We narrowed to 40,709 results for: kan
-
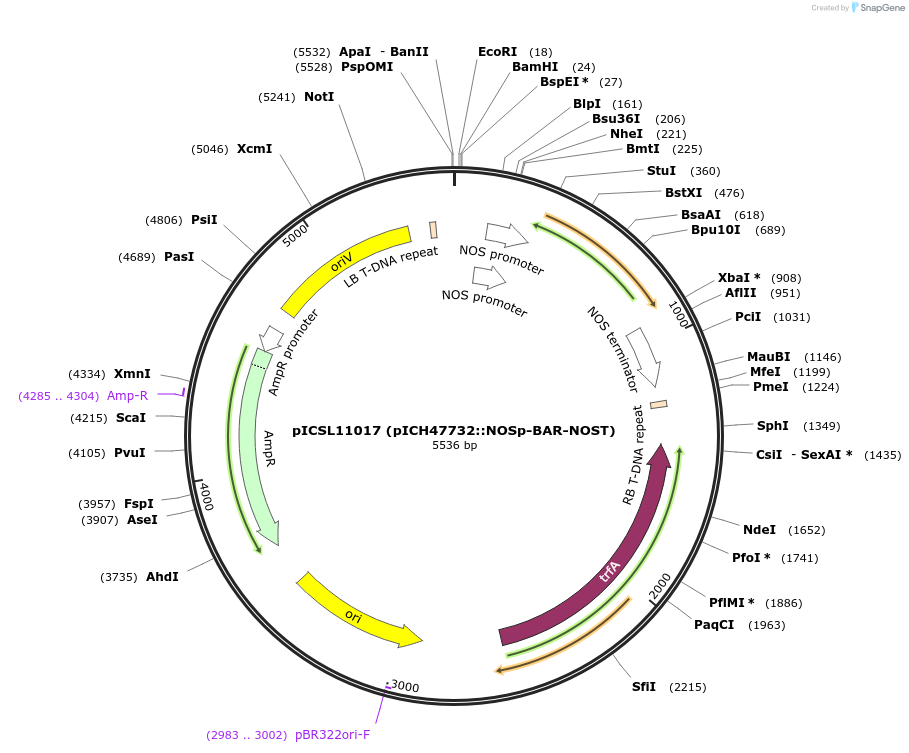
Plasmid#51145PurposeConfers Basta resistance in plantaDepositorInsertNOSp-BAR-NOST
Available SinceFeb. 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
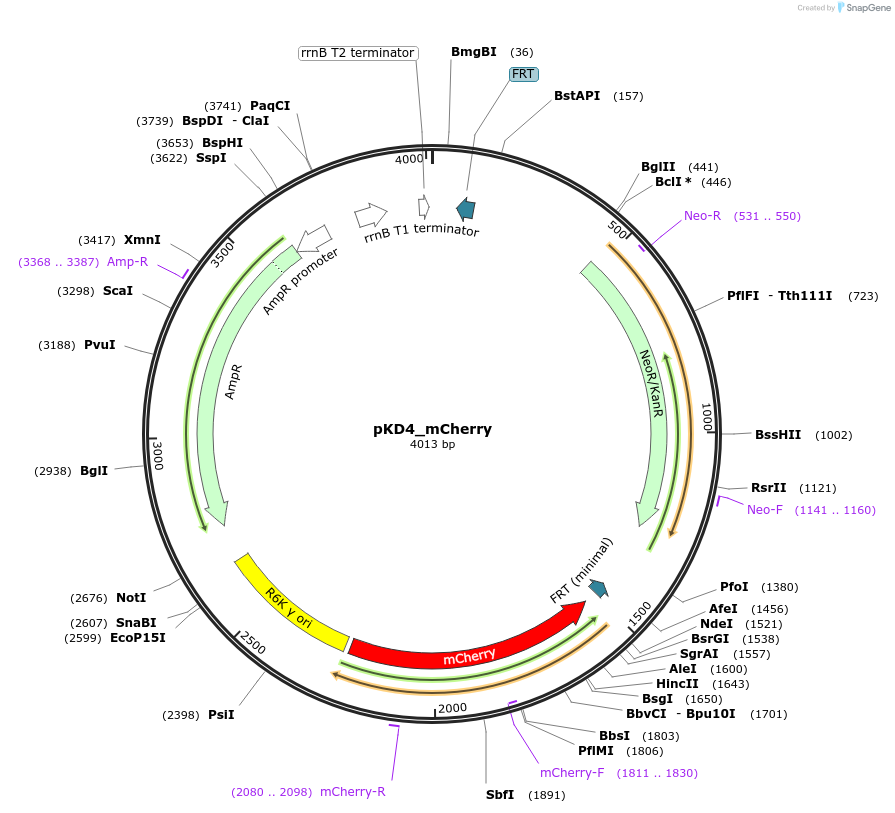
pKD4_mCherry
Plasmid#187386PurposeTo chromosomally tag genes of interest with mCherry. FRT-mCherry-kan-FRTDepositorInsertmCherry
ExpressionBacterialAvailable SinceSept. 8, 2022AvailabilityAcademic Institutions and Nonprofits only -
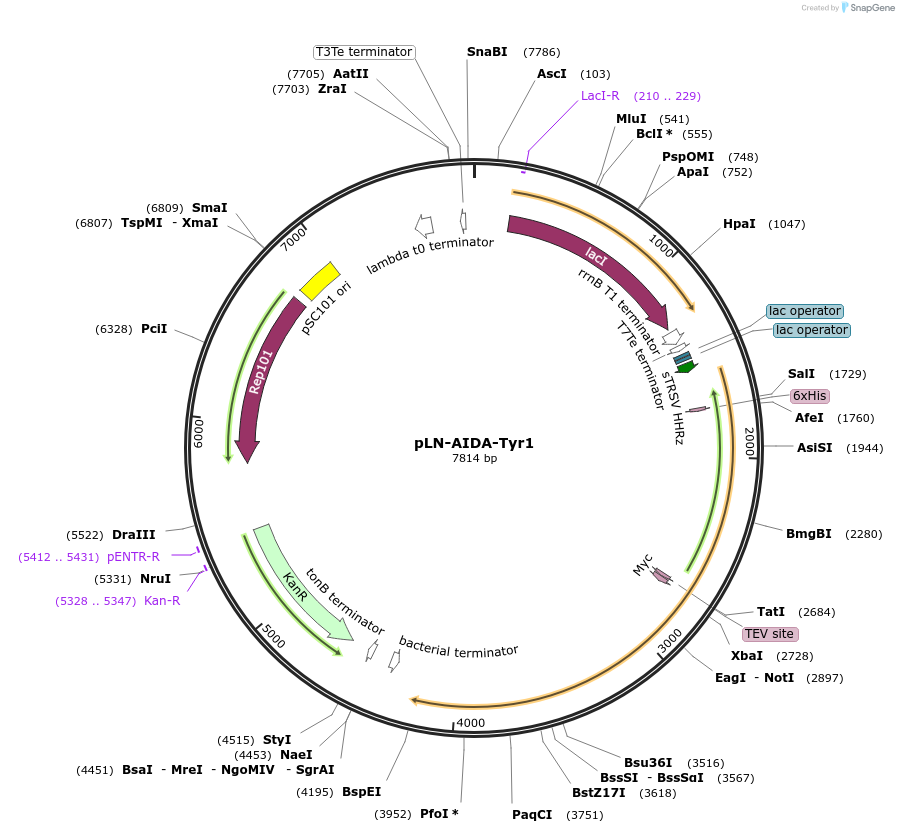
pLN-AIDA-Tyr1
Plasmid#192831PurposeIPTG inducible AIDA-Tyrosinase expressionDepositorInsertAIDA-Tyr1
ExpressionBacterialPromoterpL-lacAvailable SinceFeb. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
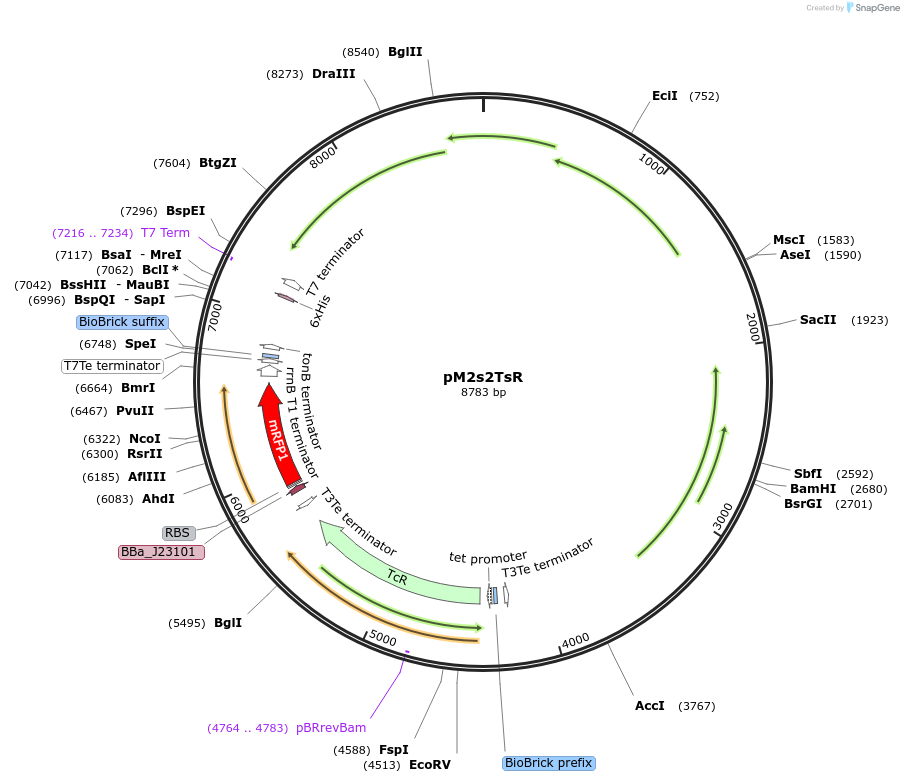
pM2s2TsR
Plasmid#137923PurposeE. coli Nissle pMUT2-derived plasmid with tetracycline resistance and constitutive RFP expressionDepositorInsertmRFP1
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterJ23101Available SinceJan. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
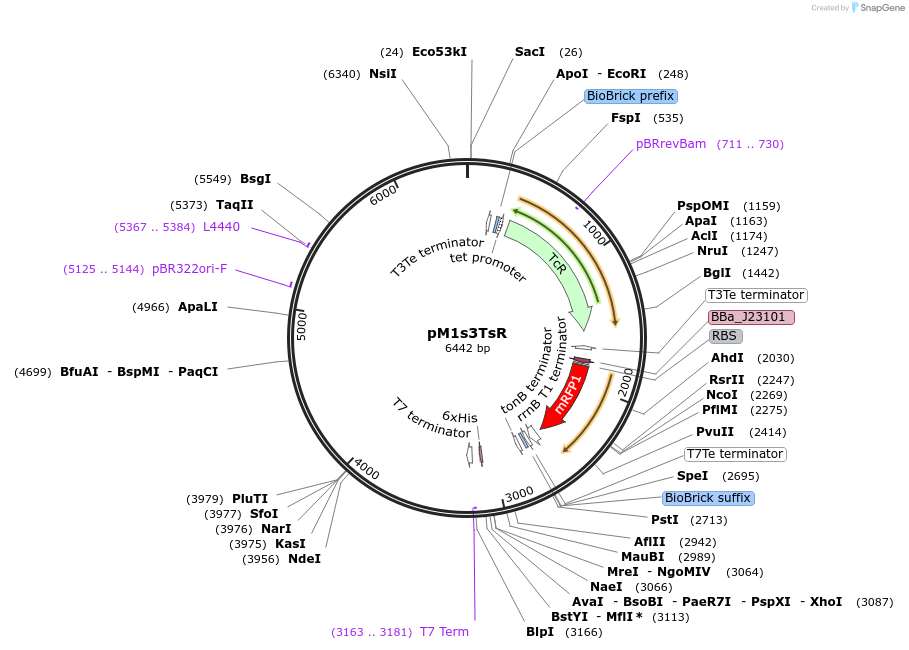
pM1s3TsR
Plasmid#137922PurposeE. coli Nissle pMUT1-derived plasmid with tetracycline resistance and constitutive RFP expressionDepositorInsertmRFP1
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterJ23101Available SinceFeb. 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
AID* Kit
Plasmid Kit#1000000120PurposePlasmid for fusion of a protein of interest to the auxin-inducible degron to control protein levels in vivo.DepositorApplicationGene Expression and LabelingVector TypeYeast ExpressionAvailable SinceJan. 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
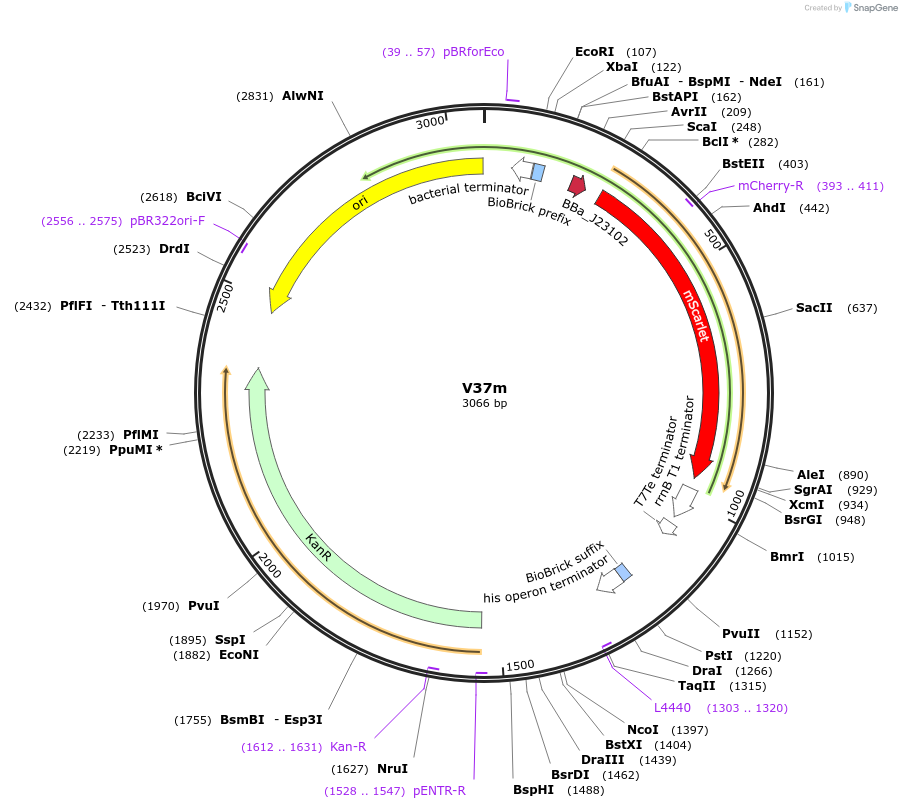
V37m
Plasmid#121043PurposeMoClo golden gate assembly AE destination vector DVCK-AE-mScar (ColE1/kan resistance destination vector with constitutive mScarlet expressing for screening.DepositorInsertKan destination vector with ColE1 origin and mScarlet selection
UseSynthetic BiologyAvailable SinceDec. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
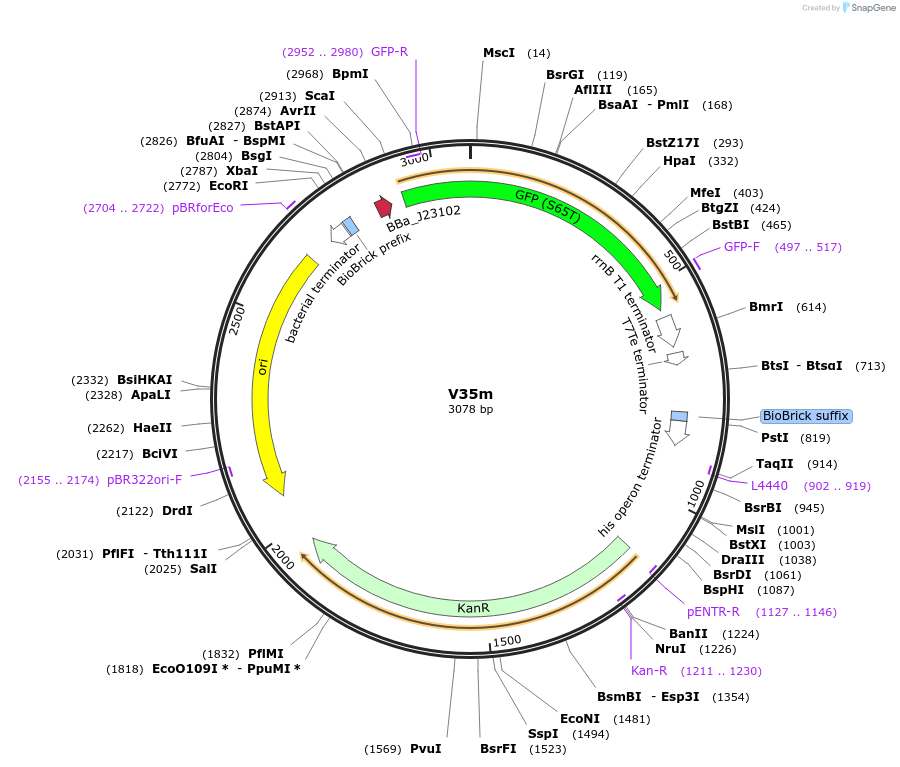
V35m
Plasmid#121042PurposeMoClo golden gate assembly AE destination vector DVCK-AE-GFP (ColE1/kan resistance destination vector with constitutive GFP expression for screening.DepositorInsertKan destination vector with ColE1 origin and GFP selection
UseSynthetic BiologyAvailable SinceDec. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
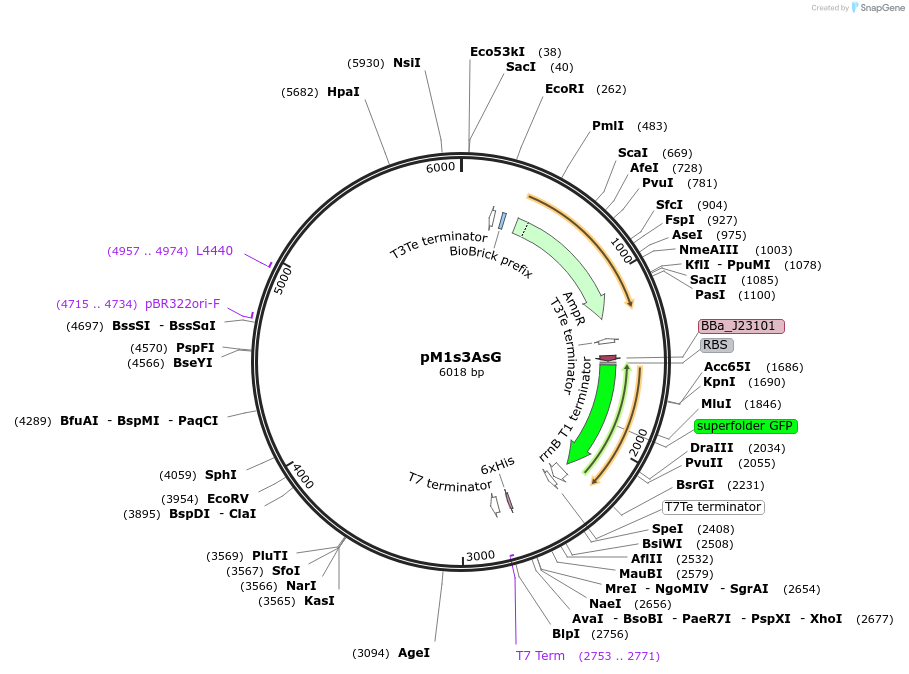
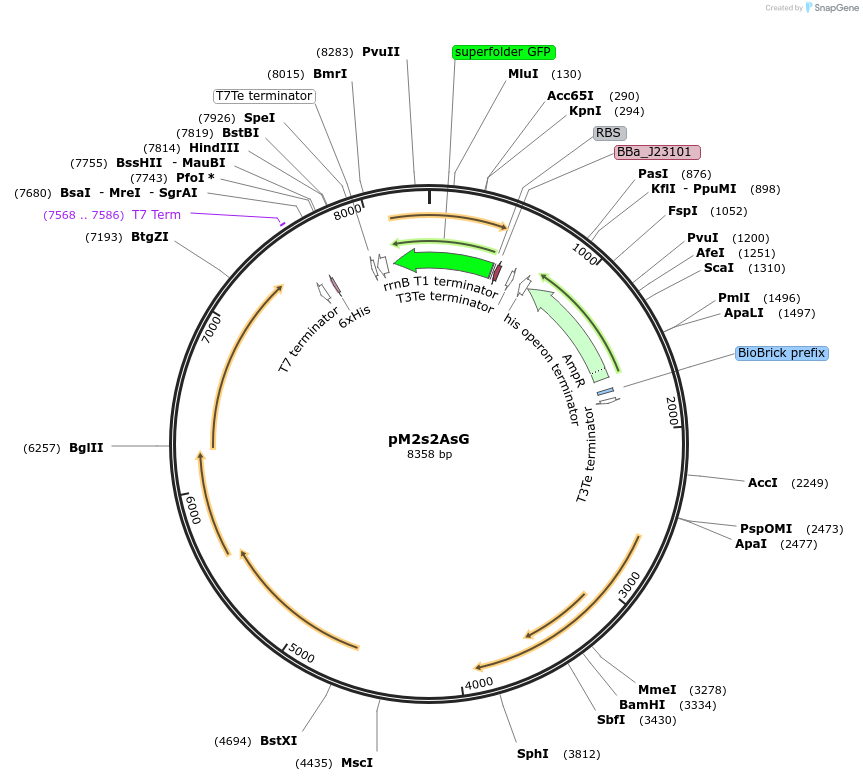
pM1s3AsG
Plasmid#137921PurposeE. coli Nissle pMUT1-derived plasmid with ampicillin resistance and constitutive GFP expressionDepositorInsertSuperfolder GFP
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterJ23101Available SinceJan. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
pM2s2AsG
Plasmid#137924PurposeE. coli Nissle pMUT2-derived plasmid with ampicillin resistance and constitutive GFP expressionDepositorInsertSuperfolder GFP
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterJ23101Available SinceMay 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
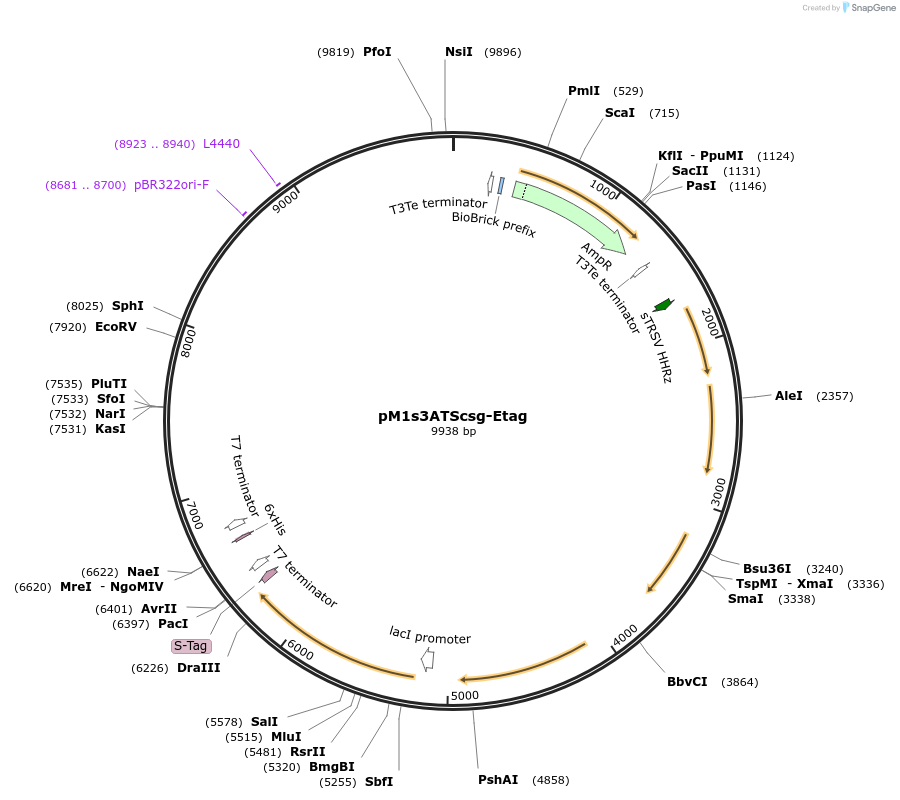
pM1s3ATScsg-Etag
Plasmid#137946PurposeE. coli Nissle pMUT1-derived plasmid with ampicillin resistance and temperature sensitive curli expressionDepositorInsertsSynthetic curli operon
Synthetic curli operon
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterpTlpA36Available SinceApril 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
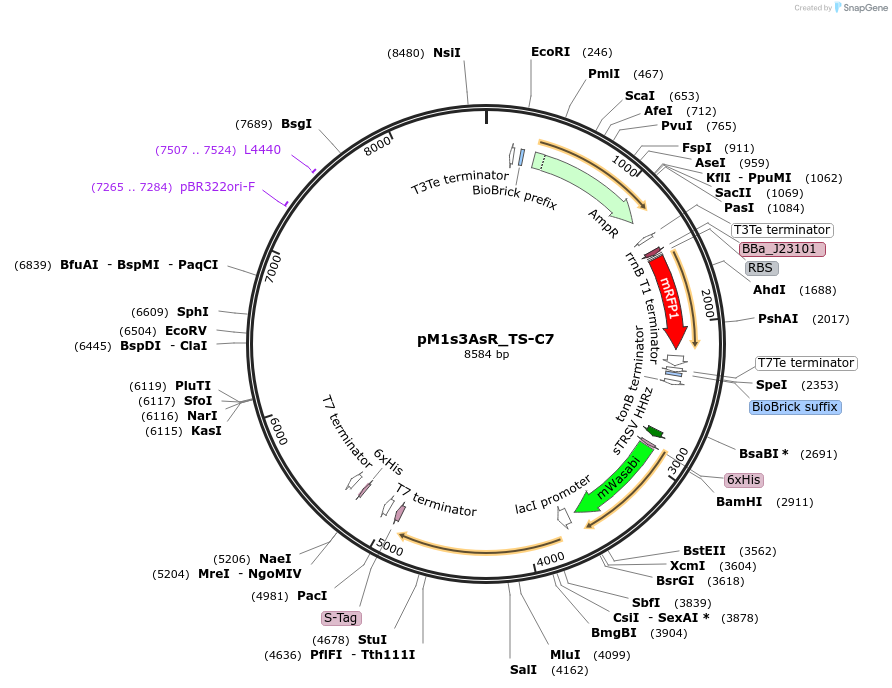
pM1s3AsR_TS-C7
Plasmid#137945PurposeE. coli Nissle pMUT1-derived plasmid with ampicillin resistance and constitutive RFP expression and strong temperature sensitive GFP expressionDepositorInsertmRFP1
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterJ23101Available SinceApril 27, 2021AvailabilityAcademic Institutions and Nonprofits only -
pM2s2AsR_TS-E10
Plasmid#137944PurposeE. coli Nissle pMUT2-derived plasmid with ampicillin resistance and constitutive RFP expression and strong temperature sensitive GFP expressionDepositorInsertsmRFP1
mWasabi
UseSynthetic Biology; For use with e. coli nissle in…ExpressionBacterialPromoterJ23101 and pTlpA-C7Available SinceMay 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
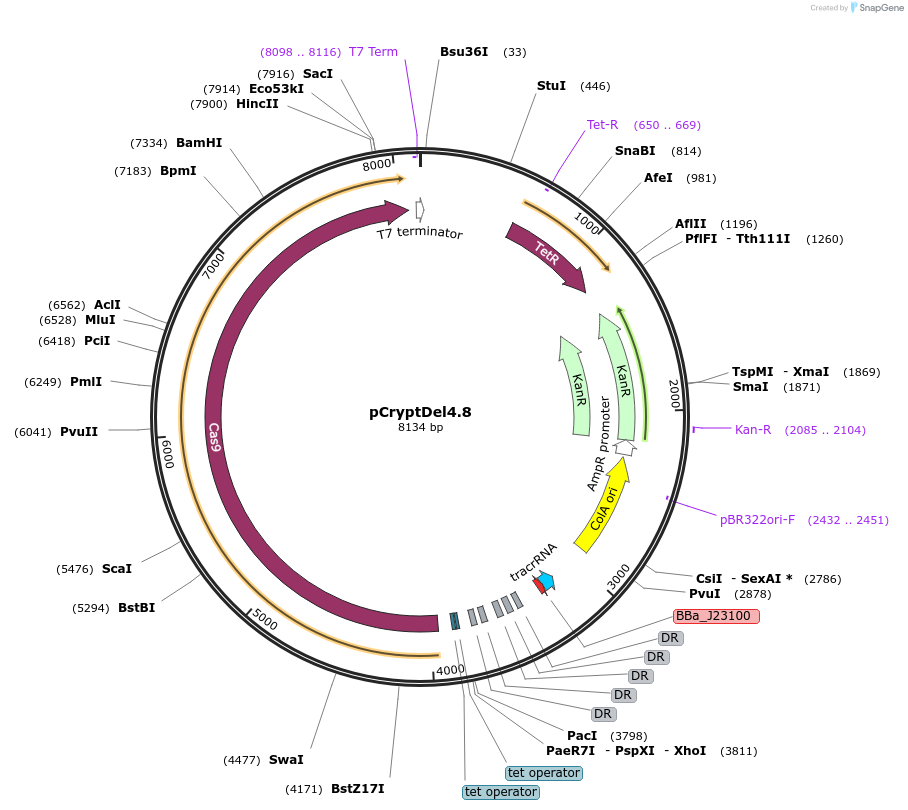
pCryptDel4.8
Plasmid#141293PurposePlasmid for curing pMUT2 in one step based on pFREE. Contains RelB antitoxin, as well as gRNA targetting pMUT2 plasmid as well as pCryptDel4.8 itself.DepositorInsertsRelB
gRNA targetting pMUT2
UseCRISPR and Synthetic BiologyExpressionBacterialPromoterNative promoter from pMUT2 relB/relE operon and P…Available SinceJan. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
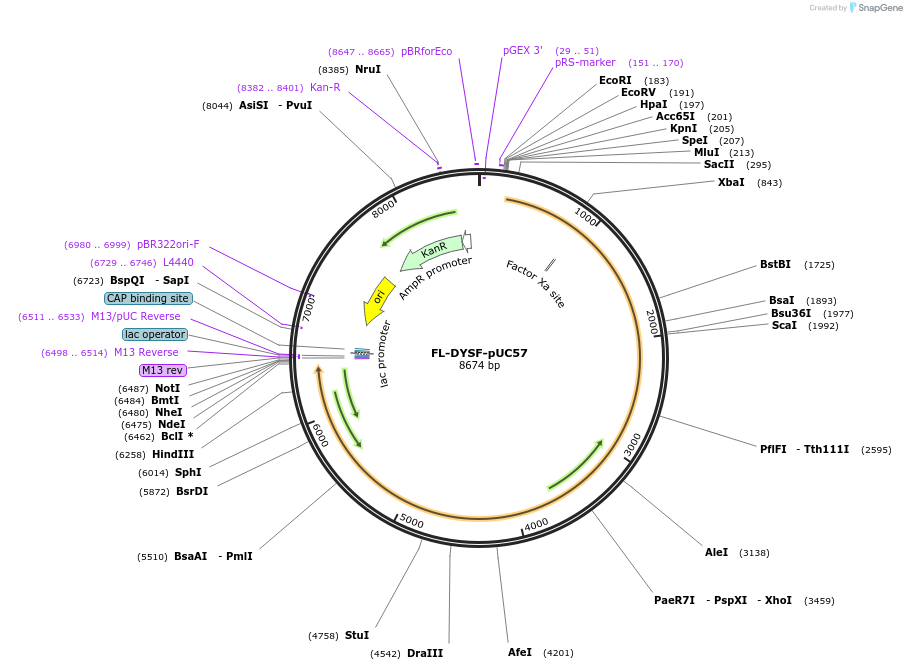
FL-DYSF-pUC57
Plasmid#60177PurposePlasmid contains full-length human dysferlin in a multiple cloning site of pUC57-Kan and can be easily cloned into other vectors.DepositorInsertDysferlin (DYSF Human)
UseCloning vectorAvailable SinceOct. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
CHLOROMODAS Kit
Plasmid Kit#1000000269PurposePlasmids for for engineering the Chlamydomonas reinhardtii chloroplast.DepositorApplicationCloning and Synthetic BiologyVector TypeAlgal ExpressionCloning TypeGolden Gate (MoClo)Available SinceNov. 7, 2025AvailabilityAcademic Institutions and Nonprofits only -
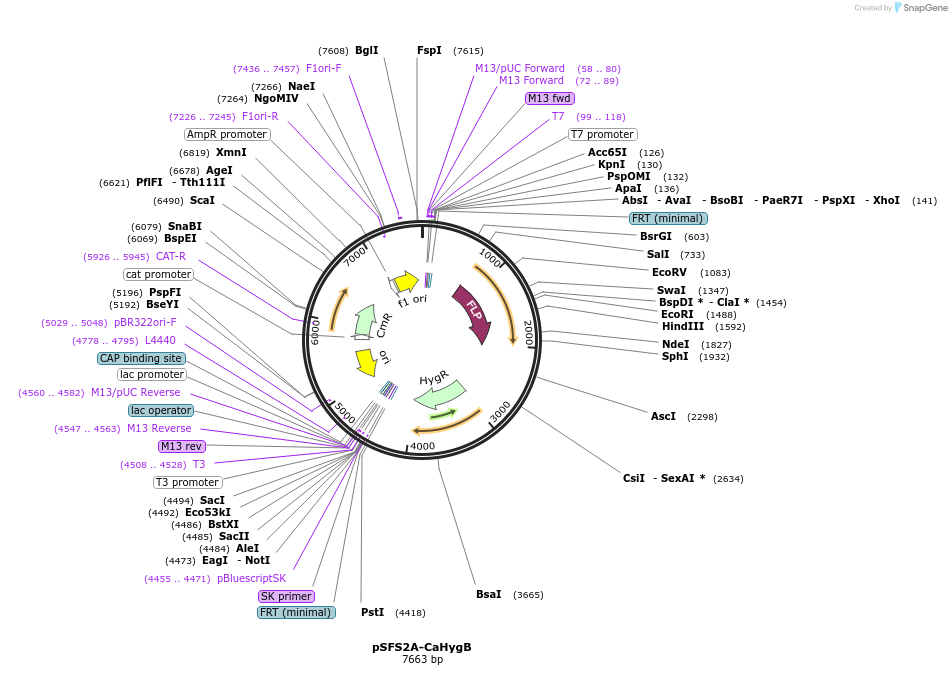
pSFS2A-CaHygB
Plasmid#189564PurposeCaHygB hygromycin resistance gene in the pSFS2A flipper backbone for genetic manipulation of Candida albicansDepositorInsertHygB
ExpressionYeastAvailable SinceAug. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
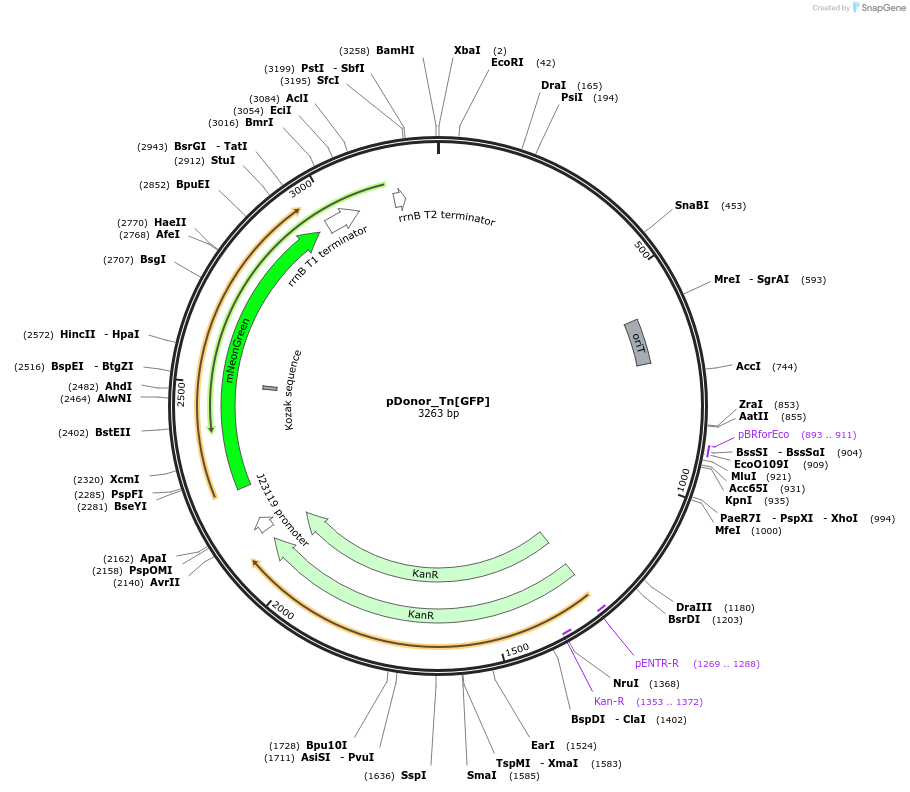
pDonor_Tn[GFP]
Plasmid#213903PurposeMariner transposon expressing mNeonGreen with kan selection casetteDepositorInsertmariner transposon with kan resistance and mNeonGreen
ExpressionBacterialAvailable SinceApril 9, 2024AvailabilityAcademic Institutions and Nonprofits only -
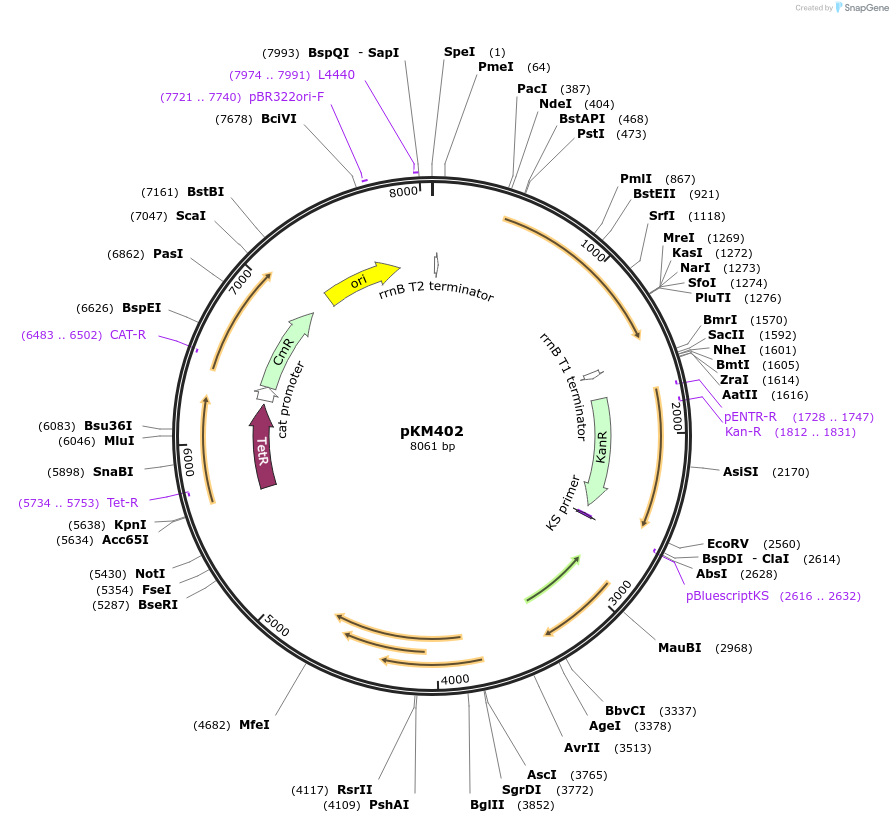
pKM402
Plasmid#107770PurposeExpression of phage Che9c RecT for oligo-mediated recombineering in mycobacteriaDepositorInsertsChe9C RecT
TetR (tet repressor)
cat
kan
ExpressionBacterialPromoterCat promoter, Ptet, kan promoter, and pTB21Available SinceMay 15, 2018AvailabilityAcademic Institutions and Nonprofits only -
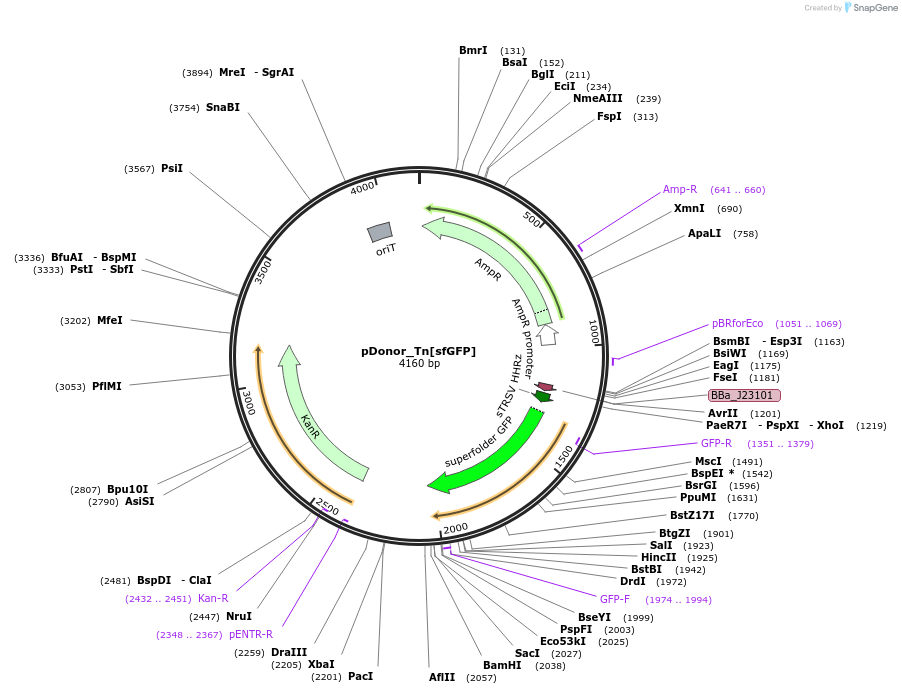
pDonor_Tn[sfGFP]
Plasmid#213902PurposeMariner transposon expressing sfGFP with kan selection casetteDepositorInsertmariner transposon with kan resistance and superfolder GFP
ExpressionBacterialAvailable SinceFeb. 14, 2024AvailabilityAcademic Institutions and Nonprofits only