We narrowed to 19,568 results for: RAN-1
-
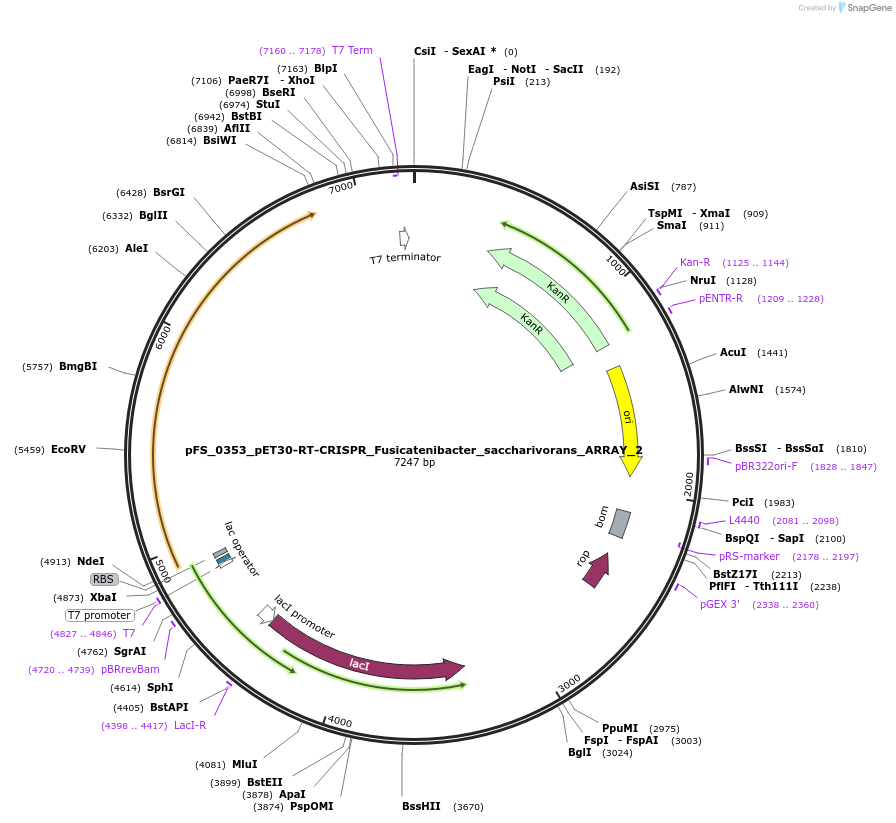
Plasmid#116965PurposeExpresses FsRT-Cas1-Cas2 under pT7lac promoter, encodes FsCRISPR array 2, compatible with SENECA acquisition readoutDepositorInsertFusicatenibacter saccharivorans RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceDec. 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
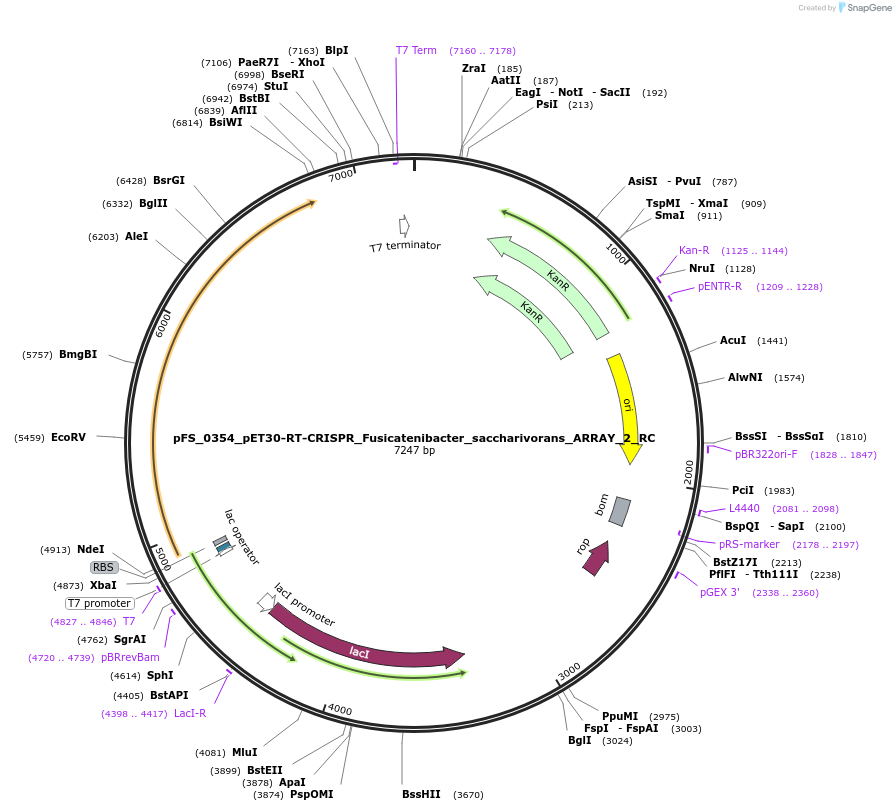
pFS_0354_pET30-RT-CRISPR_Fusicatenibacter_saccharivorans_ARRAY_2_RC
Plasmid#116966PurposeExpresses FsRT-Cas1-Cas2 under pT7lac promoter, encodes FsCRISPR array 2 RC, compatible with SENECA acquisition readoutDepositorInsertFusicatenibacter saccharivorans RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
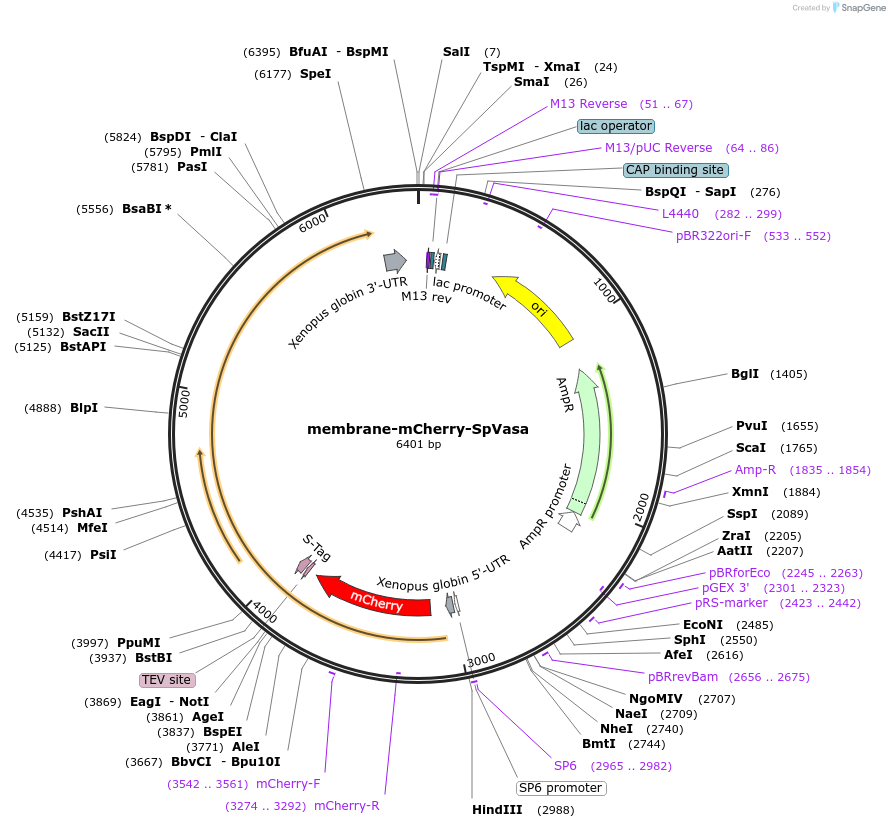
membrane-mCherry-SpVasa
Plasmid#185586Purposeto recruit Vasa to membraneDepositorInsertVasa
UseIn vitro transcription (mrna synthesis)TagsmCherry, S-Tag and membrane targeting sequence fr…Available SinceOct. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
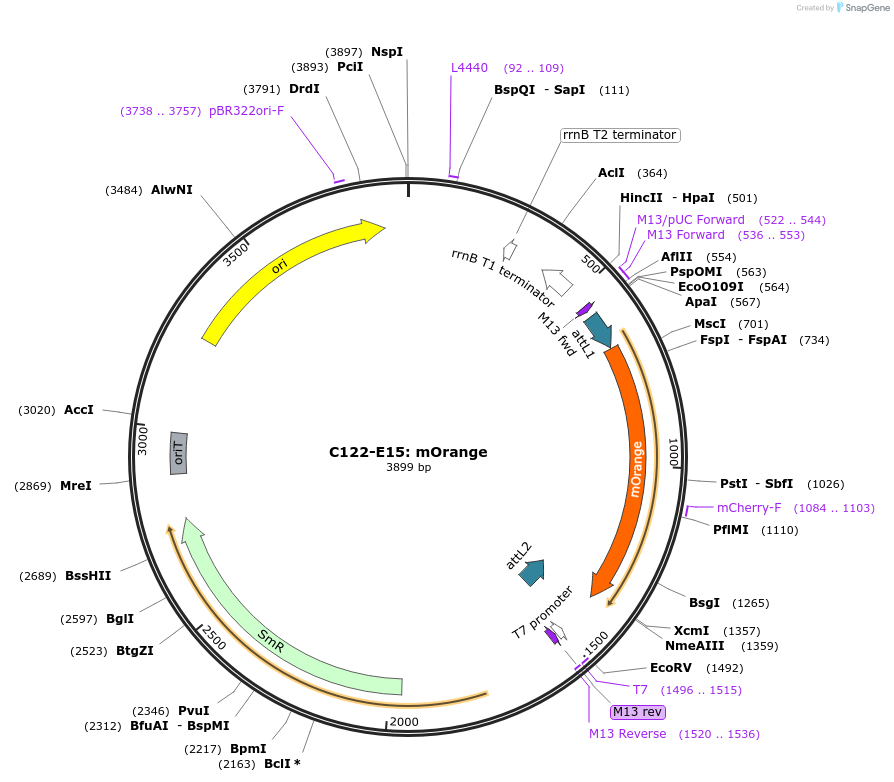
C122-E15: mOrange
Plasmid#162895PurposeStandard Gateway entry clone encoding mOrange fluorescent proteinDepositorInsertmonomeric Orange fluorescent protein
UseSynthetic BiologyAvailable SinceApril 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
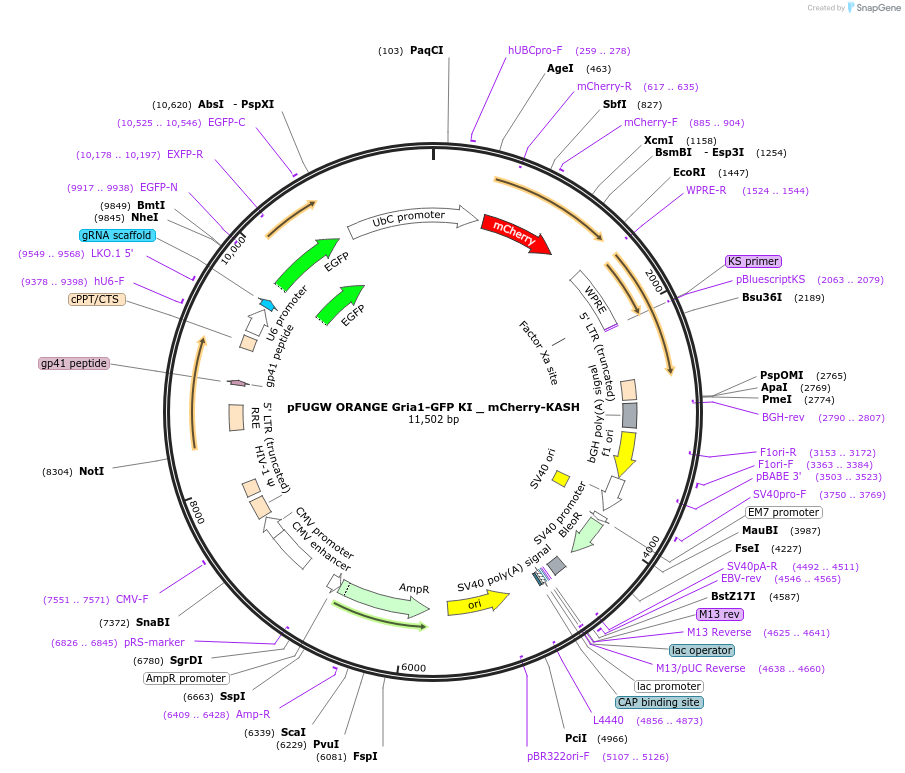
pFUGW ORANGE Gria1-GFP KI _ mCherry-KASH
Plasmid#131507PurposeEndogenous tagging of GluA1: C-terminal (amino acid position: STOP codon)DepositorUseLentiviralExpressionMammalianPromoterhUBCAvailable SinceSept. 20, 2019AvailabilityAcademic Institutions and Nonprofits only -
mOrange2-H2B-6
Plasmid#57962PurposeLocalization: Nucleus/Histones, Excitation: 549, Emission: 565DepositorInsertH2B (H2BC11 Human)
TagsmOrange2ExpressionMammalianMutationD26G and V119I in H2BPromoterCMVAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
mOrange-H2B-6
Plasmid#57942PurposeLocalization: Nucleus/Histones, Excitation: 548, Emission: 562DepositorInsertH2B (H2BC11 Human)
TagsmOrangeExpressionMammalianMutationD26G and V119I in H2BPromoterCMVAvailable SinceOct. 22, 2014AvailabilityAcademic Institutions and Nonprofits only -
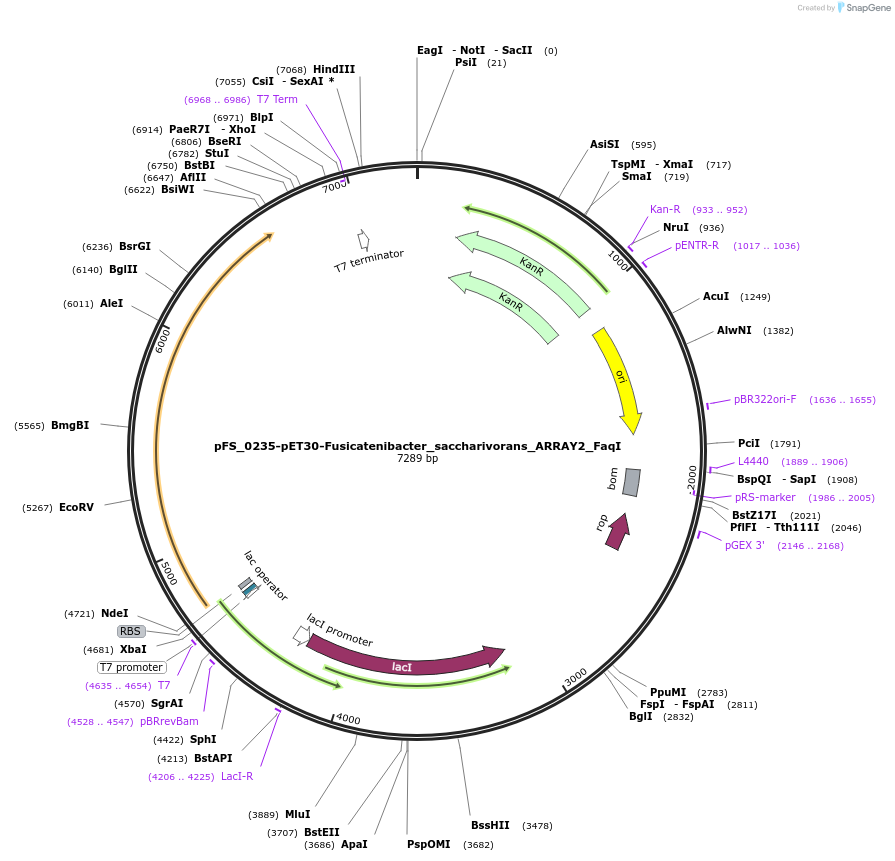
pFS_0235-pET30-Fusicatenibacter_saccharivorans_ARRAY2_FaqI
Plasmid#116954PurposeExpresses FsRT-Cas1-Cas2 under pT7lac promoter, encodes FsCRISPR array 2, compatible with SENECA acquisition readoutDepositorInsertFusicatenibacter saccharivorans RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
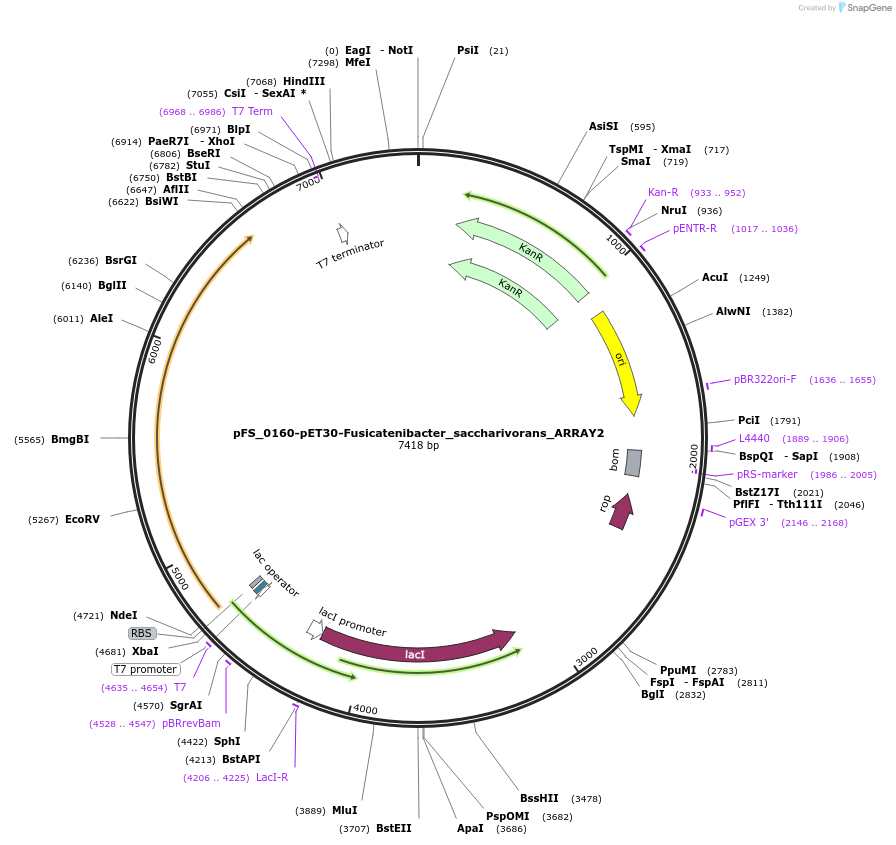
pFS_0160-pET30-Fusicatenibacter_saccharivorans_ARRAY2
Plasmid#116953PurposeExpresses FsRT-Cas1-Cas2 under pT7lac promoter, encodes FsCRISPR array 2, compatible with classic acquisition readoutDepositorInsertFusicatenibacter saccharivorans RT-Cas1-Cas2
ExpressionBacterialPromoterT7Available SinceNov. 2, 2018AvailabilityAcademic Institutions and Nonprofits only -
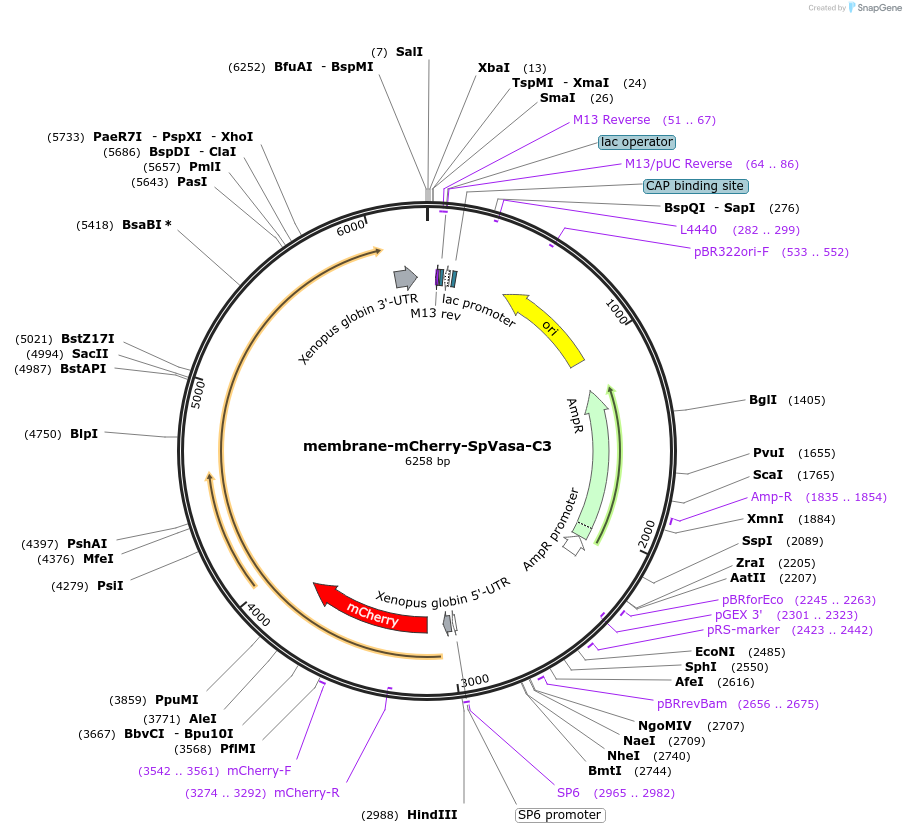
membrane-mCherry-SpVasa-C3
Plasmid#185587Purposeto recruit Vasa-C3 to membraneDepositorInsertVasa-C3
UseIn vitro transcription (mrna synthesis)TagsmCherry and membrane targeting sequence from Neur…MutationC3 mutantAvailable SinceOct. 17, 2022AvailabilityAcademic Institutions and Nonprofits only -
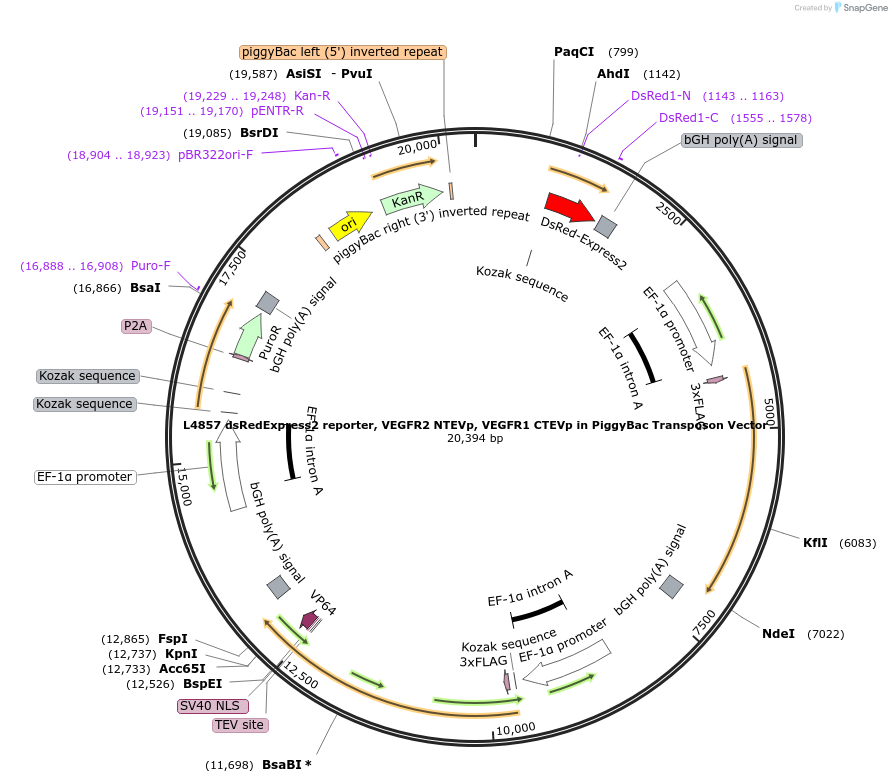
L4857 dsRedExpress2 reporter, VEGFR2 NTEVp, VEGFR1 CTEVp in PiggyBac Transposon Vector
Plasmid#244187PurposePiggyBac transposon vector for expression of dsRed-Express2 synTF promoter; constitutive expression of VEGFR2 NTEVp chain, VEGFR1 CTEVp chain, and mNeonGreen-P2A-PuroR selection markerDepositorInsertdsRed-Express2 under synTF responsive promoter; VEGFR2 NTEVp chain with WT NTEVp; VEGFR1 CTEVp chain; mNeonGreen-P2A-PuroR
UseSynthetic BiologyTags3xFLAGExpressionMammalianMutationCTEVp_190KPromoterhEF1aAvailable SinceOct. 16, 2025AvailabilityAcademic Institutions and Nonprofits only -
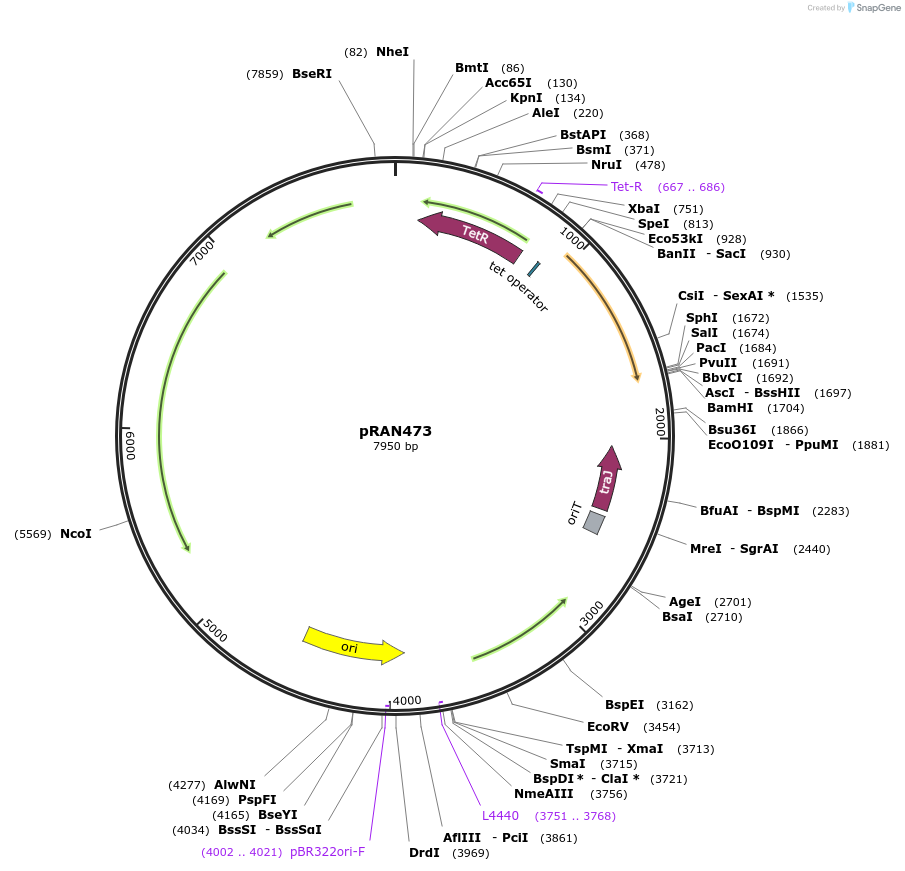
pRAN473
Plasmid#120813PurposeE. coli-C. difficile shuttle vector: tetracycline-inducible promoter (Ptet) driving expression of red fluorescent protein (mCherryOpt), multiple cloning site for fusion to N-terminus of target proteinDepositorInsertred fluorescent protein (mCherry), codon-optimized for C. difficile, multiple-cloning site for protein fusion
ExpressionBacterialPromoterPtetAvailable SinceFeb. 12, 2019AvailabilityAcademic Institutions and Nonprofits only -
pET3CTc3A(1-135)
Plasmid#63138PurposeExpression of human Tc3(1–135) bipartite domain tagged with 6xHisDepositorInsertTc3A
Tags6xHisExpressionBacterialMutation1-135 aa bipartite domainPromoterT7Available SinceApril 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
YFP Gamma 1
Plasmid#36101DepositorAvailable SinceMay 3, 2012AvailabilityAcademic Institutions and Nonprofits only -
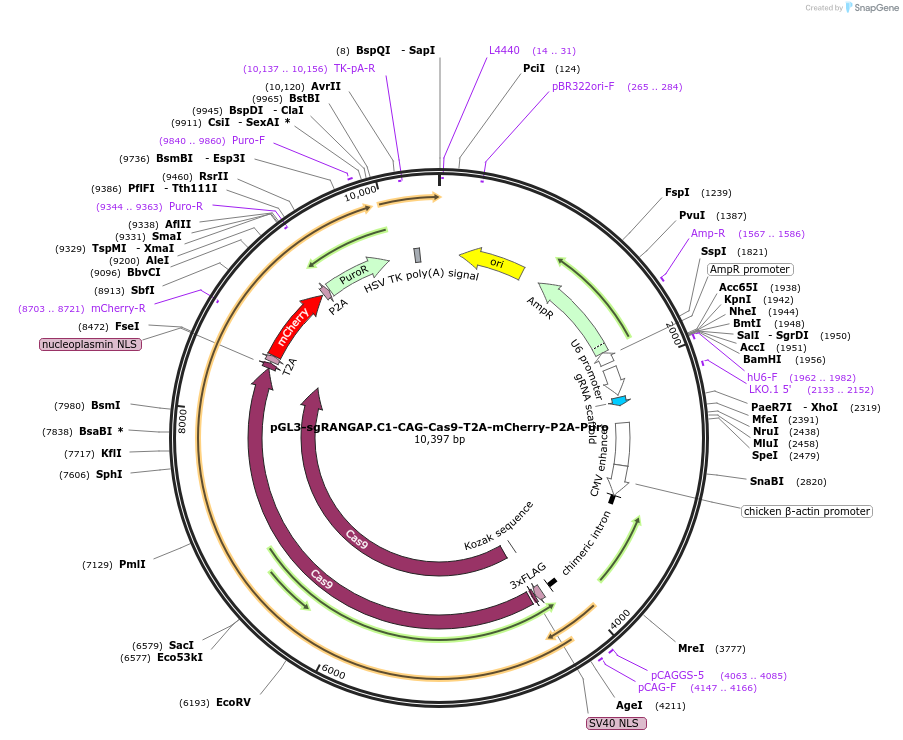
pGL3-sgRANGAP.C1-CAG-Cas9-T2A-mCherry-P2A-Puro
Plasmid#216249PurposeExpress Cas9 with CAG promoter to improve the expression in hES cells and sgRANGAP.C1 to tag endogenous RANGAP1 C-terminusDepositorInsertCas9 and sgRANGAP.C1 (RANGAP1 Human)
UseCRISPR; Endogenous taggingExpressionMammalianPromoterCAGAvailable SinceMarch 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
mcherry Beta 1
Plasmid#36111DepositorAvailable SinceMay 3, 2012AvailabilityAcademic Institutions and Nonprofits only -
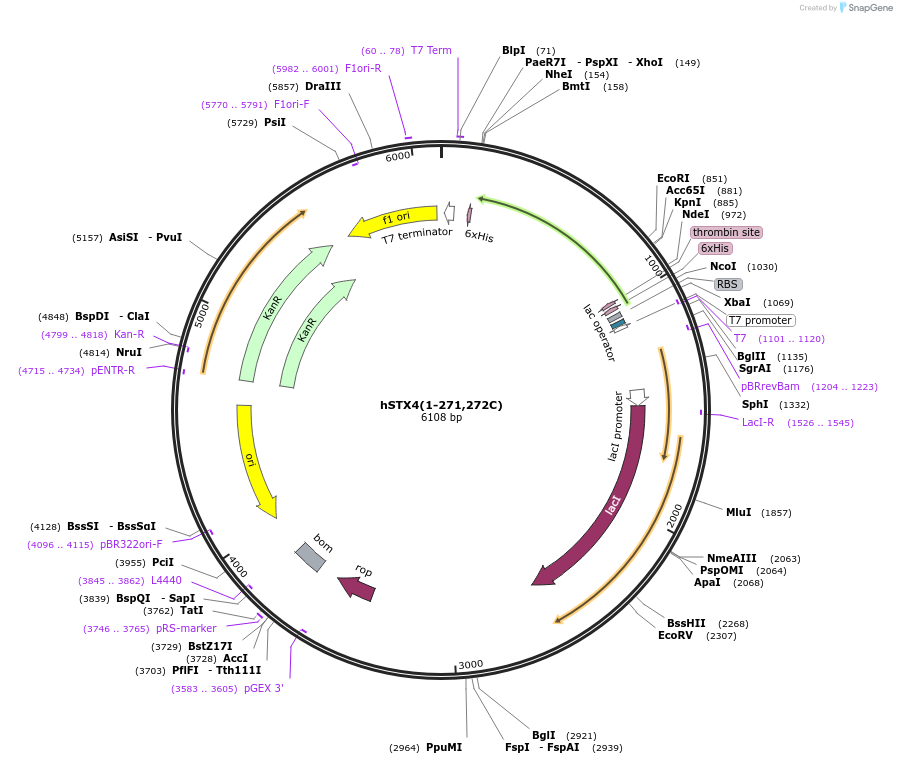
hSTX4(1-271,272C)
Plasmid#206031PurposeBacterial expression of human Syntaxin 4 without transmembrane helix and a C-terminal cysteineDepositorInsertSyntaxin-4 (STX4 Human)
Tagshis-tag (removable with thrombin)ExpressionBacterialMutationdeleted amino acids 272-297 (encoding the transme…PromoterT7Available SinceOct. 1, 2025AvailabilityAcademic Institutions and Nonprofits only -
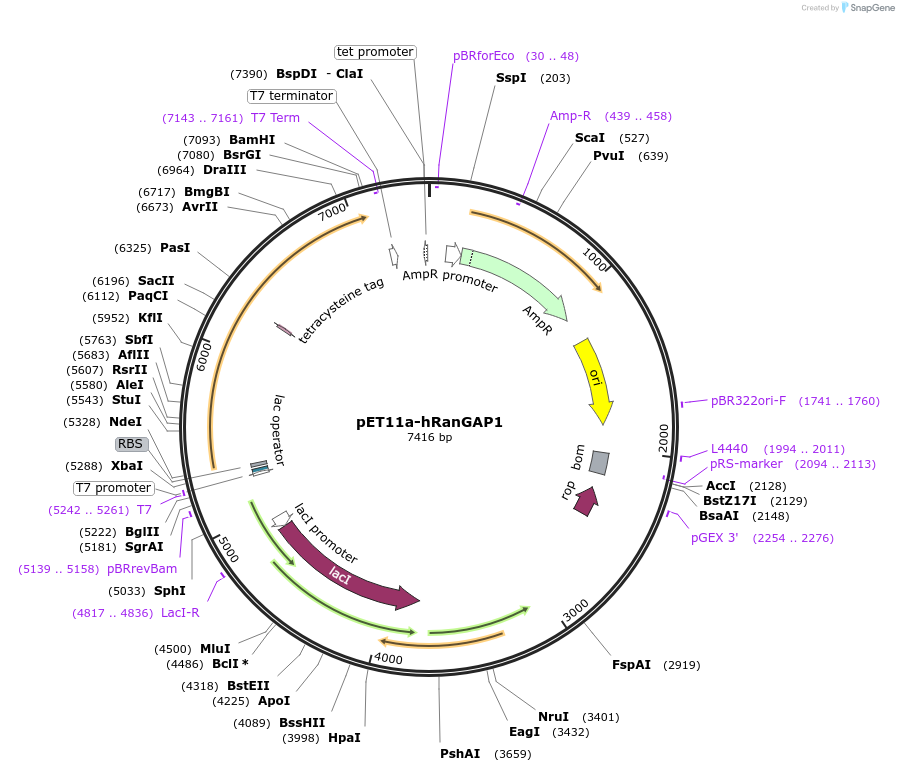
pET11a-hRanGAP1
Plasmid#50941Purposeexpression of full length untagged human RanGAP1 in E.coliDepositorAvailable SinceMay 30, 2014AvailabilityAcademic Institutions and Nonprofits only -
mOrange2-Cx43-7
Plasmid#57951PurposeLocalization: Gap Junctions, Excitation: 549, Emission: 565DepositorInsertCx43
TagsmOrange2ExpressionMammalianPromoterCMVAvailable SinceOct. 24, 2014AvailabilityAcademic Institutions and Nonprofits only -
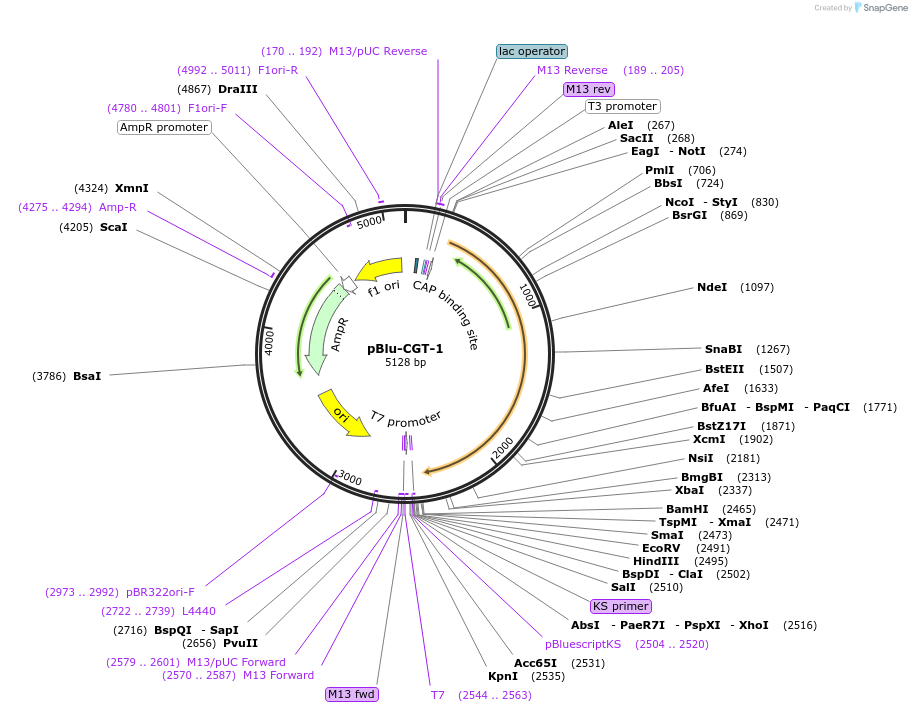
pBlu-CGT-1
Plasmid#170158PurposeExpresses bacterial Cyclomaltodextrin Glucanotransferase from Paenibacillus sp. c36 in E.coliDepositorInsertCyclomaltodextrin Glucanotransferase
ExpressionBacterialPromoterTacAvailable SinceMay 10, 2023AvailabilityAcademic Institutions and Nonprofits only