We narrowed to 556 results for: rho.2
-
Pooled Library#174296PurposeThis library contains mutations to SARS-CoV-2 Spike RBD N501Y variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only
-
SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) library E484K Tile 2 (positions 437-527)
Pooled Library#174294PurposeThis library contains mutations to SARS-CoV-2 Spike RBD E484K variant in which all surface exposed residues on S RBD (Tile 2, positions 437-527) were mutated to every other amino acid.DepositorExpressionYeastAvailable SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library A
Pooled Library#157971PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
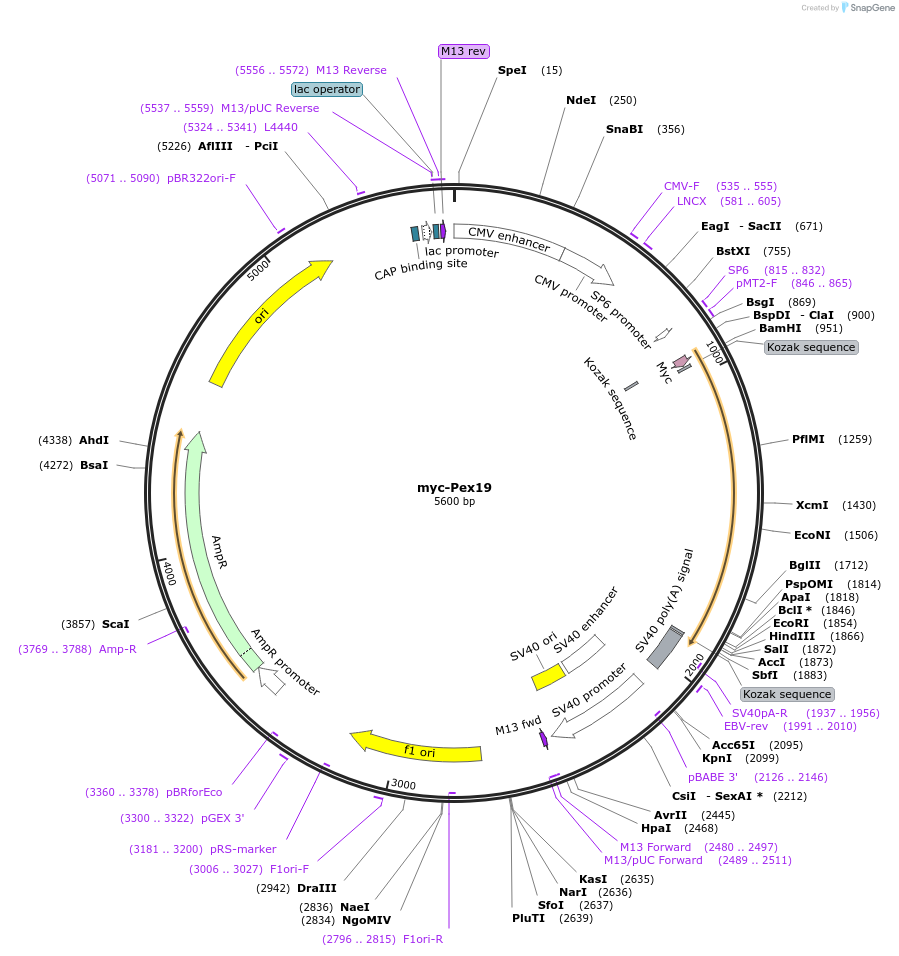
myc-Pex19
Plasmid#127625PurposeN-terminal myc-tagged mouse Pex19DepositorAvailable SinceOct. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library C
Pooled Library#157973PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
SARS-CoV-2 Spike (S) Ectodomain Library B
Pooled Library#157972PurposeEncodes Spike ectodomain mutations that stabilize 'up' prefusion conformationDepositorExpressionBacterial and YeastSpeciesSars-cov-2Available SinceSept. 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
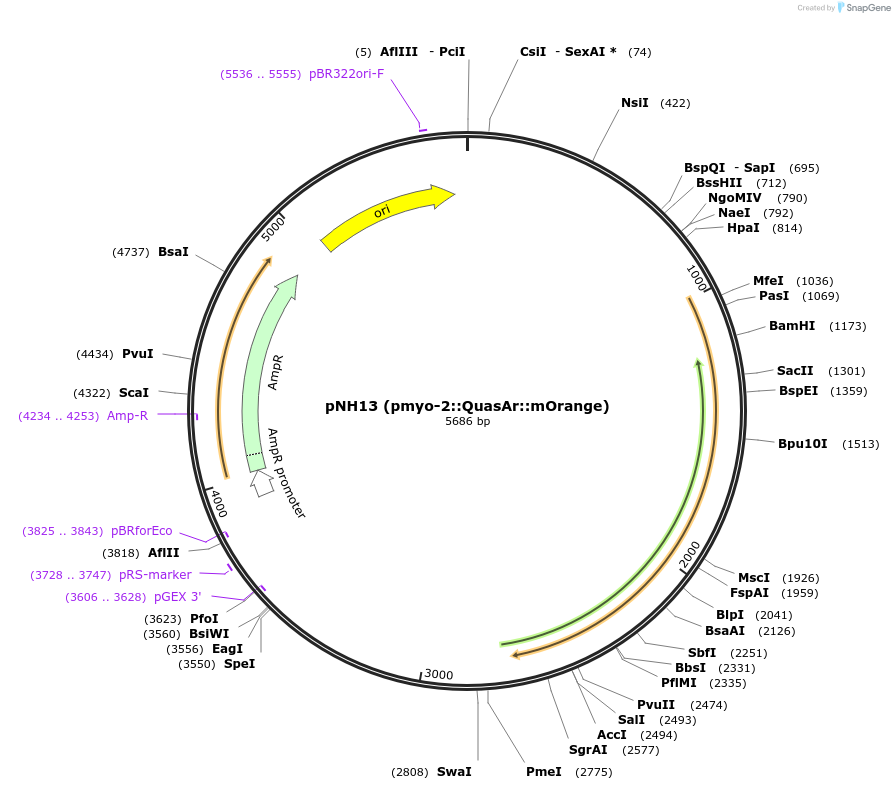
pNH13 (pmyo-2::QuasAr::mOrange)
Plasmid#130273PurposeExpresses the eFRET-based voltage sensor QuasAr::mOrange in the pharynx of C. elegans.DepositorInsertQuasAr::mOrange
TagsmOrangeExpressionWormAvailable SinceAug. 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
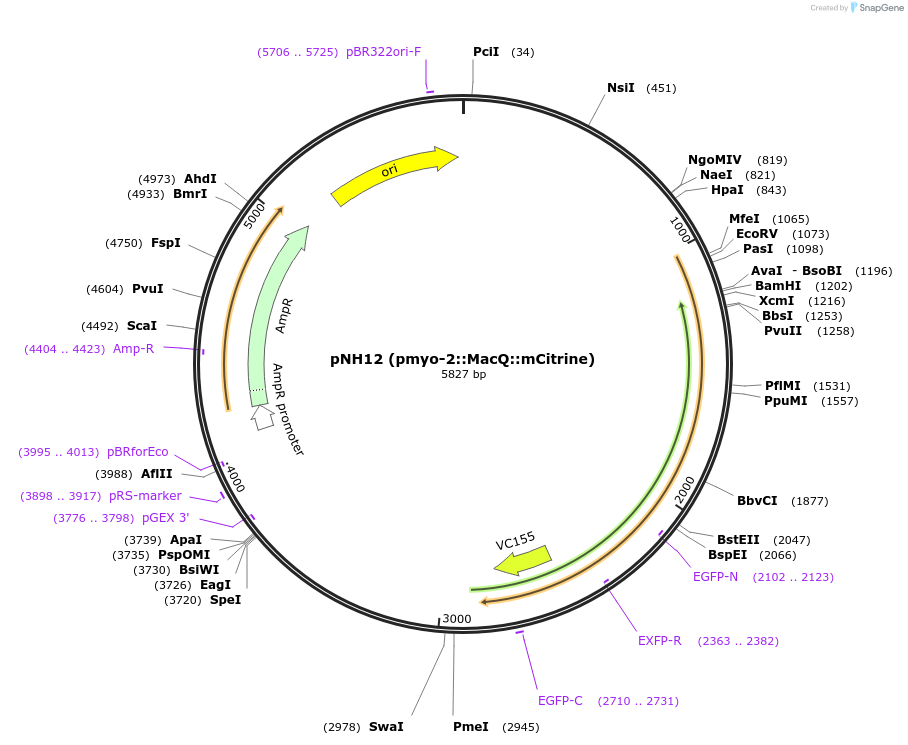
pNH12 (pmyo-2::MacQ::mCitrine)
Plasmid#130274PurposeExpresses the eFRET-based voltage sensor MacQ::mCitrine in the pharynx of C. elegans.DepositorInsertMacQ::mCitrine
TagsmCitrineExpressionWormAvailable SinceAug. 27, 2019AvailabilityAcademic Institutions and Nonprofits only -
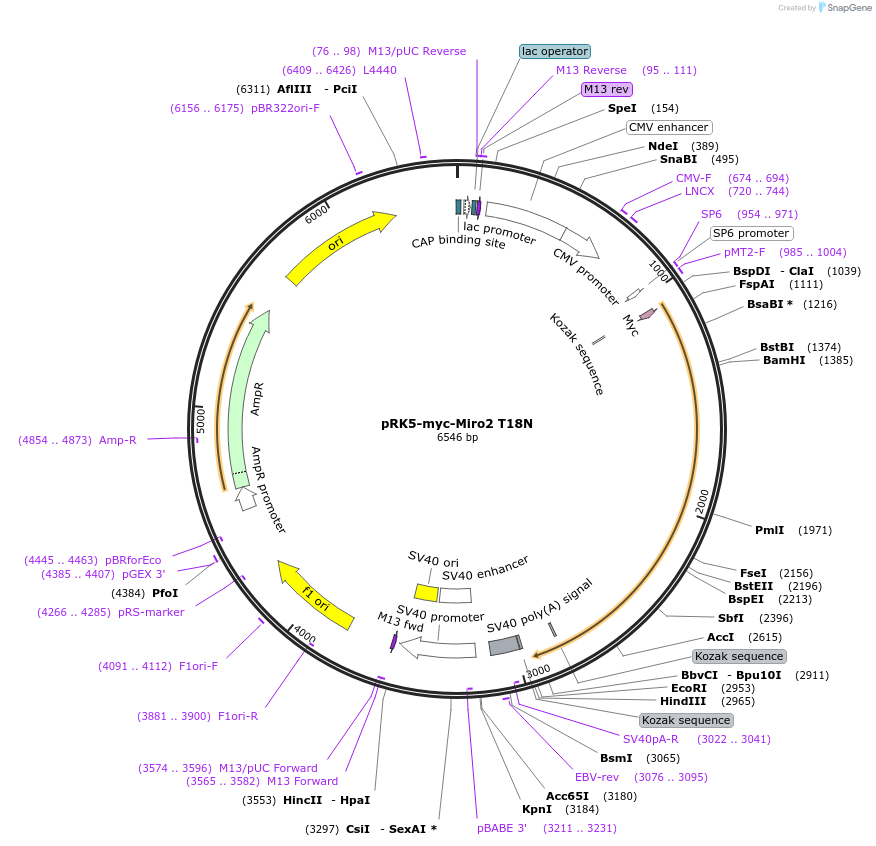
pRK5-myc-Miro2 T18N
Plasmid#47897PurposeExpresses myc tagged Miro2 T18N dominant negative mutantDepositorInsertMiro2 T18N (RHOT2 Human)
TagsmycExpressionMammalianMutationT18N, dominant negative mutantPromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pRK5-myc-Miro2 A13V
Plasmid#47896PurposeExpresses myc tagged Miro2 A13V constitutively active mutantDepositorInsertMiro2 A13V (RHOT2 Human)
TagsmycExpressionMammalianMutationA13V, constitutively active mutantPromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pRK5-myc-Miro2 E208K/E328K
Plasmid#47900PurposeExpresses myc tagged Miro2 V208K/E328K mutantDepositorInsertMiro2 E208K/E328K (RHOT2 Human)
TagsmycExpressionMammalianMutationE208K/E328K, abolishes calcium bindingPromoterCMVAvailable SinceSept. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
pRK5-myc-Miro2 R425V
Plasmid#47898PurposeExpresses myc tagged Miro2 R425V constitutively active mutantDepositorInsertMiro2 R425V (RHOT2 Human)
TagsmycExpressionMammalianMutationR425V, constitutively active mutantPromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pRK5-myc-Miro2 S430N
Plasmid#47899PurposeExpresses myc tagged Miro2 S430N dominant negative mutantDepositorInsertMiro2 S430N (RHOT2 Human)
TagsmycExpressionMammalianMutationS430N, dominant negative mutantPromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
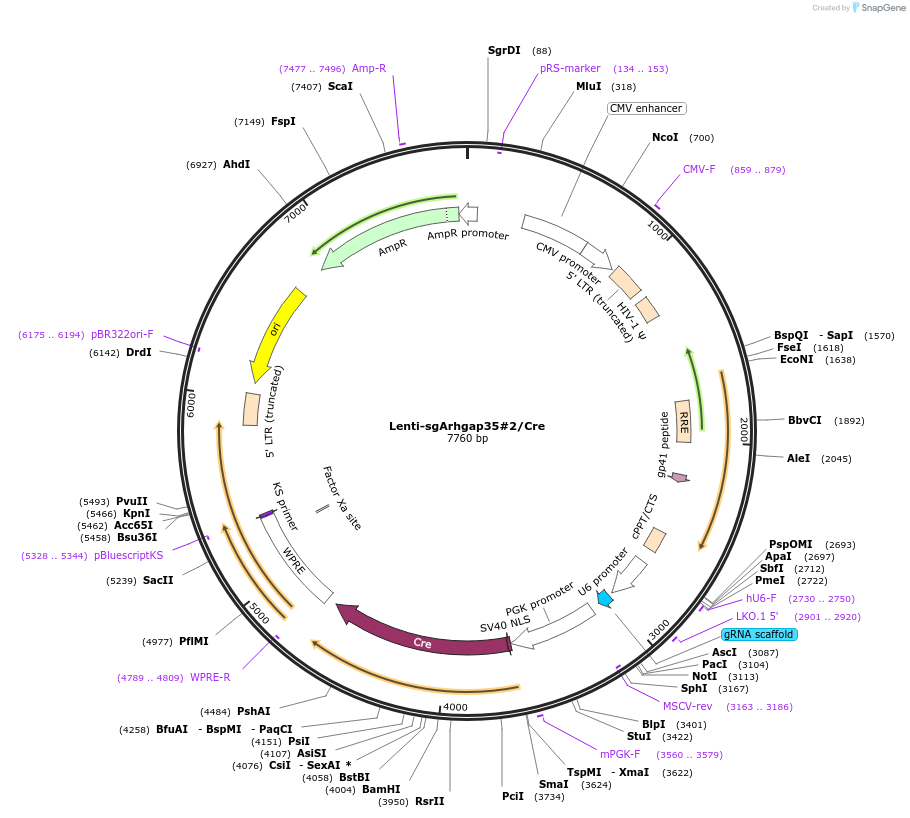
Lenti-sgArhgap35#2/Cre
Plasmid#173570PurposeExpresses a Arhgap35-targeting gRNA and Cre-recombinaseDepositorInsertgRNA targeting Arhgap35 (Grlf1 Mouse)
UseCRISPR, Cre/Lox, and LentiviralAvailable SinceApril 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
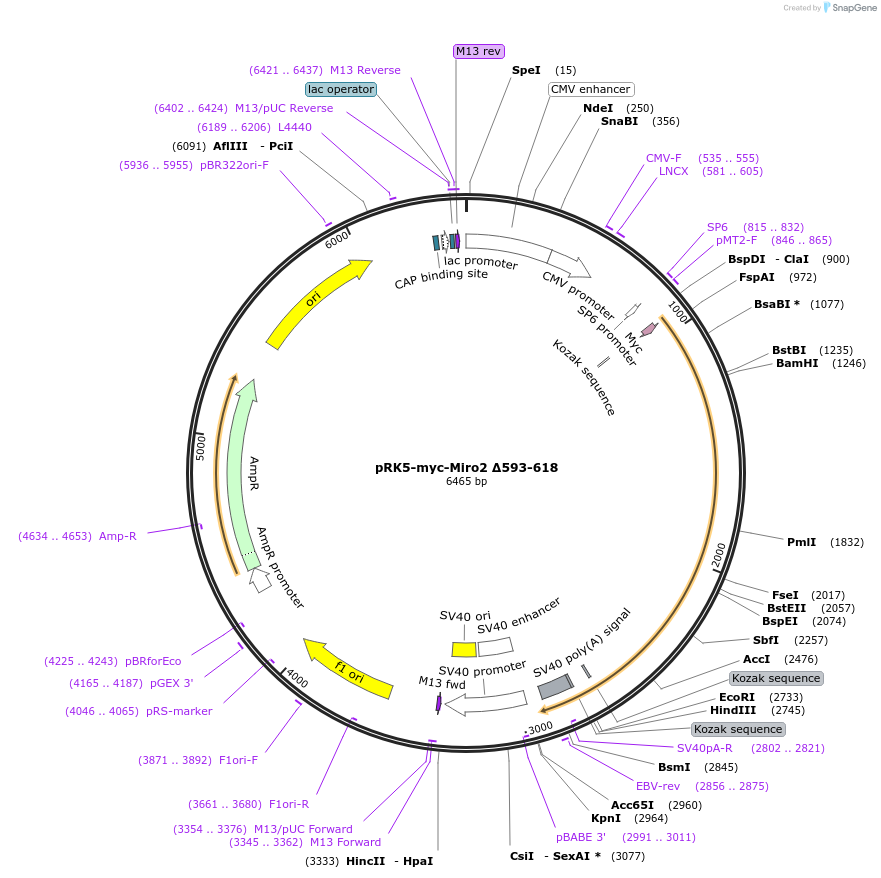
pRK5-myc-Miro2 Δ593-618
Plasmid#47901PurposeExpresses myc tagged Miro2 lacking transmembrane domainDepositorInsertMiro2 Δ593-618 (RHOT2 Human)
TagsmycExpressionMammalianMutationΔ593-618, lacks transmembrane domain. Also see De…PromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
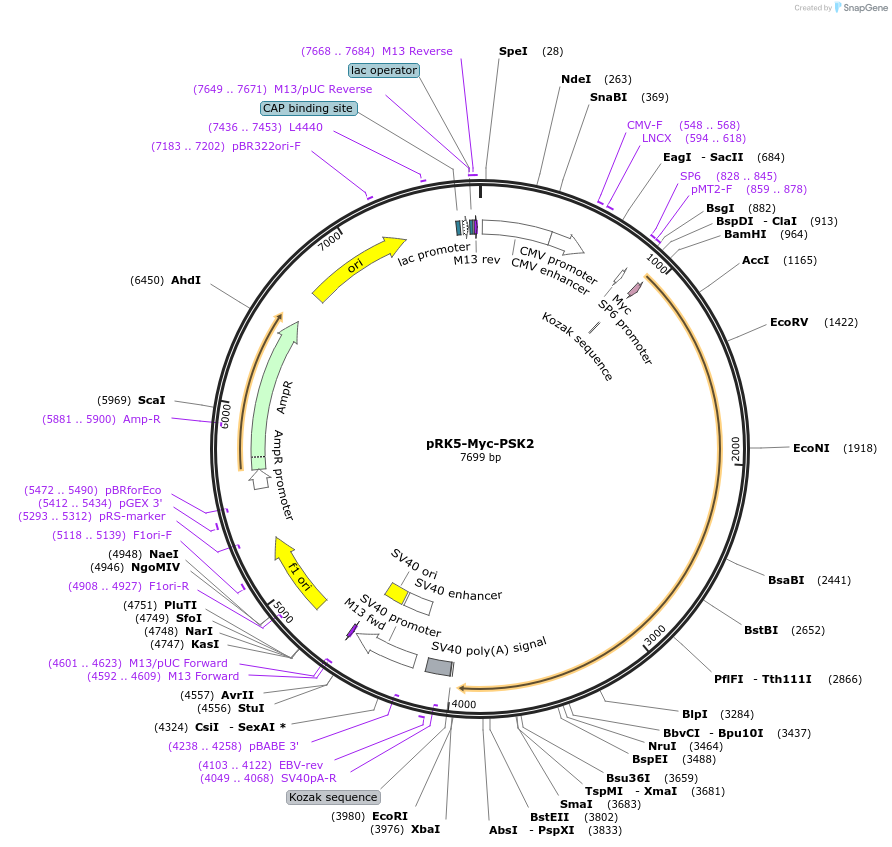
pRK5-Myc-PSK2
Plasmid#197117PurposeExpression of Myc-tagged PSK2 / TAOK1 in mammalian cellsDepositorAvailable SinceMarch 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
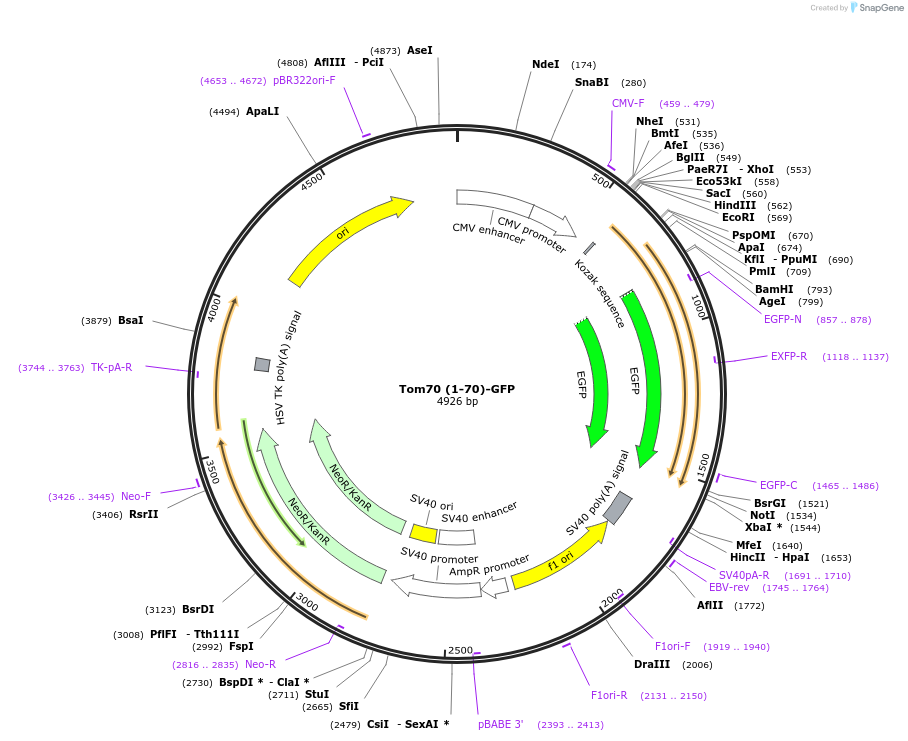
Tom70 (1-70)-GFP
Plasmid#127633PurposeMitochondrial targeting sequence of Tom70 fused to GFPDepositorAvailable SinceOct. 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
pRK5-myc-Miro1 K427V
Plasmid#47892PurposeExpresses myc tagged Miro1 K427 constitutively active mutantDepositorInsertMiro1 K427V (RHOT1 Human)
TagsmycExpressionMammalianMutationK427V, constitutively active mutantPromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
pRK5-myc-Miro1 S432N
Plasmid#47893PurposeExpresses myc tagged Miro1 S432N dominant negative mutantDepositorInsertMiro1 S432N (RHOT1 Human)
TagsmycExpressionMammalianMutationS432N, dominant negative mutantPromoterCMVAvailable SinceSept. 12, 2013AvailabilityAcademic Institutions and Nonprofits only -
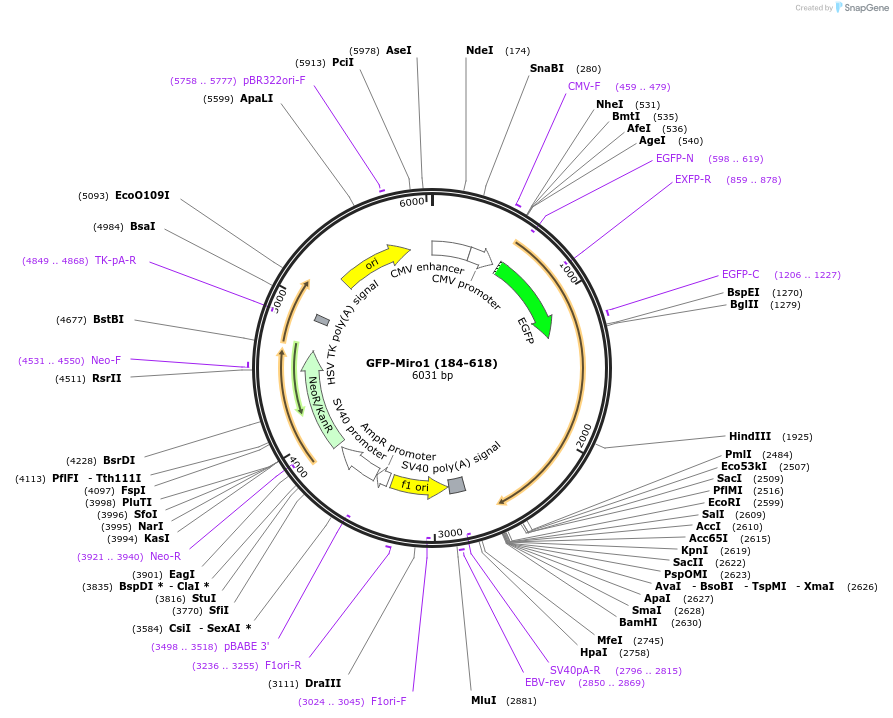
GFP-Miro1 (184-618)
Plasmid#127626PurposeHuman Miro1 lacking the first GTPase domainDepositorAvailable SinceOct. 20, 2020AvailabilityAcademic Institutions and Nonprofits only