We narrowed to 3,004 results for: PHI-1;
-
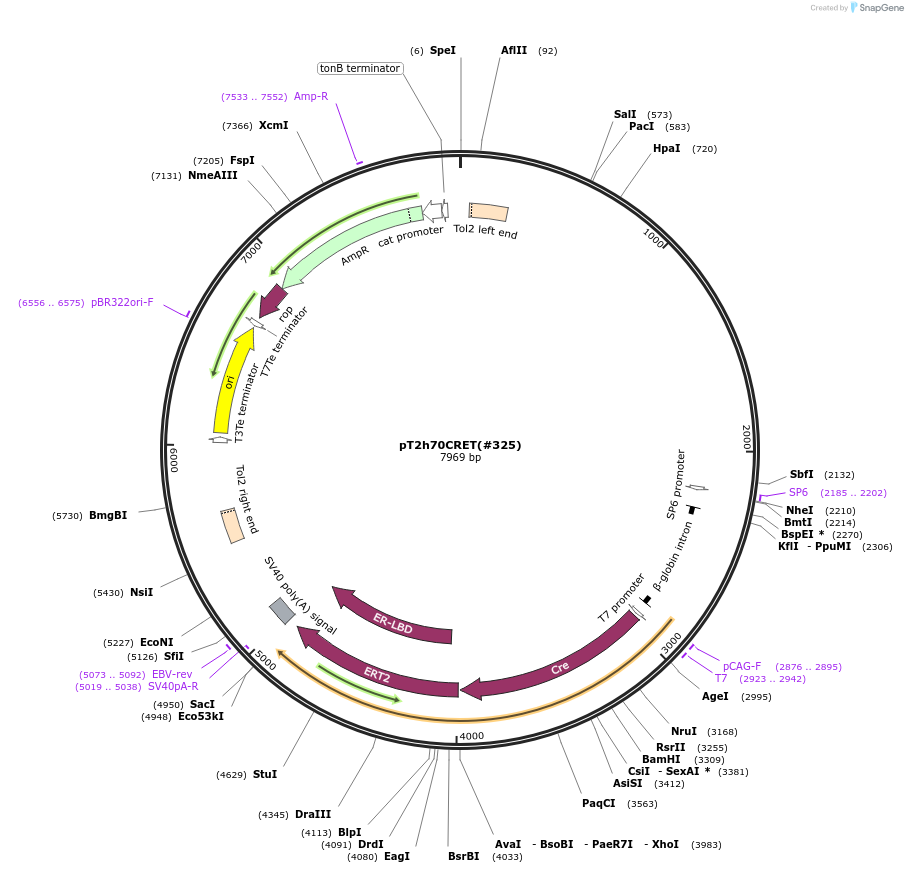
Plasmid#184060Purposecyclofen-inducible CRE activation in zebrafish transient transgenicDepositorInsertCRE-ERT2
UseZebrafish transient transgenesisMutationERT2: G400V, M543A, L544A, deleted 1-281;PromoterZf-Hsp70-4Available SinceJuly 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
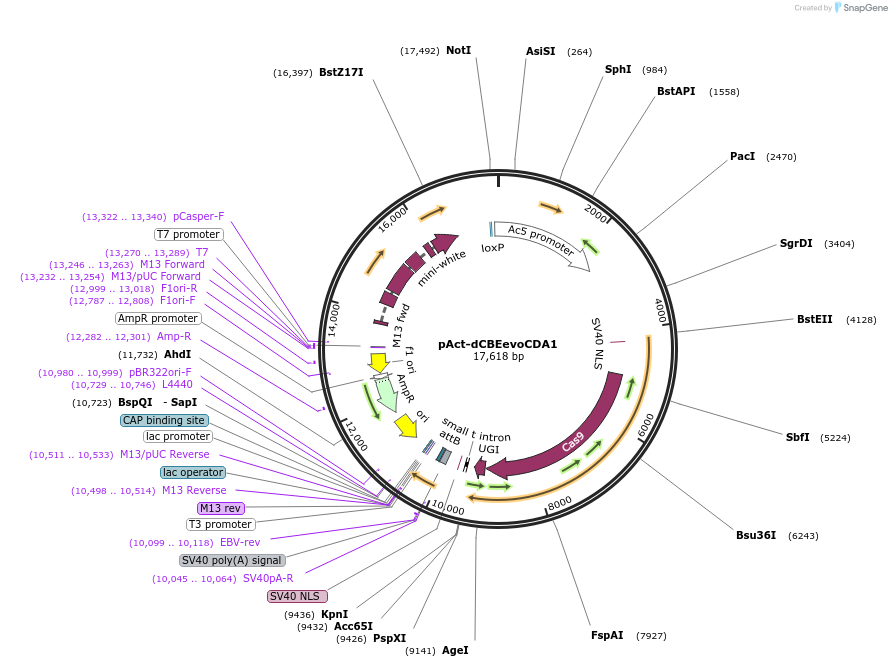
pAct-dCBEevoCDA1
Plasmid#195515PurposeUbiquitous expression of dCBEevoCDA1 in DrosophilaDepositorInsertdCBE-evoCDA1
UseCRISPRExpressionInsectPromoteract5CAvailable SinceSept. 12, 2023AvailabilityAcademic Institutions and Nonprofits only -
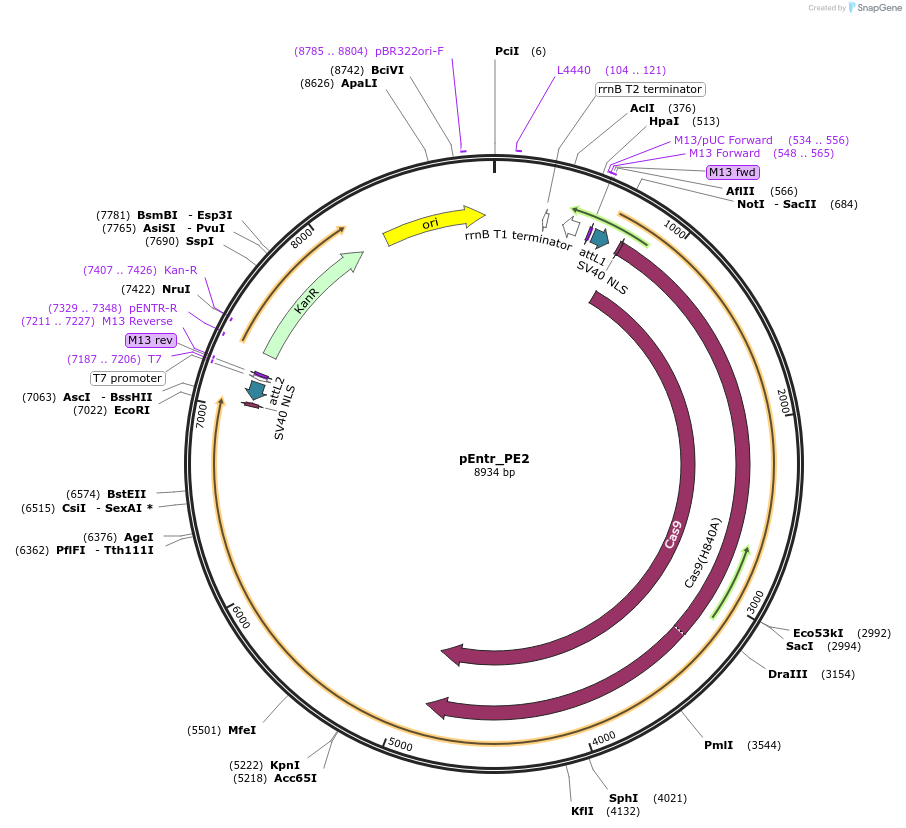
pEntr_PE2
Plasmid#149548PurposeGateway entry clone with PE2 enzyme (with stop codon)DepositorInsertPE2
UseCRISPR; Gateway shuttle vectorAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
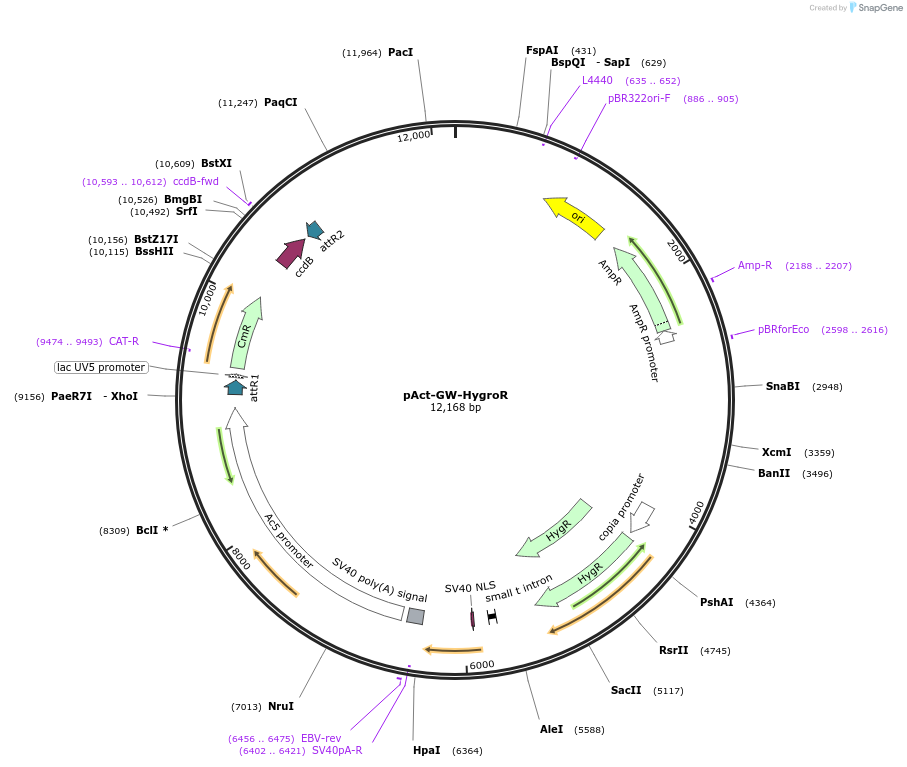
pAct-GW-HygroR
Plasmid#149610PurposeGateway compatible expression plasmid for Drosophila and establishing stable cell lines by Hygromycin selection. Actin5c ubiquitous promoter.DepositorInsertGateway cassette (ccdb + Chlor.R.)
ExpressionInsectPromoterAct5cAvailable SinceAug. 6, 2020AvailabilityAcademic Institutions and Nonprofits only -
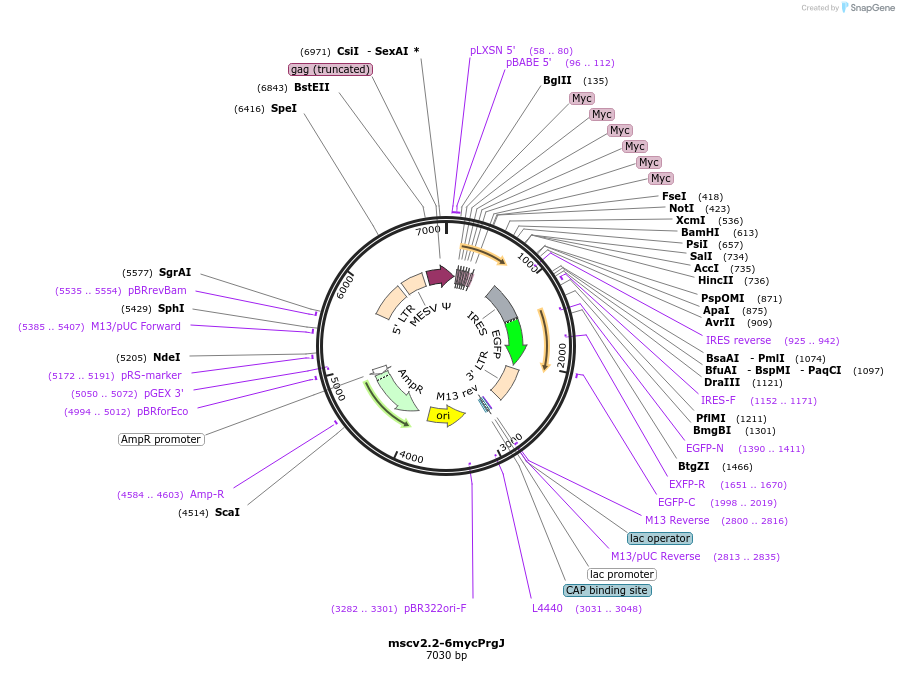
mscv2.2-6mycPrgJ
Plasmid#60204Purpose6myc-tagged PrgJ (SPI-1 rod protein) from Salmonella typhimurium, under the constitutive, moderate-level retroviral LTR promoter for mammalian cell expression; IRES-GFP reporter downstream of PrgJDepositorInsertPrgJ
UseRetroviralTags6xMYCExpressionMammalianAvailable SinceMarch 18, 2015AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut G
Plasmid#11320DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation G (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut I
Plasmid#11322DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation I (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut A
Plasmid#11314DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation A (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut B
Plasmid#11315DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation B (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut C
Plasmid#11316DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation C (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut D
Plasmid#11317DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation D (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut E
Plasmid#11318DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation E (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut F
Plasmid#11319DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation F (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut H
Plasmid#11321DepositorInsert4 bulged binding sites for CXCR4 siRNA antisense
UseLuciferaseTagsRr-lucMutationMutation H (see paper) on inner 2 binding sites.Available SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
pRL-TK 4x mut GFP
Plasmid#11323DepositorInsert2 bulged binding sites for CXCR4 siRNA antisense, flanking 2 binding sites for GFP
UseLuciferaseTagsRr-lucAvailable SinceFeb. 15, 2006AvailabilityAcademic Institutions and Nonprofits only -
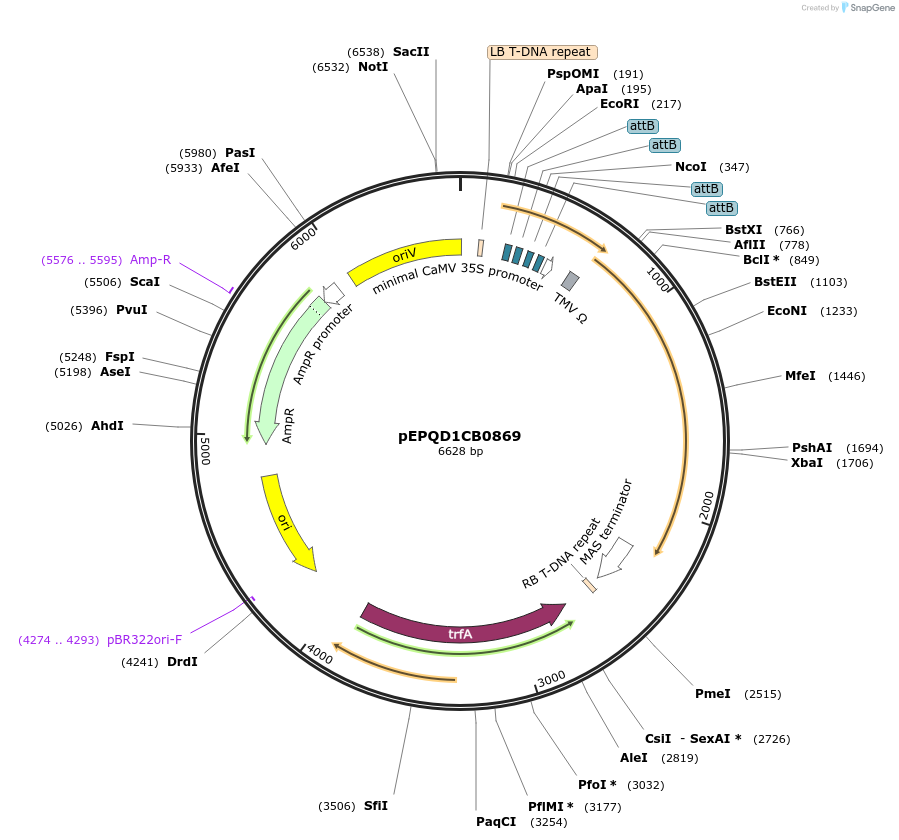
pEPQD1CB0869
Plasmid#177086PurposeMoClo Level 1, position 5, transcriptional unit for transient expression of 7-deoxyloganic acid hydroxylase (Cr7-DLH) from Catharanthus roseus promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsert7-deoxyloganic acid hydroxylase (Cr7-DLH) from Catharanthus roseus
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
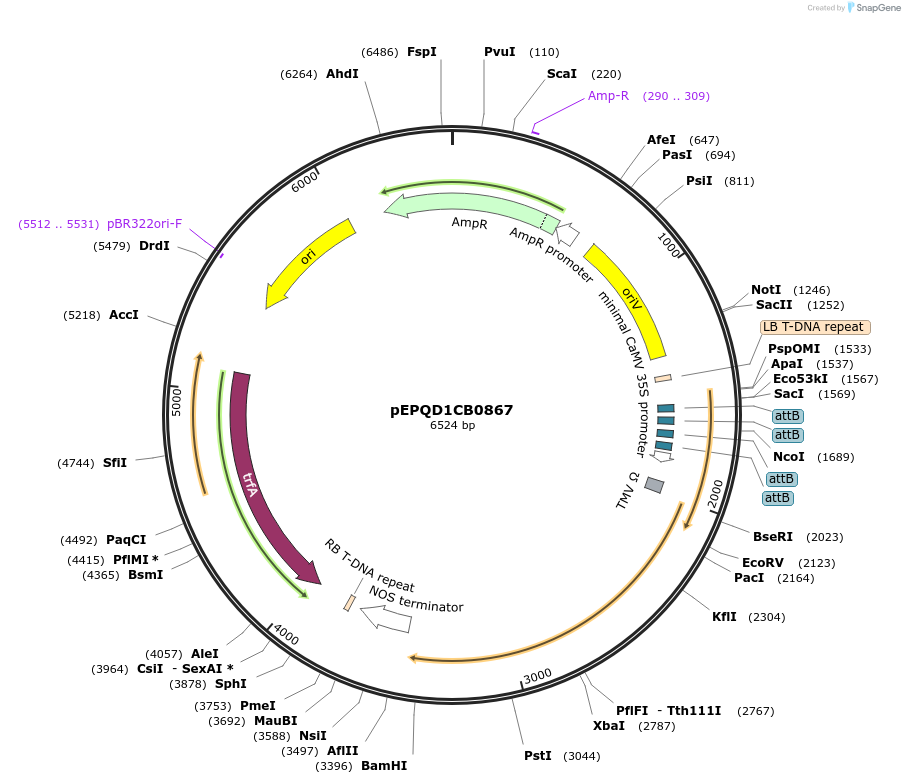
pEPQD1CB0867
Plasmid#177085PurposeMoClo Level 1, position 3, transcriptional unit for transient expression of 7-deoxyloganetic acid glucosyl transferase (Cr7-DLGT) from Catharanthus roseus promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsert7-deoxyloganetic acid glucosyl transferase (Cr7-DLGT) from Catharanthus roseus
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
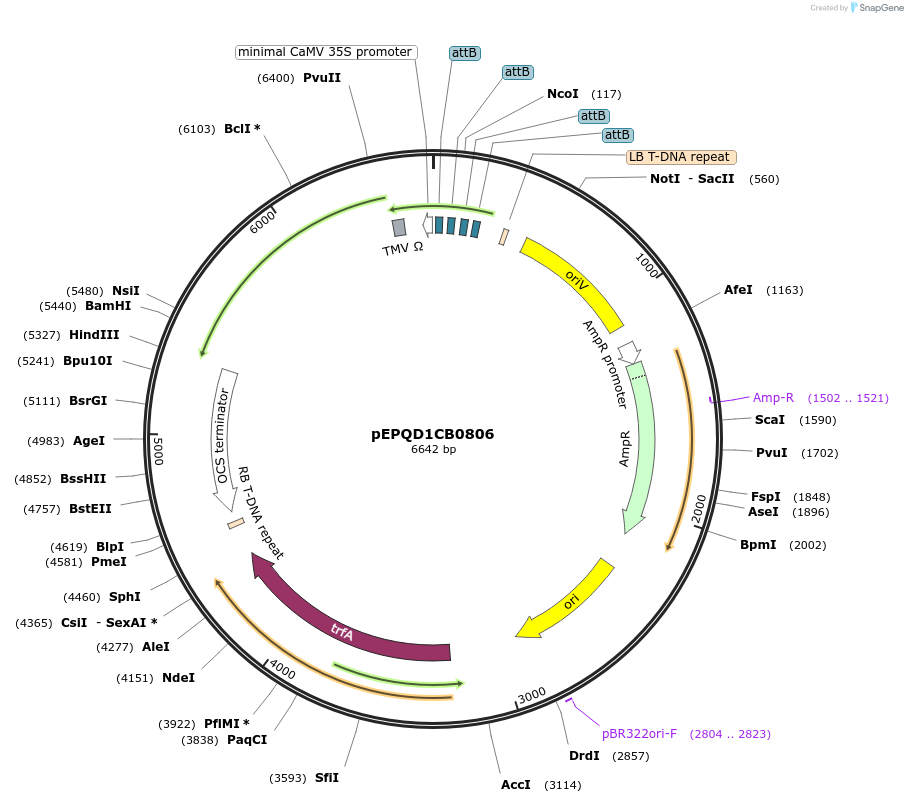
pEPQD1CB0806
Plasmid#177080PurposeMoClo Level 1, position 3, transcriptional unit for transient expression of loganic acid O-methyltransferase (CrLAMT) from Catharanthus roseus promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsertloganic acid O-methyltransferase (CrLAMT) from Catharanthus roseus
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
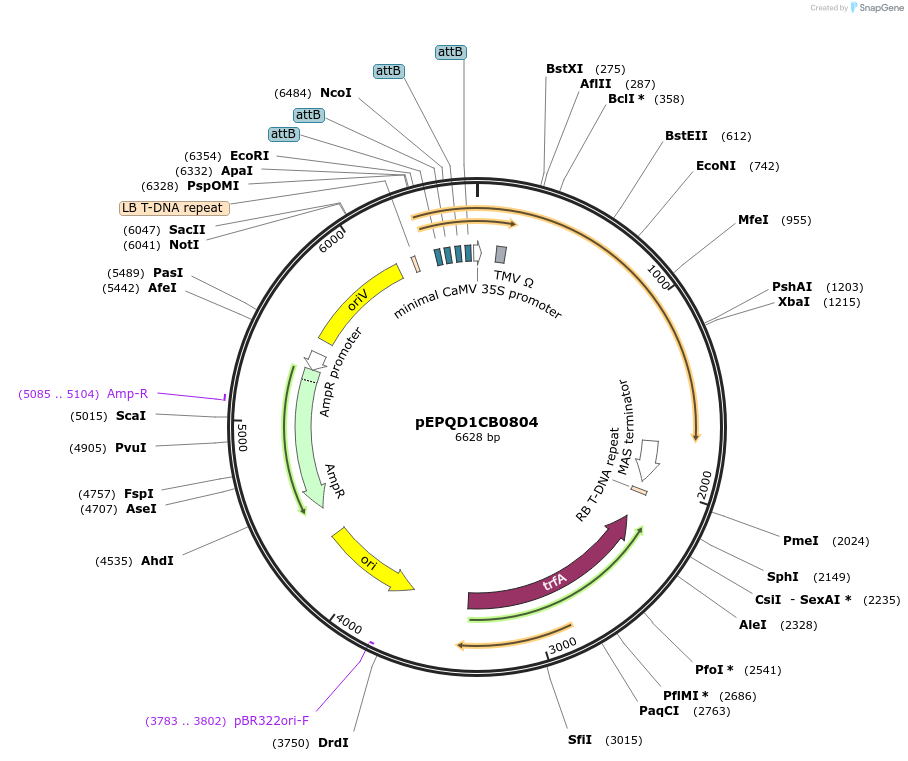
pEPQD1CB0804
Plasmid#177079PurposeMoClo Level 1, position 2, transcriptional unit for transient expression of 7-deoxyloganic acid hydroxylase (Cr7-DLH) from Catharanthus roseus promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsert7-deoxyloganic acid hydroxylase (Cr7-DLH) from Catharanthus roseus
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
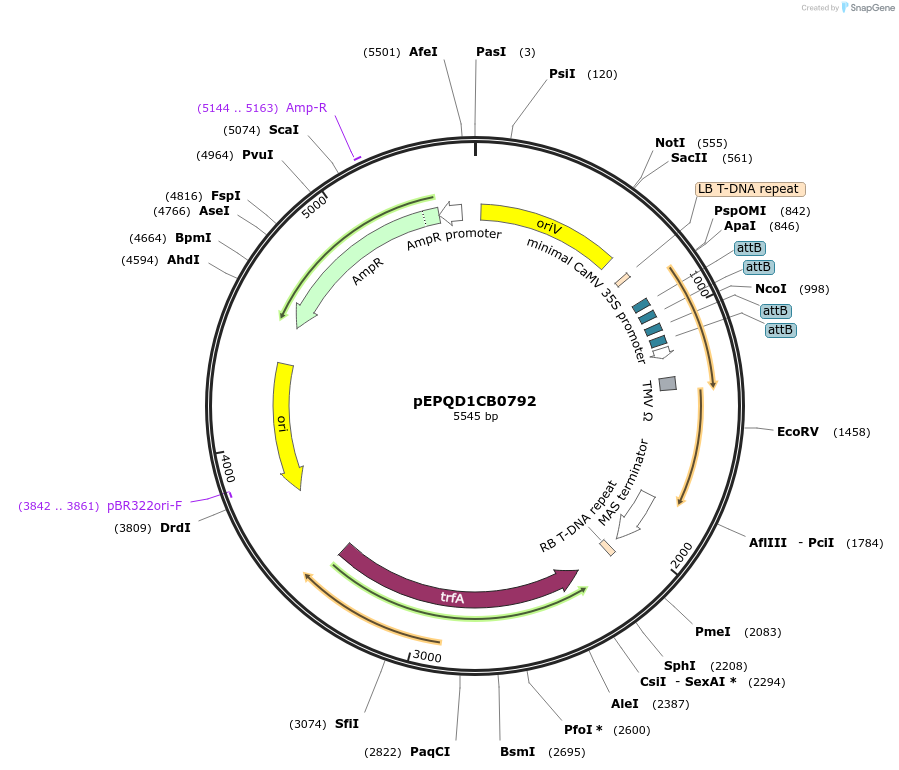
pEPQD1CB0792
Plasmid#177076PurposeMoClo Level 1, position 2, transcriptional unit for transient expression of major latex protein-like (NmMLPL) from Nepeta mussinii promoter regions contains 4x attB sites for recruitment fo gal4AD-phiC31DepositorInsertmajor latex protein-like (NmMLPL) from Nepeta mussinii
UseSynthetic BiologyAvailable SinceDec. 6, 2021AvailabilityAcademic Institutions and Nonprofits only