We narrowed to 8,660 results for: gal
-
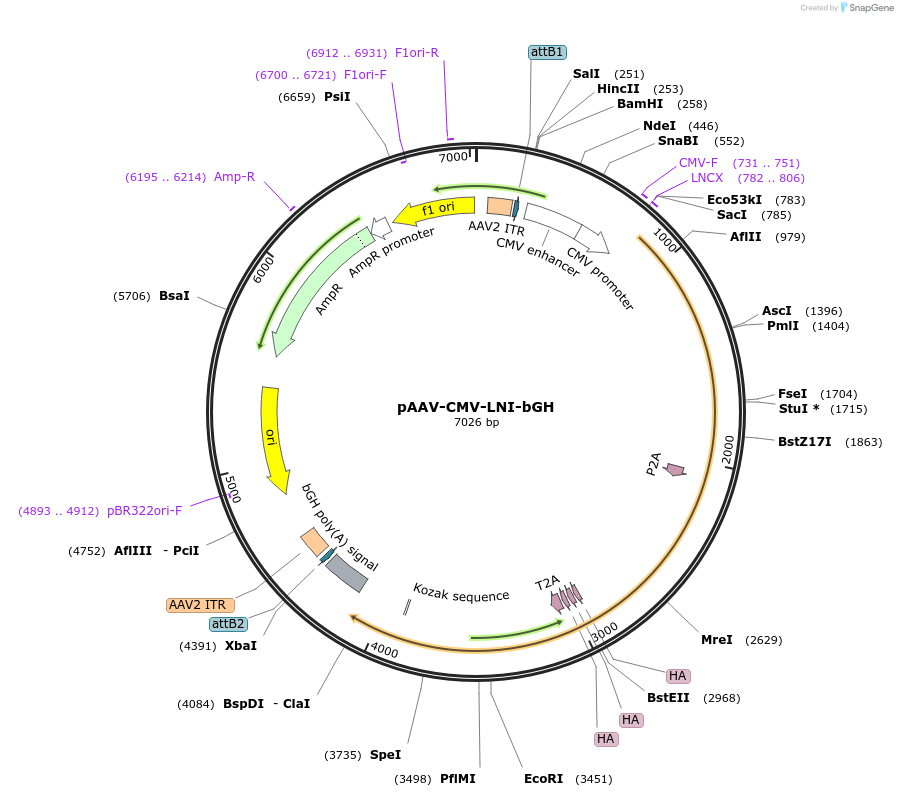
Plasmid#233203PurposeAAV expression of Lhx3, Ngn2x3HA, and Isl1 for motor neuron reprogrammingDepositorAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only
-
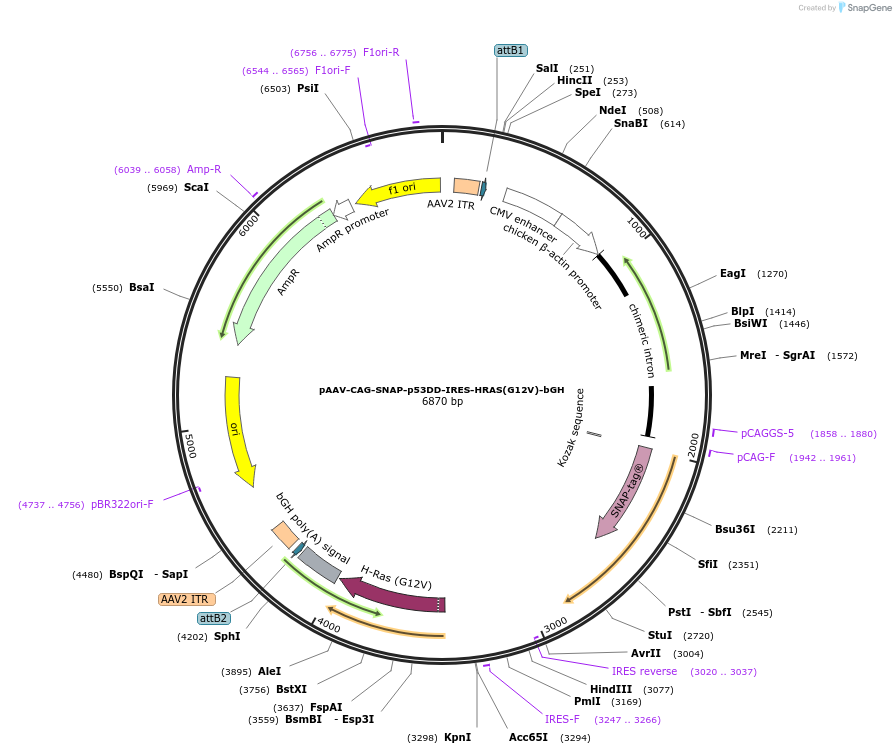
pAAV-CAG-SNAP-p53DD-IRES-HRAS(G12V)-bGH
Plasmid#233204PurposeAAV expression of SNAP-p53DD and HRAS(G12V)DepositorUseAAVExpressionMammalianMutationG12VAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
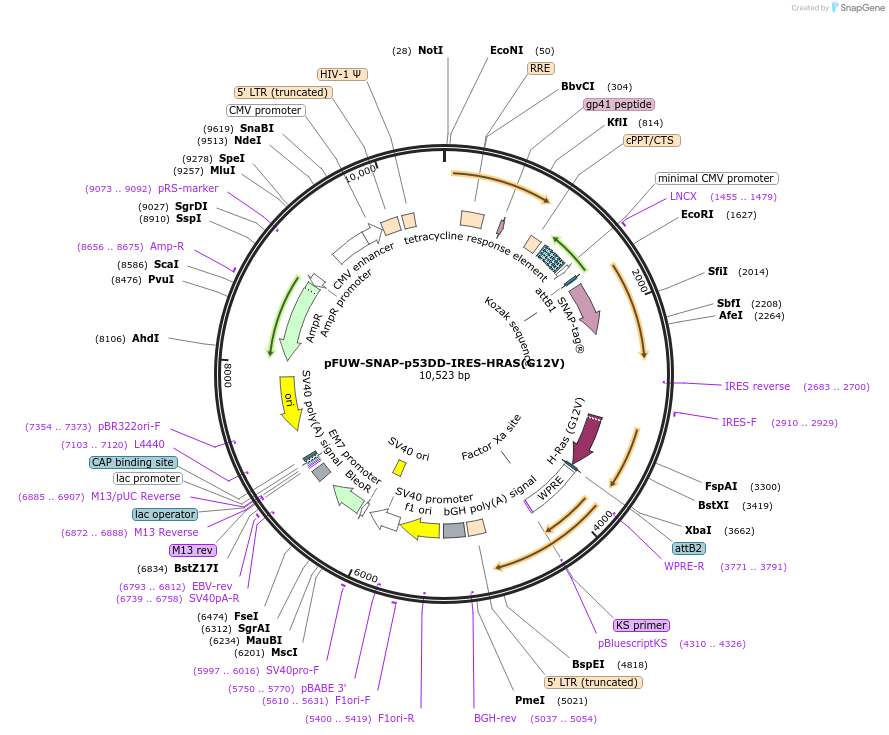
pFUW-SNAP-p53DD-IRES-HRAS(G12V)
Plasmid#233206PurposeLentiviral dox-inducible expression of SNAP-p53DD and HRAS(G12V)DepositorUseLentiviralExpressionMammalianMutationG12VAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
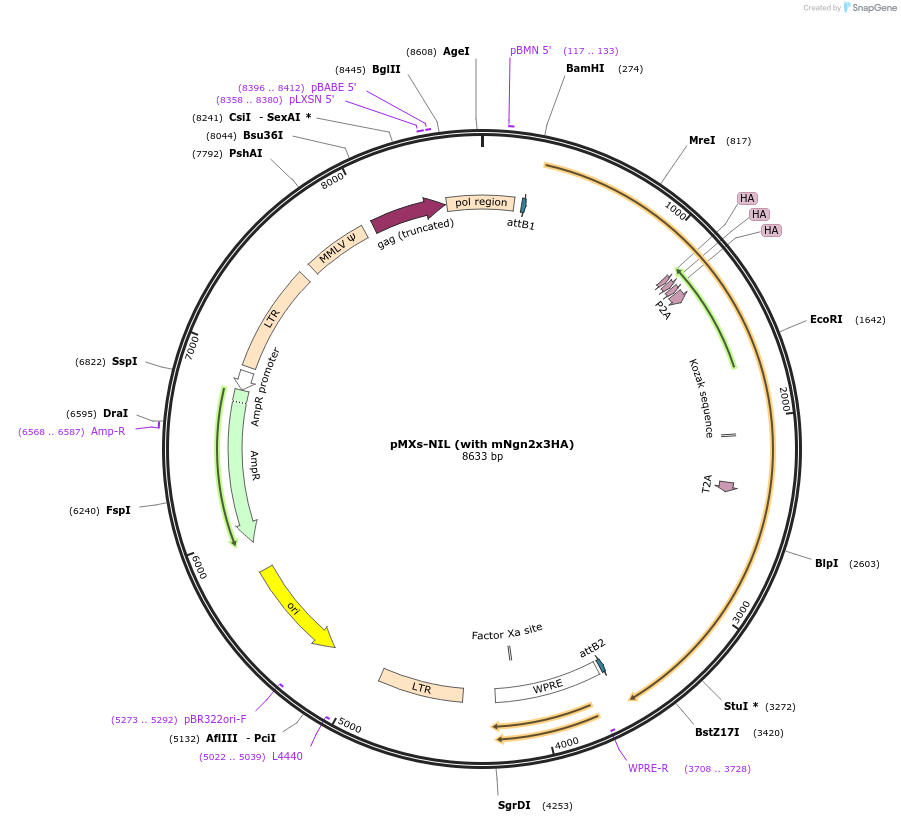
pMXs-NIL (with mNgn2x3HA)
Plasmid#233192PurposeRetroviral expression of Ngn2x3HA, Isl1, and Lhx3 for motor neuron reprogrammingDepositorUseRetroviralTagsx3HAExpressionMammalianAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
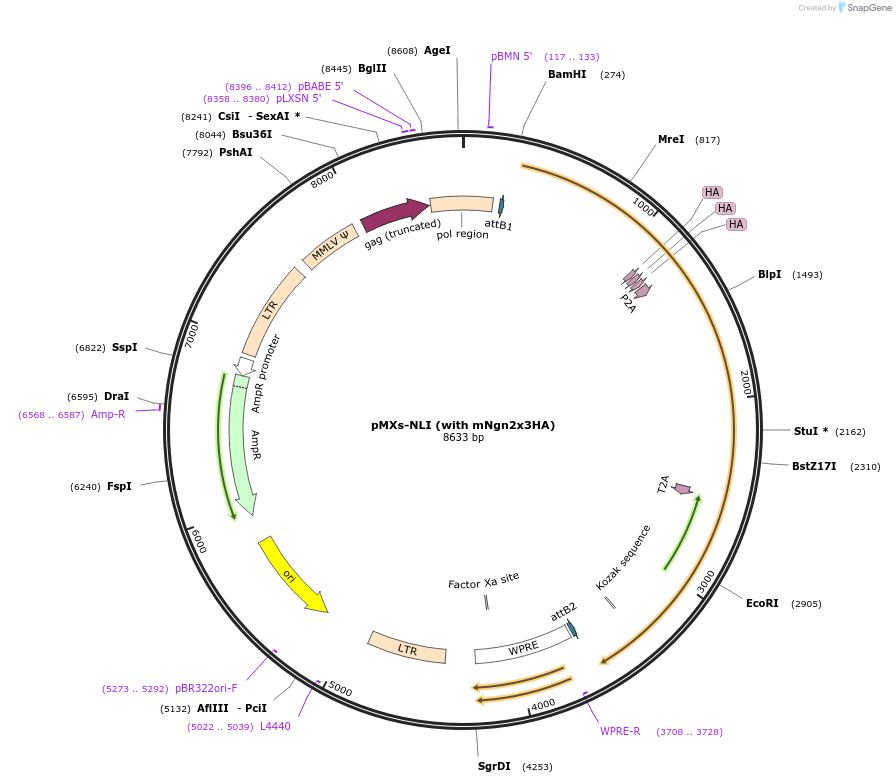
pMXs-NLI (with mNgn2x3HA)
Plasmid#233193PurposeRetroviral expression of Ngn2x3HA, Lhx3, and Isl1 for motor neuron reprogrammingDepositorUseRetroviralTagsx3HAExpressionMammalianAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
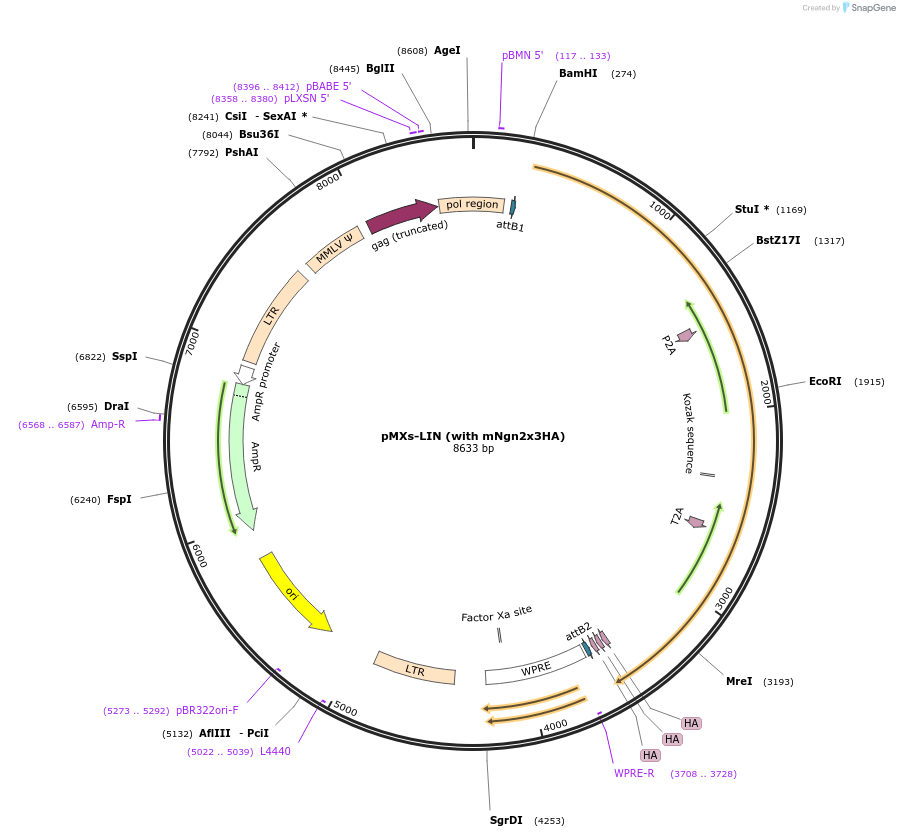
pMXs-LIN (with mNgn2x3HA)
Plasmid#233194PurposeRetroviral expression of Lhx3, Isl1, and Ngn2x3HA for motor neuron reprogrammingDepositorUseRetroviralTagsx3HAExpressionMammalianAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
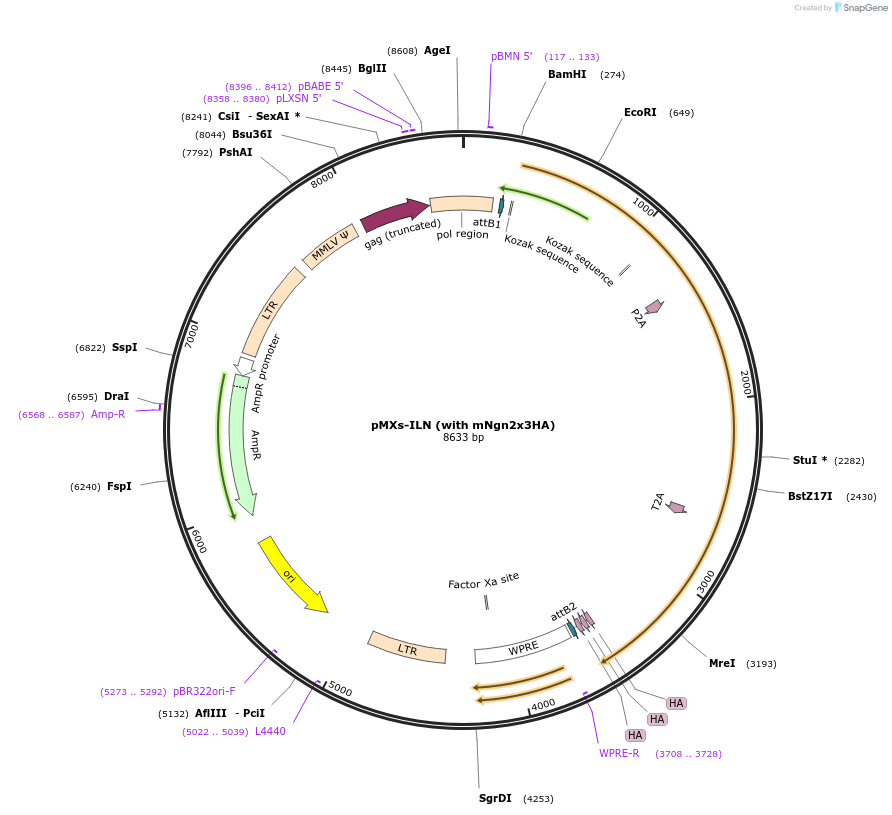
pMXs-ILN (with mNgn2x3HA)
Plasmid#233196PurposeRetroviral expression of Isl1, Lhx3, and Ngn2x3HA for motor neuron reprogrammingDepositorUseRetroviralTagsx3HAExpressionMammalianAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
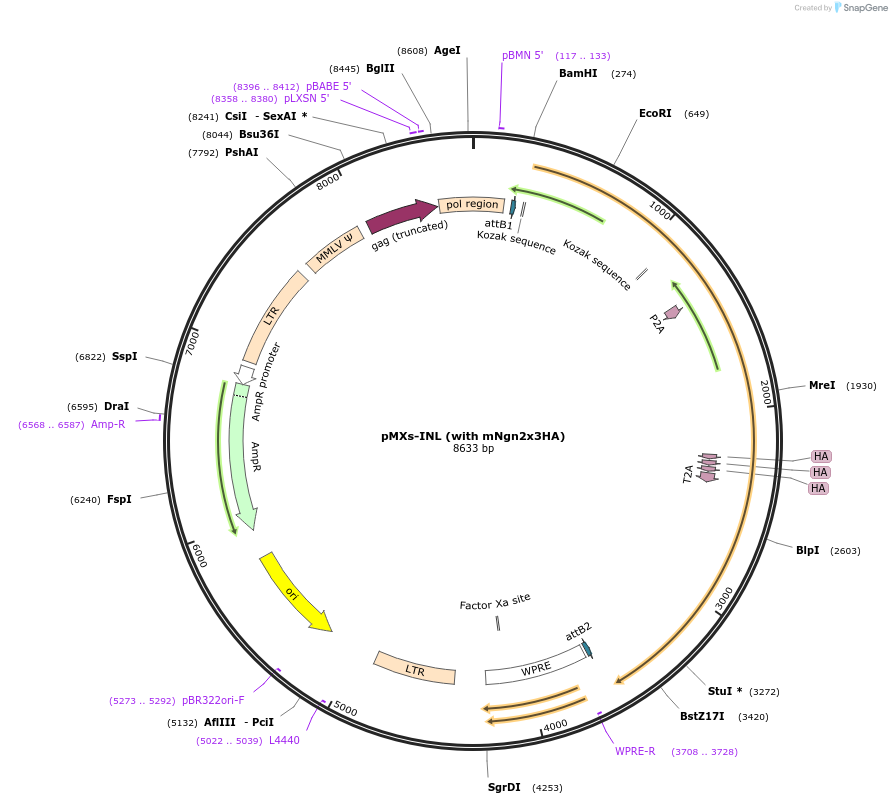
pMXs-INL (with mNgn2x3HA)
Plasmid#233197PurposeRetroviral expression of Isl1, Ngn2x3HA, and Lhx3 for motor neuron reprogrammingDepositorUseRetroviralTagsx3HAExpressionMammalianAvailable SinceMarch 13, 2025AvailabilityAcademic Institutions and Nonprofits only -
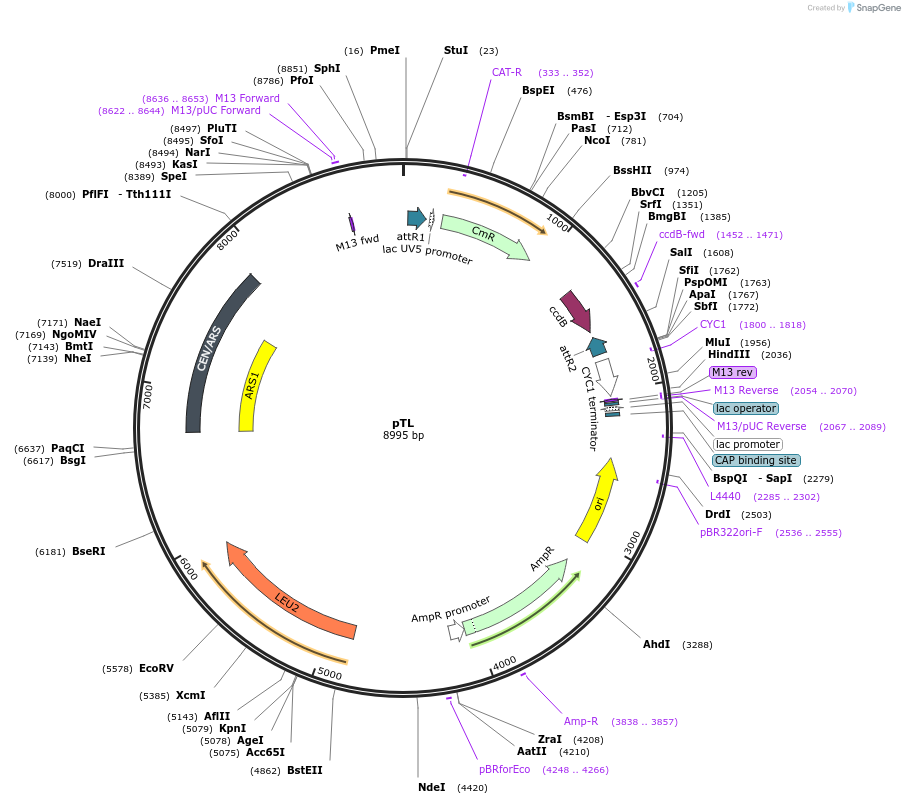
pTL
Plasmid#203176PurposeCentromeric yeast expression vector, leucine selectionDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 29, 2023AvailabilityAcademic Institutions and Nonprofits only -
pT3-EF1A-MYC-IRES-lucOS
Plasmid#129776PurposeTransposon based vector that expresses human MYC followed by IRES and firefly luciferase and the antigens SIYRYYGL (SIY), SIINFEKL (SIN, OVA257-264), and OVA323-339DepositorInserthuman MYC (MYC Human)
TagsOS: SIYRYYGL (SIY), SIINFEKL (SIN, OVA257-264), a…ExpressionMammalianPromoterEF1AAvailable SinceFeb. 13, 2020AvailabilityAcademic Institutions and Nonprofits only -
Antibody#199805-rAbPurposeAnti-HA chimeric recombinant antibody. The hybridoma-derived heavy chain variable sequence and kappa light chain are fused to a chicken IgY Fc.DepositorRecommended ApplicationsWestern BlotSource SpeciesChickenIsotypeIgYTrial SizeAvailable to purchaseAvailable SinceApril 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits

-
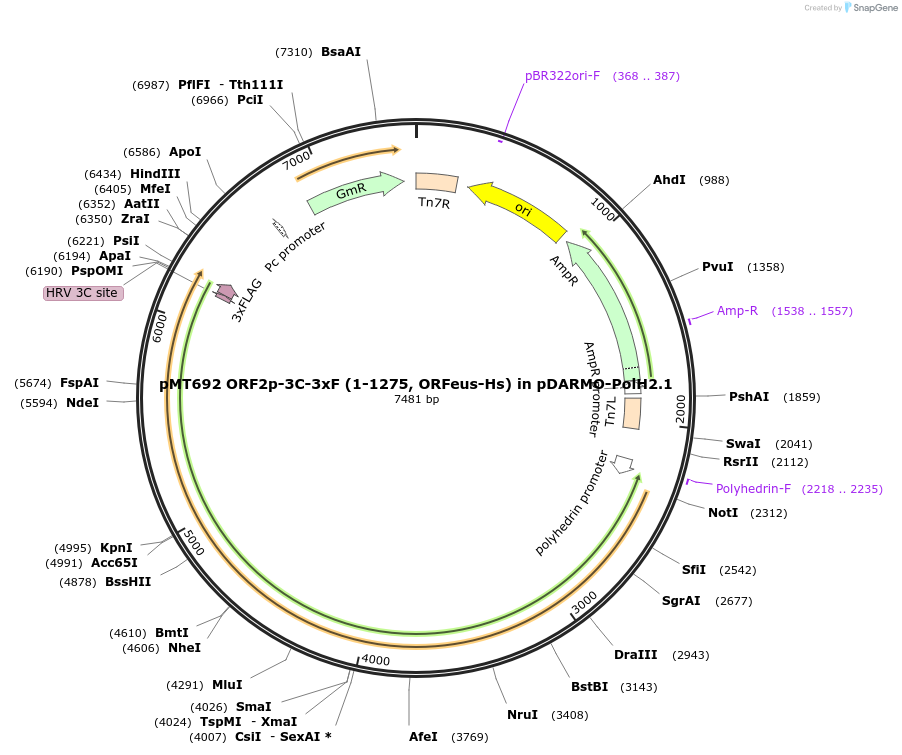
pMT692 ORF2p-3C-3xF (1-1275, ORFeus-Hs) in pDARMO-PolH2.1
Plasmid#213025PurposeExpresses full length human LINE-1 ORF2p Core (1-1275) with a C-terminal 3xFlag tag for baculovirus production in insect cellsDepositorInsertLINE-1 ORF2p (1-1275)
Tags3C-3xFlag (ORF2p)ExpressionInsectPromoterPolyhedrinAvailable SinceMay 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
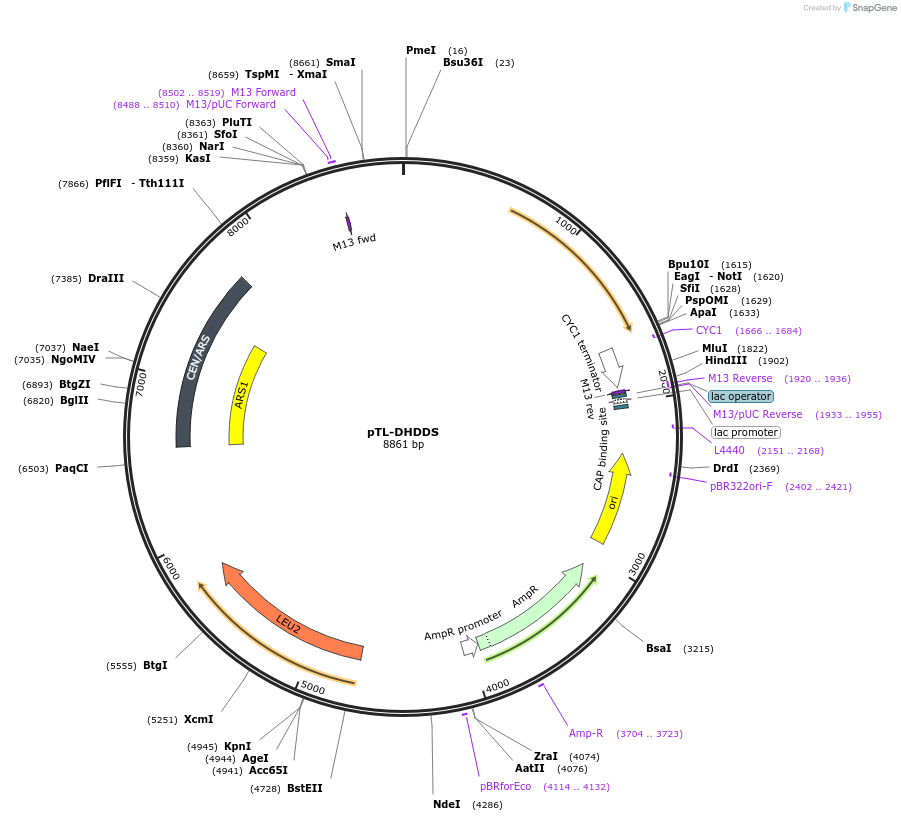
pTL-DHDDS
Plasmid#203177PurposeCentromeric yeast expression vector for hDHDDSDepositorAvailable SinceAug. 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
Antibody#194520-rAbPurposeAnti-c-Myc chimeric recombinant antibody. The hybridoma-derived heavy chain variable sequence and kappa light chain are fused to a chicken IgY Fc.DepositorRecommended ApplicationsWestern BlotReactivityHumanSource SpeciesChickenIsotypeIgYTrial SizeAvailable to purchaseAvailable SinceFeb. 9, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits

-
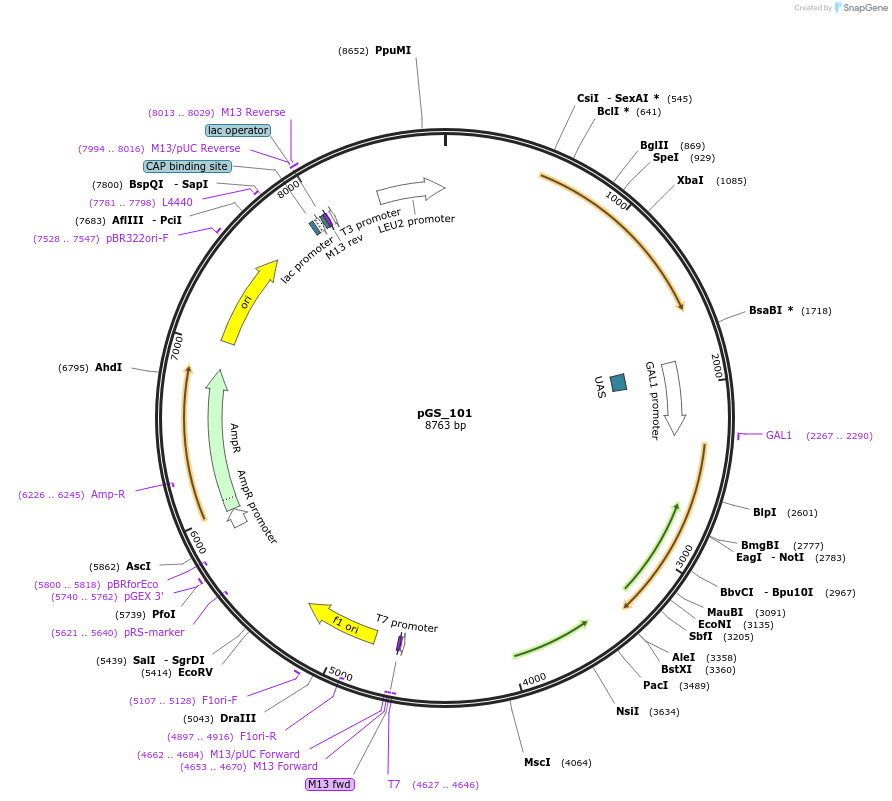
pGS_101
Plasmid#160541PurposeExpresses Human APOE4 with a yeast secretory signal peptide under the control of the yeast Gal promoterDepositorInsertApolipoprotein E e4 (APOE Human)
TagsKar2 signal sequenceExpressionYeastMutatione4 allelePromoterGAL1Available SinceJan. 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
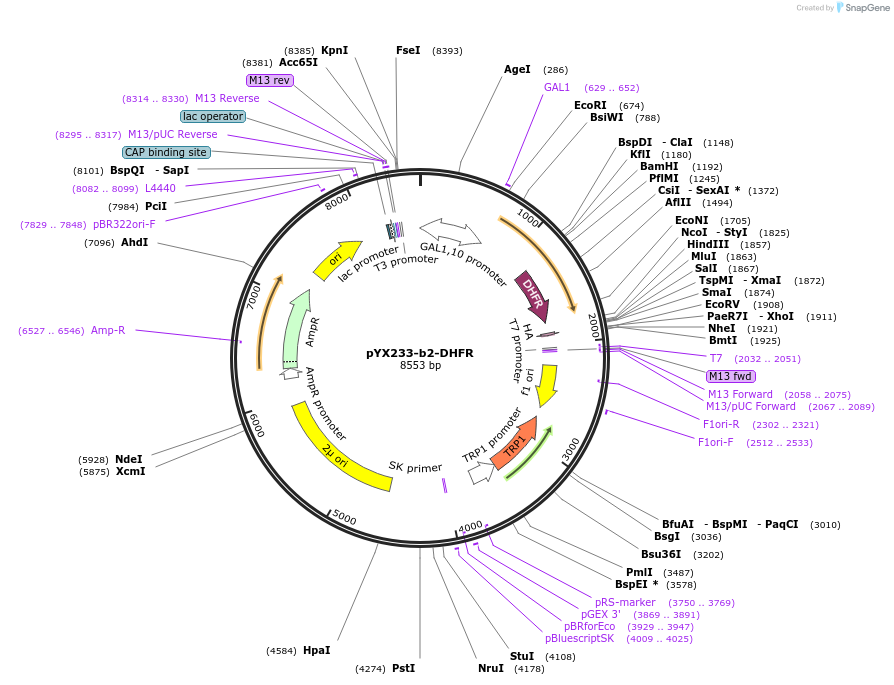
pYX233-b2-DHFR
Plasmid#163759PurposeGal-inducible yeast expression of the mitochondrial "clogger" protein b2(167)-DHFRDepositorInsertb2-DHFR (Dhfr Mouse, Budding Yeast)
ExpressionYeastMutationAmino acids 1-167 of S.c. Cytochrome b2 (Cyb2) fu…PromoterGAL1Available SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
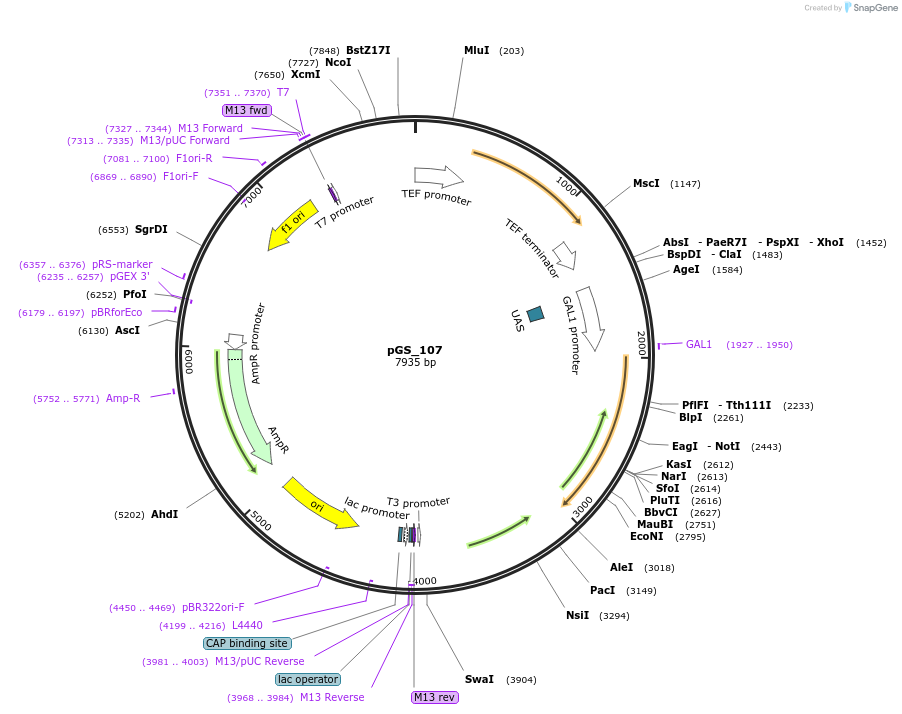
pGS_107
Plasmid#160650PurposeExpresses Human APOE4 with a yeast secretory signal peptide under the control of the yeast Gal promoterDepositorInsertApolipoprotein E e4 (APOE Human)
TagsKar2 signal sequenceExpressionYeastMutatione4 allelePromoterGAL1Available SinceJan. 4, 2023AvailabilityAcademic Institutions and Nonprofits only -
Antibody#198594-rAbPurposeAnti-PSD-95 (Human) chimeric recombinant antibody. The hybridoma-derived heavy chain variable sequence and kappa light chain are fused to a chicken IgY Fc.DepositorRecommended ApplicationsWestern BlotReactivityHumanSource SpeciesChickenIsotypeIgYTrial SizeAvailable to purchaseAvailable SinceApril 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits

-
Antibody#198267-rAbPurposeAnti-GFP chimeric recombinant antibody. The hybridoma-derived heavy chain variable sequence and kappa light chain are fused to a chicken IgY Fc.DepositorRecommended ApplicationsWestern BlotReactivitySyntheticSource SpeciesChickenIsotypeIgYTrial SizeNot available to purchaseAvailable SinceApril 3, 2023AvailabilityIndustry, Academic Institutions, and Nonprofits

-
Antibody#213675-rAbPurposeAnti-Integrin αM (Mouse) recombinant rat monoclonal antibody; binds αM subunit. For more data, see Antibody 213665.DepositorRecommended ApplicationsFlow CytometryReactivityHuman and MouseSource SpeciesRatIsotypeIgG2aTrial SizeNot available to purchaseAvailable SinceMarch 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits

-
Antibody#213665-rAbPurposeAnti-Integrin αM (Mouse) chimeric recombinant antibody with fused rat variable and rabbit constant domains; binds αM subunit.DepositorRecommended ApplicationsFlow CytometryReactivityHuman and MouseSource SpeciesRabbitIsotypeIgGTrial SizeNot available to purchaseAvailable SinceMarch 27, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits

-
-
Gaq-LSS-SGFP2
Plasmid#112951PurposeGalphaq tagged with LSS-SGFP2DepositorInsertGalphaq
TagsLSS-SGFP2ExpressionMammalianPromoterCMVAvailable SinceSept. 12, 2018AvailabilityAcademic Institutions and Nonprofits only -
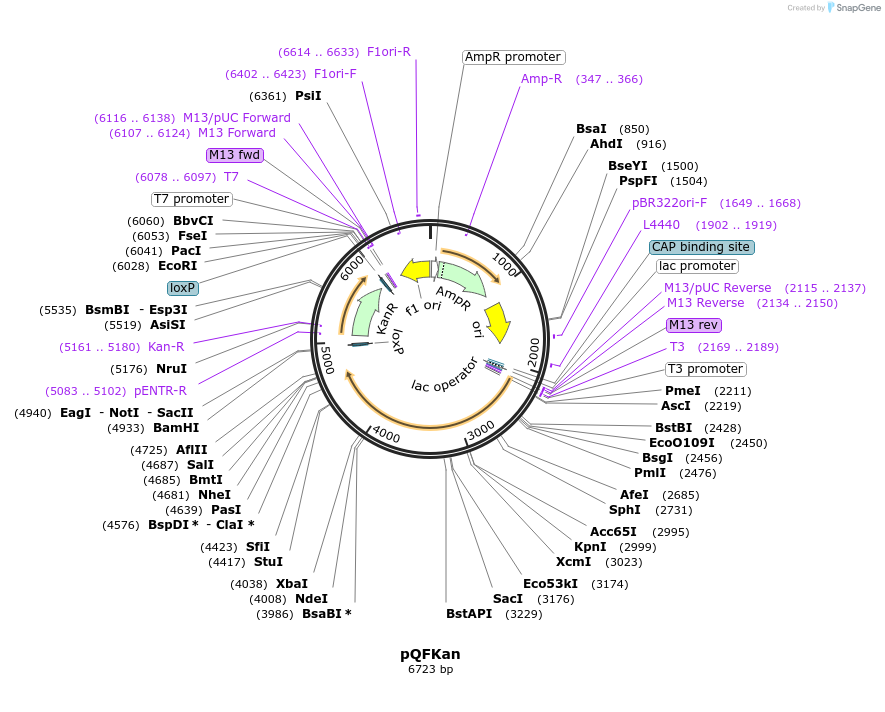
pQFKan
Plasmid#32321DepositorInsertQF Kan
Available SinceSept. 15, 2011AvailabilityAcademic Institutions and Nonprofits only -
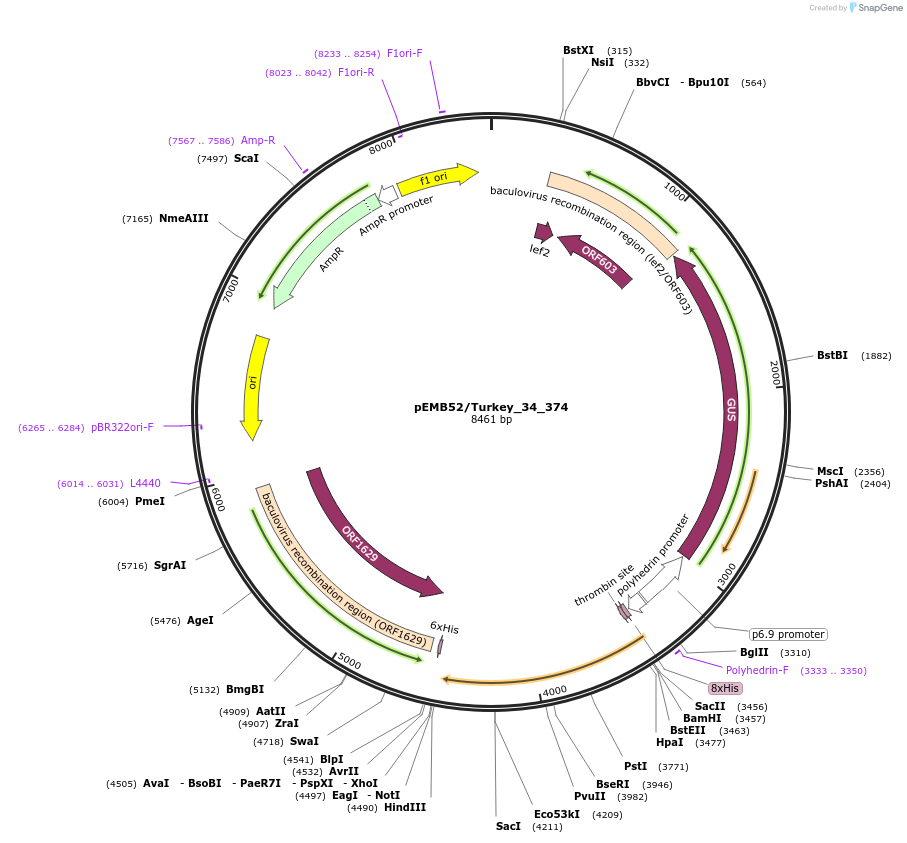
pEMB52/Turkey_34_374
Plasmid#40545DepositorInsertLRRK2
UseBaculovirusTags6xHisMutationWTPromoterpolHAvailable SinceOct. 1, 2012AvailabilityIndustry, Academic Institutions, and Nonprofits -
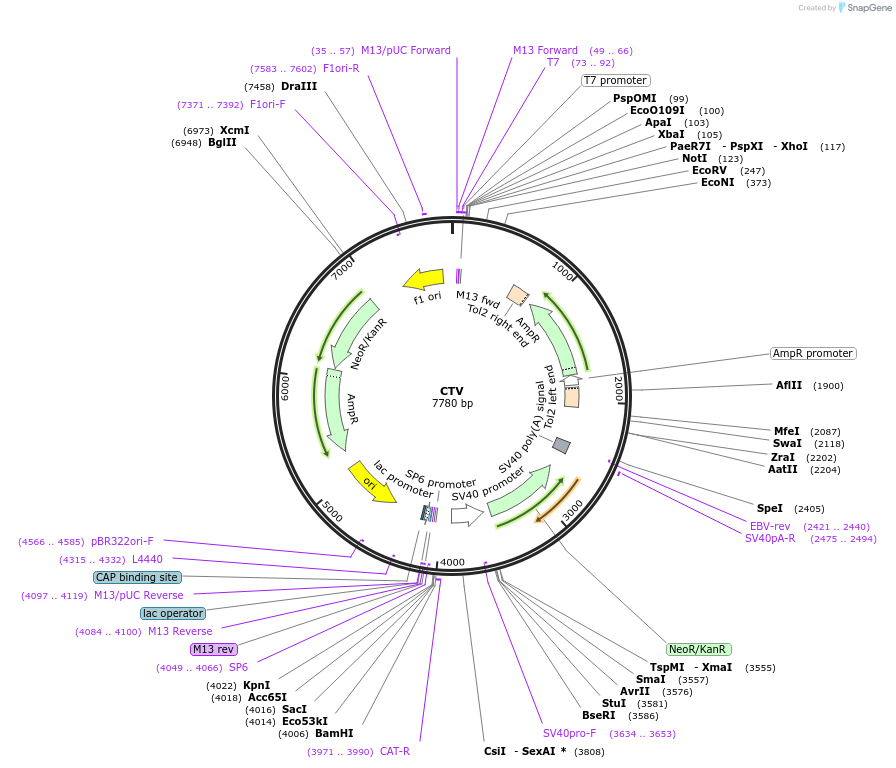
CTV
Plasmid#131590PurposePlasmid for the targeting of chloramphenicol geneDepositorInsertNeomycin resistance gene
ExpressionBacterialAvailable SinceSept. 18, 2019AvailabilityAcademic Institutions and Nonprofits only -
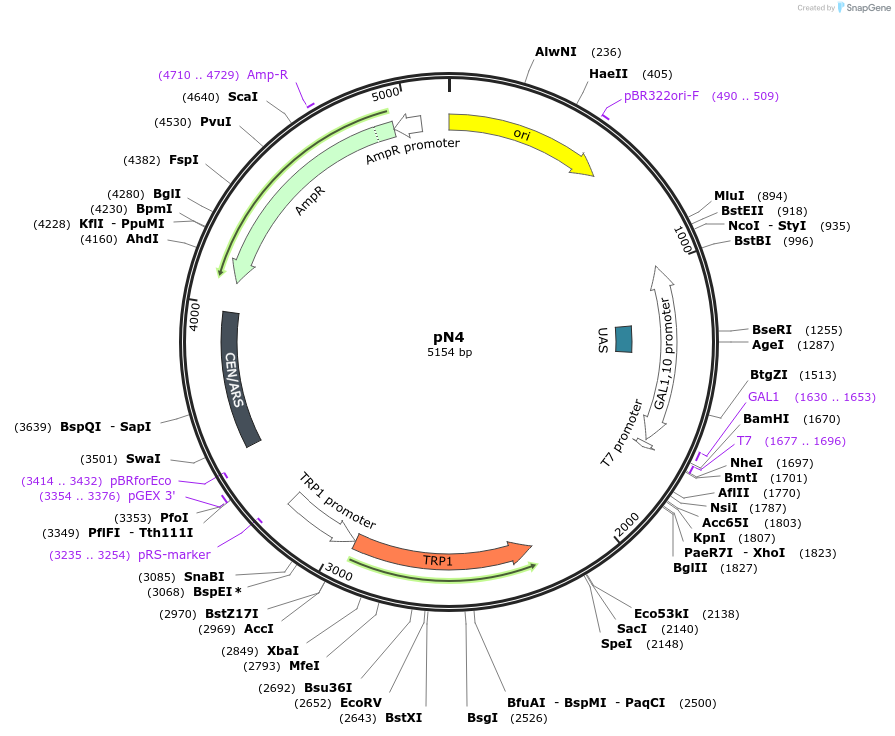
pN4
Plasmid#233521PurposeNb Vector Plasmid (under galactose induction), AmpR (E. coli) and TRP1 (yeast), Cen6/ARSDepositorTypeEmpty backboneExpressionBacterial and YeastPromoterpGAL1-10Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
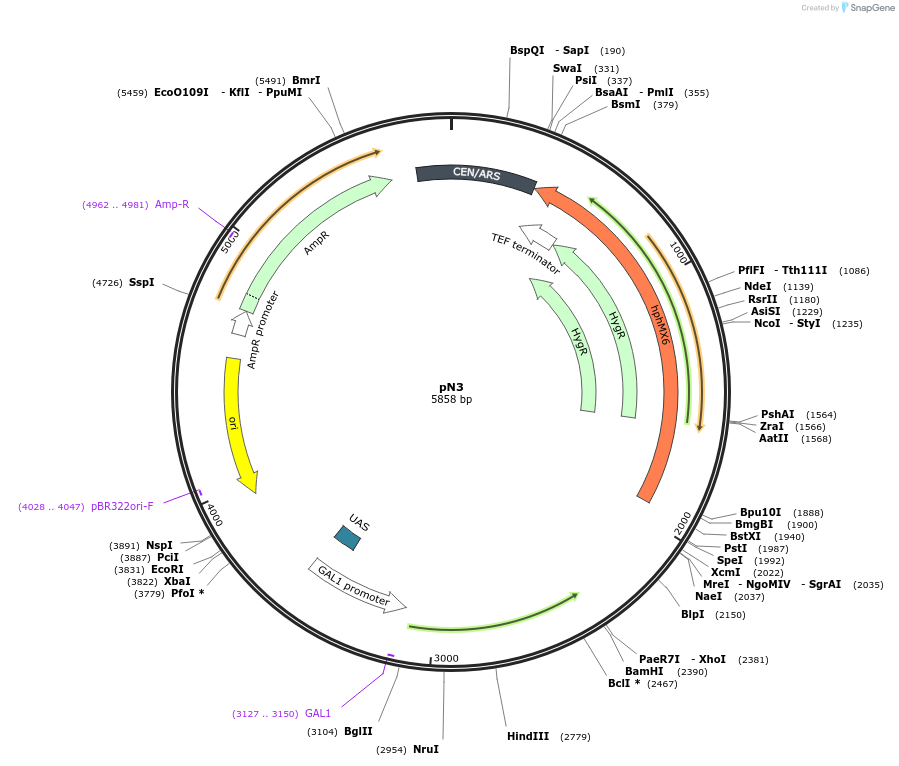
pN3
Plasmid#233520PurposeNb Vector Plasmid (under galactose induction), AmpR (E. coli) and HygroR (yeast), Cen6/ARSDepositorTypeEmpty backboneExpressionBacterial and YeastPromoterpGAL1Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
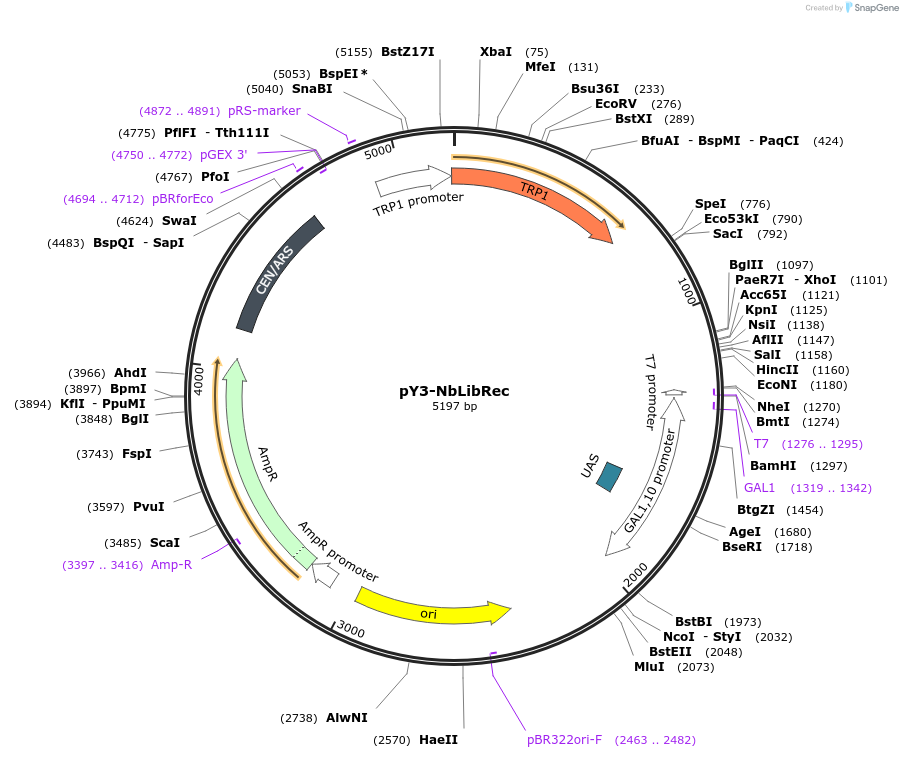
pY3-NbLibRec
Plasmid#233522PurposeNb Library Receiver Vector (under galactose induction), AmpR (E. coli) and TRP1 (yeast), Cen6/ARSDepositorTypeEmpty backboneExpressionBacterial and YeastPromoterpGAL1-10Available SinceJuly 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
mEmerald-SiT-N-15
Plasmid#54255PurposeLocalization: Golgi TGN, Excitation: 487, Emission: 509DepositorAvailable SinceJuly 25, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
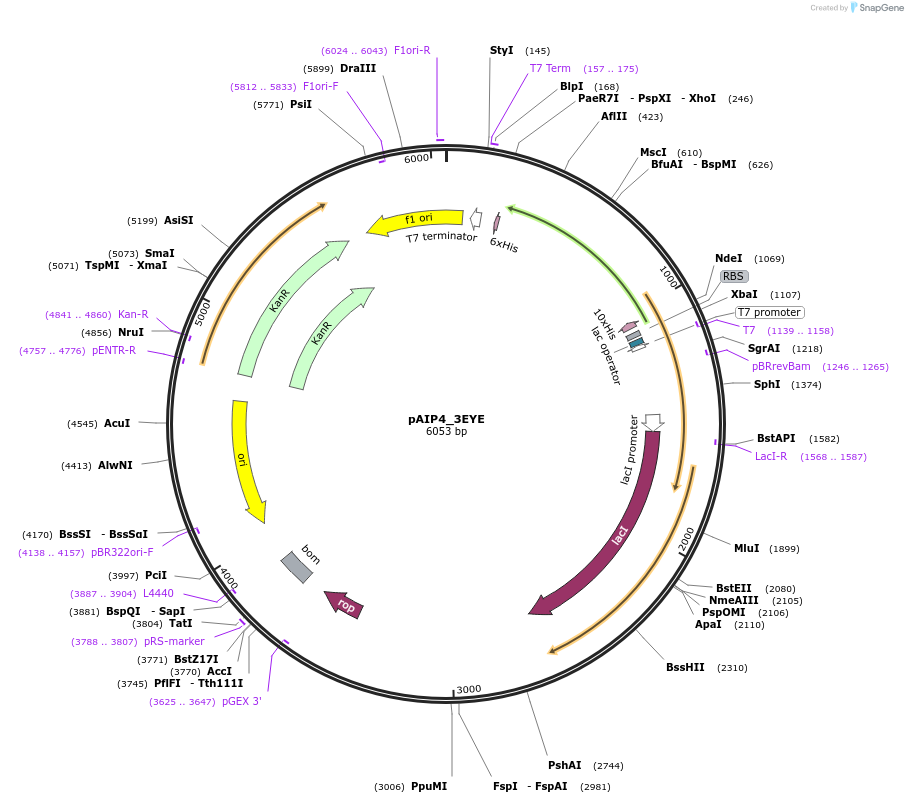
pAIP4_3EYE
Plasmid#234176PurposeHeterologous protein expression of N-acetylgalactosamine-specific enzyme IIB component of PTS in Escherichia coliDepositorInsert3EYE
Tags10x HisTag-Smt3ExpressionBacterialPromoterT7Available SinceMarch 6, 2025AvailabilityIndustry, Academic Institutions, and Nonprofits -
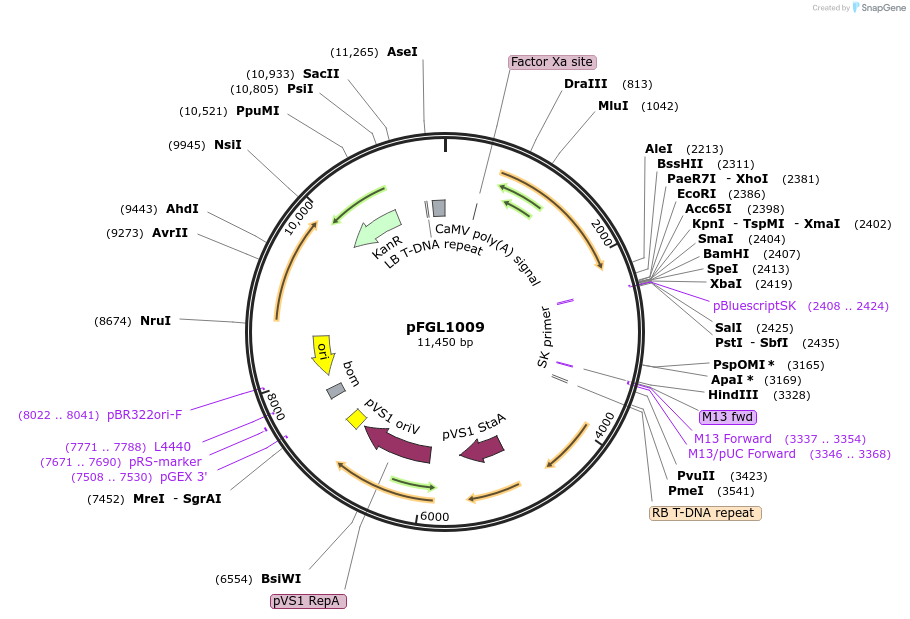
pFGL1009
Plasmid#119080PurposeIntroducing ectopic integration at the defined fungal ILV2-locus, based on SRR (Sulfonylurea Resistance Reconstitution).DepositorTypeEmpty backboneUseFungal expressionAvailable SinceAug. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
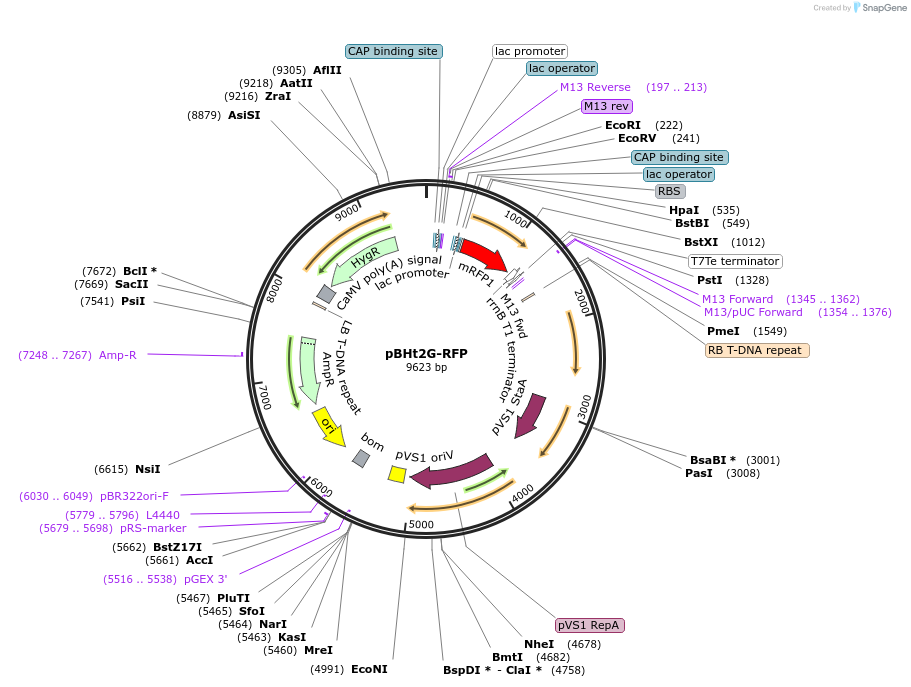
pBHt2G-RFP
Plasmid#107162PurposeGolden-Gate compatible fungal transformation vector carrying Hygromycin Resistance w RFP screening cassette (RFP knocked out by GG reaction, allowing for red-white screening)DepositorTypeEmpty backboneUseFungal grobacterium transformation plasmid, teste…Available SinceMarch 19, 2018AvailabilityAcademic Institutions and Nonprofits only -
mCherry-SiT-N-15
Plasmid#55133PurposeLocalization: Golgi TGN, Excitation: 587, Emission: 610DepositorAvailable SinceSept. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
mVenus-SiT-N-15
Plasmid#56336PurposeLocalization: Golgi TGN, Excitation: 515, Emission: 528DepositorAvailable SinceDec. 18, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
mApple-SiT-N-15
Plasmid#54948PurposeLocalization: Golgi TGN, Excitation: 568, Emission: 592DepositorAvailable SinceAug. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
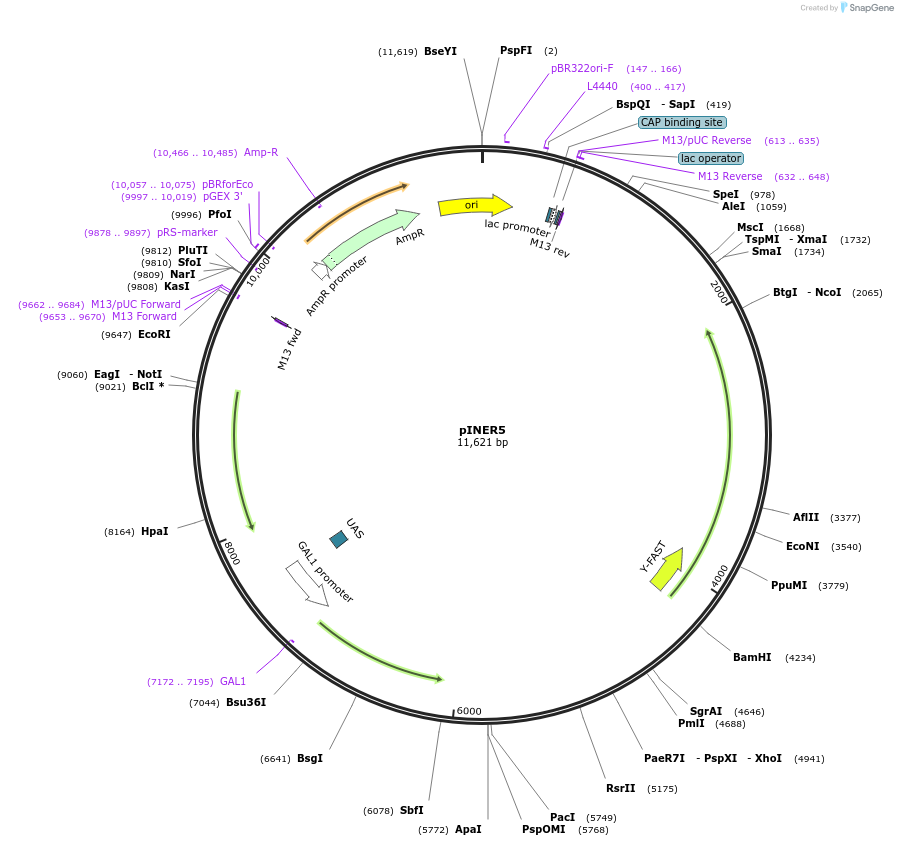
pINER5
Plasmid#185895PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (15 copy).DepositorInsertPRPL25(Arm1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm3)-ARS305-PSk.GAL2>YFAST-EVBR1.2A-AcNES1>TRPL41B-PPDA1>RPL25(part;Arm2
ExpressionYeastMutationNoneAvailable SinceAug. 25, 2022AvailabilityIndustry, Academic Institutions, and Nonprofits -
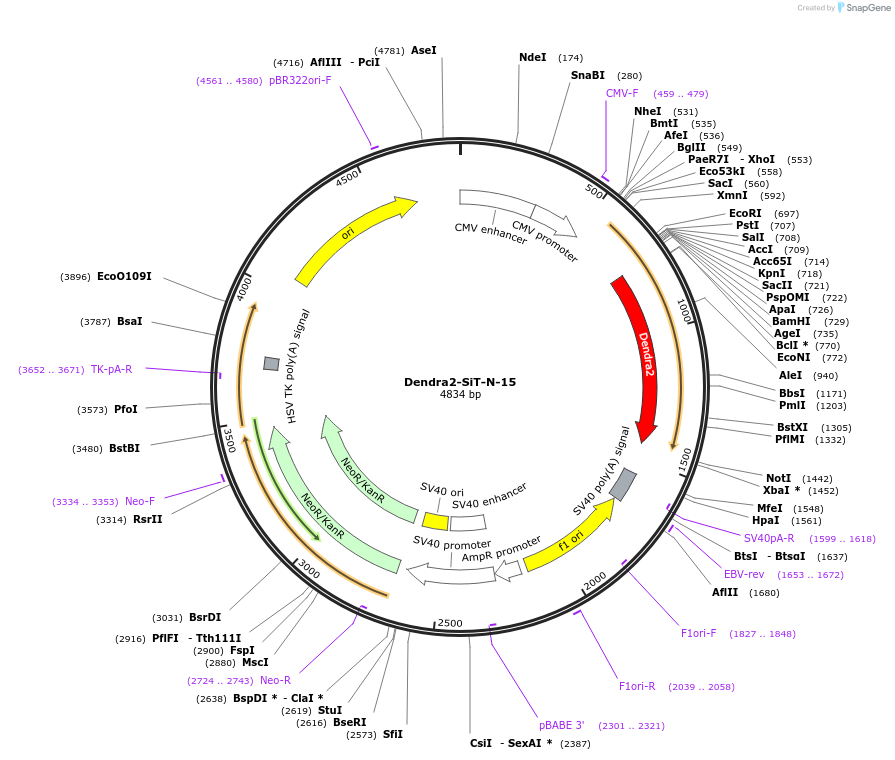
Dendra2-SiT-N-15
Plasmid#57739PurposeLocalization: Golgi TGN, Excitation: 490 / 553, Emission: 507 / 573DepositorAvailable SinceJan. 6, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
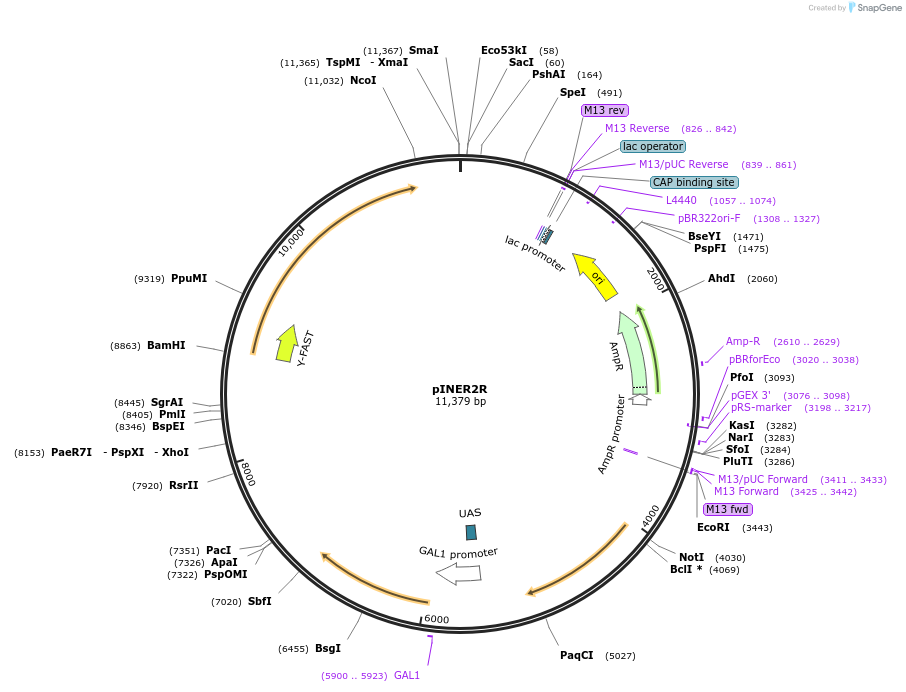
pINER2R
Plasmid#185882PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (11 copy).DepositorInsertPRPL25(Arm 1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm 3)-ARS305-PGAL2>Y.FAST-EVBR1.2A-AcNES1>TRPL41B>RPL25(partial; Arm2)
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
PA-mCherry1-SiT-N-15
Plasmid#57197PurposeLocalization: Golgi TGN, Excitation: 400 / 504, Emission: 515 / 517DepositorInsertSiT (ST6GAL1 Human)
TagsPA-mCherry1ExpressionMammalianMutationaa 1-45 of SiTPromoterCMVAvailable SinceJan. 13, 2015AvailabilityAcademic Institutions and Nonprofits only -
mEos2-SiT-N-15
Plasmid#57418PurposeLocalization: Golgi TGN, Excitation: 505 / 569, Emission: 516 / 581DepositorAvailable SinceFeb. 5, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
tdTomato-SiT-N-15
Plasmid#58131PurposeLocalization: Golgi TGN, Excitation: 554, Emission: 581DepositorAvailable SinceDec. 19, 2014AvailabilityAcademic Institutions and Nonprofits only -
Dronpa-SiT-N-15
Plasmid#57295PurposeLocalization: Golgi TGN, Excitation: 503, Emission: 518DepositorAvailable SinceJan. 13, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mCitrine-SiT-N-15
Plasmid#56318PurposeLocalization: Golgi TGN, Excitation: 516, Emission: 529DepositorAvailable SinceDec. 5, 2014AvailabilityIndustry, Academic Institutions, and Nonprofits -
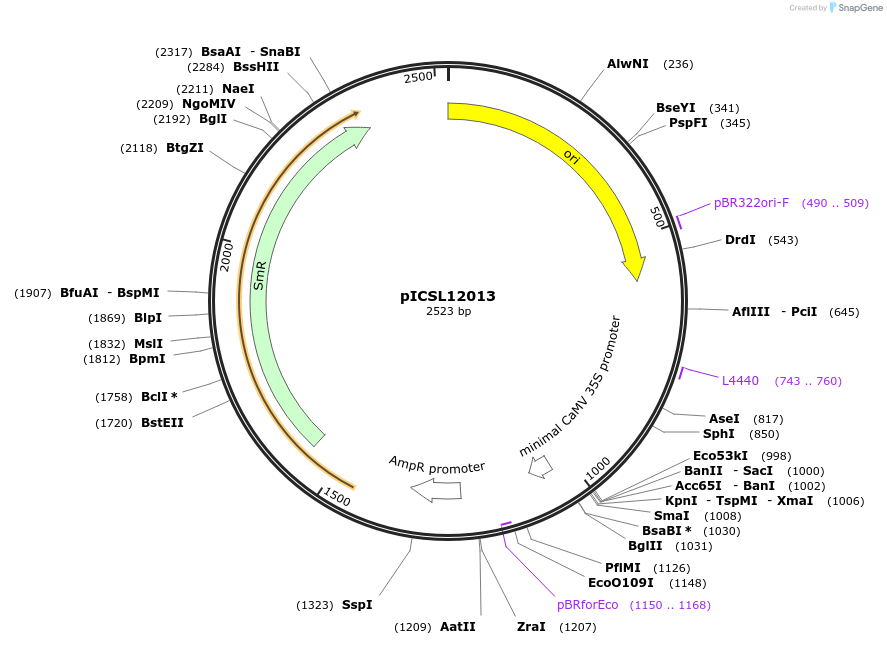
pICSL12013
Plasmid#245630PurposeLevel 0 Golden Gate part 6GAL4UAS promoter, GGAG - AATGDepositorInsert6GAL4UAS promoter
UseSynthetic BiologyMutationBsaI/BpiI sites removed by point-mutationAvailable SinceDec. 17, 2025AvailabilityAcademic Institutions and Nonprofits only -
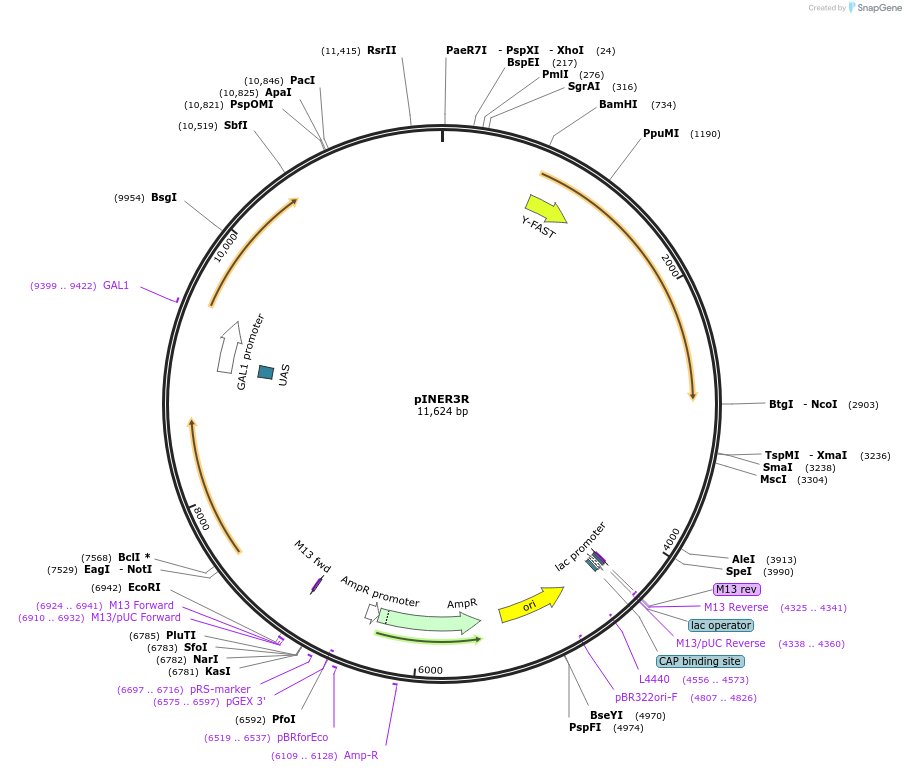
pINER3R
Plasmid#185883PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (15 copy).DepositorInsertPRPL25(Arm1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm3)-ARS305-PGAL2>Y.FAST-EVBR1.2A-AcNES1>TRPL41B-PPDA1>RPL25(part;Arm2)
ExpressionYeastMutationNoneAvailable SinceApril 16, 2024AvailabilityAcademic Institutions and Nonprofits only -
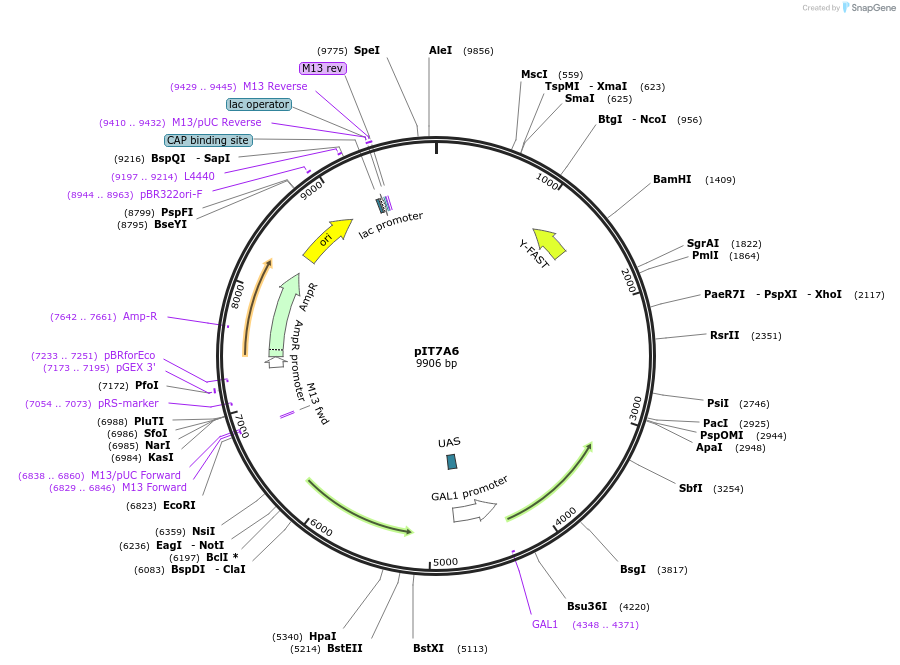
pIT7A6
Plasmid#185896PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module at RPL25 locus (15 copy).DepositorInsertPRPL25(Arm 1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm 3)-ARS305-PSk.GAL2>Y.FAST-EVBR1.2A>TRPL41B-PPDA1>RPL25(partial;Arm2)
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
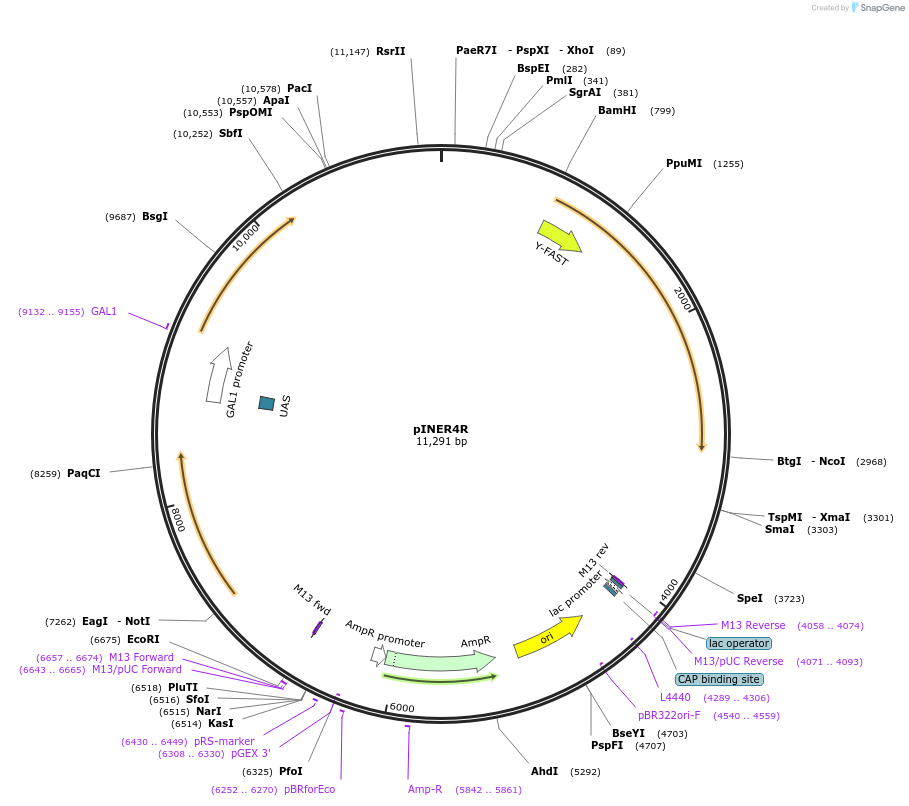
pINER4R
Plasmid#185884PurposeIn vivo gene amplification (HamAmp) of nerolidol synthetic module locus (25 copy).DepositorInsertPRPL25(Arm1)>Kl.LEU2>TKl.LEU2-PGAL1>ERG20>TRPL3-TRPL25(Arm3)-ARS305-PGAL2>Y.FAST-EVBR1.2A-AcNES1>TRPL41B-PBTS1>RPL25(part;Arm2)
ExpressionYeastMutationNoneAvailable SinceSept. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
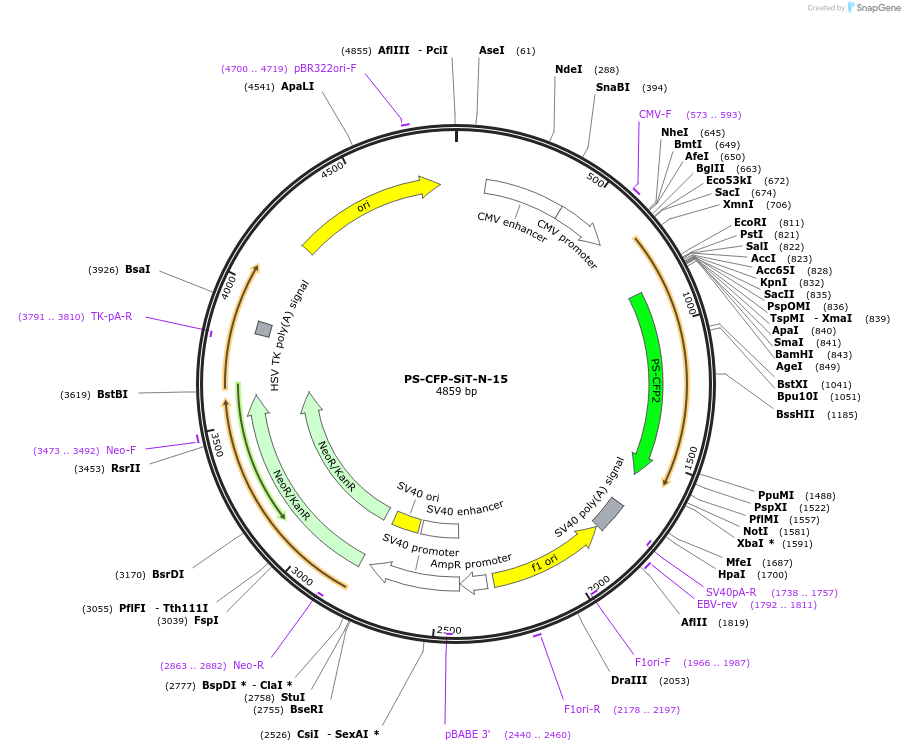
PS-CFP-SiT-N-15
Plasmid#57206PurposeLocalization: Golgi TGN, Excitation: 400 / 490, Emission: 468 / 511DepositorAvailable SinceApril 1, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
mPlum-SiT-N-15
Plasmid#55999PurposeLocalization: Golgi TGN, Excitation: 590, Emission: 649DepositorAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only