, M694A, Q695A an…PromoterT7Available SinceNov. 6, 2017AvailabilityAcademic Institutions and Nonprofits only -
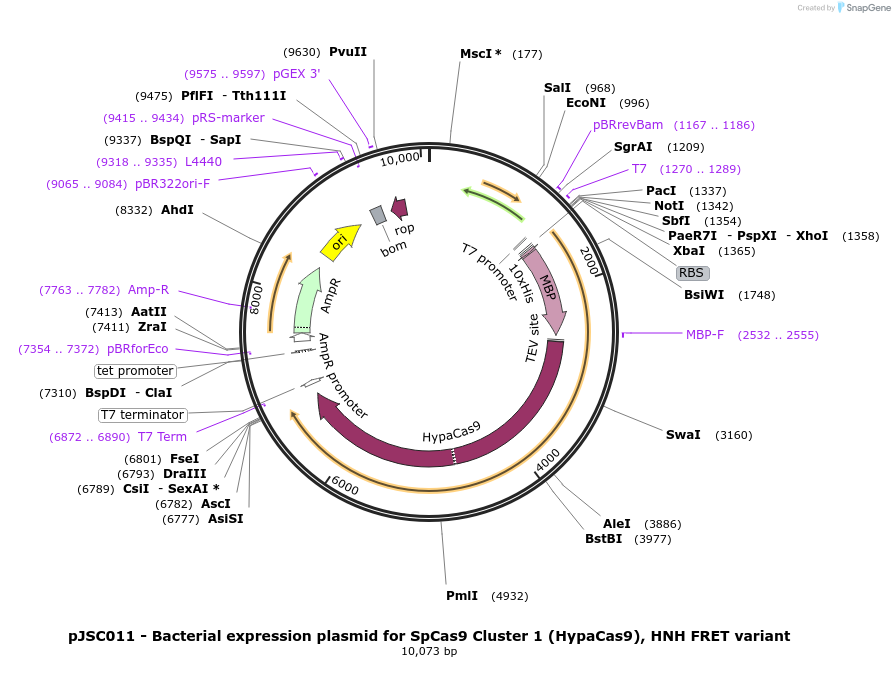
pJSC011 - Bacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), HNH FRET variant
Plasmid#101229PurposeBacterial expression plasmid for SpCas9 Cluster 1 (HypaCas9), HNH FRET variantDepositorInsertSpCas9 variant C80S/C574S/S355C/S867C/N692A/M694A/Q695A/H698A
Tags10x His, MBP, and TEV siteExpressionBacterialMutationC80S, C574S, S355C, S867C, N692A, M694A, Q695A an…PromoterT7Available SinceOct. 25, 2017AvailabilityAcademic Institutions and Nonprofits only -
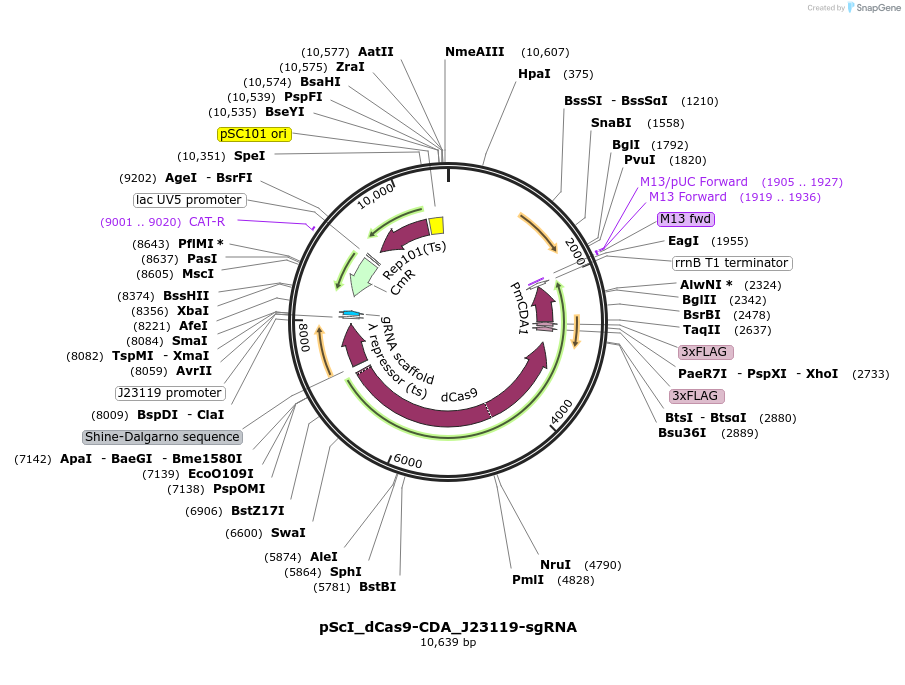
pScI_dCas9-CDA_J23119-sgRNA
Plasmid#108550PurposeBacterial Target-AID vector with sgRNADepositorInsertsdCas9-PmCDA1
Streptococcus pyogenes sgRNA
TagsFlagExpressionBacterialMutationD10A and H840A for SpCas9PromoterJ23119 and lambda ORAvailable SinceSept. 14, 2018AvailabilityAcademic Institutions and Nonprofits only -
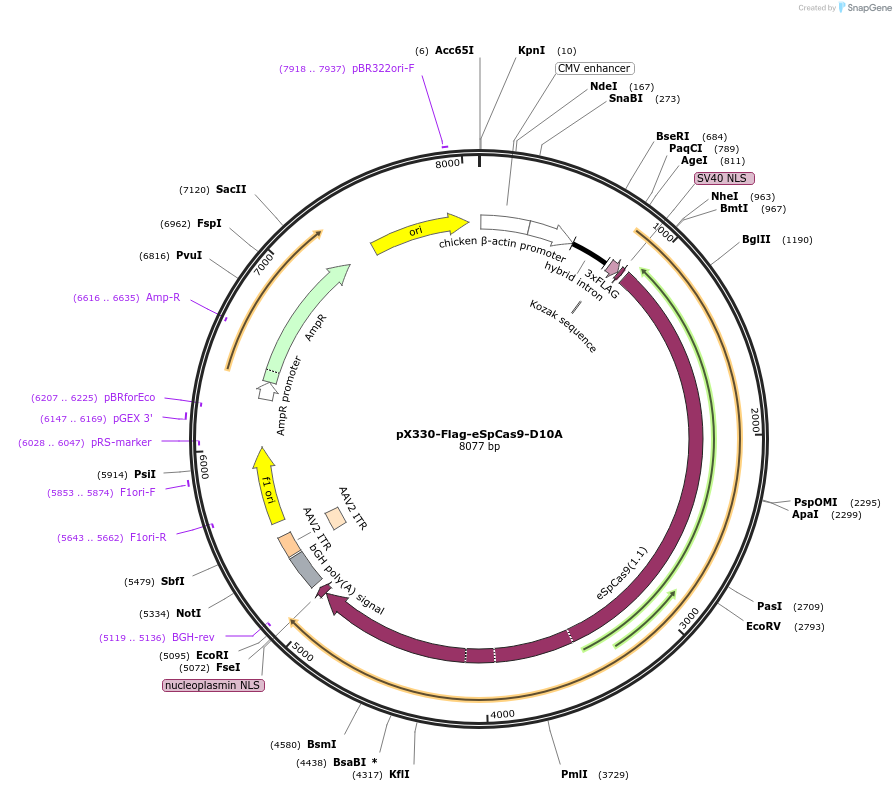
pX330-Flag-eSpCas9-D10A
Plasmid#80449Purposeenhanced SpCas9 mammalian expression vectorDepositorInsertehSpCas9_D10A
UseCRISPRTags3xFLAG and NLSExpressionMammalianPromoterCBhAvailable SinceAug. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
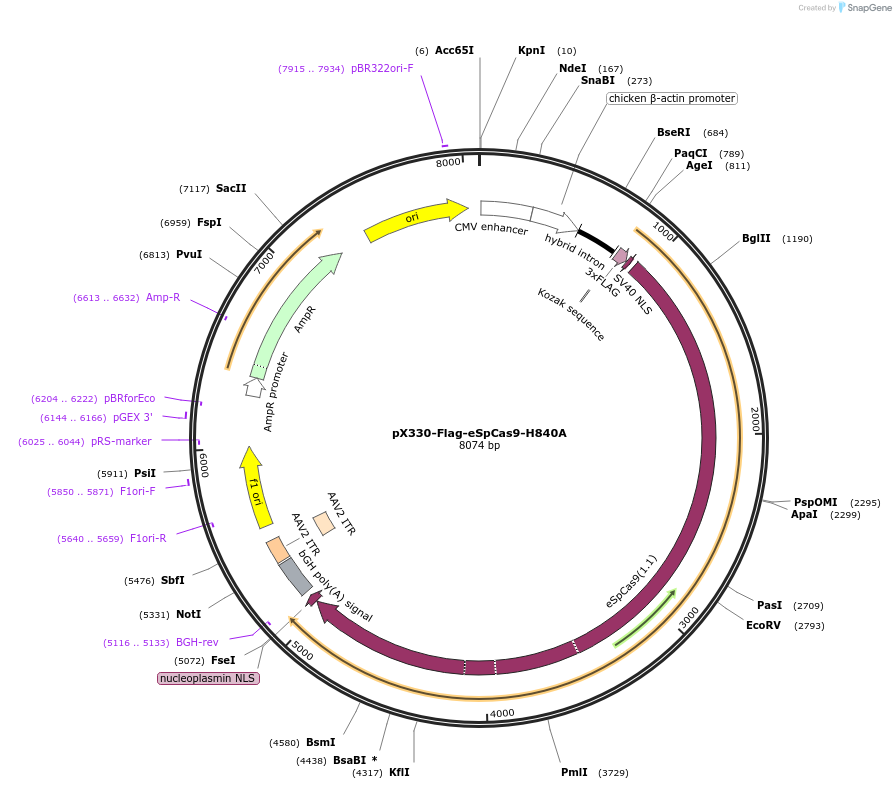
pX330-Flag-eSpCas9-H840A
Plasmid#80454Purposeenhanced SpCas9 mammalian expression vectorDepositorInsertehSpCas9_H840A
UseCRISPRTags3xFLAG and NLSExpressionMammalianPromoterCBhAvailable SinceAug. 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
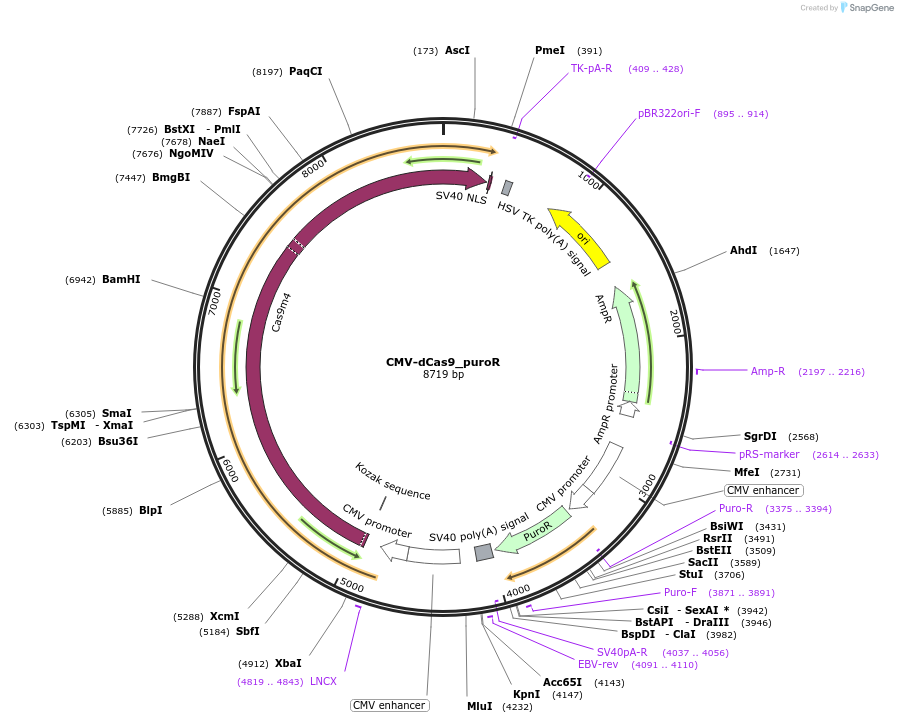
CMV-dCas9_puroR
Plasmid#219825PurposeCMV-driven expression of dCas9 with puromycin resistance (Adapted from plasmid #68416)DepositorInsertdCas9
UseCRISPRExpressionMammalianMutationD10A/H841APromoterCMVAvailable SinceJuly 31, 2024AvailabilityAcademic Institutions and Nonprofits only -
-
-
-
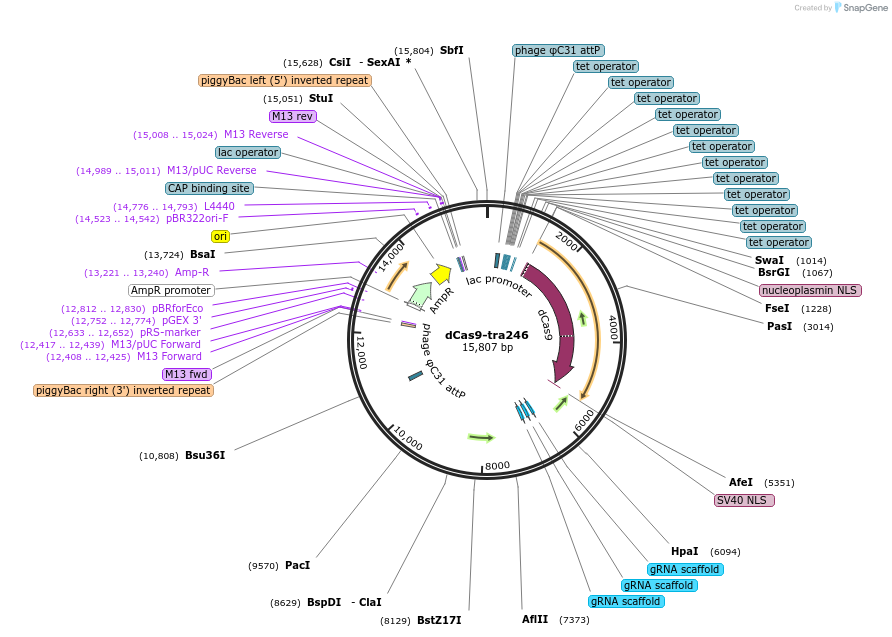
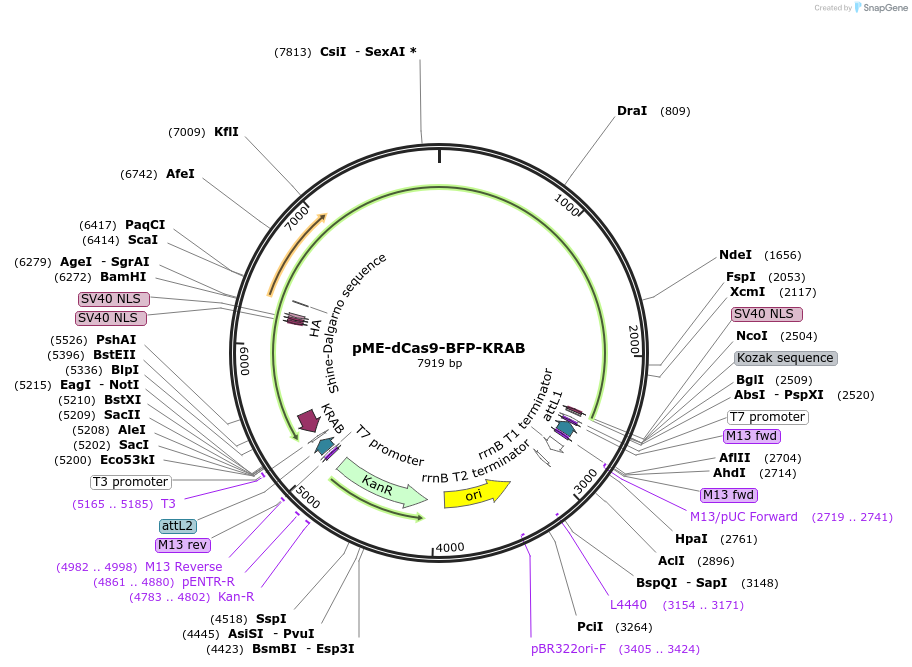
pME-dCas9-BFP-KRAB
Plasmid#221502PurposeCRISPRi machinery fish codon optimizedDepositorInsertdCas9-tagBFP-KRAB
UseFish expressionMutationD10A and H839A point mutationsAvailable SinceJuly 17, 2024AvailabilityAcademic Institutions and Nonprofits only -
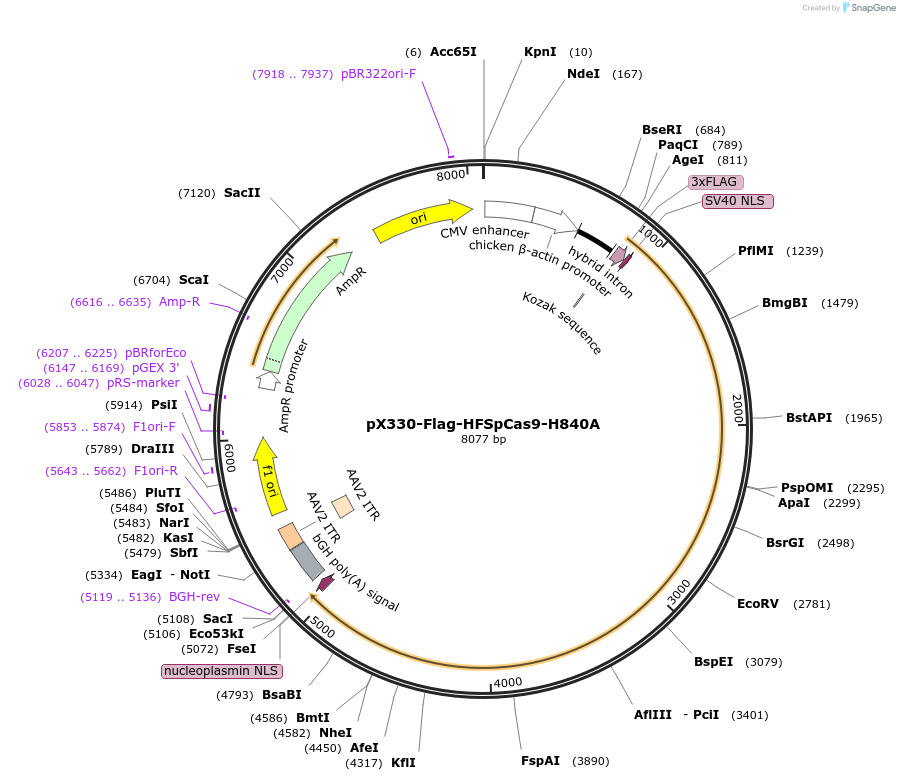
pX330-Flag-HFSpCas9-H840A
Plasmid#80455Purposehigh fidelity SpCas9 mammalian expression vectorDepositorInsertHFSpCas9_H840A
UseCRISPRTags3xFLAG and NLSExpressionMammalianPromoterCBhAvailable SinceSept. 6, 2016AvailabilityAcademic Institutions and Nonprofits only -
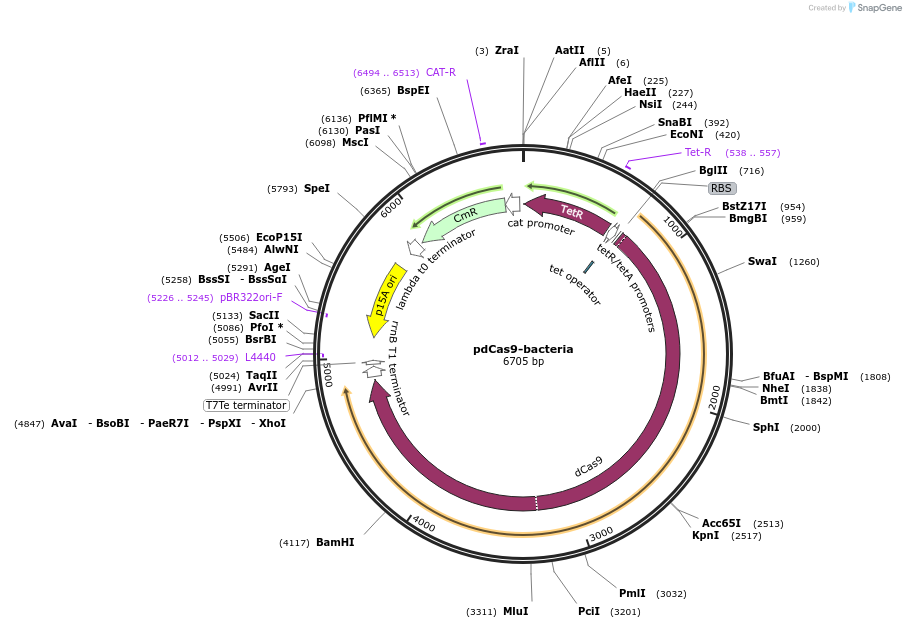
pdCas9-bacteria
Plasmid#44249PurposeaTc-inducible expression of a catalytically inactive bacterial Cas9 (S. pyogenes) for bacterial gene knockdownDepositorInsertdCas9 (bacteria)
UseCRISPRExpressionBacterialMutationD10A H840A (catalytically inactive)PromoterpLtetO-1Available SinceApril 11, 2013AvailabilityAcademic Institutions and Nonprofits only