We narrowed to 1,662 results for: MB;
-
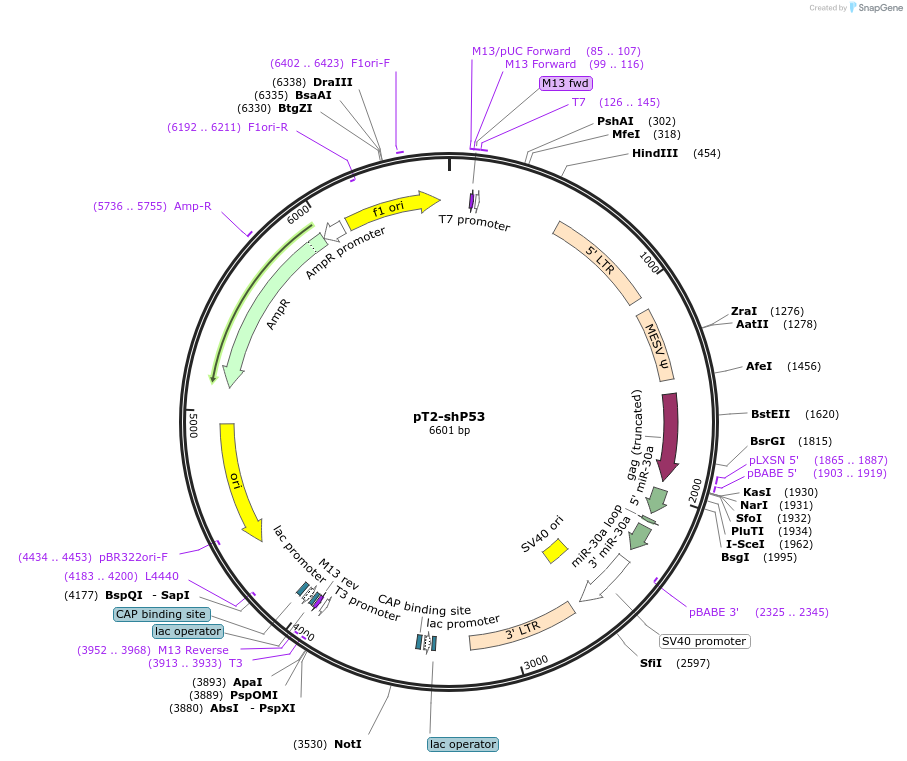
Plasmid#124261PurposeExpresses shRNA targeting P53. Construct has inverted repeats to be used in Sleeping beauty system.DepositorInsertshP53
ExpressionMammalianAvailable SinceApril 8, 2019AvailabilityIndustry, Academic Institutions, and Nonprofits -
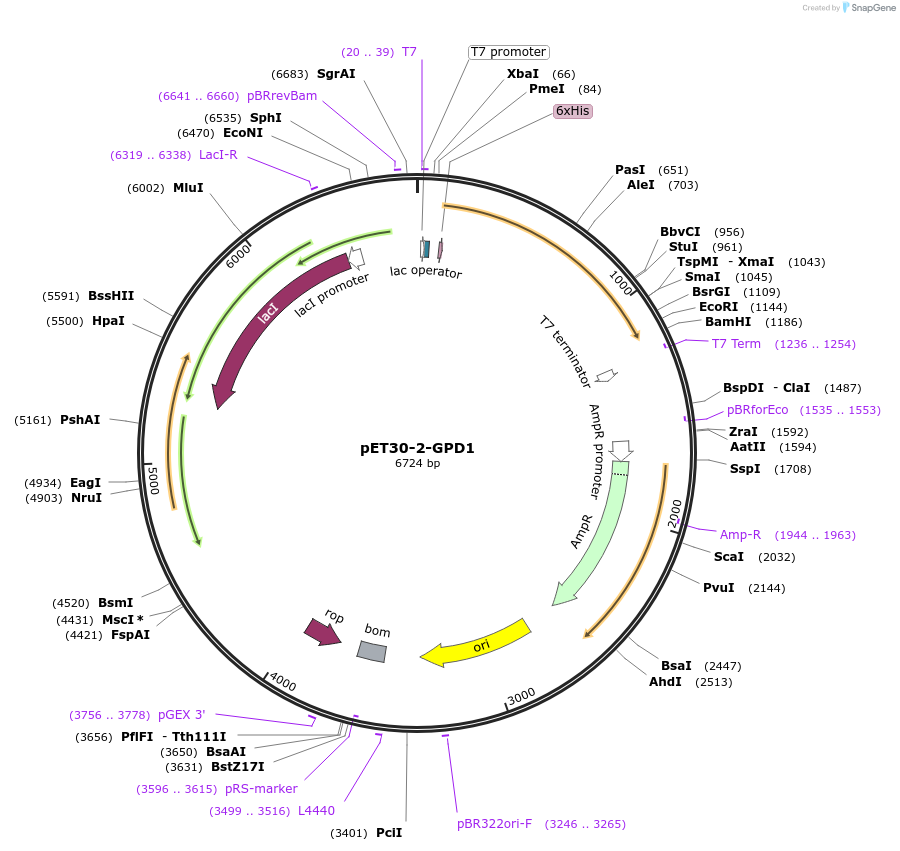
pET30-2-GPD1
Plasmid#83911Purposestable overexpressionDepositorInsertglycerol-3-phosphate dehydrogenase 1 (GPD1 Human)
Tags6x His tagsExpressionBacterialPromoterT7Available SinceOct. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
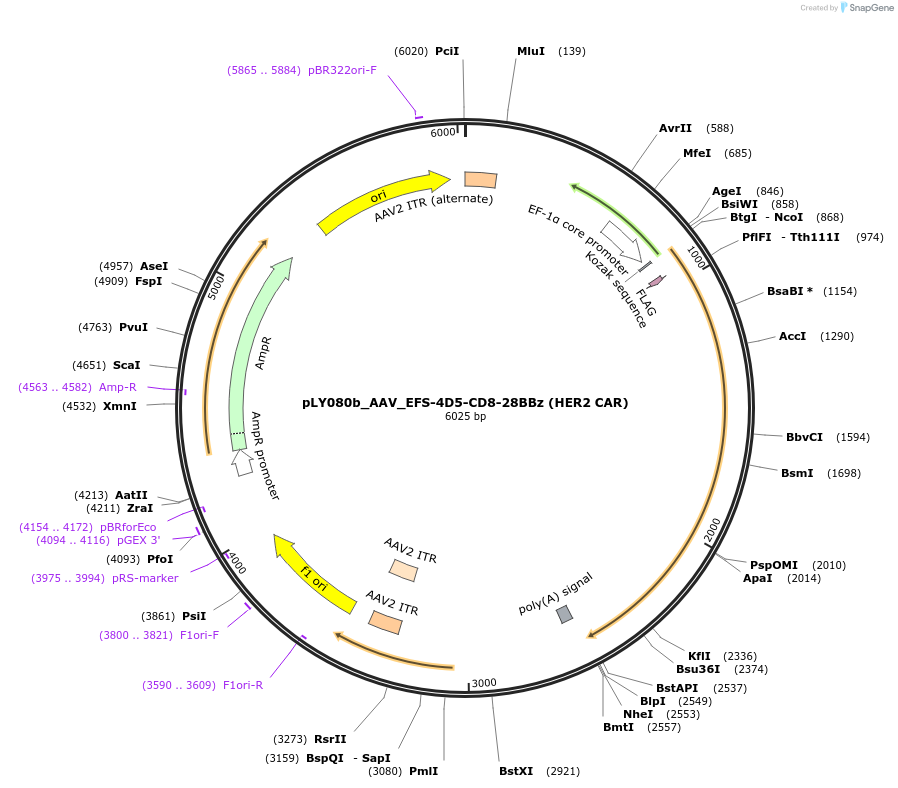
pLY080b_AAV_EFS-4D5-CD8-28BBz (HER2 CAR)
Plasmid#192190PurposeHER2 CAR AAV vector PRODH2 KI (pLY080b)DepositorInsertHER2 CAR AAV vector PRODH2 KI (pLY080b) (ERBB2 Synthetic)
UseAAV; Mammalian expressionMutationNAAvailable SinceOct. 24, 2022AvailabilityAcademic Institutions and Nonprofits only -
sgTP53_3
Plasmid#78164PurposesgTP53DepositorAvailable SinceJune 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
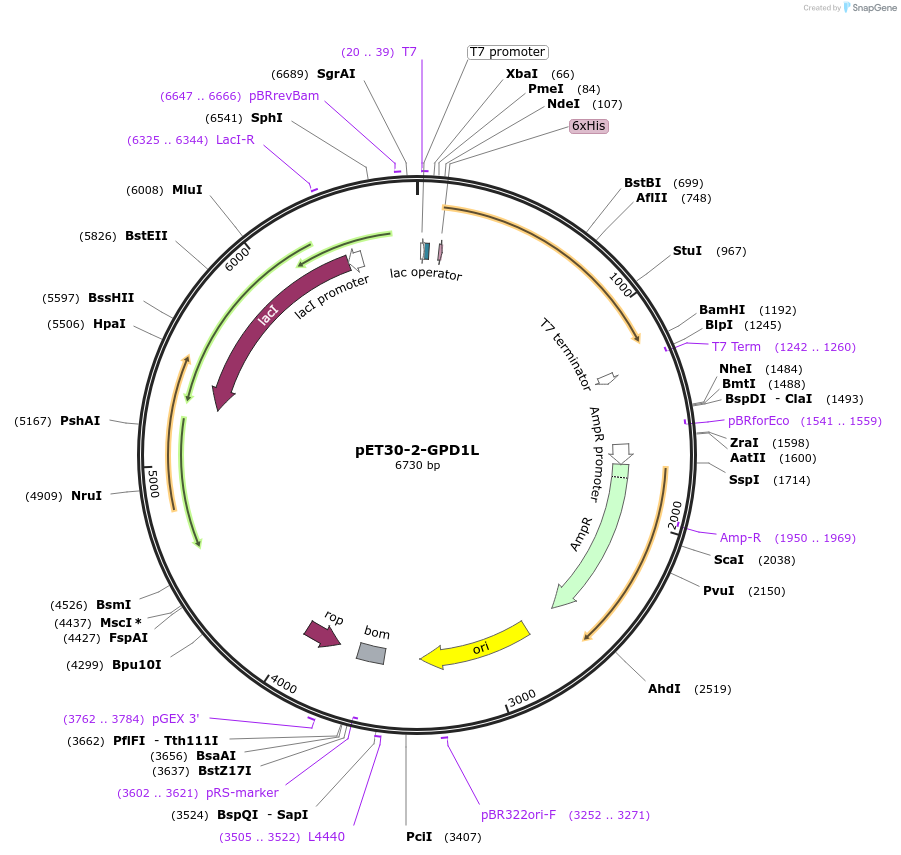
pET30-2-GPD1L
Plasmid#83912Purposestable overexpressionDepositorInsertglycerol-3-phosphate dehydrogenase 1-like (GPD1L Human)
Tags6x His tagsExpressionBacterialPromoterT7Available SinceOct. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
pET30-2-PHGDH
Plasmid#83907Purposestable overexpressionDepositorInsertPhosphoglycerate dehydrogenase (PHGDH Human)
Tags6x His tagsExpressionBacterialPromoterT7Available SinceOct. 21, 2016AvailabilityAcademic Institutions and Nonprofits only -
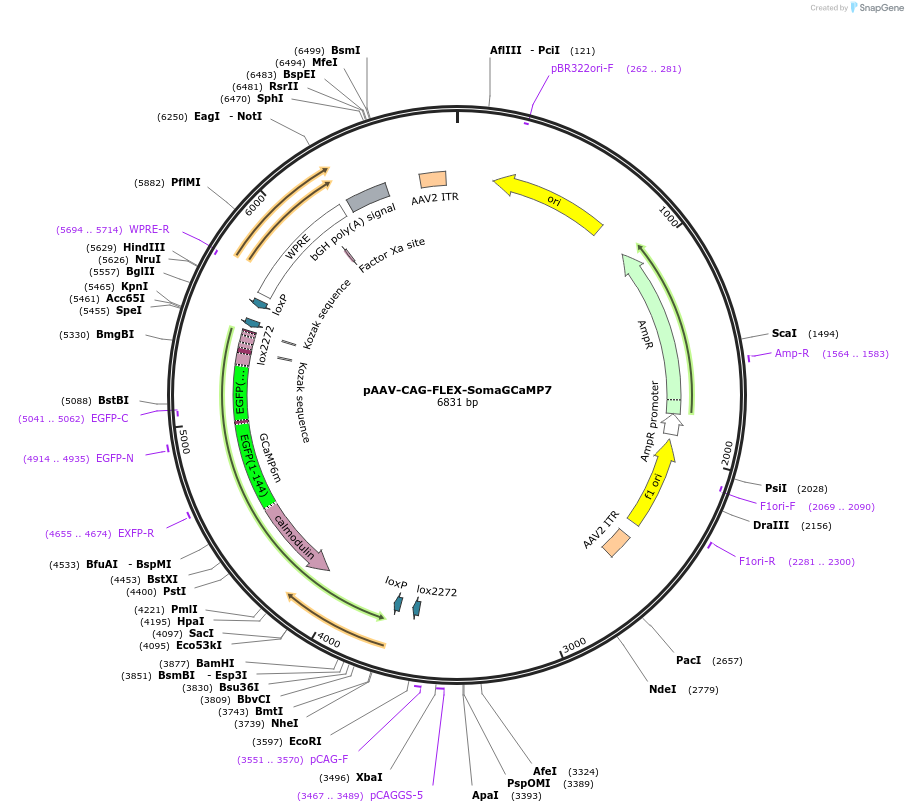
pAAV-CAG-FLEX-SomaGCaMP7
Plasmid#182942PurposeCre-dependent expression of cell body-targeted GCaMP7f, under CAG promoter. As bright as conventional GCaMP7f.DepositorInsertSoma-GCaMP7
UseAAVAvailable SinceSept. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
pLentiCRISPRv1-sgPHGDH-G5
Plasmid#83913Purposestable knockoutDepositorInsertPhosphoglycerate dehydrogenase (PHGDH Human)
UseCRISPR and LentiviralExpressionMammalianPromoterhU6Available SinceOct. 24, 2016AvailabilityAcademic Institutions and Nonprofits only -
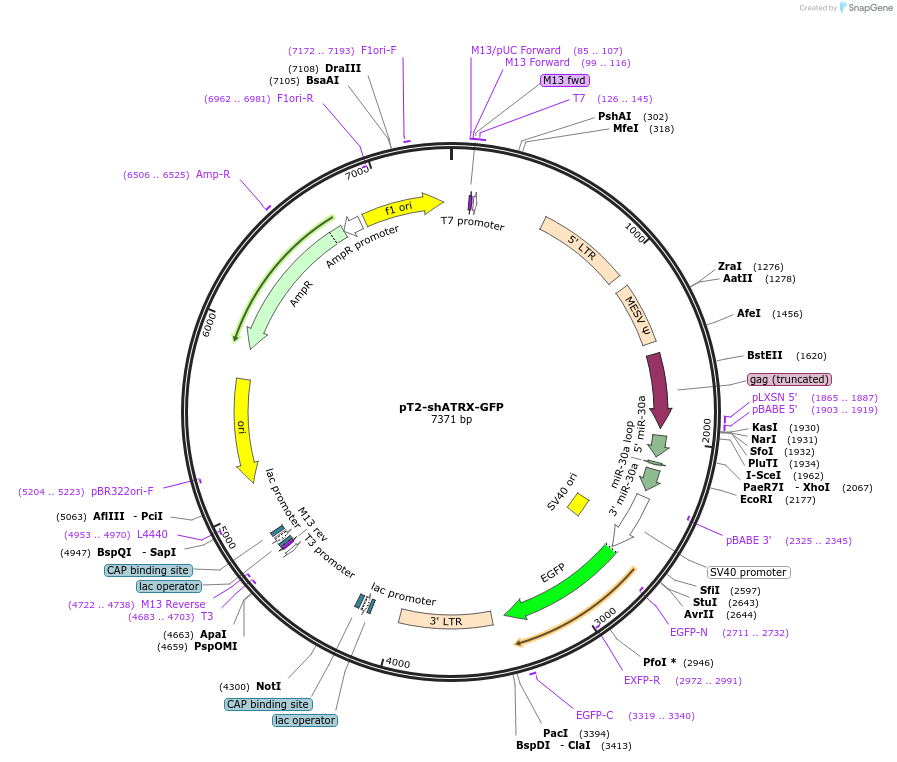
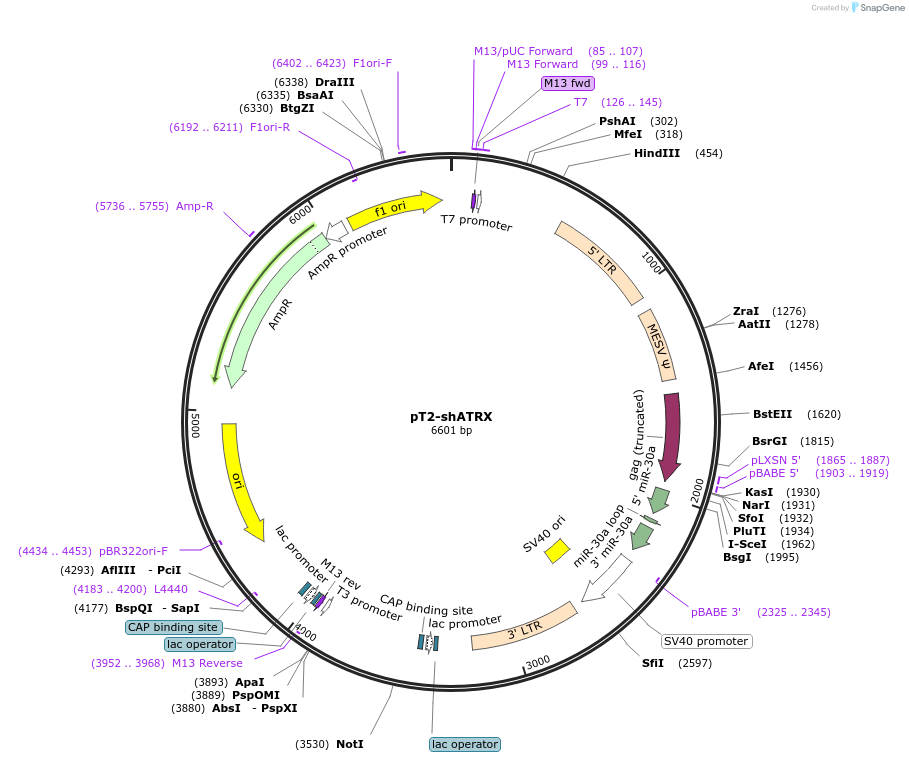
pT2-shATRX-GFP
Plasmid#124259PurposeExpresses shRNA targeting ATRX with a GFP reporter which is driven by the SV40 promoter. Construct has inverted repeats to be used in Sleeping beauty system.DepositorInsertshATRX
ExpressionMammalianAvailable SinceJuly 3, 2019AvailabilityAcademic Institutions and Nonprofits only -
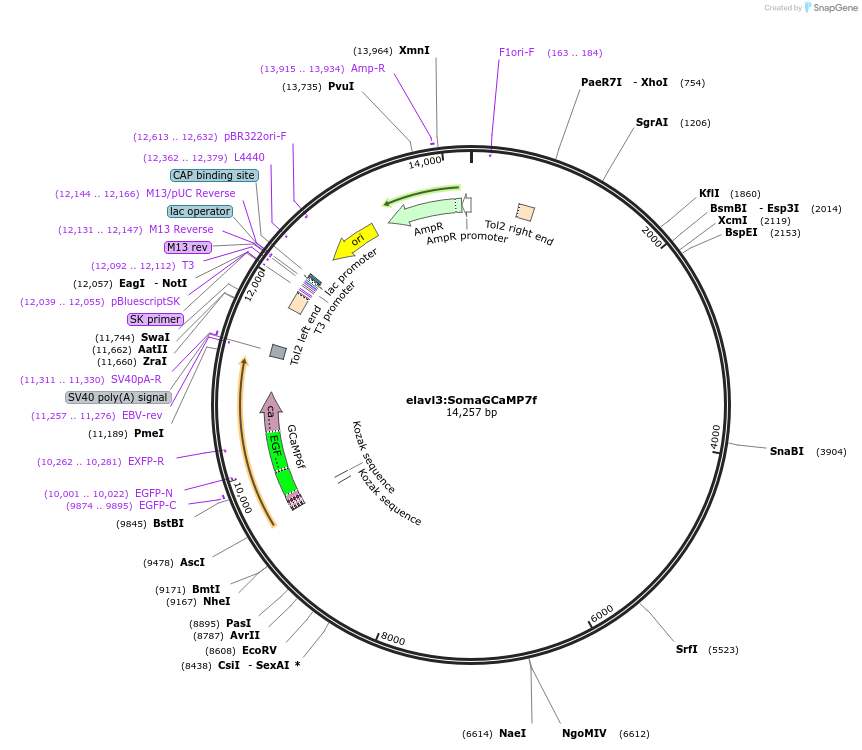
elavl3:SomaGCaMP7f
Plasmid#158760PurposeCell body-targeted GCaMP7f, under HuC promoter: GCaMP7f followed by a linker, and the EE-RR coiled coil motif (GCaMP7f-27-EE-RR). As bright as conventional GCaMP7f.DepositorInsertSomaGCaMP7
UseSynthetic BiologyPromoterHuC promoterAvailable SinceSept. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
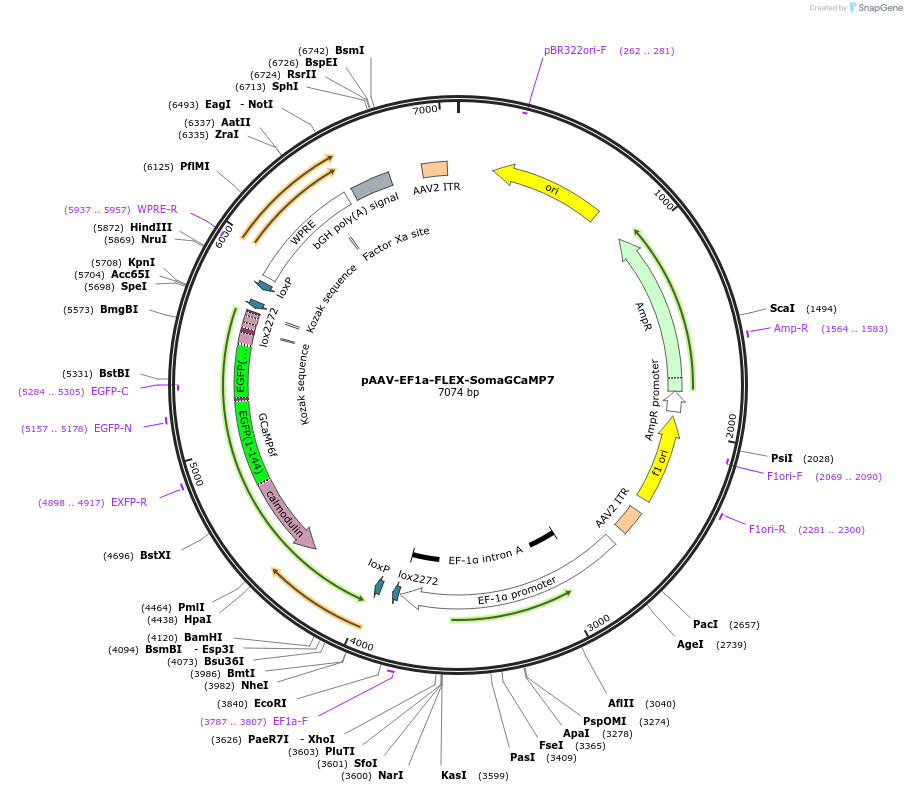
pAAV-EF1a-FLEX-SomaGCaMP7
Plasmid#182941PurposeCre-dependent expression of cell body-targeted GCaMP7f, under EF1a promoter. As bright as conventional GCaMP7f.DepositorInsertSoma-GCaMP7
UseAAVAvailable SinceSept. 23, 2022AvailabilityAcademic Institutions and Nonprofits only -
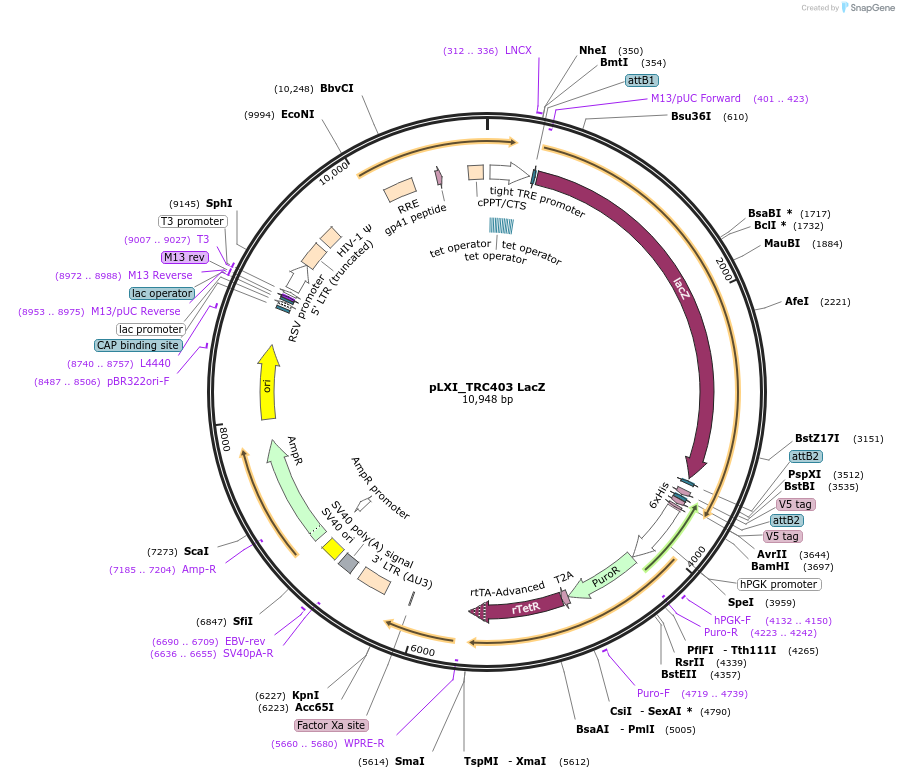
pLXI_TRC403 LacZ
Plasmid#111184PurposeControl inducible vector with LacZ (V5 tagged)DepositorInsertlacZ
UseLentiviralTagsV5PromoterTREAvailable SinceMarch 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
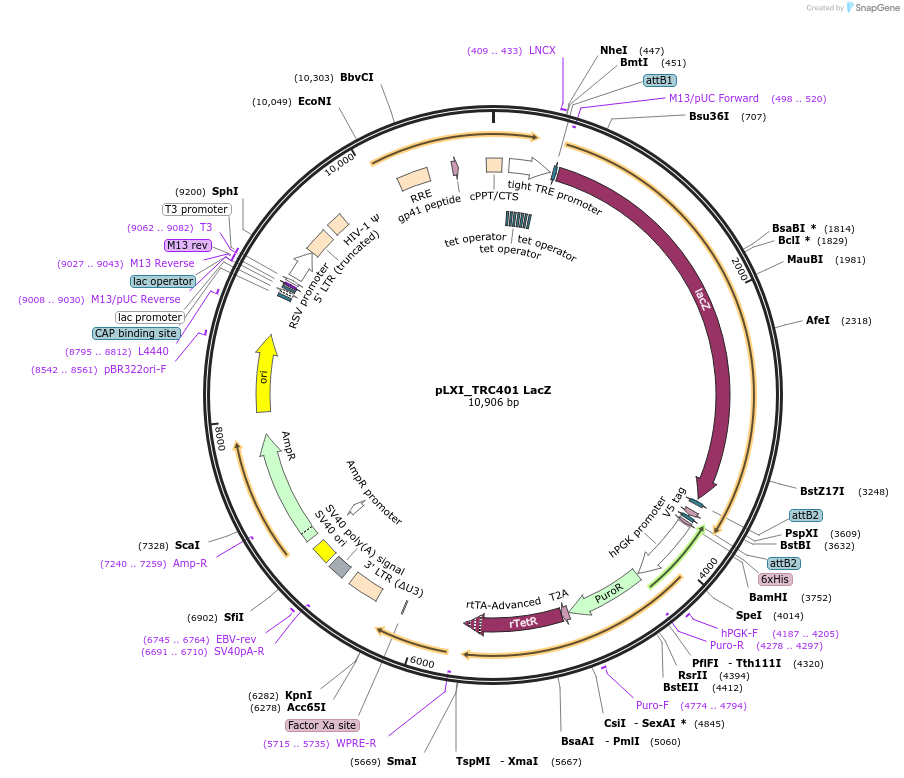
pLXI_TRC401 LacZ
Plasmid#111183PurposeControl inducible vector with LacZ (V5 tagged)DepositorInsertlacZ
UseLentiviralTagsV5PromoterTREAvailable SinceMarch 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
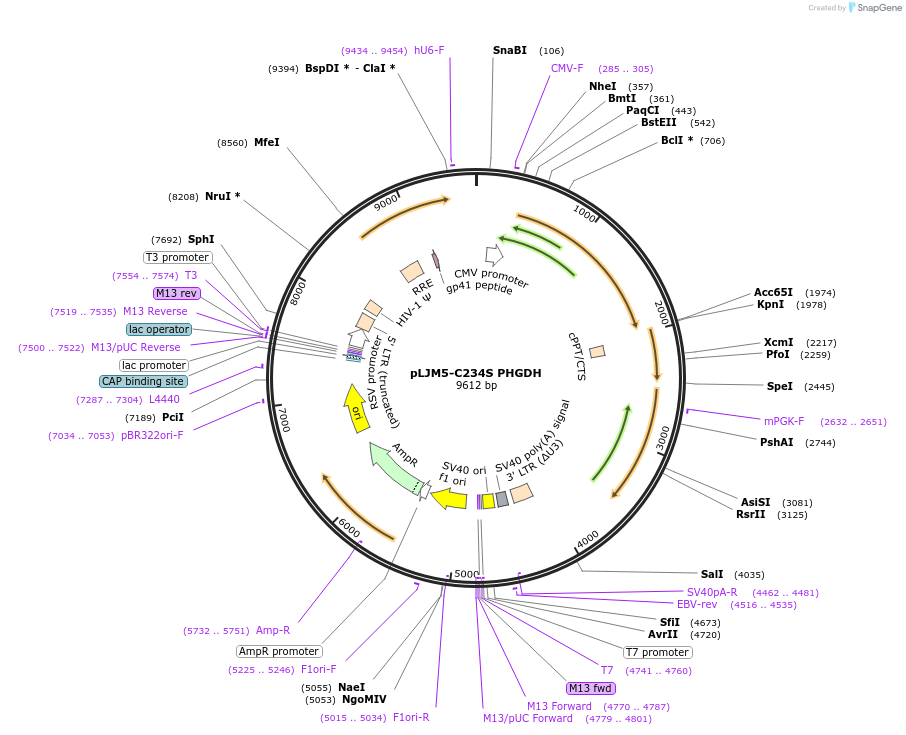
pLJM5-C234S PHGDH
Plasmid#83902Purposestable overexpressionDepositorInsertPhosphoglycerate dehydrogenase (PHGDH Human)
UseLentiviralExpressionMammalianMutationquikchanged cysteine 234 to serinePromoterCMVAvailable SinceOct. 20, 2016AvailabilityAcademic Institutions and Nonprofits only -
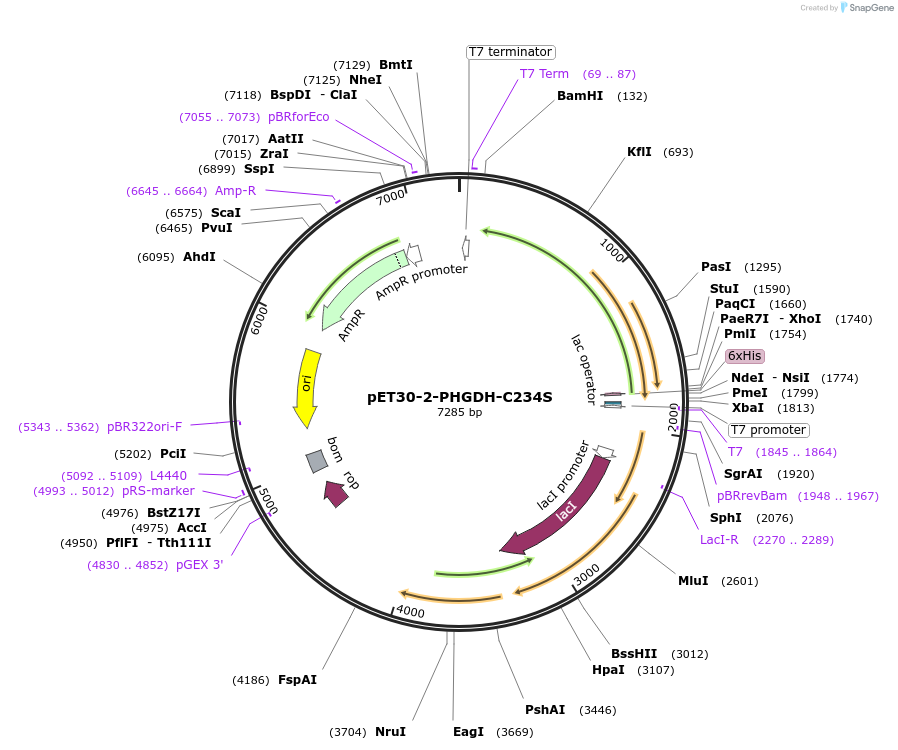
pET30-2-PHGDH-C234S
Plasmid#107724PurposeExpression of PHGDH C234S in bacteriaDepositorAvailable SinceApril 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
pT2-shATRX
Plasmid#124258PurposeExpresses shRNA targeting ATRX. Construct has inverted repeats to be used in Sleeping beauty system.DepositorInsertshATRX
ExpressionMammalianAvailable SinceApril 8, 2019AvailabilityAcademic Institutions and Nonprofits only -
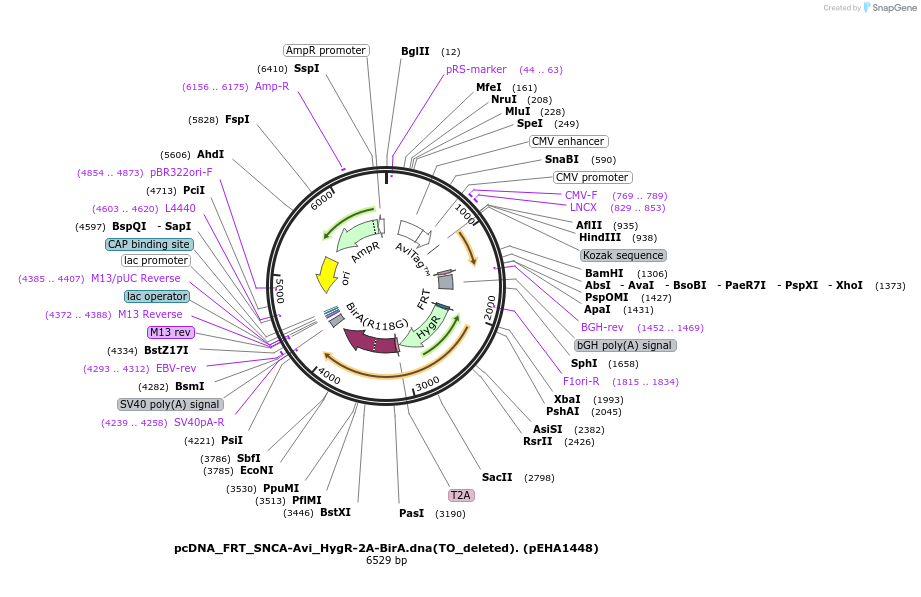
pcDNA_FRT_SNCA-Avi_HygR-2A-BirA.dna(TO_deleted). (pEHA1448)
Plasmid#209083PurposeSNCA (Avi tagged) expression along with BirA ligaseDepositorInsertsTagsAviExpressionMammalianMutationhuman codon optimizedPromoterCMVAvailable SinceOct. 27, 2023AvailabilityAcademic Institutions and Nonprofits only -
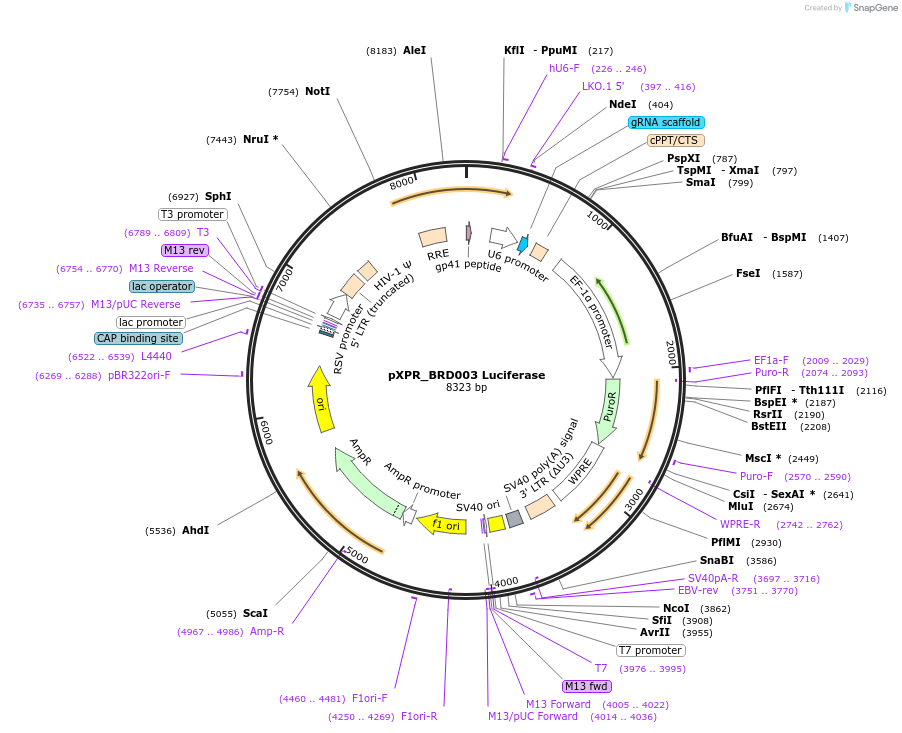
pXPR_BRD003 Luciferase
Plasmid#117072Purposesingle guide RNA targeting LuciferaseDepositorInsertLuciferase
UseCRISPRPromoterhU6Available SinceMarch 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
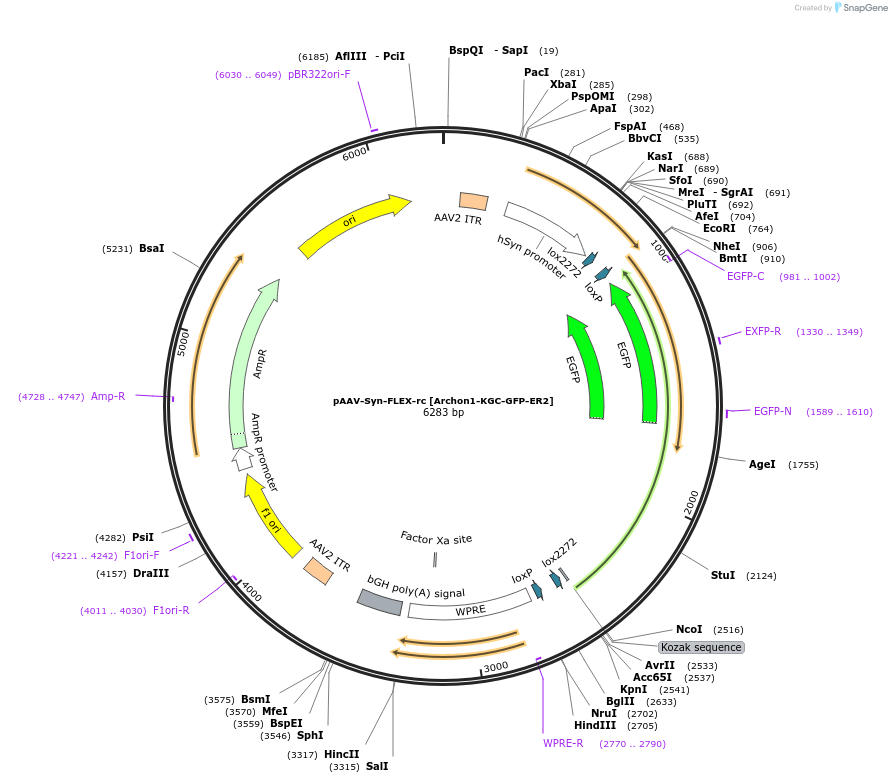
pAAV-Syn-FLEX-rc [Archon1-KGC-GFP-ER2]
Plasmid#115893PurposeAAV-mediated expression of Archon1-KGC-GFP-ER2 under the Syn promoter, in floxed/reversed (Cre-dependent) manner. Using bGHpA signal.DepositorHas ServiceAAV8InsertArchon1-KGC-EGFP-ER2
UseAAVTagsER2, GFP, and KGCExpressionMammalianPromoterSynAvailable SinceNov. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
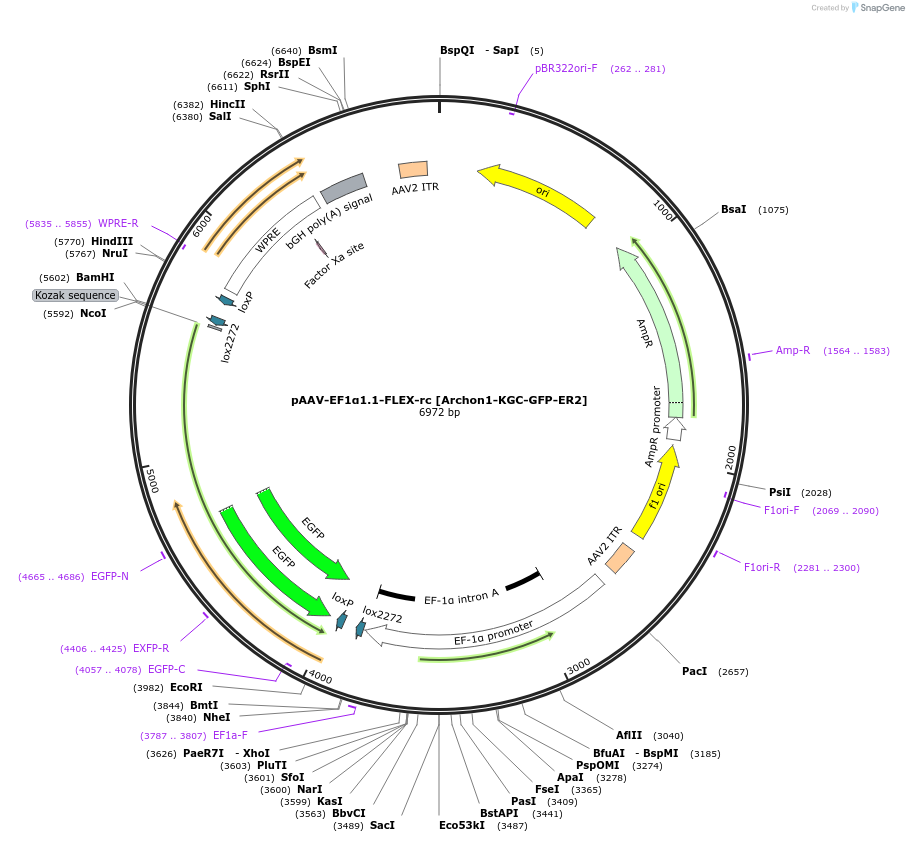
pAAV-EF1α1.1-FLEX-rc [Archon1-KGC-GFP-ER2]
Plasmid#115891PurposeAAV-mediated expression of Archon1-KGC-GFP-ER2 under the EF1α promoter (1.1kb short version), in floxed/reversed (Cre-dependent) manner. Using bGHpA signal.DepositorInsertArchon1-KGC-GFP-ER2
UseAAVTagsER2, GFP, and KGCExpressionMammalianPromoterEF1α promoter (1.1kb short version)Available SinceJuly 2, 2019AvailabilityAcademic Institutions and Nonprofits only