8,173 results
-
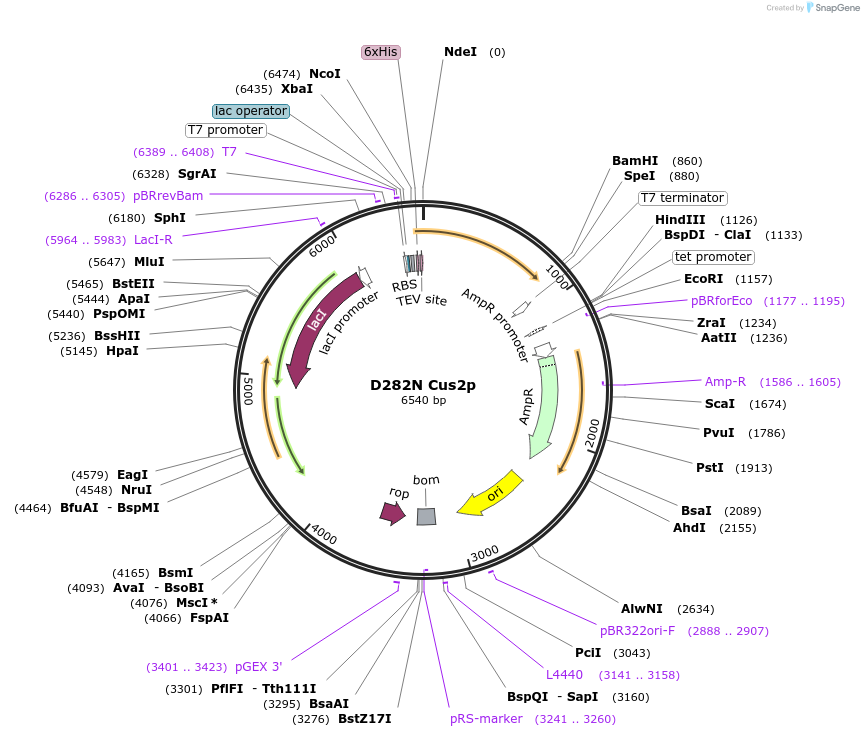
Plasmid#73060PurposeS. cerevisiae Cus2p gene with D282N MutationDepositorArticleInsertCUS2p (CUS2 Budding Yeast)
Tags6x His tag fused to TEV cleavage siteExpressionBacterial and YeastMutationAsp 282 to AsnPromoterT7AvailabilityAcademic Institutions and Nonprofits only -
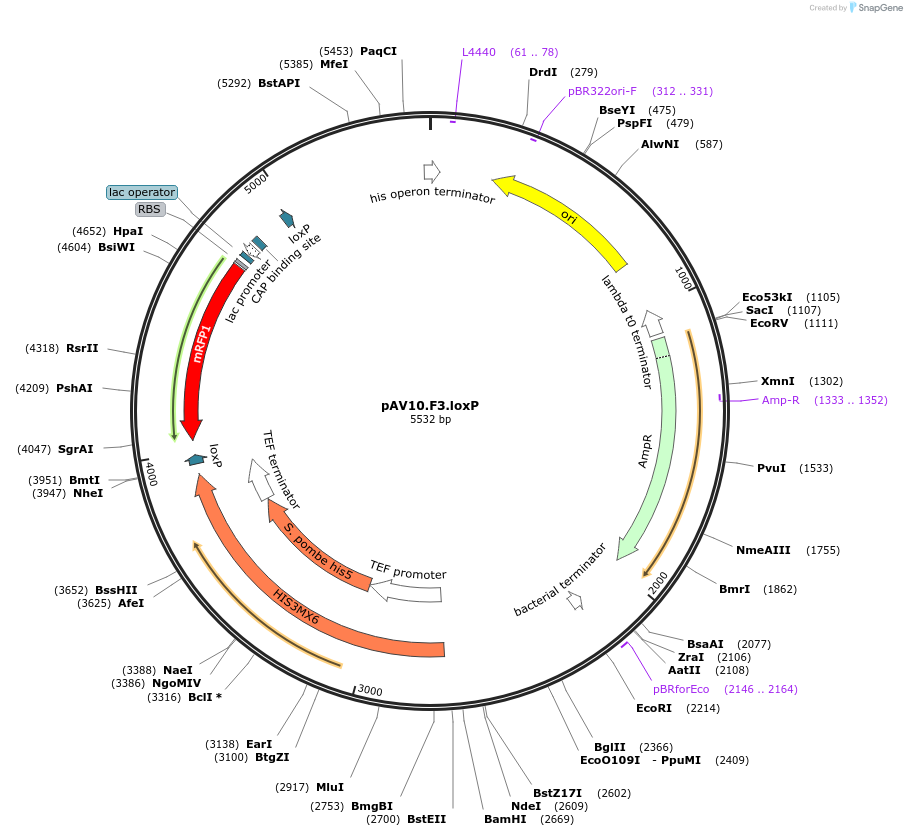
pAV10.F3.loxP
Plasmid#63200PurposepCACS backbone (Amp), SpHIS5, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into chrVI: 97873-98803 (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
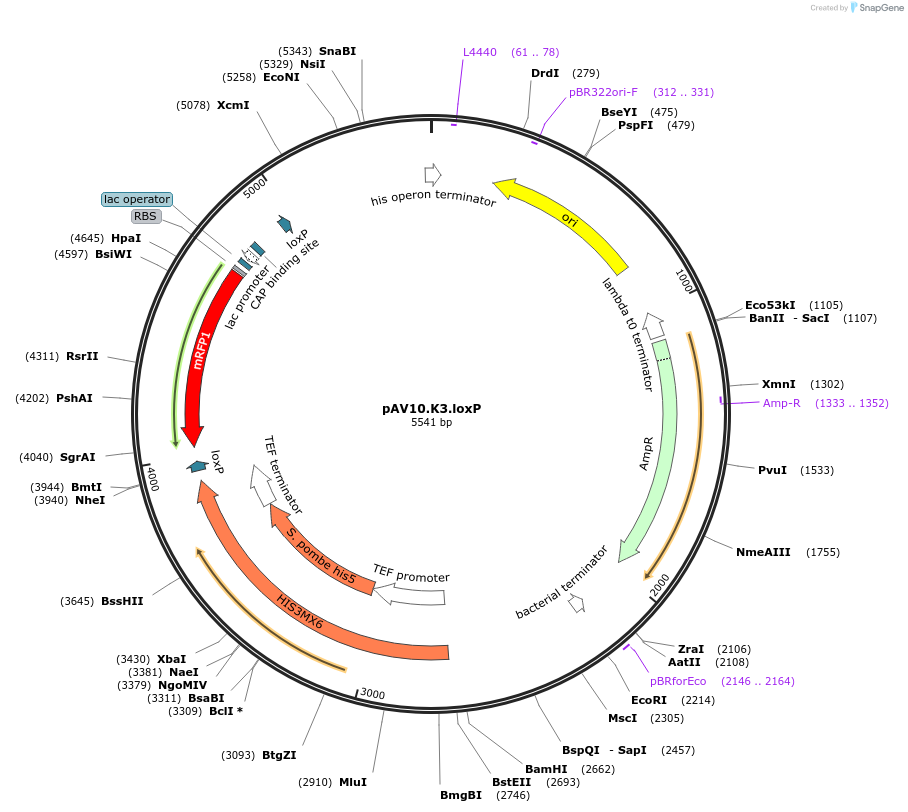
pAV10.K3.loxP
Plasmid#63202PurposepCACS backbone (Amp), SpHIS5, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into YKL162C-A (NotI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
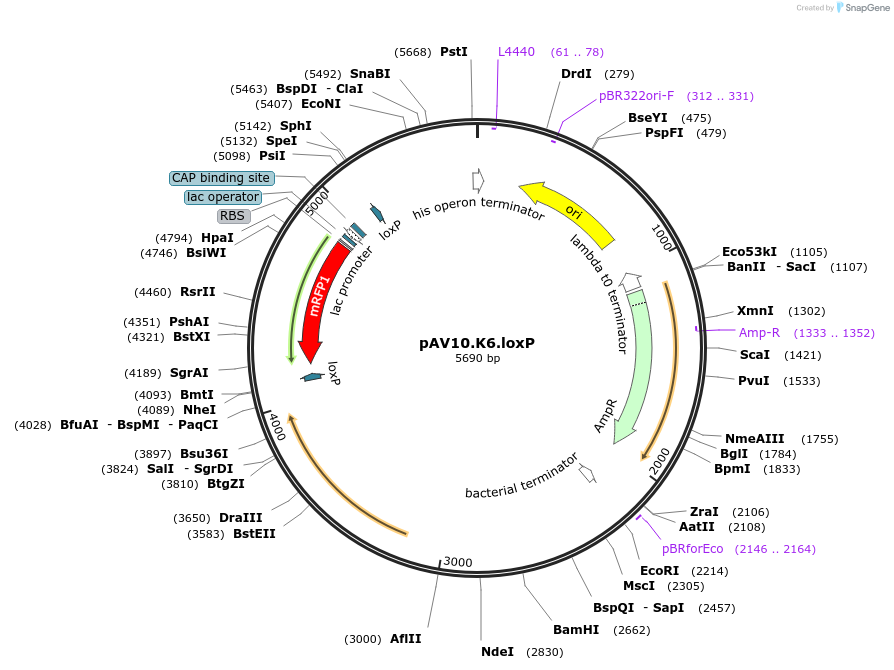
pAV10.K6.loxP
Plasmid#63203PurposepCACS backbone (Amp), KlURA3, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into YKL162C-A (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
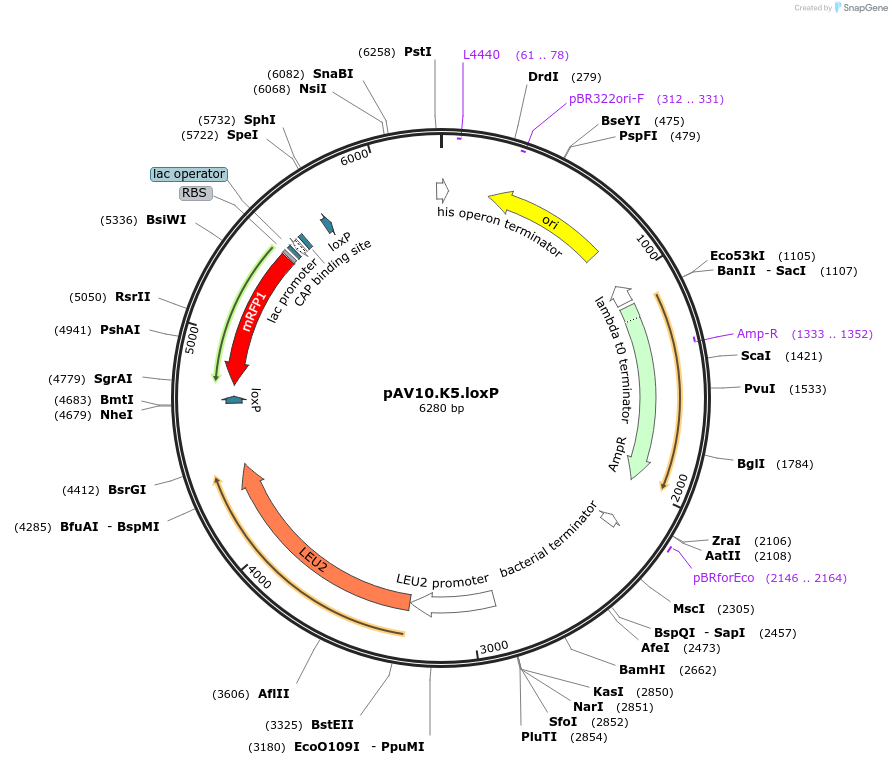
pAV10.K5.loxP
Plasmid#63204PurposepCACS backbone (Amp), LEU2, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into YKL162C-A (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
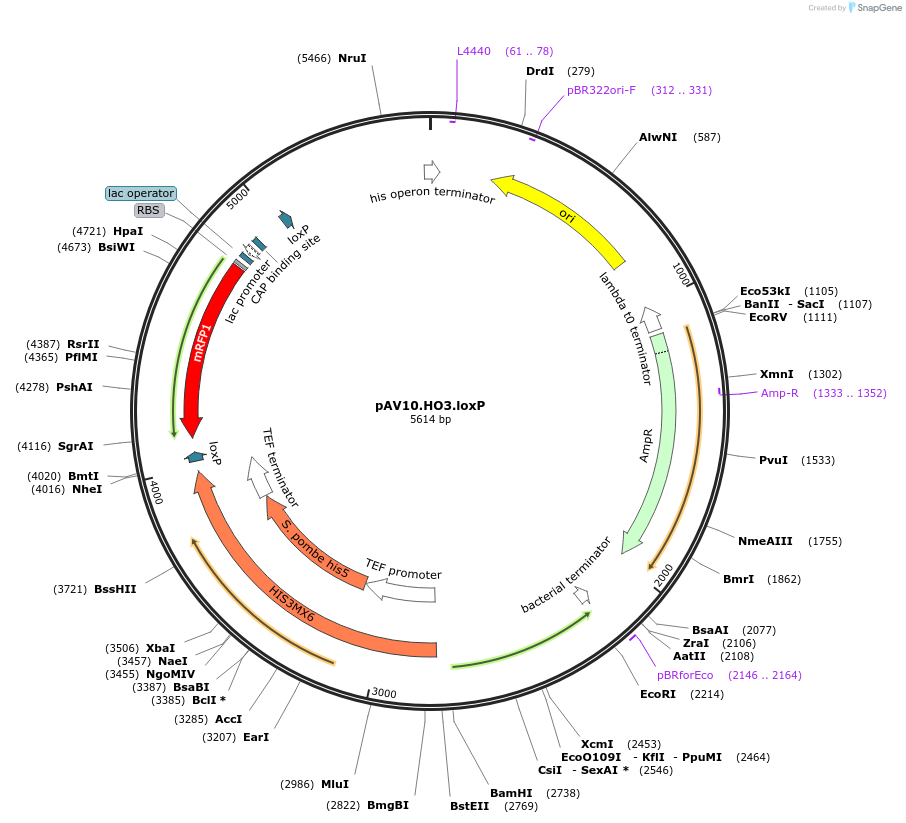
pAV10.HO3.loxP
Plasmid#63205PurposepCACS backbone (Amp), SpHIS5, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into HO locus (NotI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
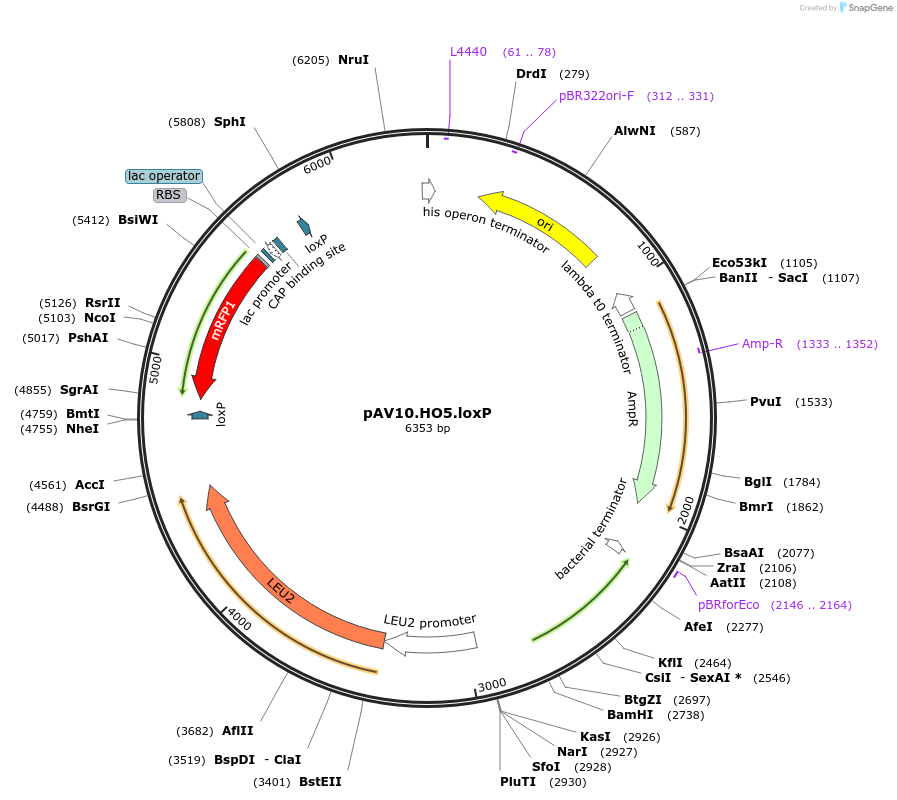
pAV10.HO5.loxP
Plasmid#63206PurposepCACS backbone (Amp), LEU2, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. LoxP flanked TU. Integration into HO locus (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
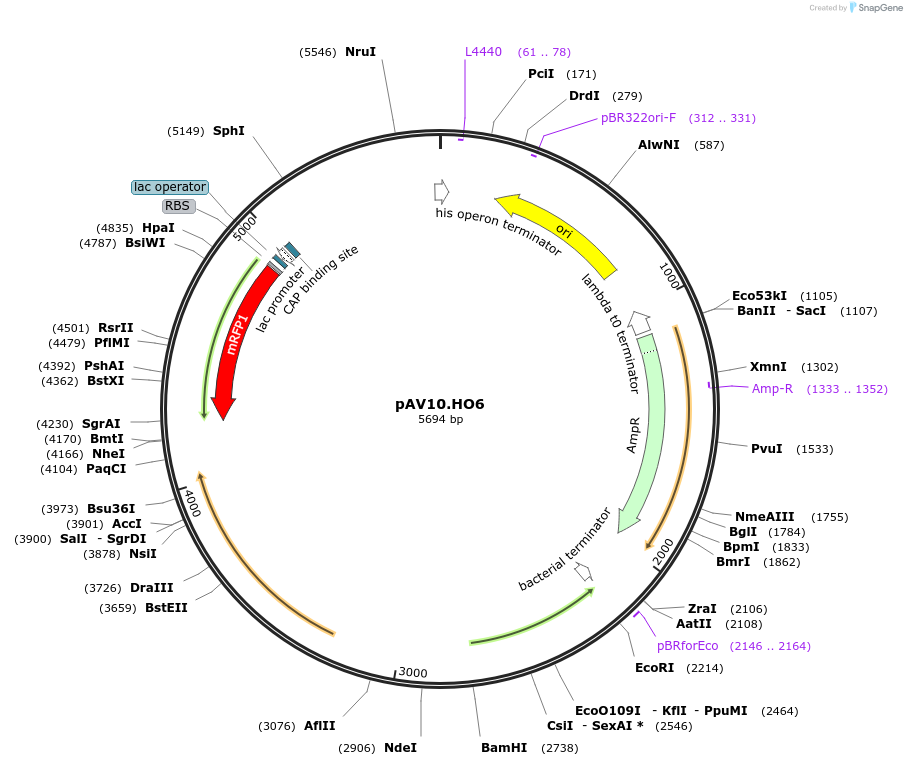
pAV10.HO6
Plasmid#63211PurposepCACS backbone (Amp), KlURA3, contains an RFP flanked by BsaI sites with CAGT & TTTT overhangs for TU assembly using yGG. Integration into HO locus (NotI or BciVI).DepositorTypeEmpty backboneUseSynthetic BiologyExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
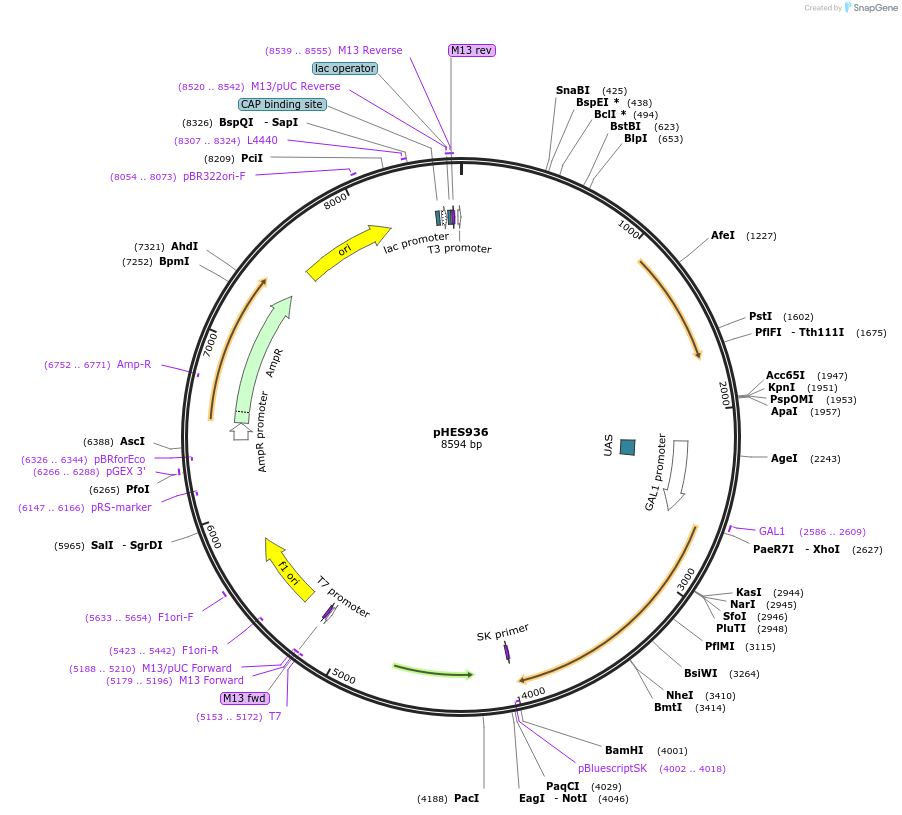
pHES936
Plasmid#89199PurposeExpresses DIG1 under control of GAL1 promoterDepositorAvailabilityAcademic Institutions and Nonprofits only -
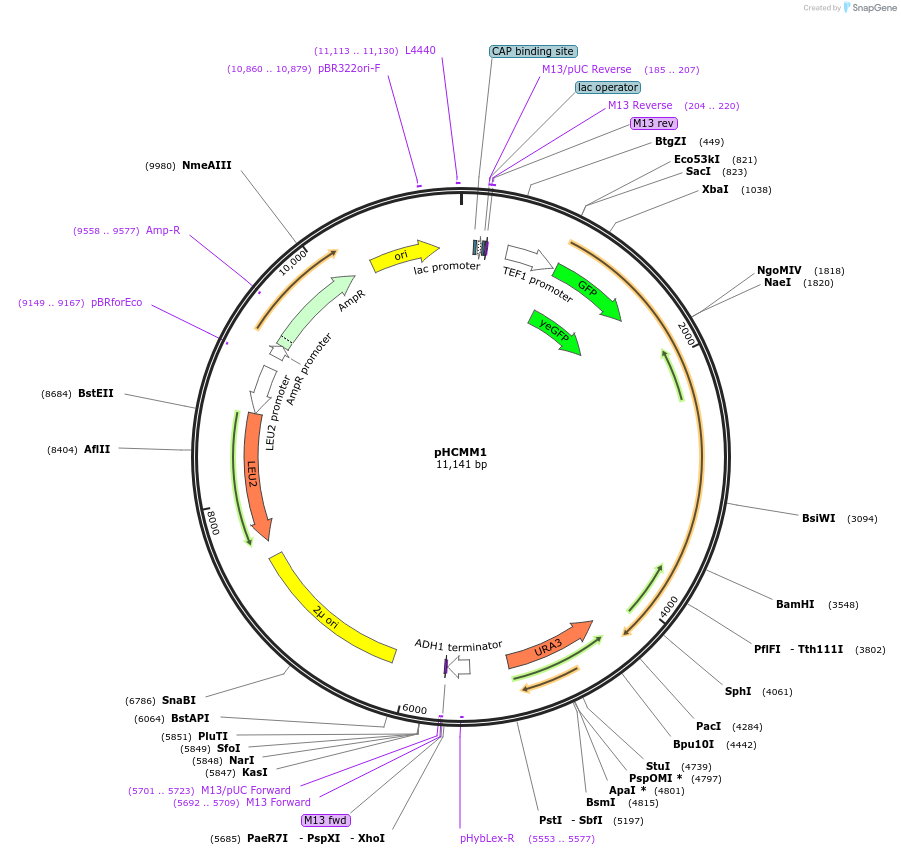
pHCMM1
Plasmid#167452PurposeExpresses C.elegans PUP-2 linked to RNA-recognition motifs (RRMs) of yeast poly(A)-binding protein [PUP alone (+PAB)] from the TEF1 promoterDepositorInsertPTEF1_yeGFP_scPAB14RRMS_cePUP-2_URA3_TADH1
ExpressionYeastPromoterTEF1AvailabilityAcademic Institutions and Nonprofits only -
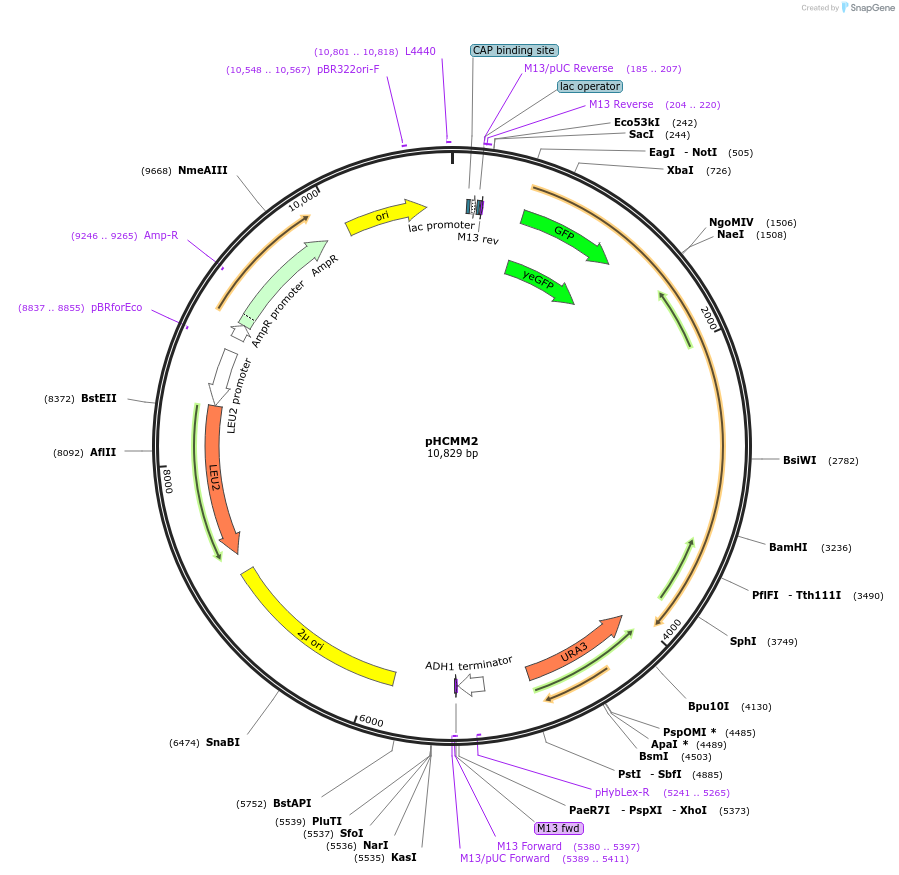
pHCMM2
Plasmid#167453PurposeExpresses C.elegans PUP-2 linked to RNA-recognition motifs (RRMs) of yeast poly(A)-binding protein [PUP alone (+PAB)] from the Sec63 promoterDepositorInsertPSEC63_yeGFP_scPAB14RRMS_cePUP-2_URA3_TADH1
ExpressionYeastPromoterPSEC63AvailabilityAcademic Institutions and Nonprofits only -
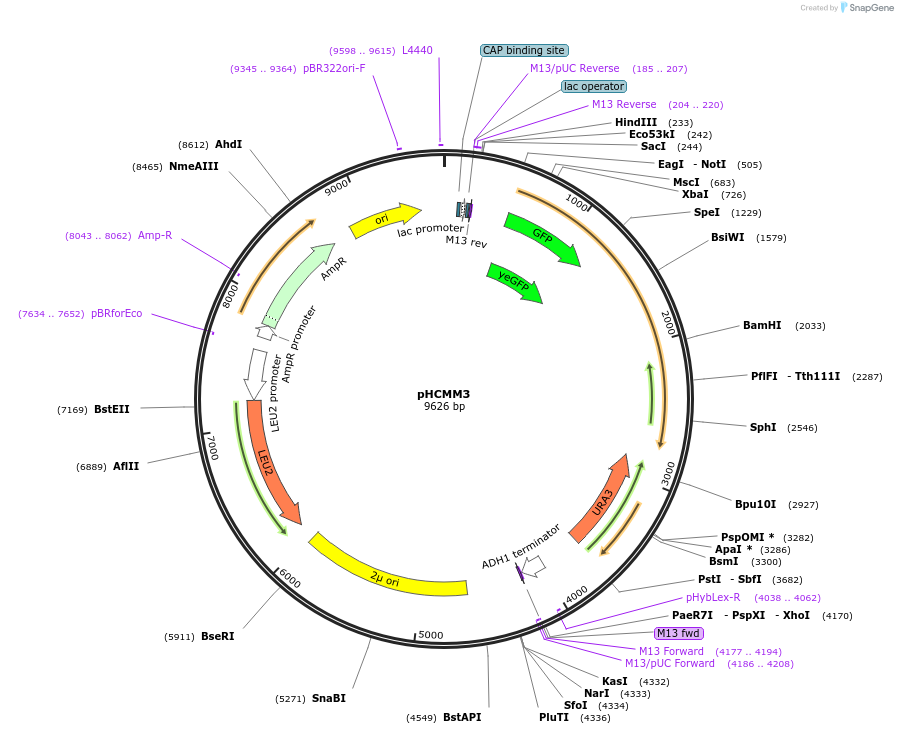
pHCMM3
Plasmid#167454PurposeA control chimera; expresses C. elegans PUP-2 from the Sec63 promoter without the RNA-recognition motifs (RRMs) of yeast poly(A)-binding protein [PUP alone (-PAB)]DepositorInsertPSEC63_yeGFP_cePUP-2_URA3_TADH1
ExpressionYeastPromoterPSEC63AvailabilityAcademic Institutions and Nonprofits only -
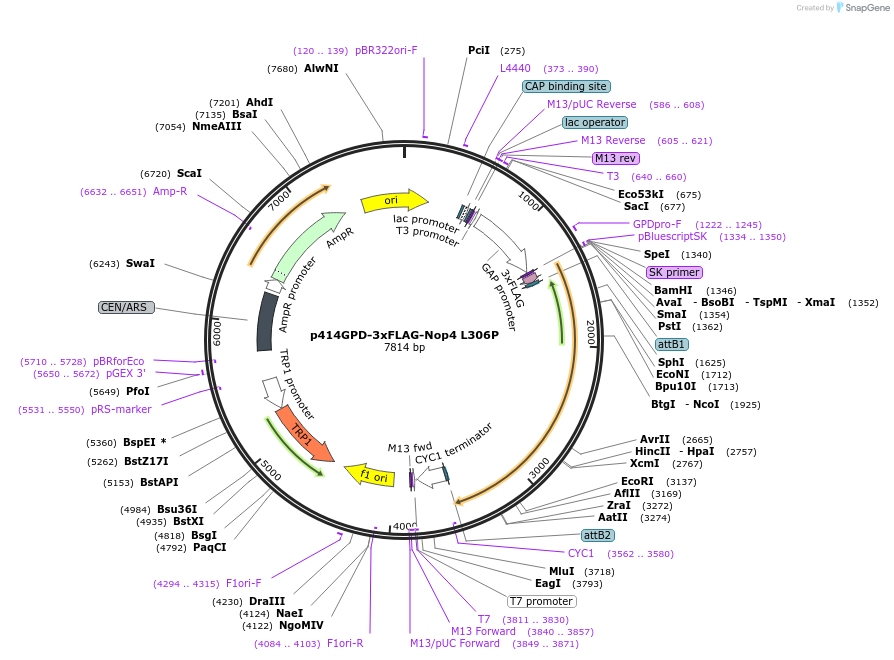
p414GPD-3xFLAG-Nop4 L306P
Plasmid#169262PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-Nop4 L306PDepositorInsertNop4 L306P (NOP4 Budding Yeast)
Tags3xFLAGExpressionYeastMutationp.(Leu306Pro)PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
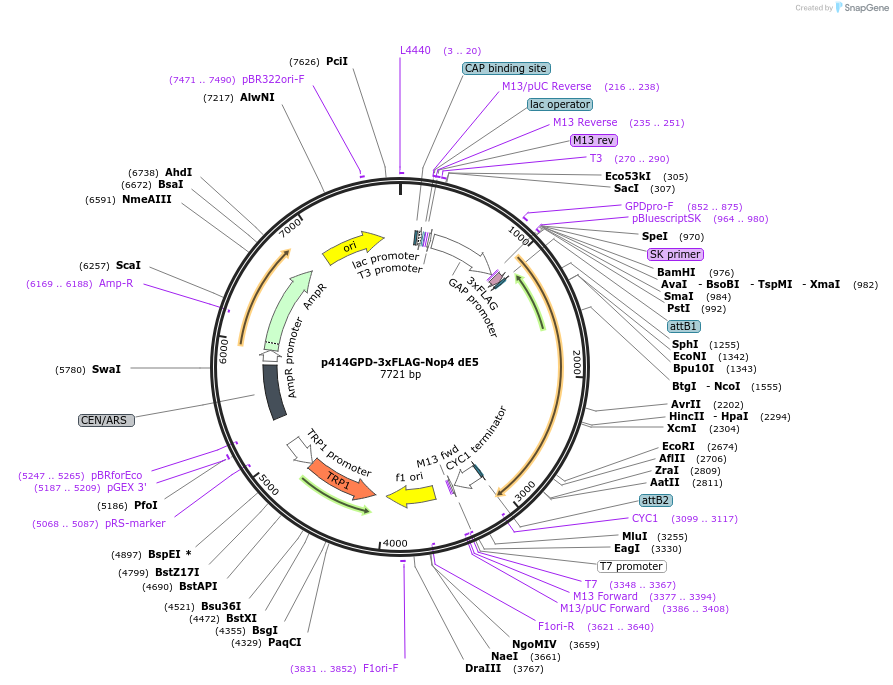
p414GPD-3xFLAG-Nop4 dE5
Plasmid#169263PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-Nop4 delta Exon 5DepositorInsertNop4 delta E5 (NOP4 Budding Yeast)
Tags3xFLAGExpressionYeastMutationdeleted region corresponding to human Exon 5PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
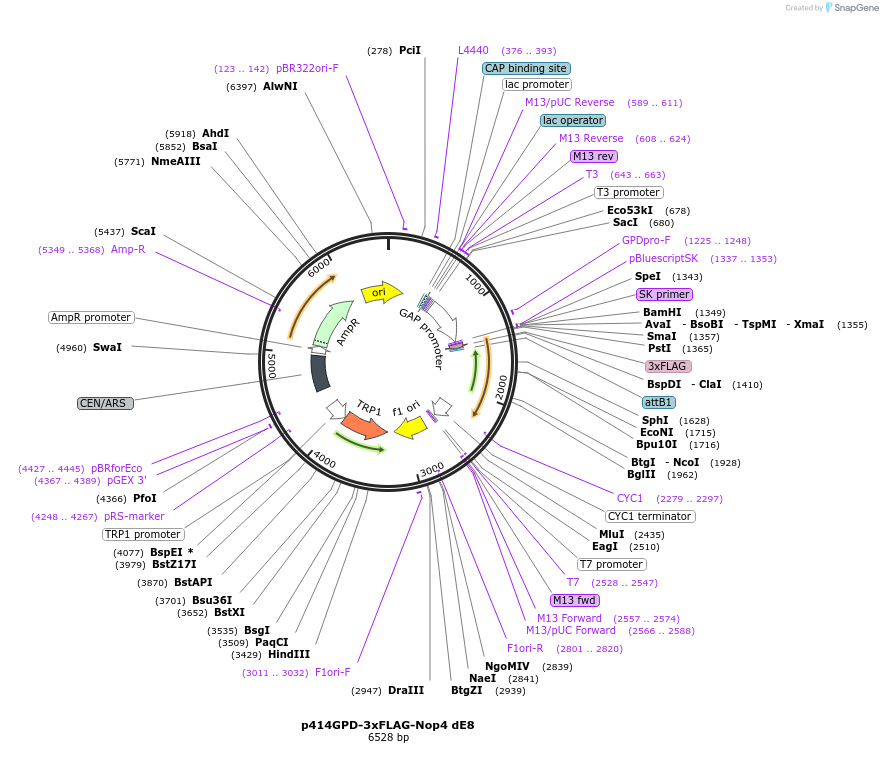
p414GPD-3xFLAG-Nop4 dE8
Plasmid#169264PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-Nop4 delta Exon 8DepositorInsertNop4 delta E8 (NOP4 Budding Yeast)
Tags3xFLAGExpressionYeastMutationdeleted region corresponding to human Exon 8PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
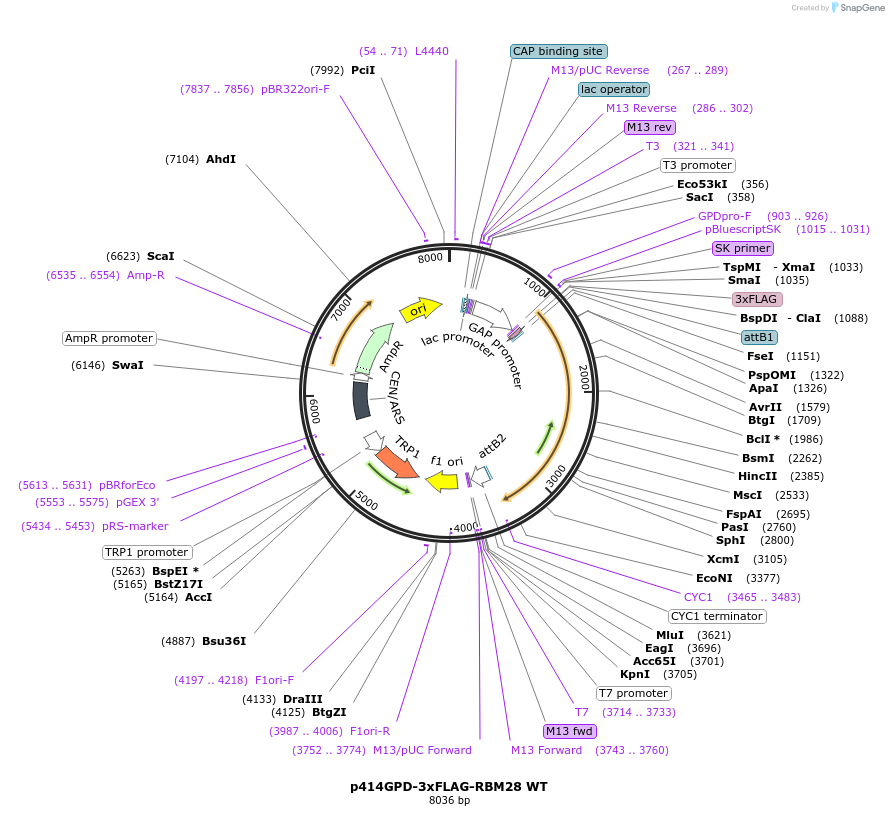
p414GPD-3xFLAG-RBM28 WT
Plasmid#169265PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-RBM28 WTDepositorAvailabilityAcademic Institutions and Nonprofits only -
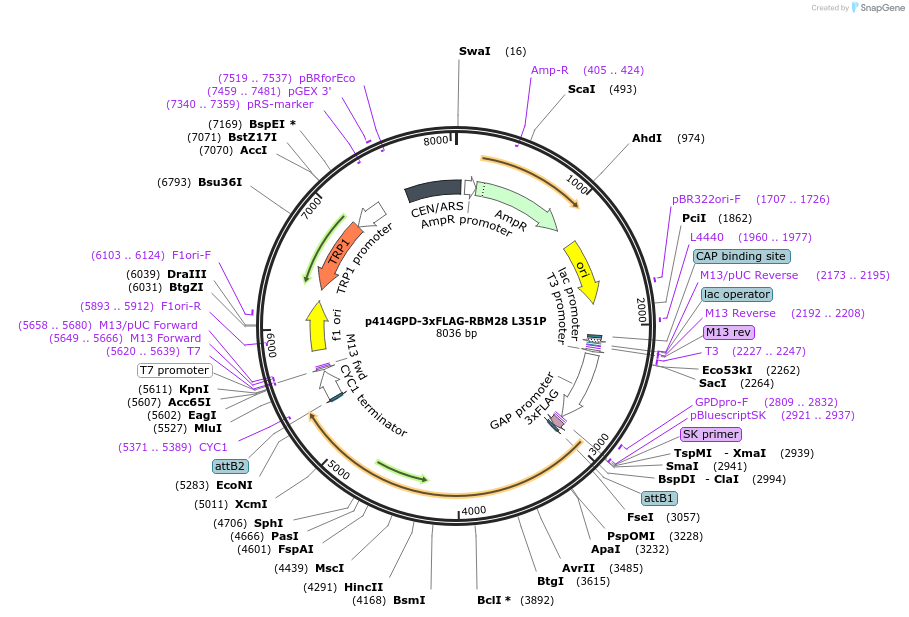
p414GPD-3xFLAG-RBM28 L351P
Plasmid#169266PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-RBM28 L351PDepositorAvailabilityAcademic Institutions and Nonprofits only -
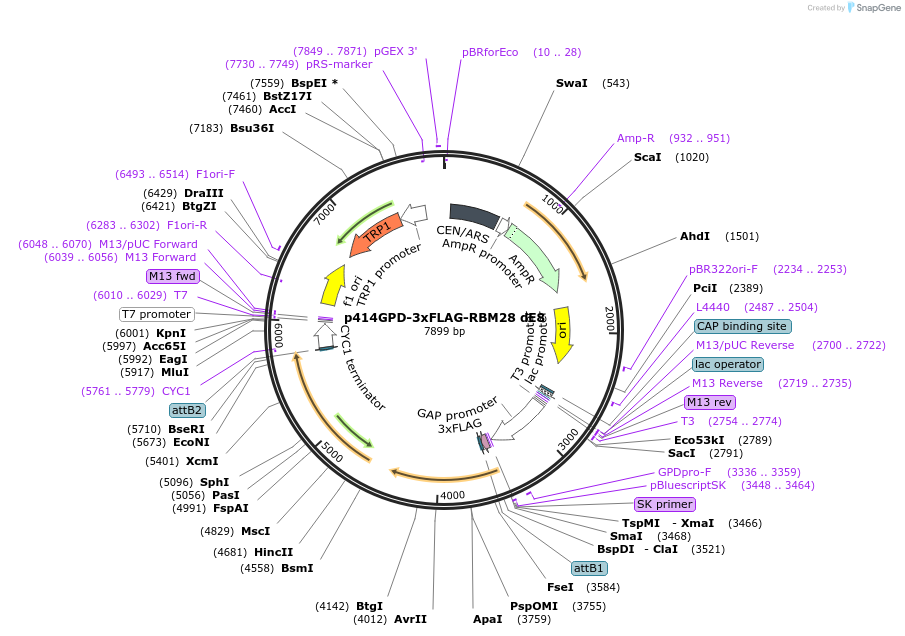
p414GPD-3xFLAG-RBM28 dE5
Plasmid#169267PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-RBM28 delta Exon 5DepositorInsertRBM28 delta E5 (RBM28 Human)
Tags3xFLAGExpressionYeastMutationdeleted Exon 5PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
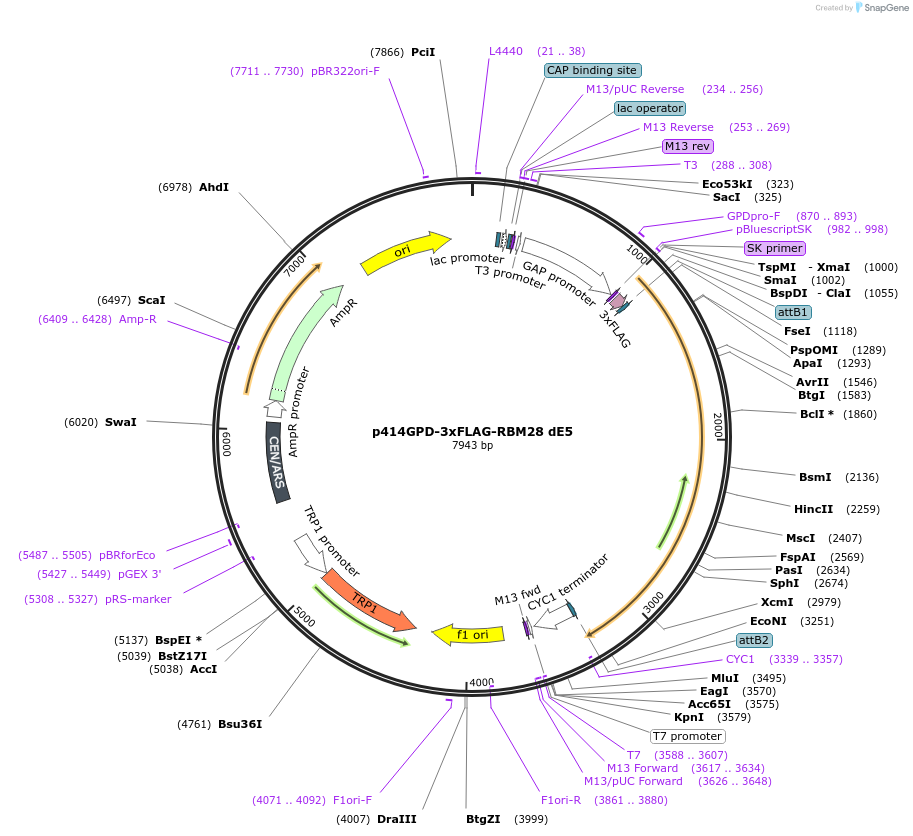
p414GPD-3xFLAG-RBM28 dE8
Plasmid#169268PurposeS. cerevisiae (yeast) vector for constitutive expression of 3xFLAG-RBM28 delta Exon 8DepositorInsertRBM28 delta E8 (RBM28 Human)
Tags3xFLAGExpressionYeastMutationdeleted Exon 8PromoterGPDAvailabilityAcademic Institutions and Nonprofits only -
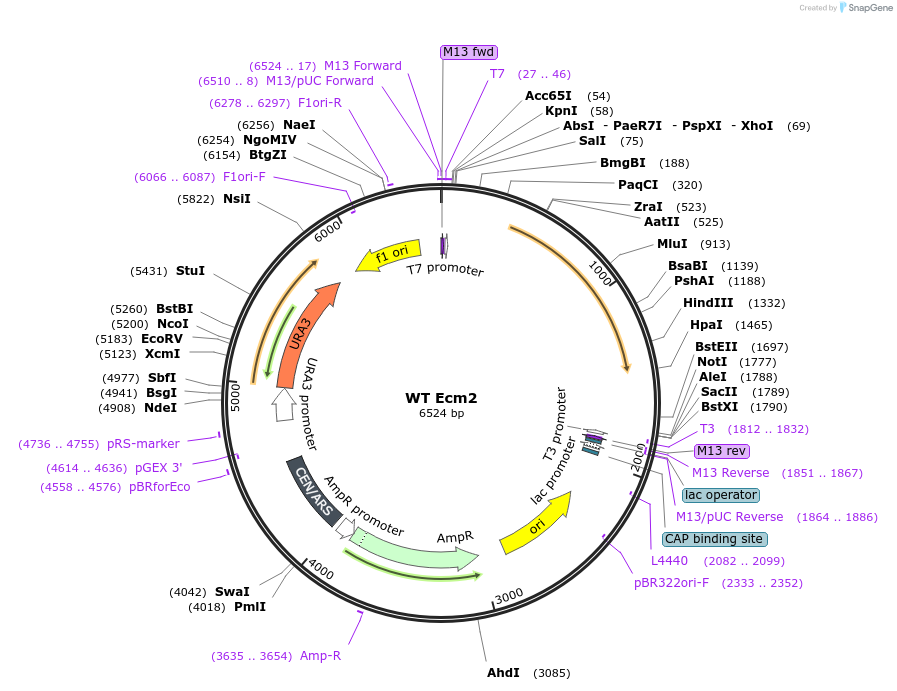
WT Ecm2
Plasmid#169922PurposeEcm2 +/-300bp of up- and downstream seq. cloned into the NotI and SalI sites of pRS416DepositorInsertEcm2
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
Ecm2 1-143
Plasmid#169923PurposeWT Ecm2 mutated to introduce stop codon after AA 143 of Ecm2DepositorInsertEcm2 1-143
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
Ecm2 1-265
Plasmid#169925PurposeWT Ecm2 mutated to introduce stop codon after AA 265 of Ecm2DepositorInsertEcm2 1-265
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
Ecm2 1-325
Plasmid#169926PurposeWT Ecm2 mutated to introduce stop codon after AA 325 of Ecm2DepositorInsertEcm2 1-325
ExpressionBacterial and YeastAvailabilityAcademic Institutions and Nonprofits only -
pICE ADE2-MET17
Plasmid#122885PurposeADE2-MET17 ICE plasmid used to facilitate yeast recombination cloning and stable genomic Integration after CEN Excision.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
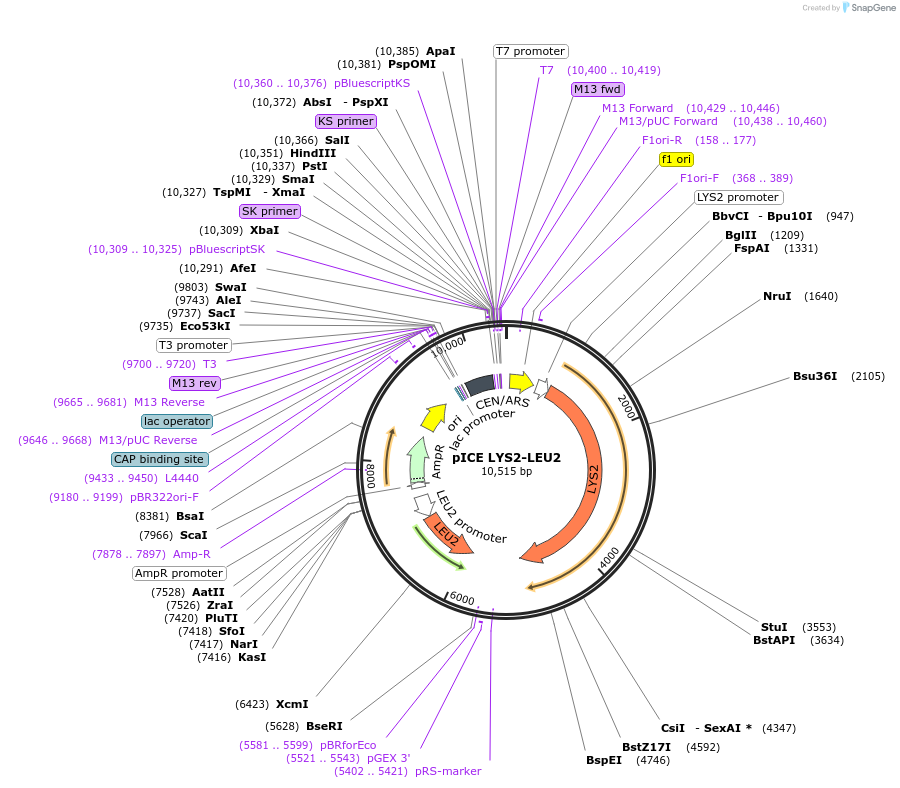
pICE LYS2-LEU2
Plasmid#122888PurposeLYS2-LEU2 ICE plasmid used to facilitate yeast recombination cloning and stable genomic Integration after CEN Excision.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
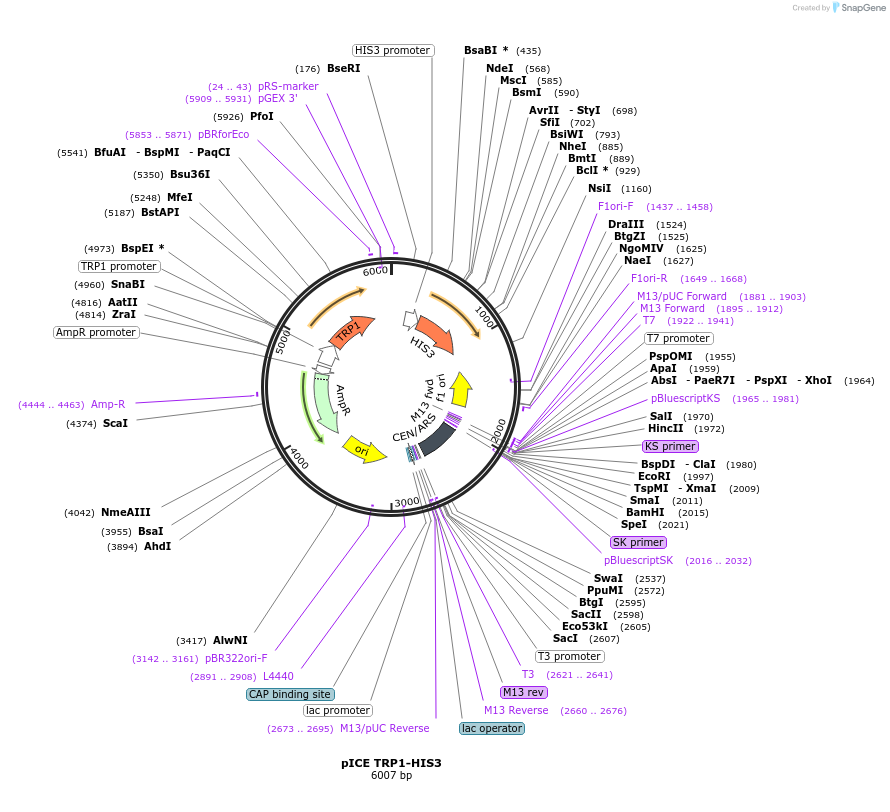
pICE TRP1-HIS3
Plasmid#122891PurposeTRP1-HIS3 ICE plasmid used to facilitate yeast recombination cloning and stable genomic Integration after CEN Excision.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
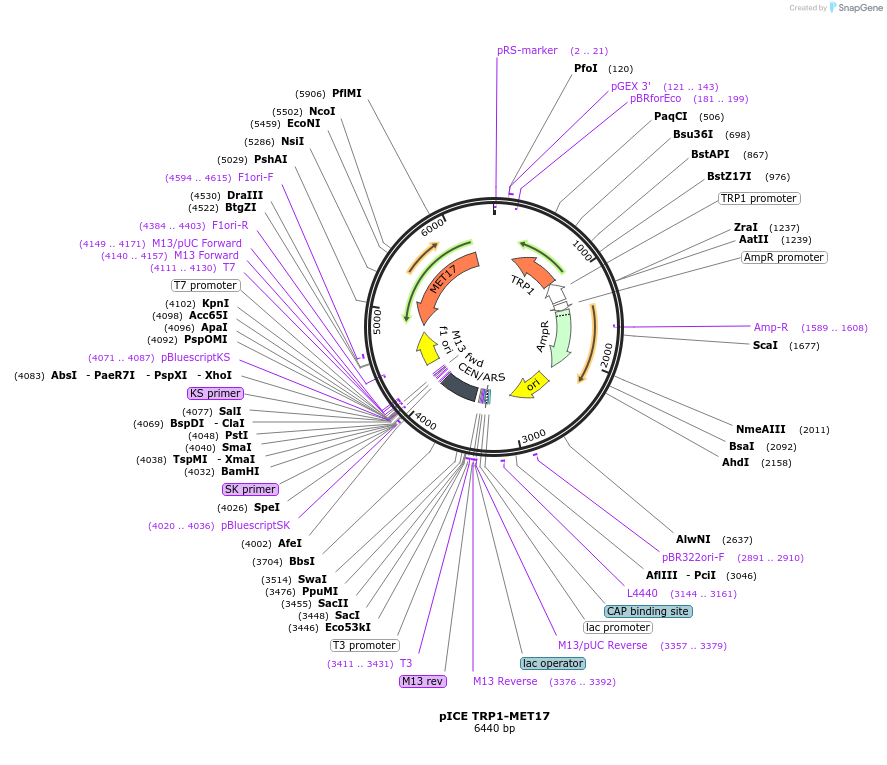
pICE TRP1-MET17
Plasmid#122893PurposeTRP1-MET17 ICE plasmid used to facilitate yeast recombination cloning and stable genomic Integration after CEN Excision.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
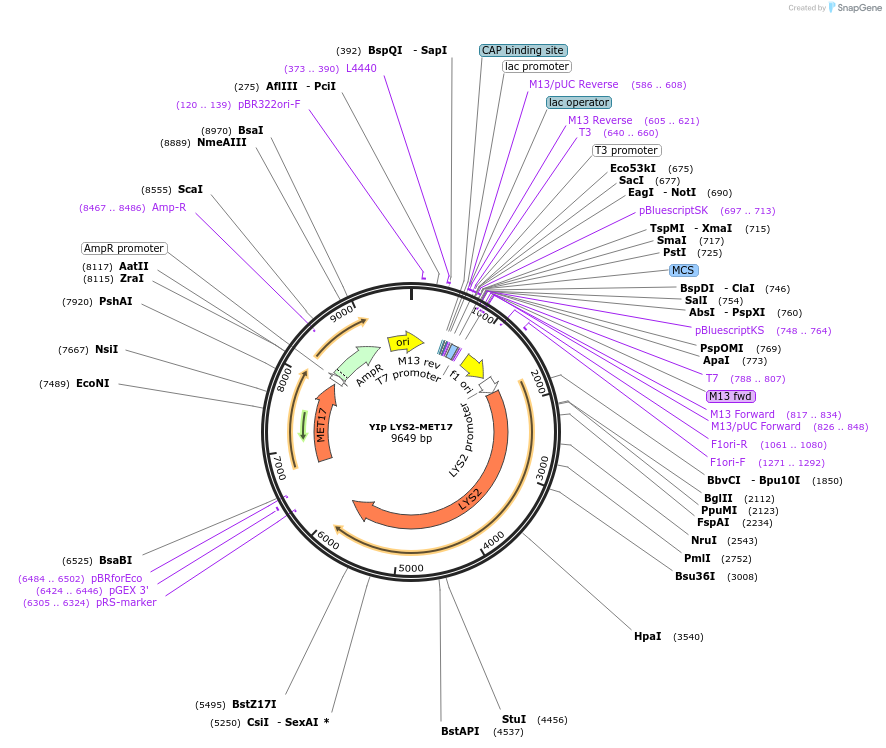
YIp LYS2-MET17
Plasmid#122910PurposeLYS2-MET17 integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
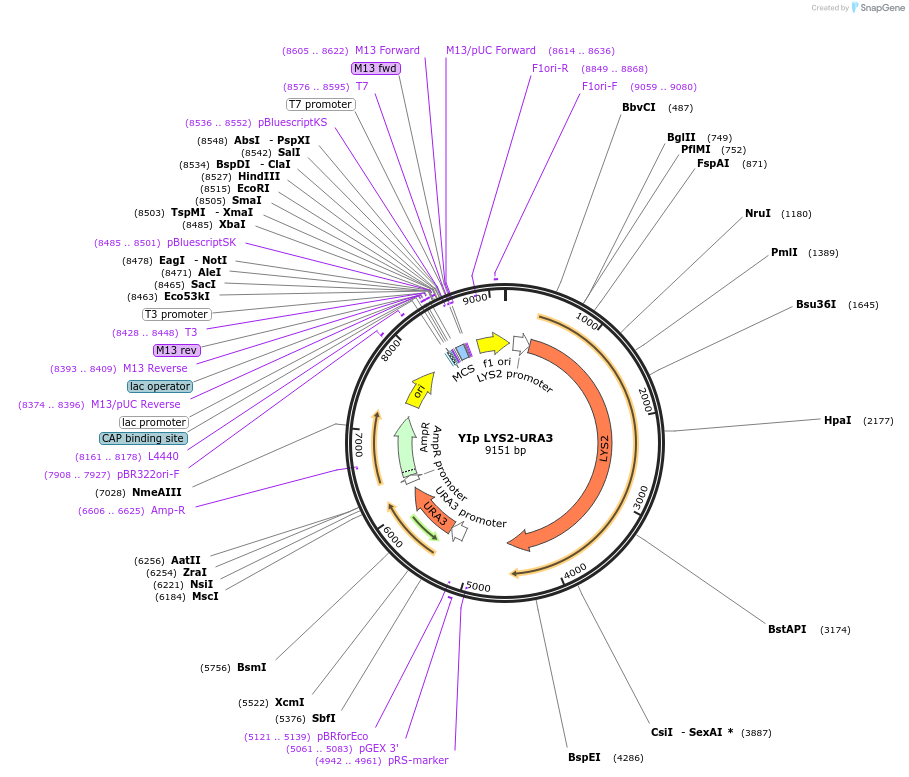
YIp LYS2-URA3
Plasmid#122911PurposeLYS2-URA3 integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
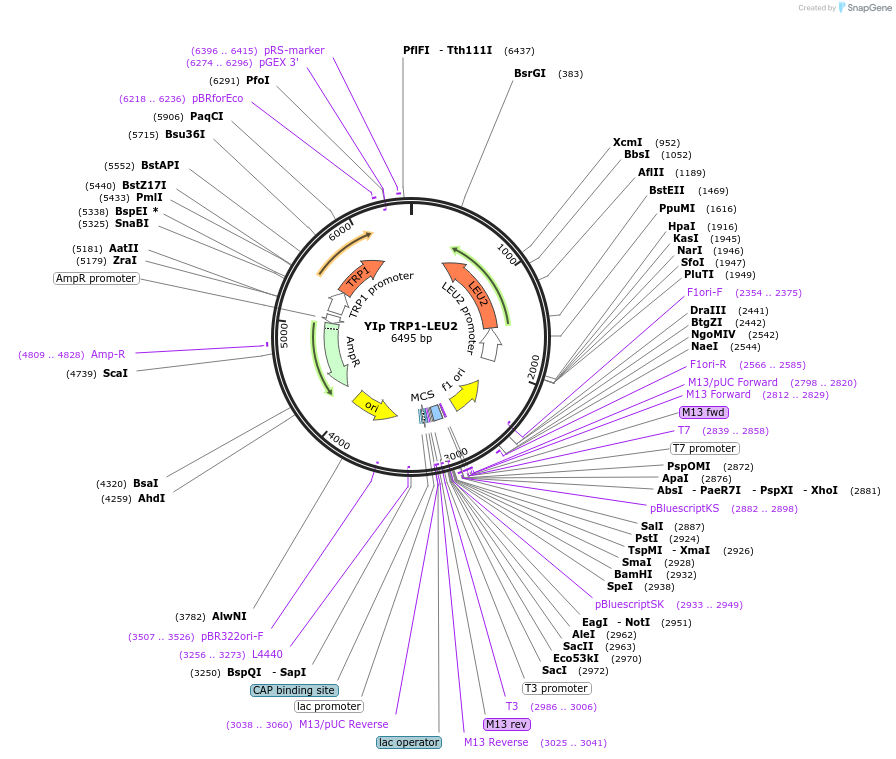
YIp TRP1-LEU2
Plasmid#122913PurposeTRP1-LEU2 integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
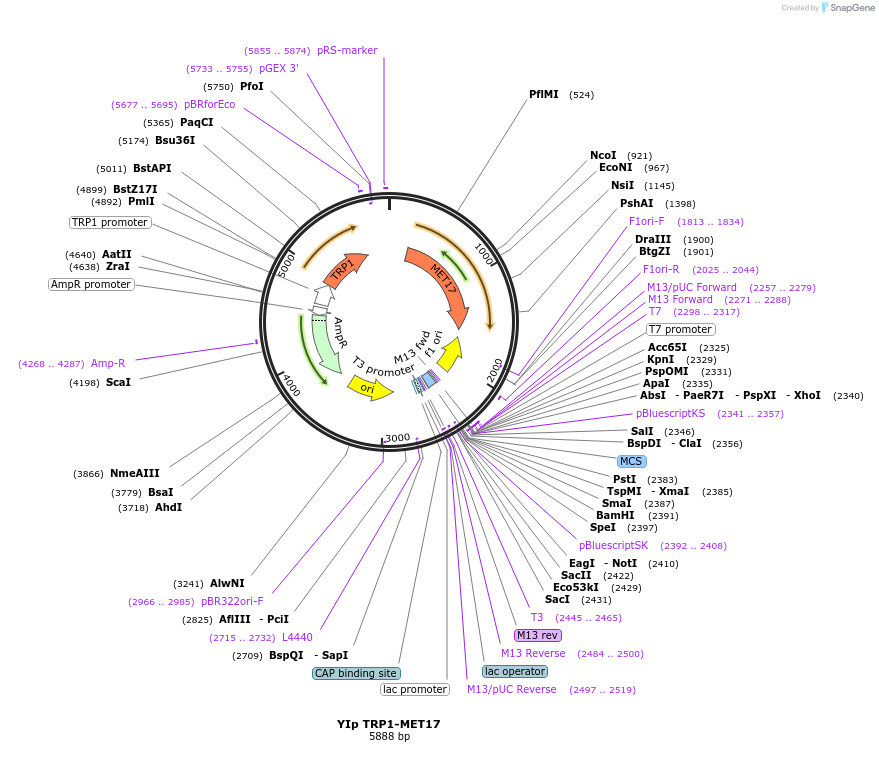
YIp TRP1-MET17
Plasmid#122914PurposeTRP1-MET17 integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
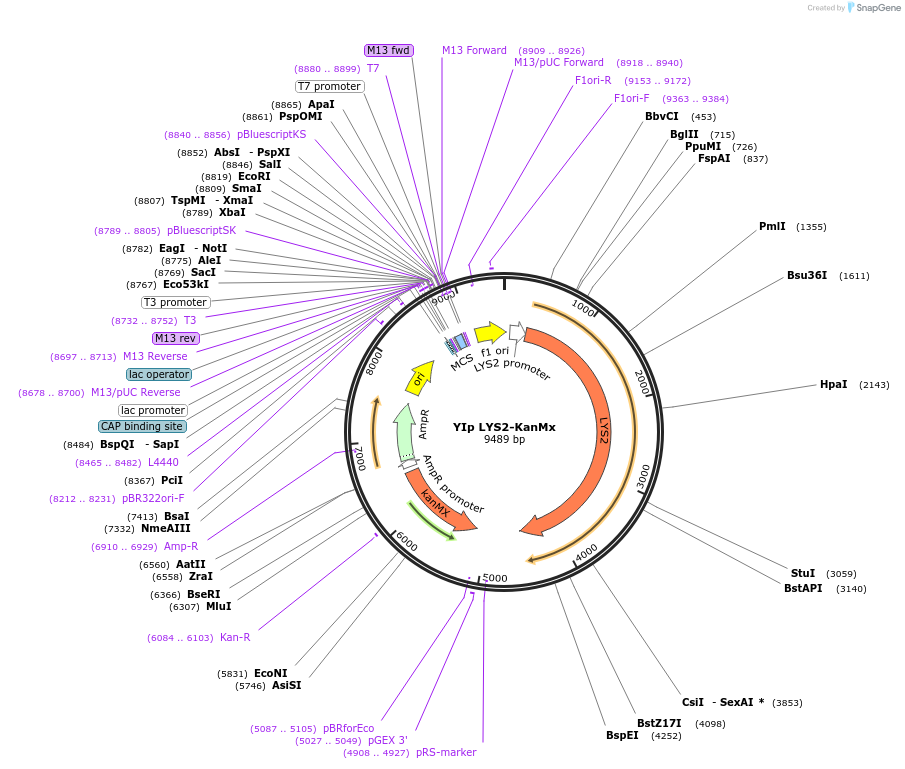
YIp LYS2-KanMx
Plasmid#122920PurposeLYS2-KanMx integrating (YIp) vector for stable genomic integration in the BY4741 background.DepositorTypeEmpty backboneUseYeast recombination cloningExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
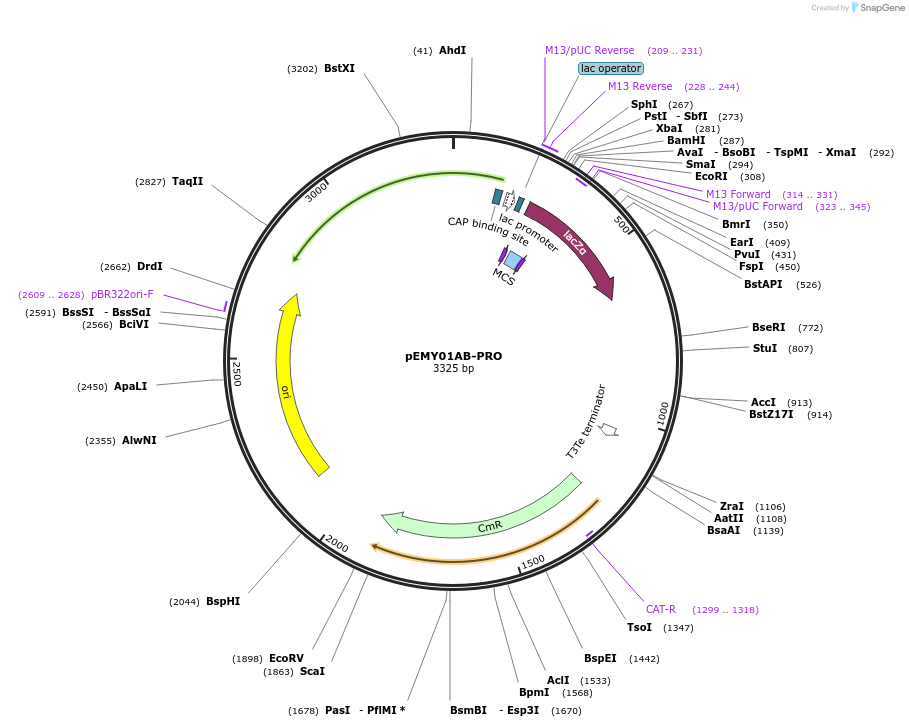
pEMY01AB-PRO
Plasmid#127714PurposeIntegrative yeast promoter characterization plasmid. Clone in with BpiI. Has ChrXV homology upstream and an mVenus fragment downstream.DepositorTypeEmpty backboneUseSynthetic BiologyExpressionYeastAvailabilityAcademic Institutions and Nonprofits only -
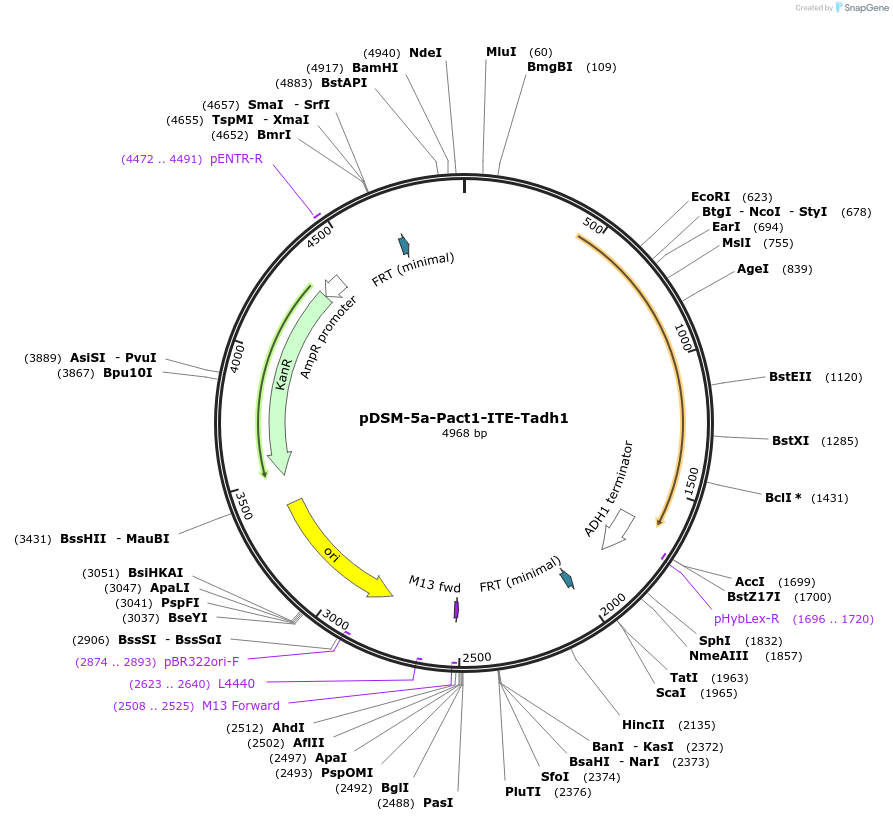
pDSM-5a-Pact1-ITE-Tadh1
Plasmid#127723PurposeYeast pathway position 1. ITE transcription unit with the ACT1 promoter and ADH1 terminator.DepositorInsertitaconate transporter from A. terreus
UseSynthetic BiologyExpressionYeastPromoterPact1AvailabilityAcademic Institutions and Nonprofits only -
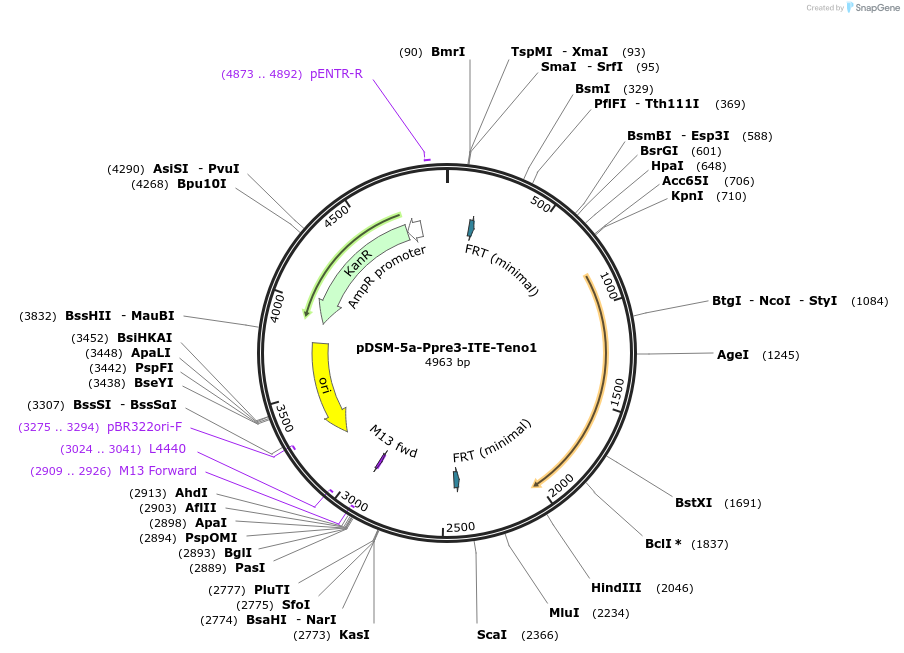
pDSM-5a-Ppre3-ITE-Teno1
Plasmid#127724PurposeYeast pathway position 1. ITE transcription unit with the PRE3 promoter and ENO1 terminator.DepositorInsertitaconate transporter from A. terreus
UseSynthetic BiologyExpressionYeastPromoterPpre3AvailabilityAcademic Institutions and Nonprofits only -
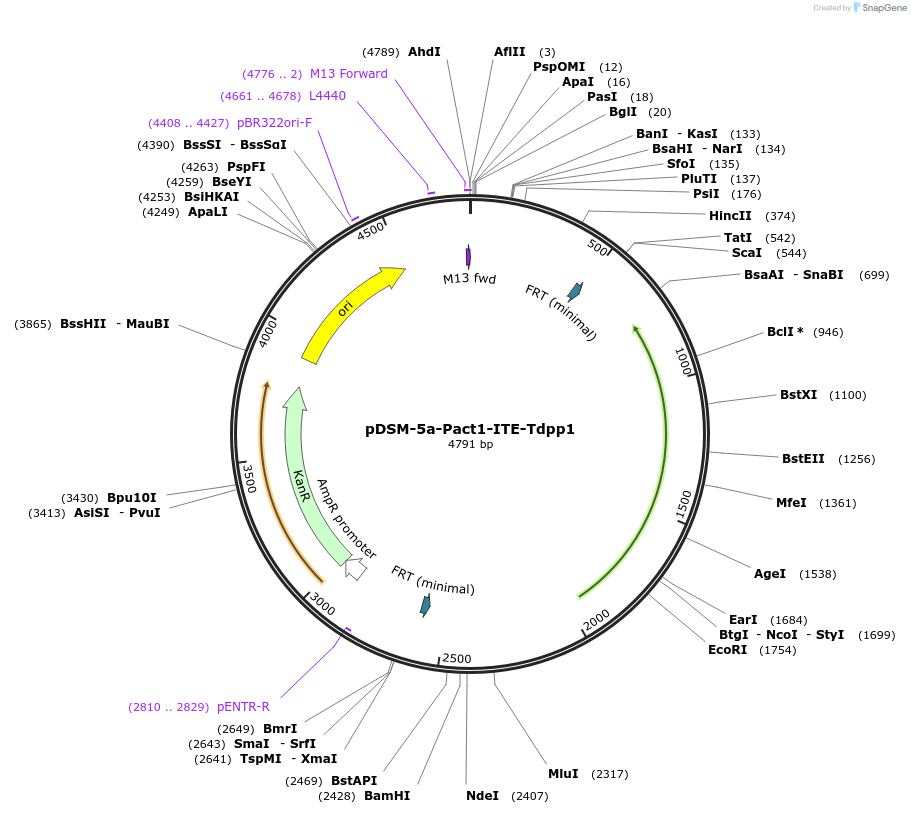
pDSM-5a-Pact1-ITE-Tdpp1
Plasmid#127725PurposeYeast pathway position 1. ITE transcription unit with the ACT1 promoter and DPP1 terminator.DepositorInsertitaconate transporter from A. terreus
UseSynthetic BiologyExpressionYeastPromoterPact1AvailabilityAcademic Institutions and Nonprofits only -
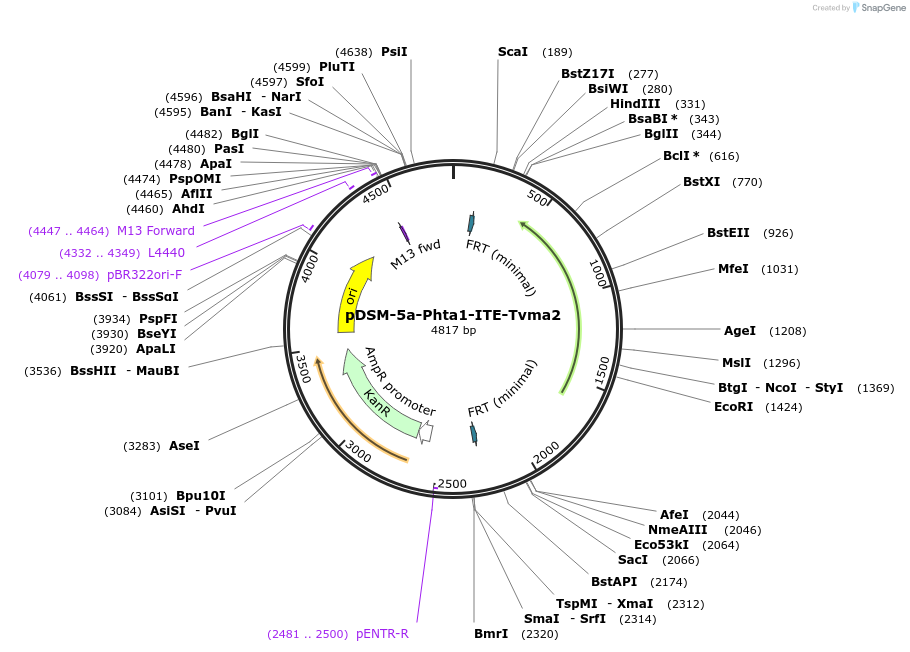
pDSM-5a-Phta1-ITE-Tvma2
Plasmid#127726PurposeYeast pathway position 1. ITE transcription unit with the HTA1 promoter and VMA2 terminator.DepositorInsertitaconate transporter from A. terreus
UseSynthetic BiologyExpressionYeastPromoterPhta1AvailabilityAcademic Institutions and Nonprofits only -
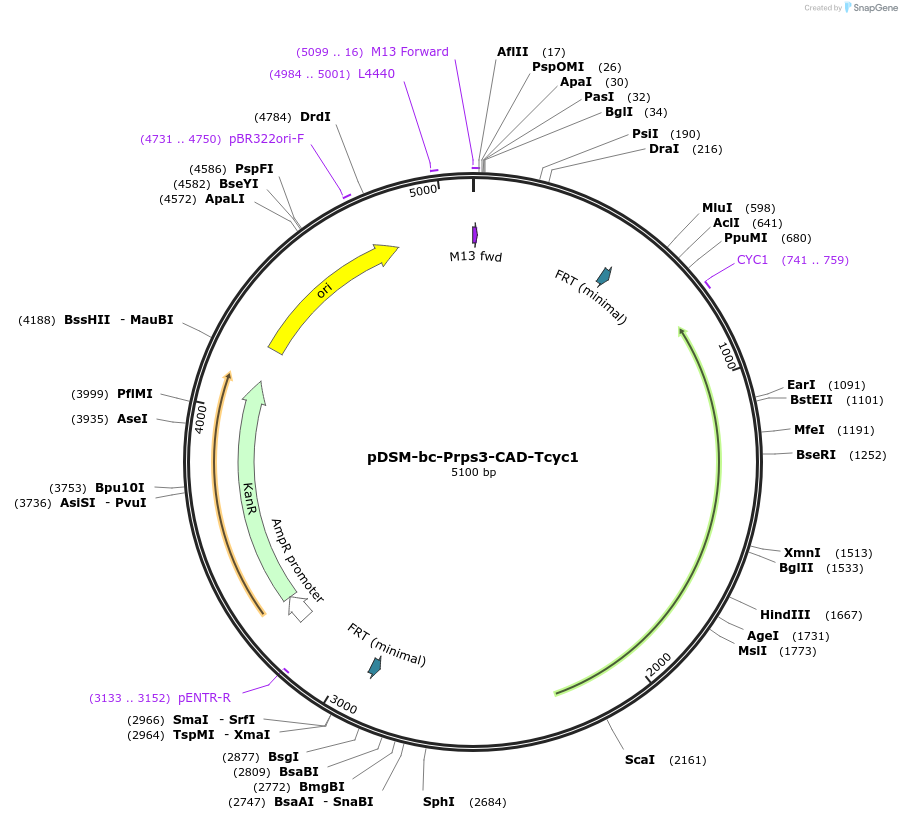
pDSM-bc-Prps3-CAD-Tcyc1
Plasmid#127728PurposeYeast pathway position 3. CAD transcription unit with the RPS3 promoter and CYC1 terminator.DepositorInsertcis-aconitate decarboxylase
UseSynthetic BiologyExpressionYeastPromoterPrps3AvailabilityAcademic Institutions and Nonprofits only -
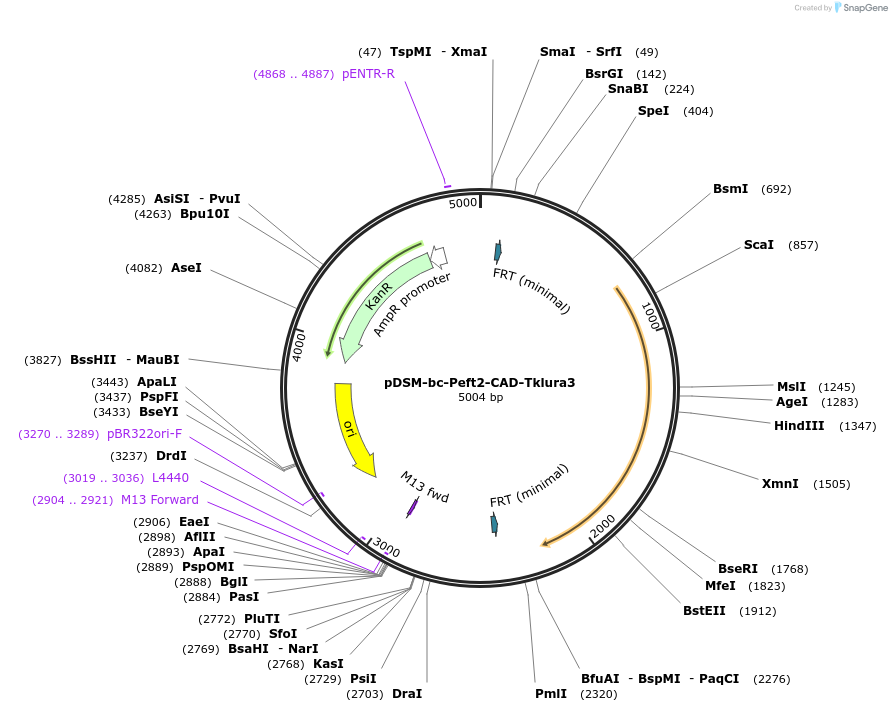
pDSM-bc-Peft2-CAD-Tklura3
Plasmid#127729PurposeYeast pathway position 3. CAD transcription unit with the EFT2 promoter and K. lactis URA3 terminator.DepositorInsertcis-aconitate decarboxylase
UseSynthetic BiologyExpressionYeastPromoterPeft2AvailabilityAcademic Institutions and Nonprofits only -
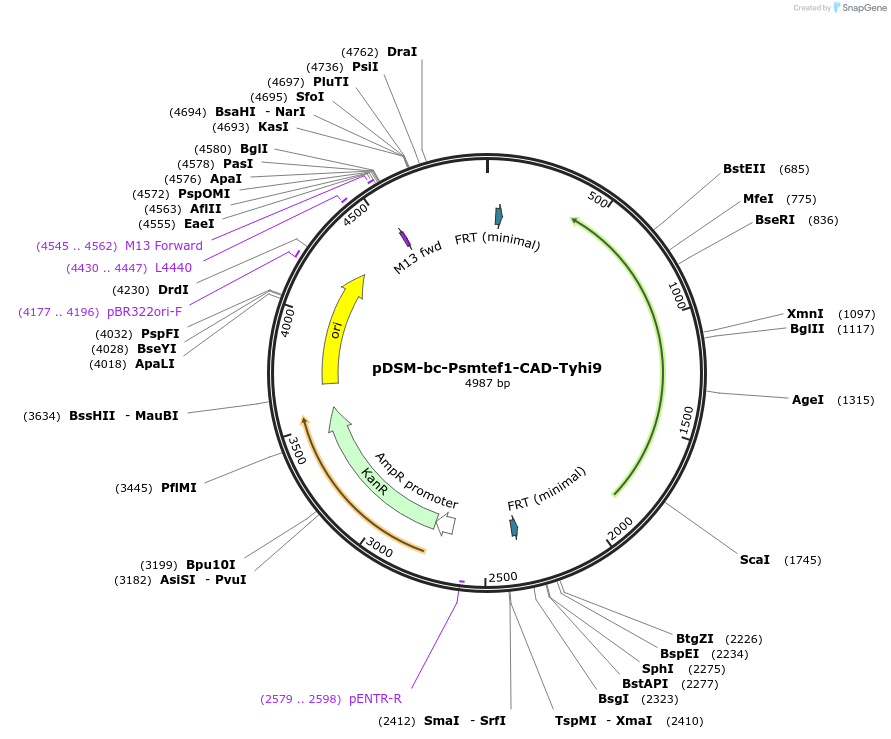
pDSM-bc-Psmtef1-CAD-Tyhi9
Plasmid#127730PurposeYeast pathway position 3. CAD transcription unit with the S. mikatae TEF1 promoter and YHI9 terminator.DepositorInsertcis-aconitate decarboxylase
UseSynthetic BiologyExpressionYeastPromoterPsmtef1AvailabilityAcademic Institutions and Nonprofits only -
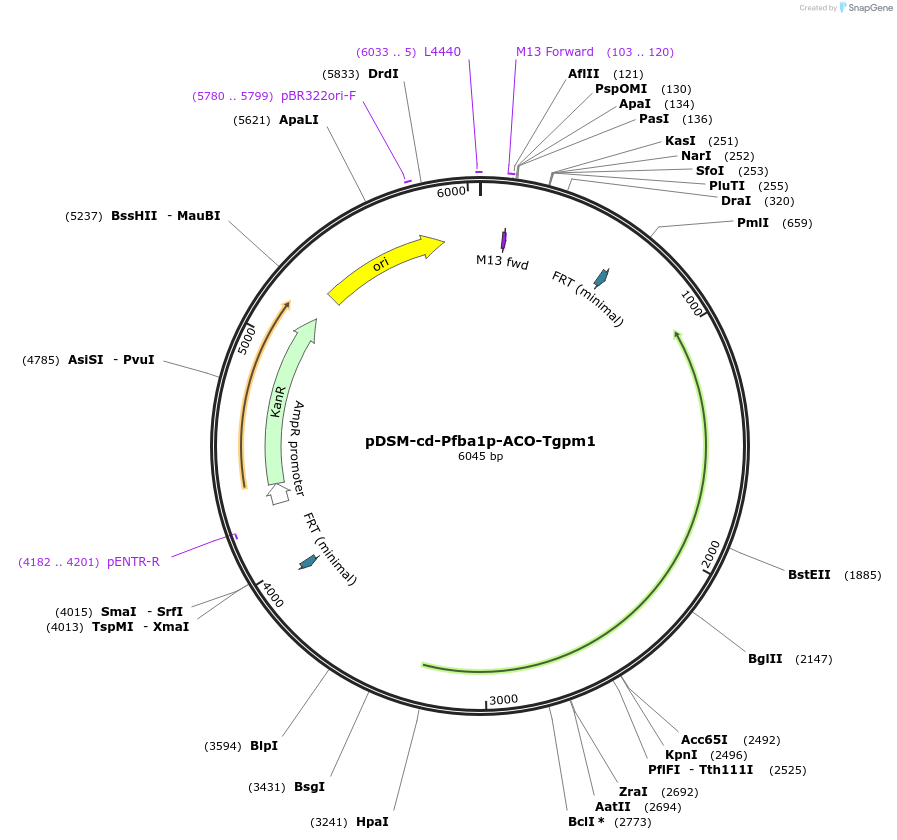
pDSM-cd-Pfba1p-ACO-Tgpm1
Plasmid#127731PurposeYeast pathway position 4. ACO transcription unit with the FBA1 promoter and GPM1 terminator.DepositorInsertaconitase
UseSynthetic BiologyExpressionYeastPromoterPfba1AvailabilityAcademic Institutions and Nonprofits only -
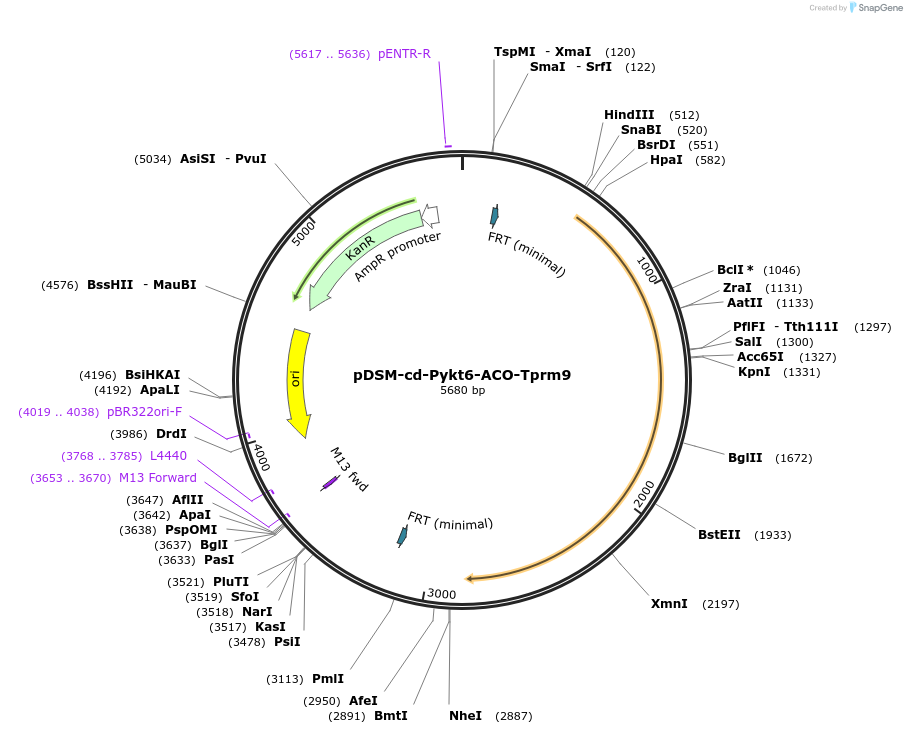
pDSM-cd-Pykt6-ACO-Tprm9
Plasmid#127732PurposeYeast pathway position 4. ACO transcription unit with the YKT6 promoter and PRM9 terminator.DepositorInsertaconitase
UseSynthetic BiologyExpressionYeastPromoterPykt6AvailabilityAcademic Institutions and Nonprofits only -
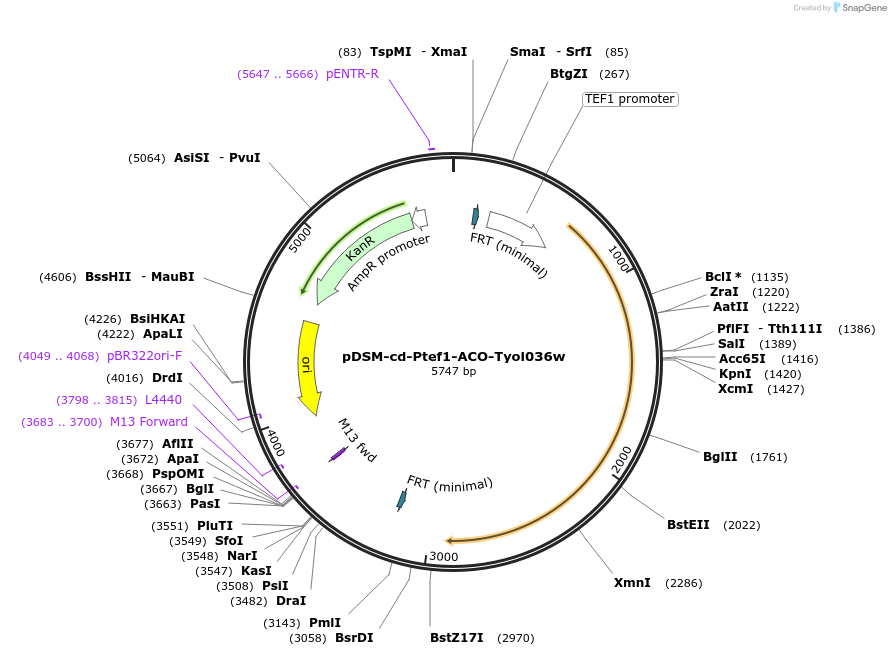
pDSM-cd-Ptef1-ACO-Tyol036w
Plasmid#127733PurposeYeast pathway position 4. ACO transcription unit with the TEF1 promoter and YOL036w terminator.DepositorInsertaconitase
UseSynthetic BiologyExpressionYeastPromoterPtef1AvailabilityAcademic Institutions and Nonprofits only -
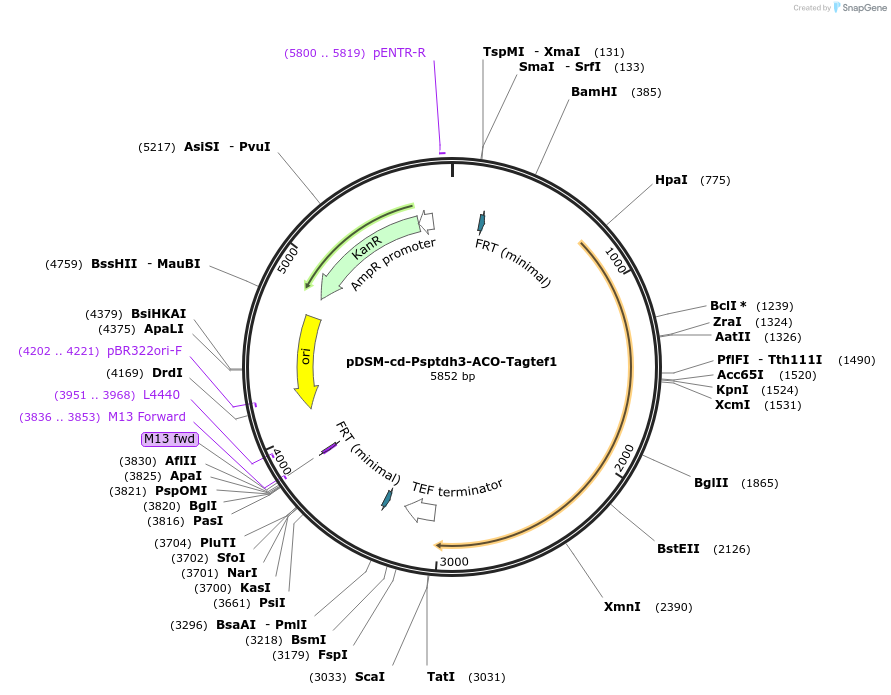
pDSM-cd-Psptdh3-ACO-Tagtef1
Plasmid#127734PurposeYeast pathway position 4. ACO transcription unit with the S. paradoxus TDH3 promoter and A. gossypii TEF1 terminator.DepositorInsertaconitase
UseSynthetic BiologyExpressionYeastPromoterPsptdh3AvailabilityAcademic Institutions and Nonprofits only -
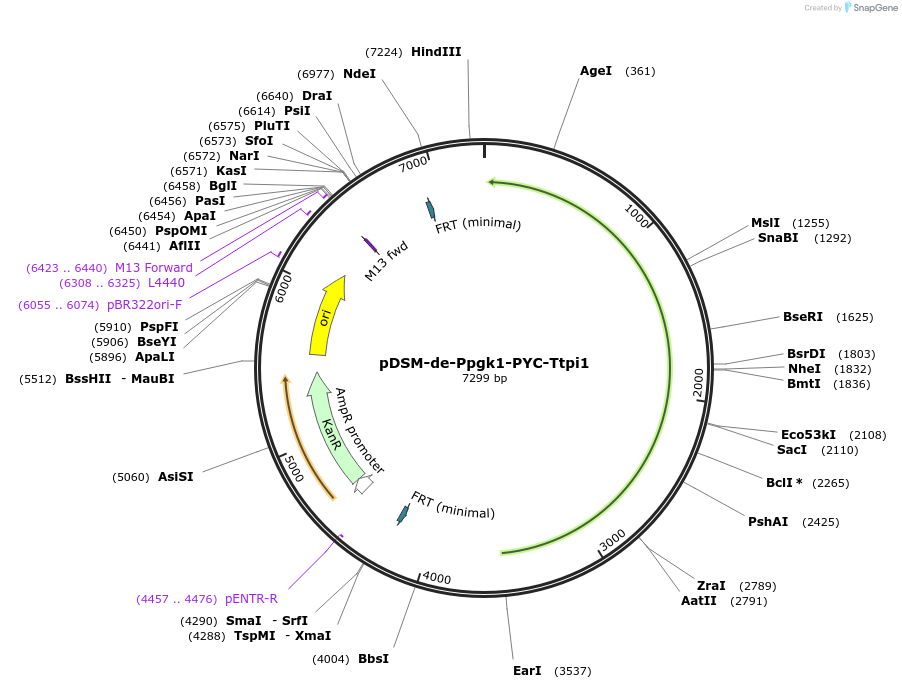
pDSM-de-Ppgk1-PYC-Ttpi1
Plasmid#127735PurposeYeast pathway position 5. PYC transcription unit with the PGK1 promoter and TPI1 terminator.DepositorInsertpyruvate carboxylase (PYC1 Budding Yeast, Synthetic)
UseSynthetic BiologyExpressionYeastPromoterPpgk1AvailabilityAcademic Institutions and Nonprofits only -
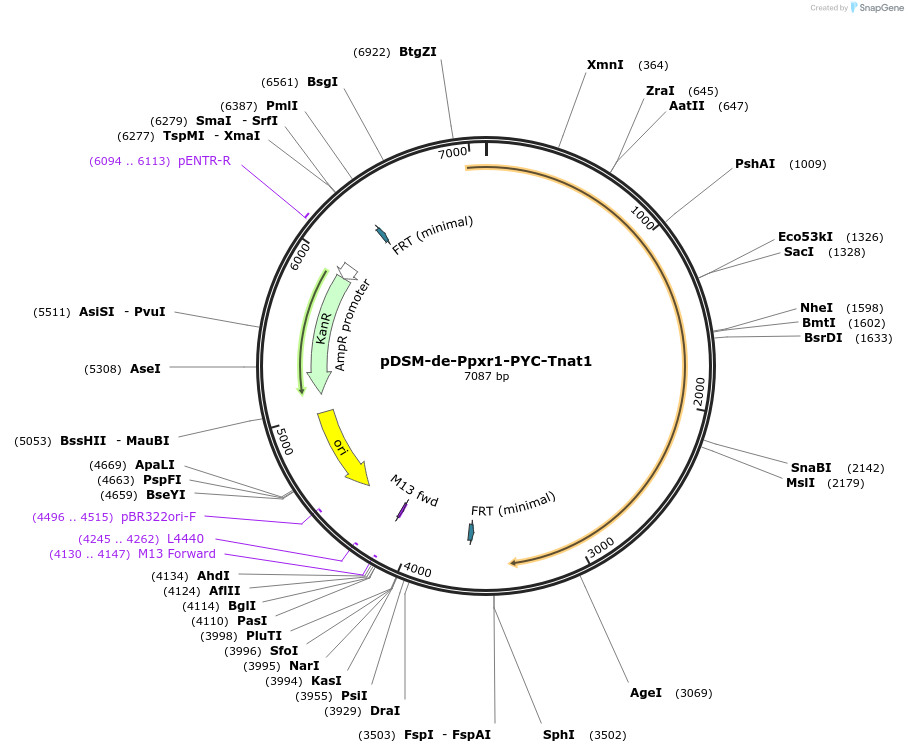
pDSM-de-Ppxr1-PYC-Tnat1
Plasmid#127736PurposeYeast pathway position 5. PYC transcription unit with the PXR1 promoter and NAT1 terminator.DepositorInsertpyruvate carboxylase (PYC1 Budding Yeast, Synthetic)
UseSynthetic BiologyExpressionYeastPromoterPpxr1AvailabilityAcademic Institutions and Nonprofits only -
pDSM-ef-Ptef1-CSC-Tpdc1
Plasmid#127739PurposeYeast pathway position 6. CSC transcription unit with the TEF1 promoter and PDC1 terminator.DepositorInsertcitrate synthase
UseSynthetic BiologyExpressionYeastPromoterPfba1AvailabilityAcademic Institutions and Nonprofits only -
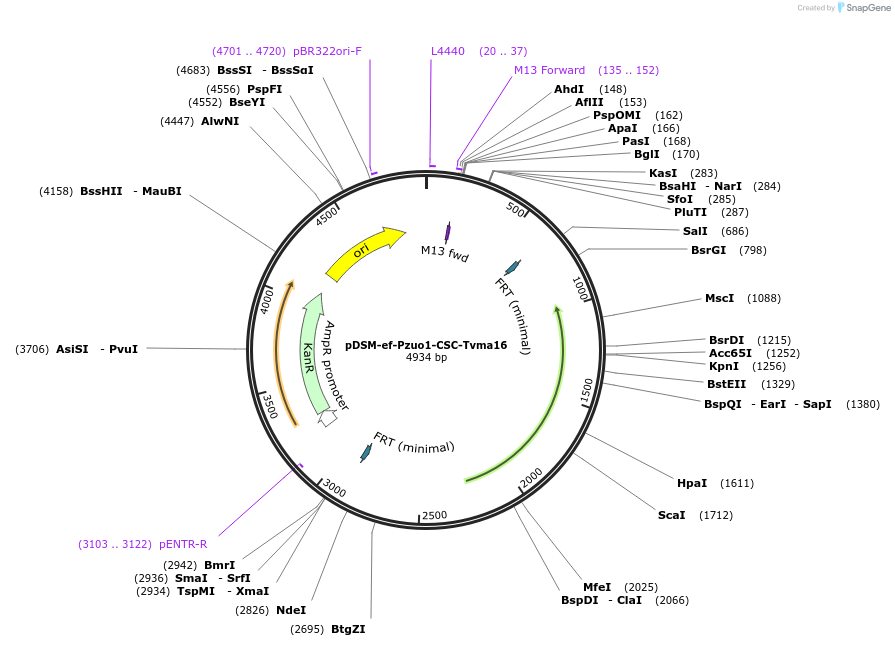
pDSM-ef-Pzuo1-CSC-Tvma16
Plasmid#127740PurposeYeast pathway position 6. CSC transcription unit with the ZUO1 promoter and VMA16 terminator.DepositorInsertcitrate synthase
UseSynthetic BiologyExpressionYeastPromoterPzuo1AvailabilityAcademic Institutions and Nonprofits only -
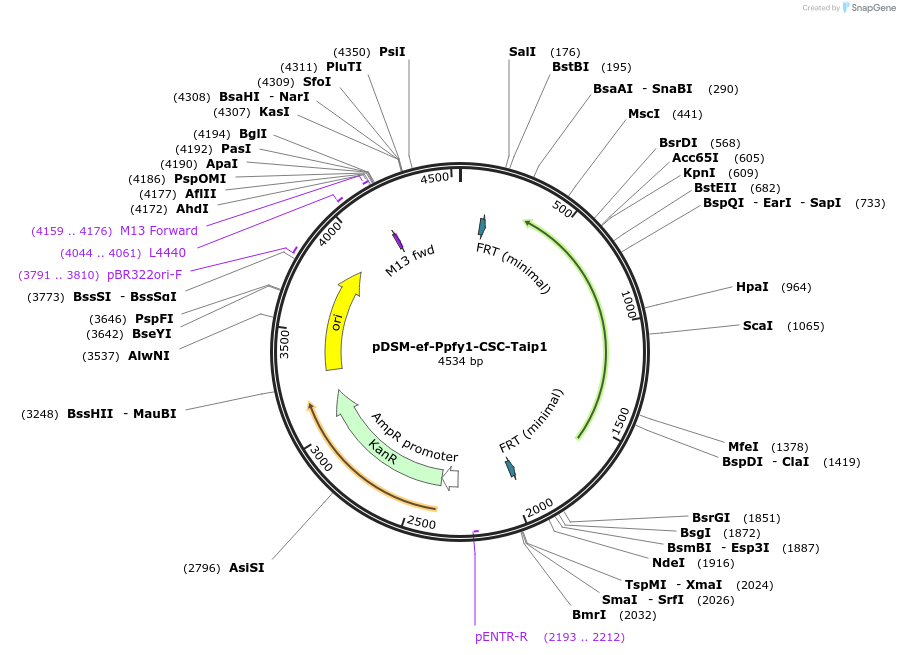
pDSM-ef-Ppfy1-CSC-Taip1
Plasmid#127741PurposeYeast pathway position 6. CSC transcription unit with the PFY1 promoter and AIP1 terminator.DepositorInsertcitrate synthase
UseSynthetic BiologyExpressionYeastPromoterPpfy1AvailabilityAcademic Institutions and Nonprofits only -
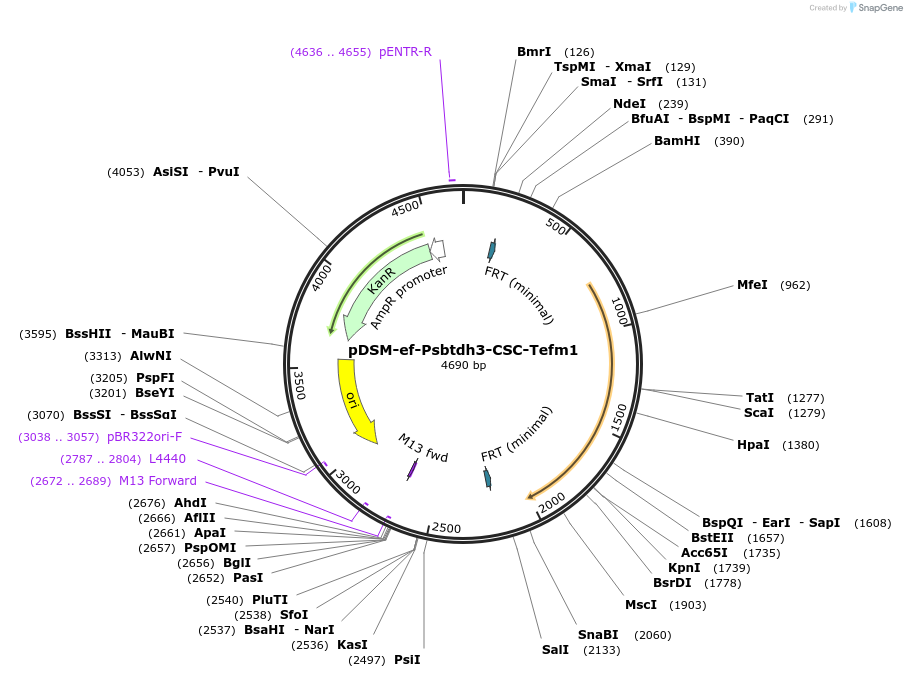
pDSM-ef-Psbtdh3-CSC-Tefm1
Plasmid#127742PurposeYeast pathway position 6. CSC transcription unit with the S. bayanus TDH3 promoter and EFM1 terminator.DepositorInsertcitrate synthase
UseSynthetic BiologyExpressionYeastPromoterPsbtdh3AvailabilityAcademic Institutions and Nonprofits only