We narrowed to 10,180 results for: yeast
-
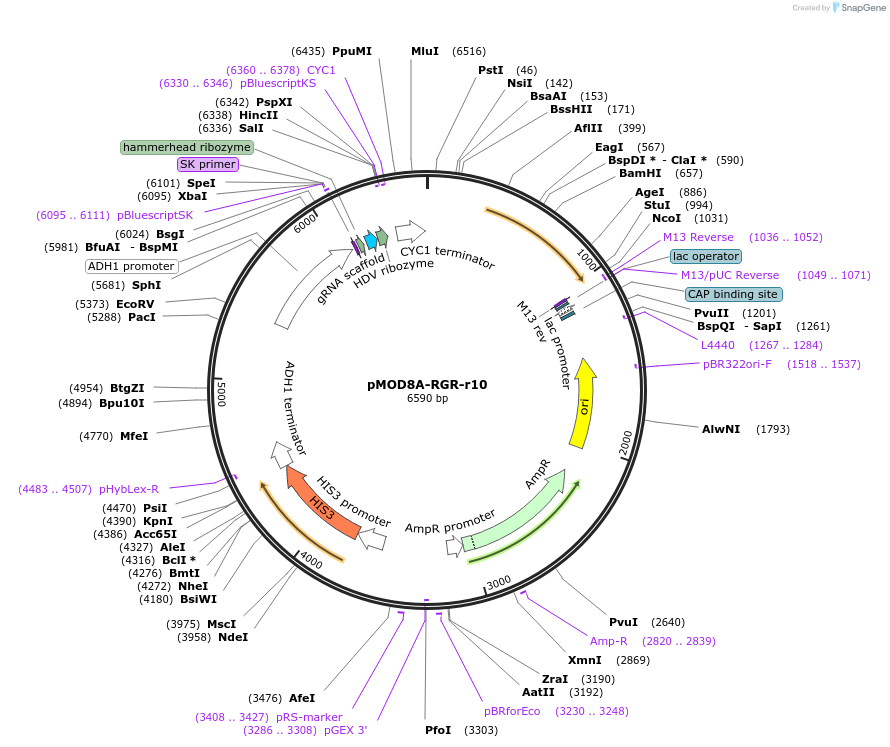
Plasmid#74370PurposeThis plasmid constitutively expresses gRNA-r10 from an AHD1 promoter using the RGR design. Integrates in the W303 HIS locus and restores HIS function.DepositorArticleInsertRGR-r10
UseCRISPR and Synthetic BiologyExpressionYeastPromoterADH1Available SinceApril 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
PP324
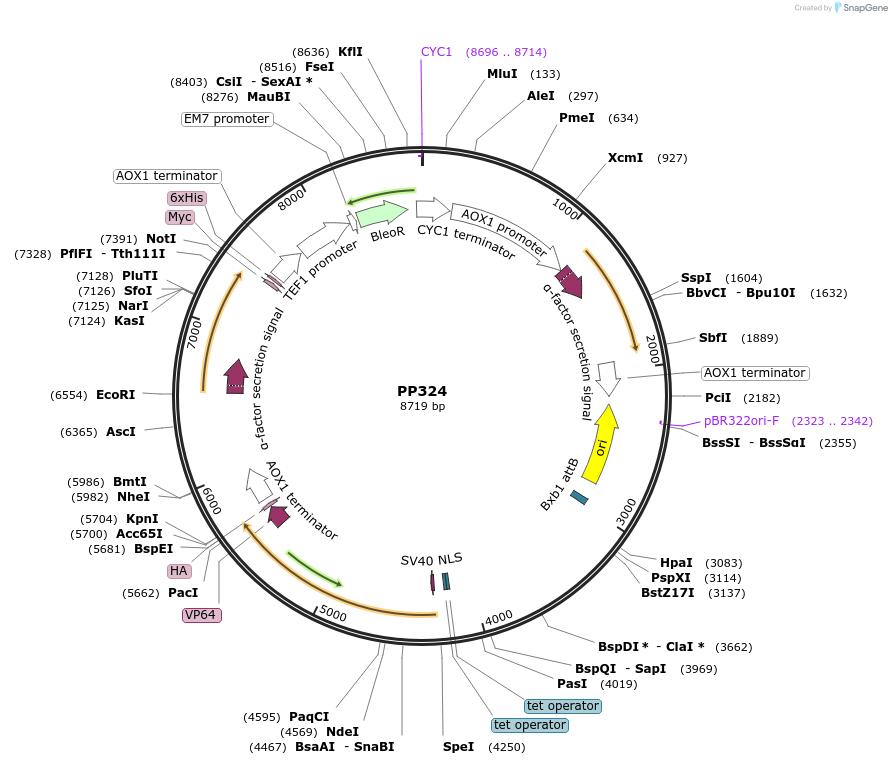
Plasmid#78982Purpose246B. Promoter: GAP6, minimal Promoter: AOX1. Figure 5. Expresses rHGH and IFNa2b.DepositorInsertsrHGH
IFNa2b
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
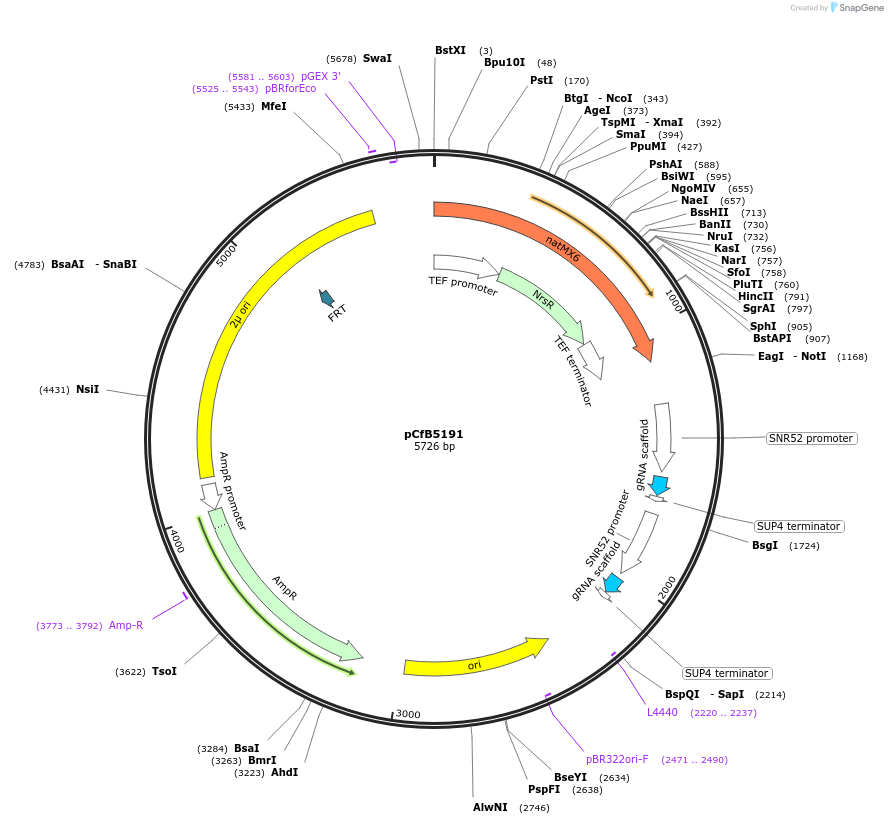
pCfB5191
Plasmid#126911PurposePlasmid containing gRNA expression cassettes targeting the X-4 and XII-5 integration sites in Saccharomyces cerevisiaeDepositorInsertX-4 and XII-5 gRNA
UseCRISPRAvailable SinceDec. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
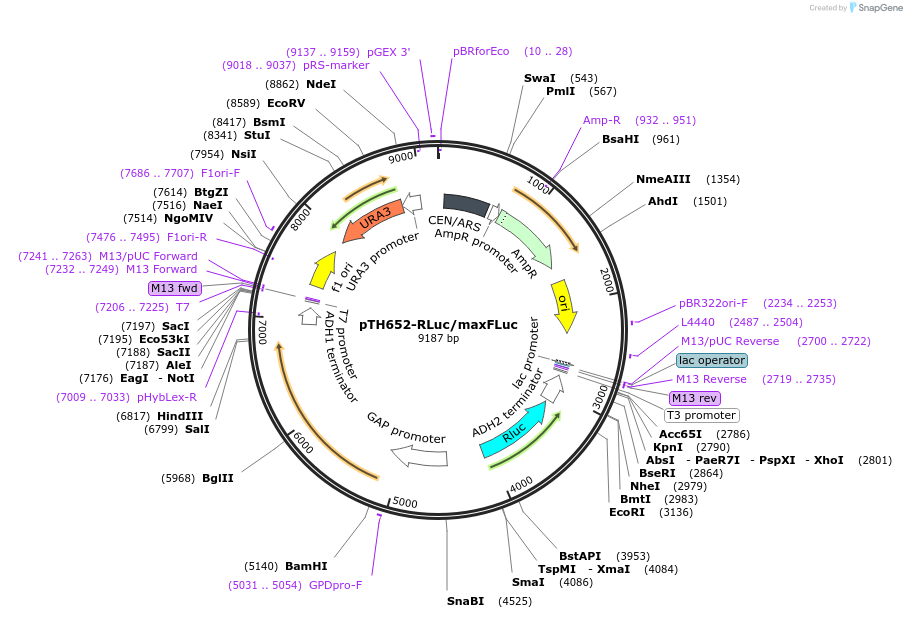
pTH652-RLuc/maxFLuc
Plasmid#29698DepositorInsertFirefly Luciferase
ExpressionYeastMutationAll codons of the original FLuc gene were exchang…Available SinceSept. 8, 2011AvailabilityAcademic Institutions and Nonprofits only -
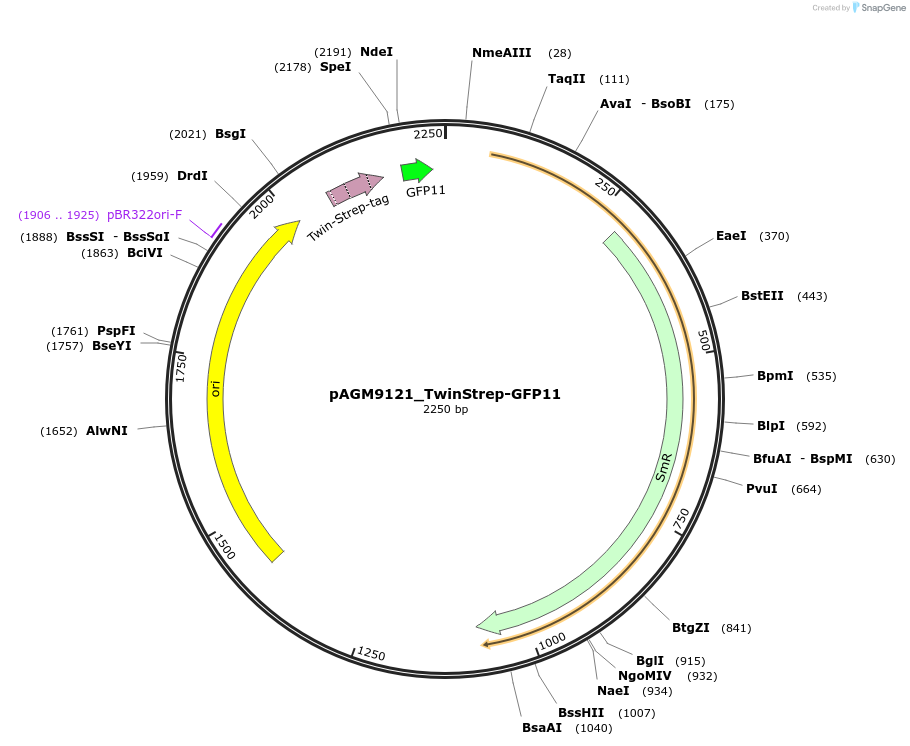
pAGM9121_TwinStrep-GFP11
Plasmid#153514PurposeTwinStrep-GFP11 C-terminal Tag Donor PlasmidDepositorInsertTwinStrep-GFP11
UseSynthetic BiologyAvailable SinceJan. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
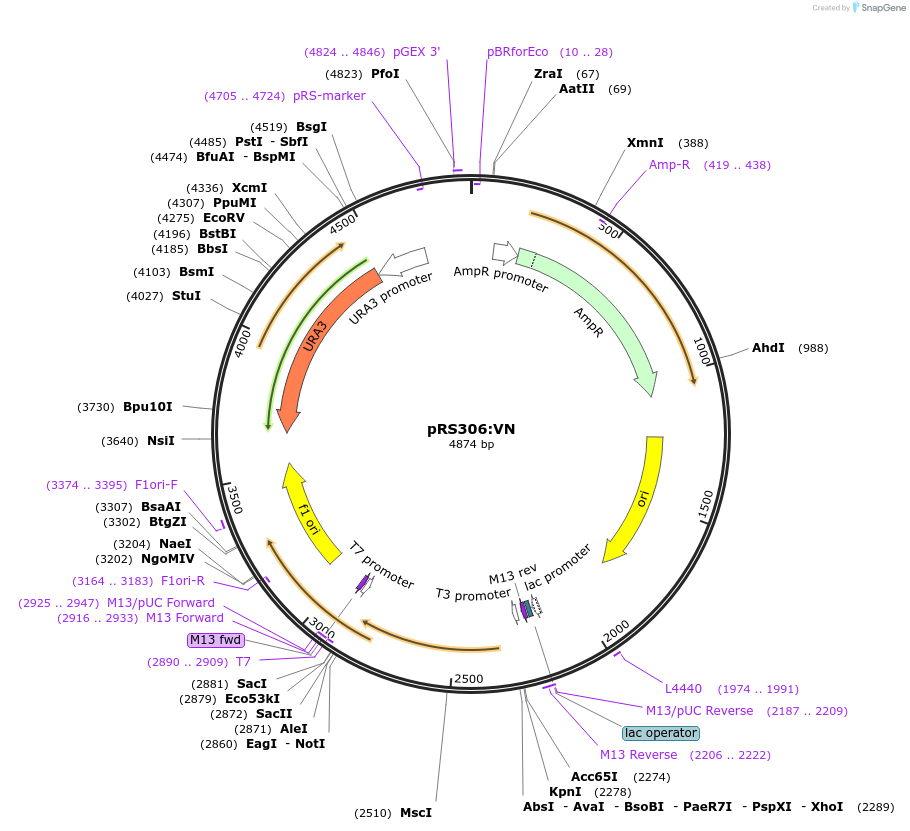
pRS306:VN
Plasmid#37555DepositorInsertVenus N-term (aa 1-272)
ExpressionBacterialAvailable SinceJuly 12, 2012AvailabilityAcademic Institutions and Nonprofits only -
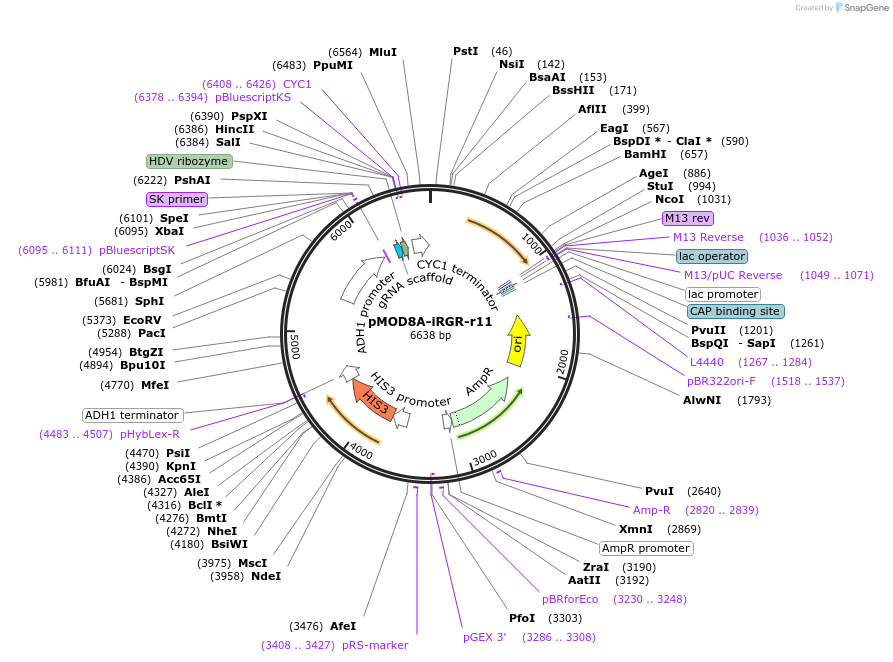
pMOD8A-iRGR-r11
Plasmid#74369PurposeThis plasmid constitutively expresses gRNA-r11 from an AHD1 promoter using the insulated RGR design. Integrates in the W303 HIS locus and restores HIS function.DepositorArticleInsertRGR-r11
UseCRISPR and Synthetic BiologyExpressionYeastPromoterAHD1Available SinceApril 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
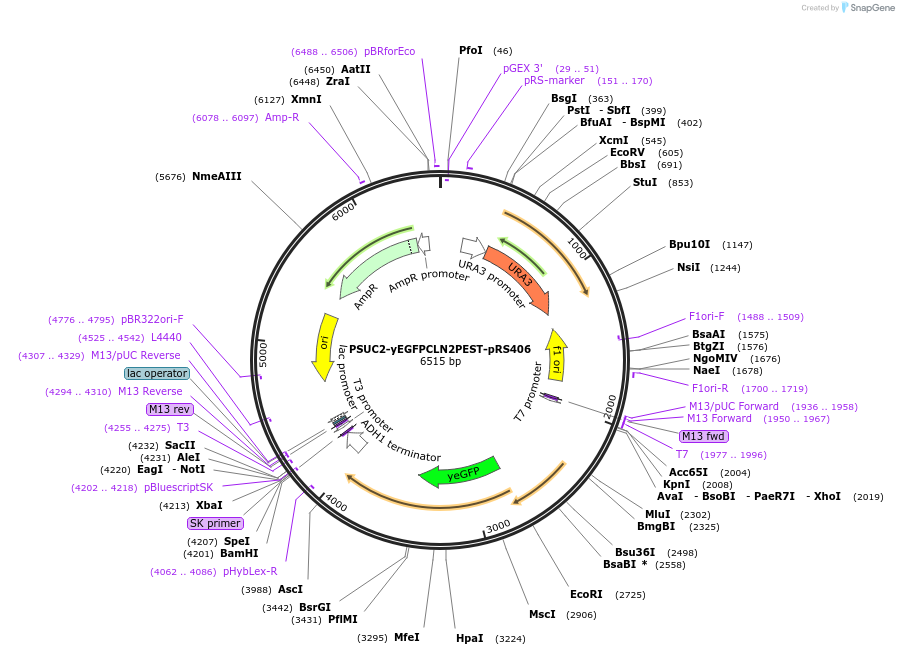
PSUC2-yEGFPCLN2PEST-pRS406
Plasmid#64405PurposeSUC2 promoter driven GFP expressionDepositorInsertSUC2 promoter (SUC2 Budding Yeast)
TagsyEGFPCLN2PEST (destabilized yeast EGFP)ExpressionYeastPromoterSUC2Available SinceMay 19, 2015AvailabilityAcademic Institutions and Nonprofits only -
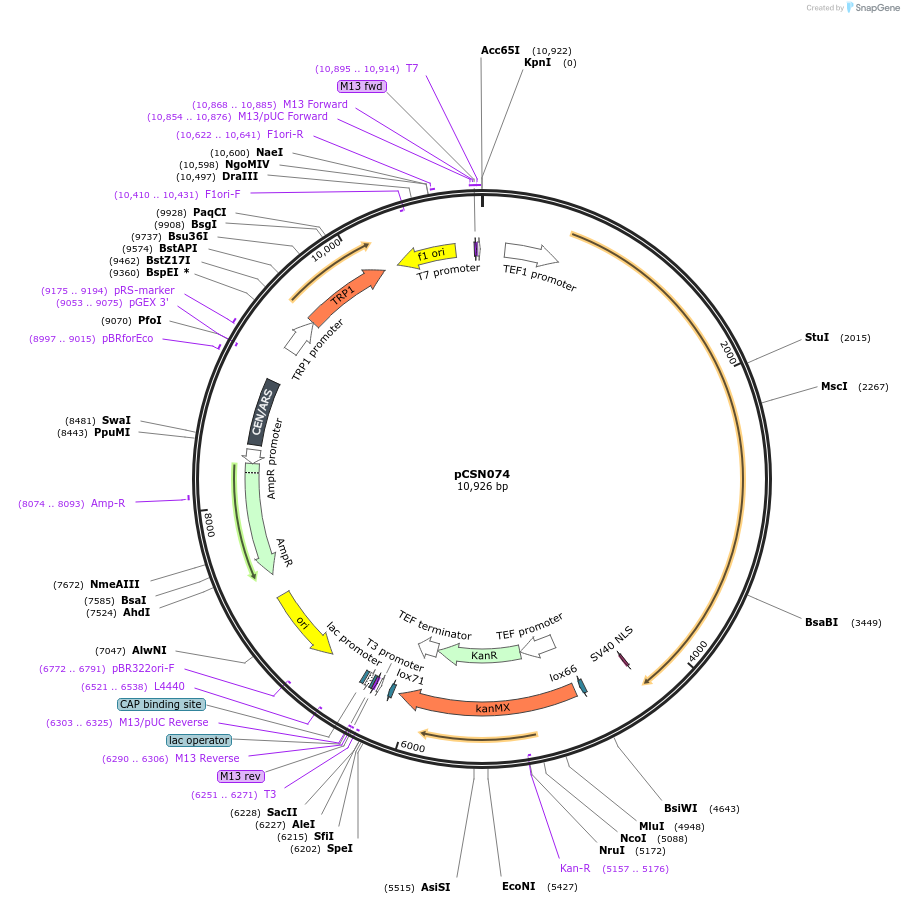
pCSN074
Plasmid#166731PurposeSingle copy yeast plasmid expressing DNase-dead dLbCas12a E925A fused to NLS, codon optimized for Saccharomyces cerevisiaeDepositorInsertdCas12a-NLS
UseCRISPRTagsSV40 NLSExpressionYeastMutationE925APromoterS. cerevisiae TEF1Available SinceJune 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
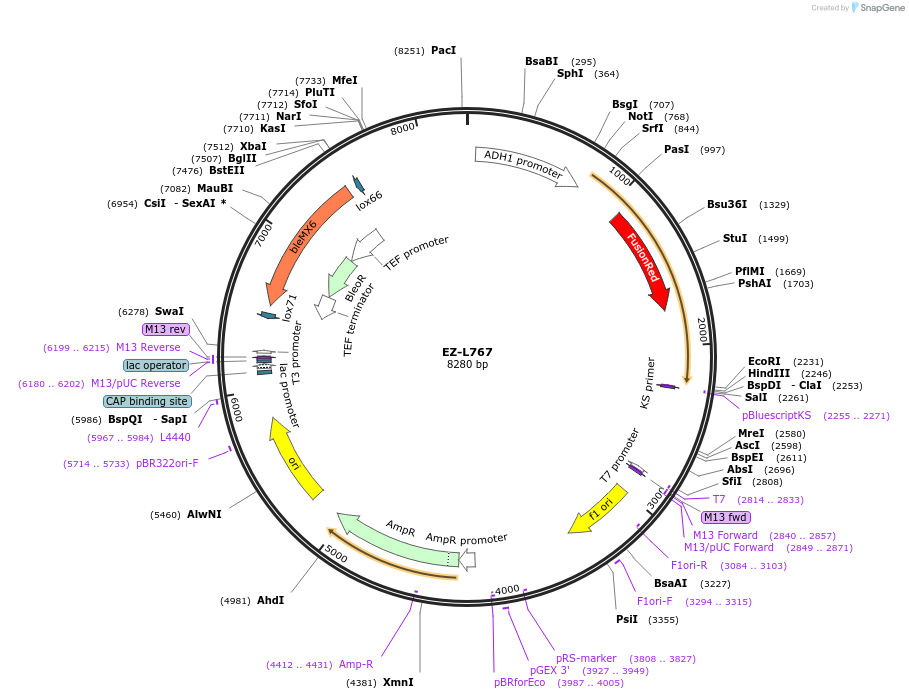
EZ-L767
Plasmid#131773PurposePADH1_sFUS_Fusion Red_PixD_TACT1DepositorInsertPADH1_sFUS_Fusion Red_PixD_TACT1
UseIntegration into δ-sitesExpressionYeastPromoterPADH1Available SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
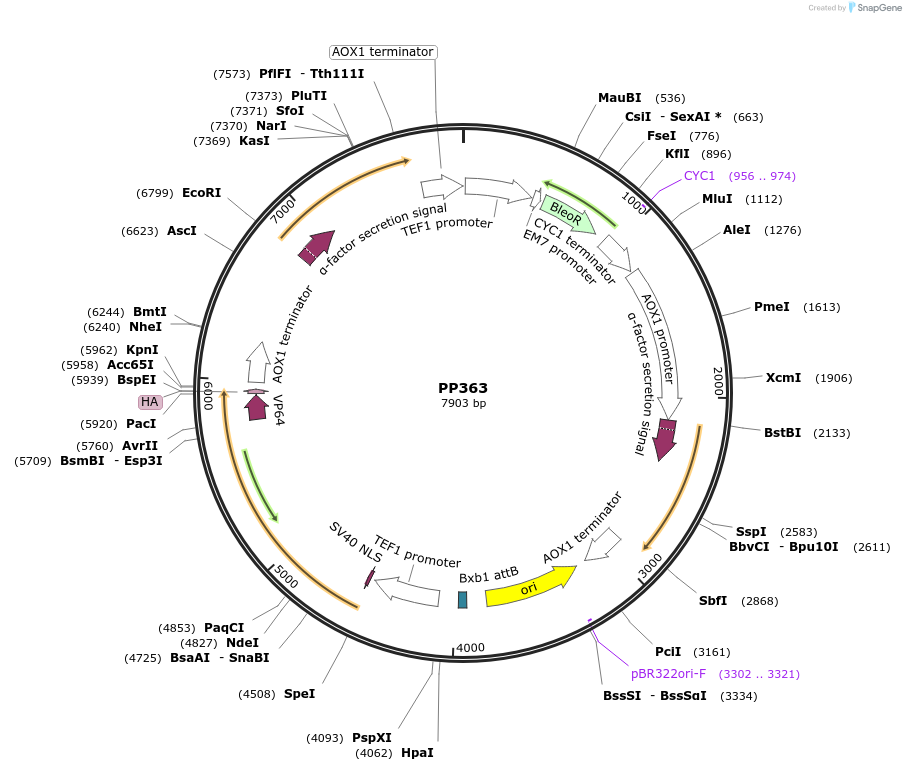
PP363
Plasmid#78984Purpose255B. Promoter: scTEF1, minimal Promoter: GAP. Figure 5. Expresses rHGH and IFNa2b.DepositorInsertsrHGH
IFNa2b
ExpressionYeastAvailable SinceDec. 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
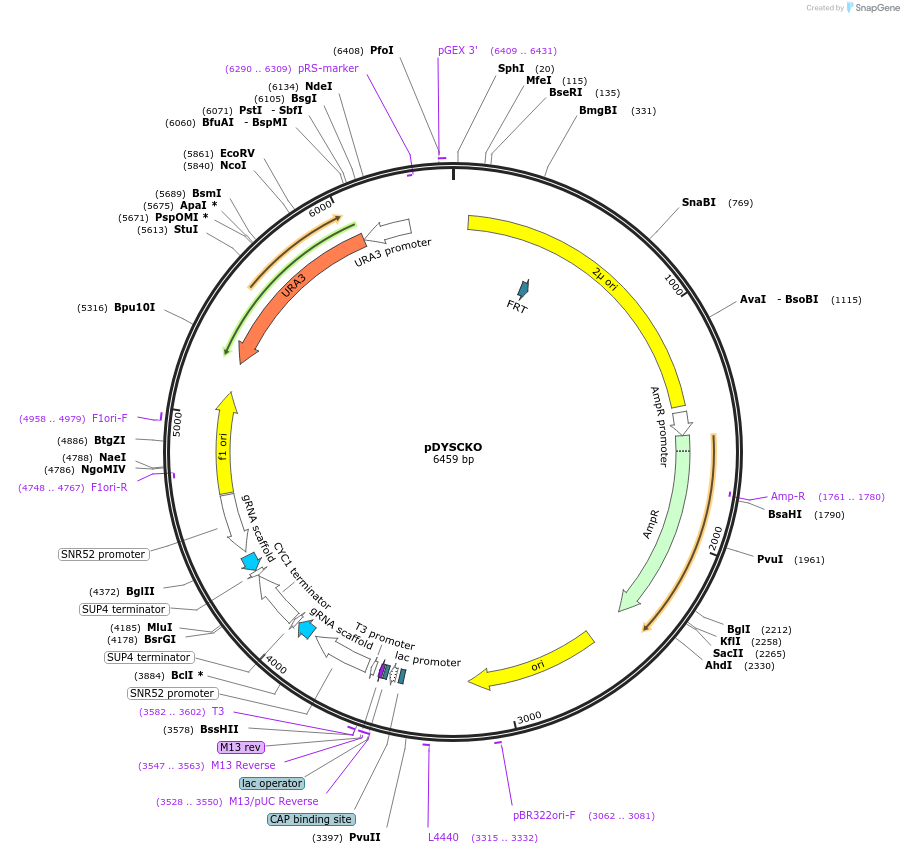
pDYSCKO
Plasmid#120263PurposeEmpty backbone for cloning any gRNA to be used with the canavanine co-selection systemDepositorInsertDYSCKO cassette
UseCRISPR and Synthetic BiologyPromoterSNR52Available SinceJan. 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
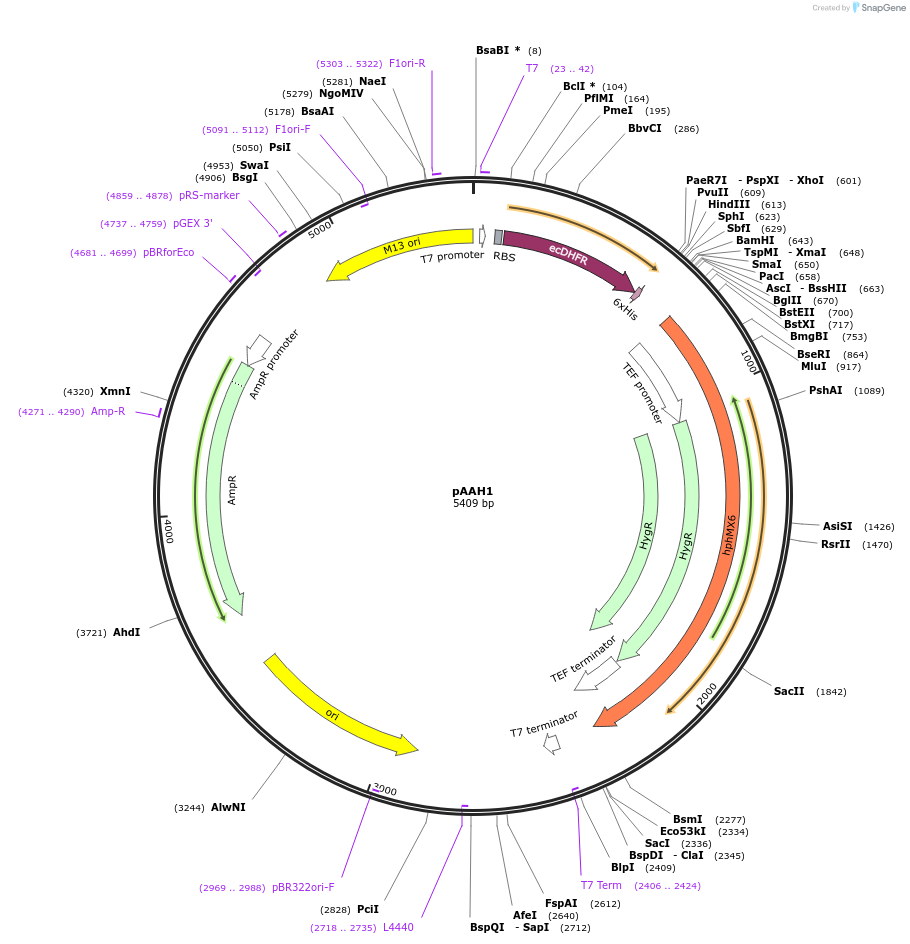
pAAH1
Plasmid#41825PurposeUsed as a PCR template for tagging yeast proteins with the DHFR tag (c-terminal histag) by homologous recombination and selection for hygromycin resistance.DepositorTypeEmpty backboneUsePcr templateExpressionBacterial and YeastPromoterT7, TEFAvailable SinceOct. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
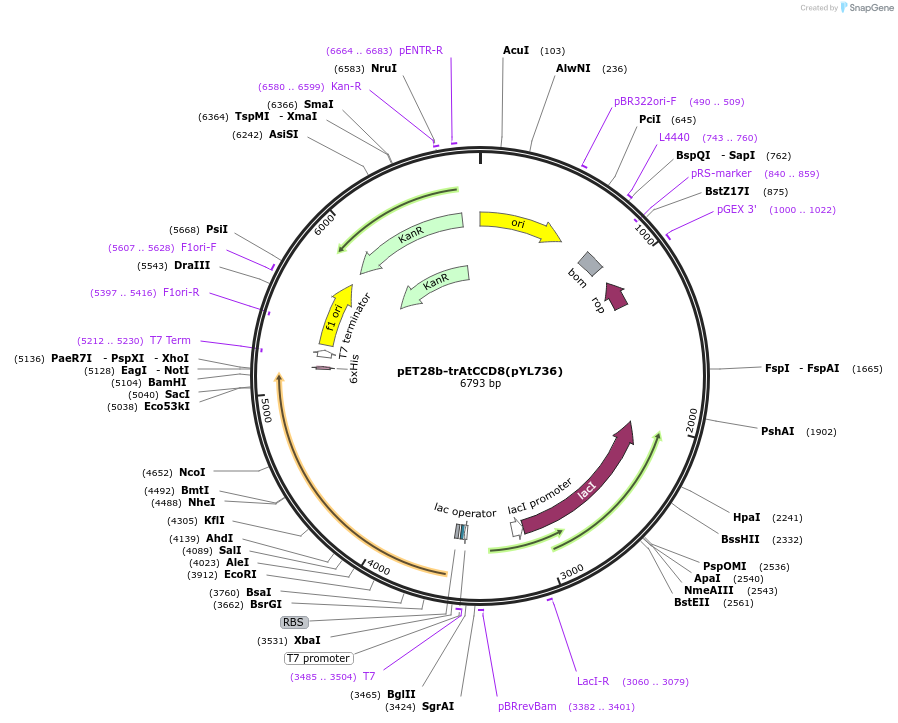
pET28b-trAtCCD8(pYL736)
Plasmid#178287PurposeExpresses trAtCCD8 in E. coli. pET28b carrying t N-terminus 56 amino-acid truncated CCD8 from ArabidopsisDepositorInserttrAtCCD8
ExpressionBacterialPromoterT7Available SinceSept. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
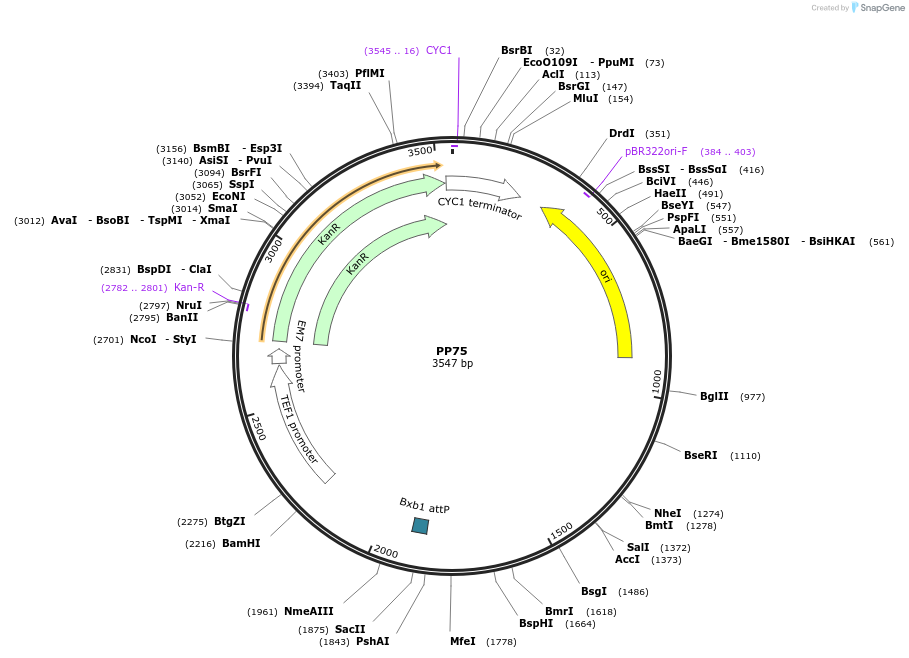
PP75
Plasmid#78945PurposeSupplemental Figure 3. Integration Site 2.DepositorInsertIntegration Site 2
ExpressionYeastAvailable SinceFeb. 9, 2017AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F53
Plasmid#23077DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Lysine 9 to Arginine, H3 K9RAvailable SinceApril 12, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F30
Plasmid#23066DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed lysine 9 to Glutamine H3 K9QAvailable SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
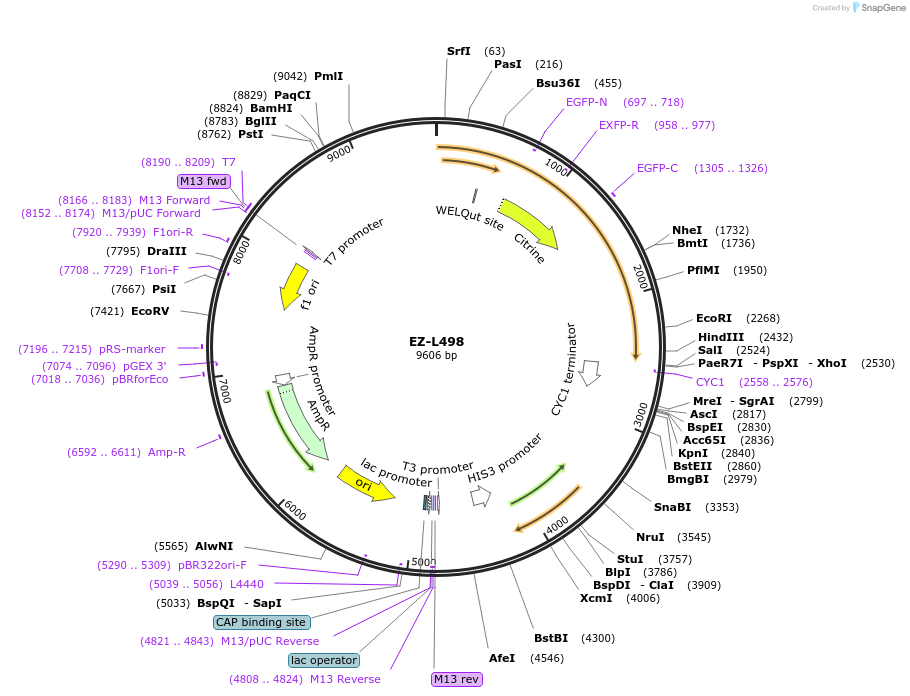
EZ-L498
Plasmid#131771PurposePPGK1_FUSN_Citrine _PixE_TCYC1DepositorInsertPPGK1_FUSN_Citrine _PixE_TCYC1
UseIntegration into his3 locusExpressionYeastPromoterPGKAvailable SinceSept. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
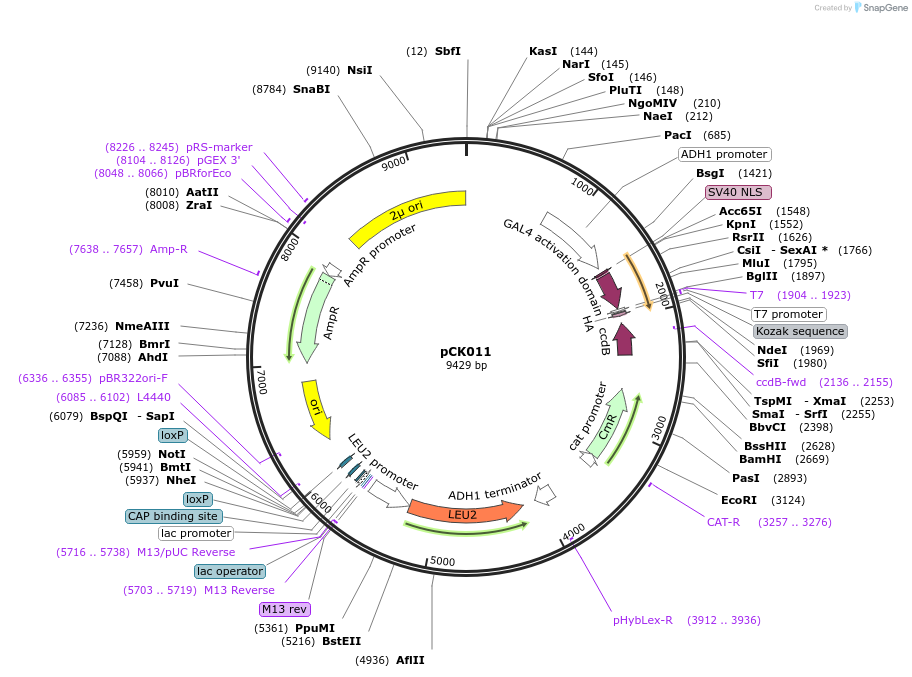
pCK011
Plasmid#105343PurposeYeast two hybridDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
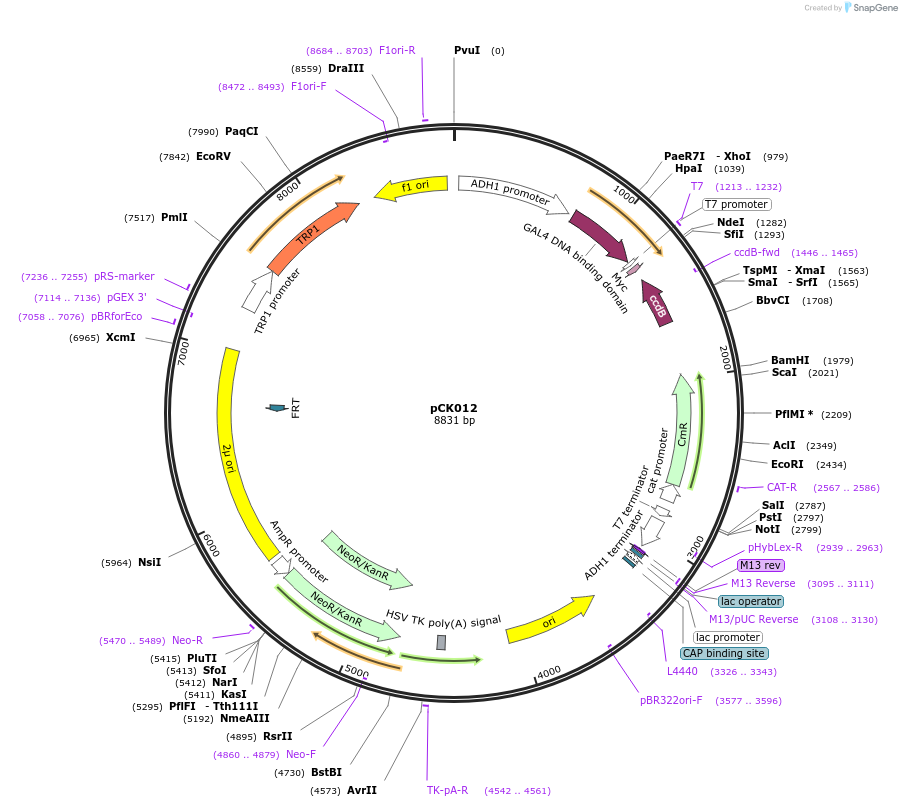
pCK012
Plasmid#105344PurposeYeast two hybridDepositorTypeEmpty backboneExpressionYeastAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F31
Plasmid#23067DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed lysine 14 to Glutamine K14QAvailable SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
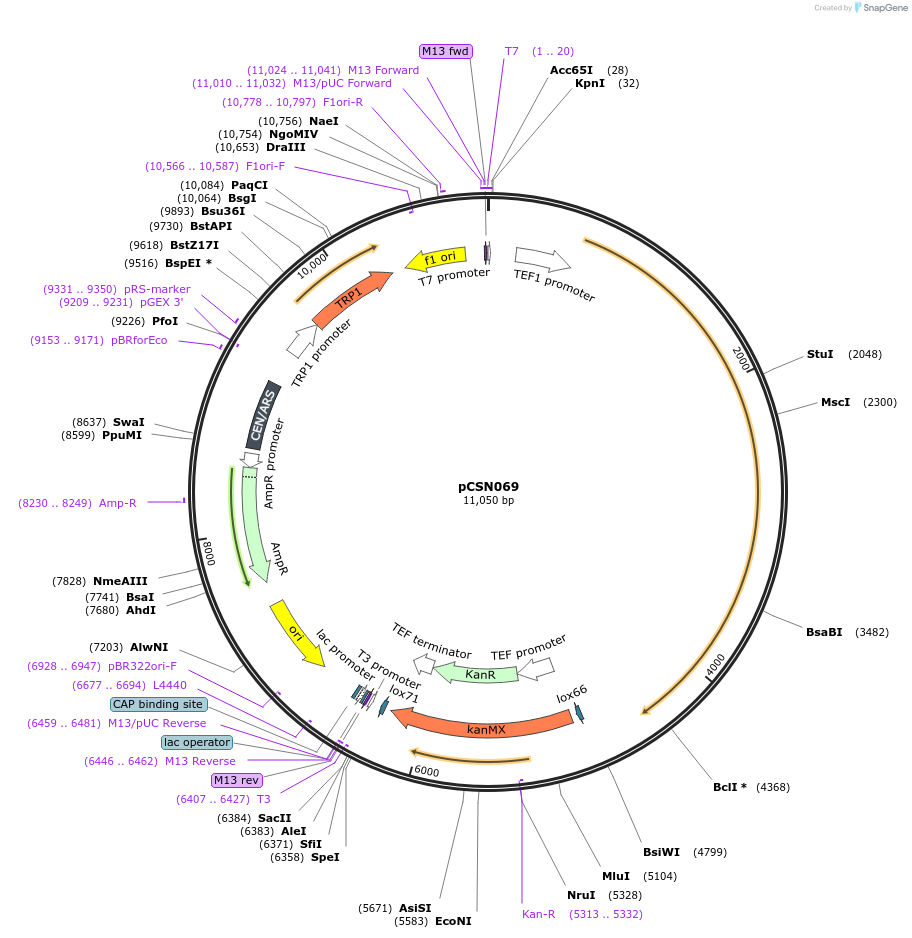
pCSN069
Plasmid#166728PurposeSingle copy yeast plasmid expressing DNase-dead dLbCas12a E925A fused to Mxi1, codon optimized for Saccharomyces cerevisiaeDepositorInsertdCas12a-Mxi1
UseCRISPRTagsMxi1ExpressionYeastMutationE925APromoterS. cerevisiae TEF1Available SinceJune 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
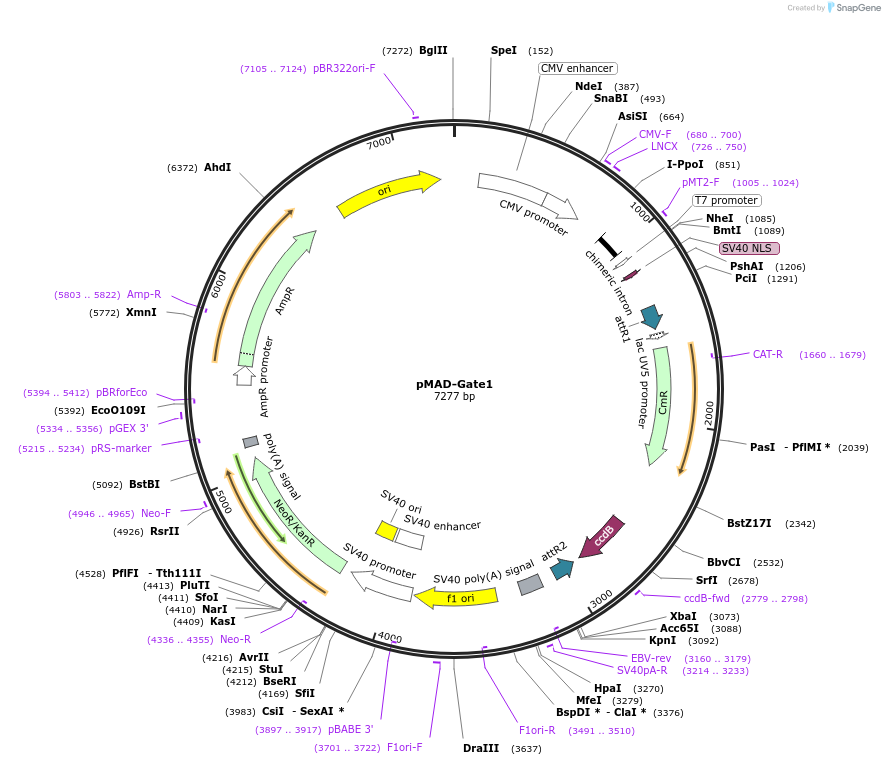
pMAD-Gate1
Plasmid#25916DepositorInsert
TagsVP16 Fusion ProteinExpressionMammalianAvailable SinceAug. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
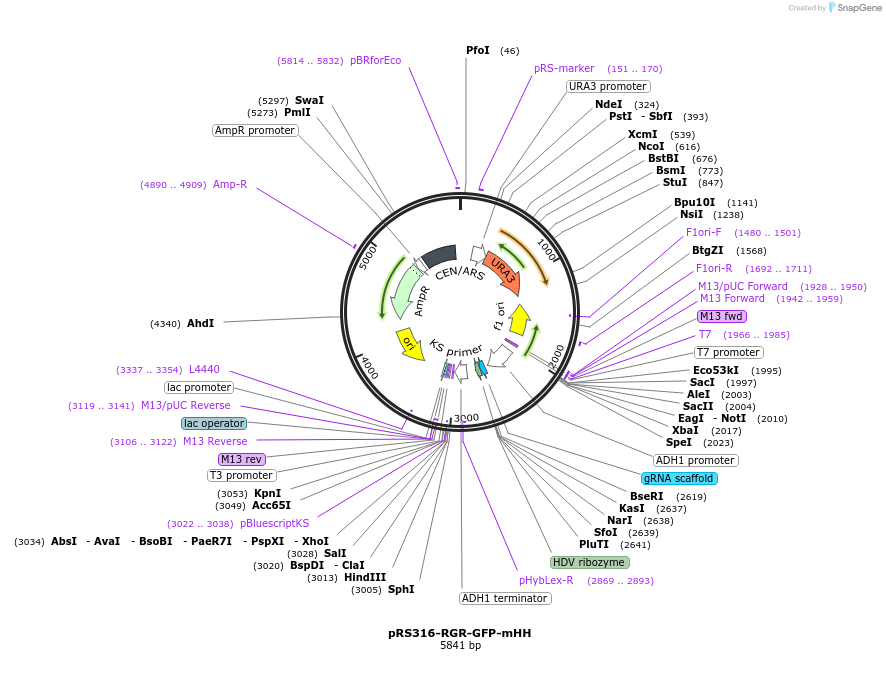
pRS316-RGR-GFP-mHDV
Plasmid#51058PurposeSame as pRS316-RGR-GFP except for the HDV ribozyme region which has a 15 bp deletion at its 3' end.DepositorInsertRGR-GFP
UseCRISPRExpressionBacterial and YeastPromoterYeast ADH1 promoterAvailable SinceFeb. 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
pRS316-RGR-GFP-mHH
Plasmid#51057PurposeSame as pRS316-RGR-GFP except for the HH ribozyme region which has a 13 bp deletion at its 5' end.DepositorInsertRGR-GFP
UseCRISPRExpressionBacterial and YeastPromoterYeast ADH1 promoterAvailable SinceFeb. 11, 2014AvailabilityAcademic Institutions and Nonprofits only -
pRS306:Met3p-mCherry-Tub1
Plasmid#37554DepositorInsertMet3p-mCherry-Tub1
ExpressionBacterialAvailable SinceAug. 2, 2012AvailabilityAcademic Institutions and Nonprofits only -
pFA3
Plasmid#46790PurposeTo express ATPase inactive Fun30 in budding yeastDepositorInsertFUN30 (FUN30 Budding Yeast)
TagsV5ExpressionYeastMutationATP-binding site mutated replaced lysin 603 by ar…PromoterPGAL1Available SinceAug. 7, 2013AvailabilityAcademic Institutions and Nonprofits only -
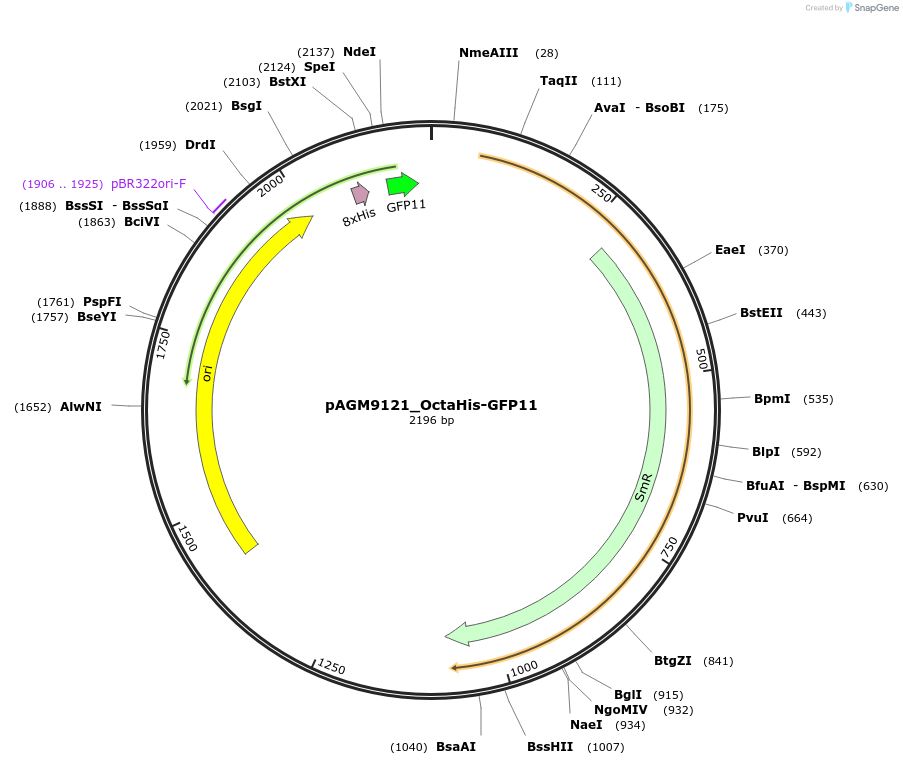
pAGM9121_OctaHis-GFP11
Plasmid#153509PurposeOctaHis-GFP11 C-terminal Tag Donor PlasmidDepositorInsertOctaHis-GFP11
UseSynthetic BiologyAvailable SinceJan. 28, 2021AvailabilityAcademic Institutions and Nonprofits only -
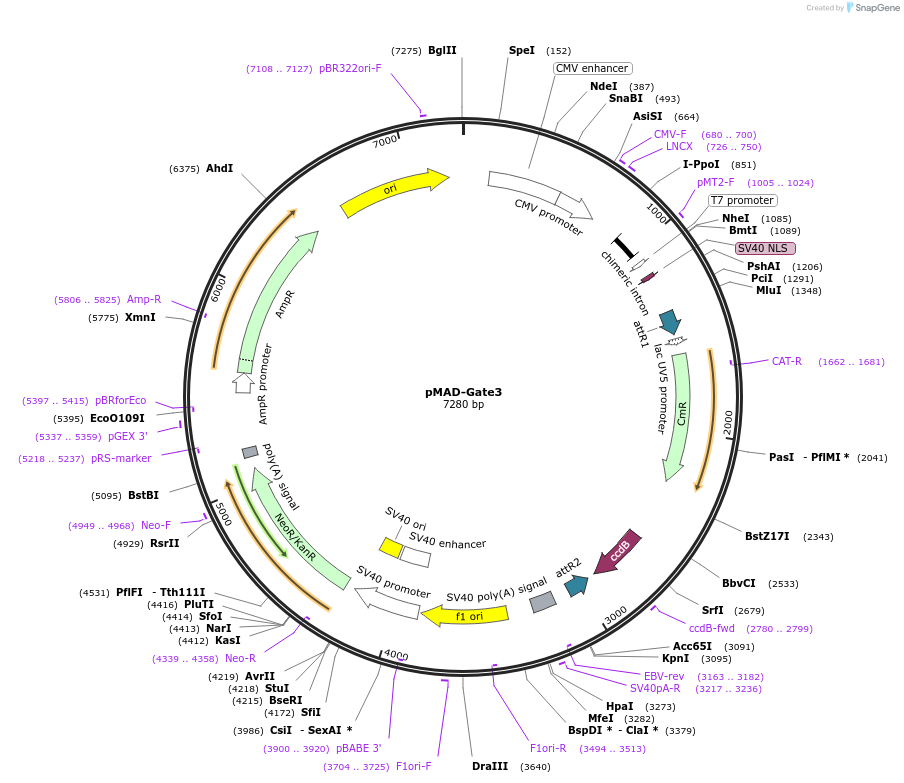
pMAD-Gate3
Plasmid#25942DepositorInsert
TagsVP16 Fusion ProteinExpressionMammalianAvailable SinceAug. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
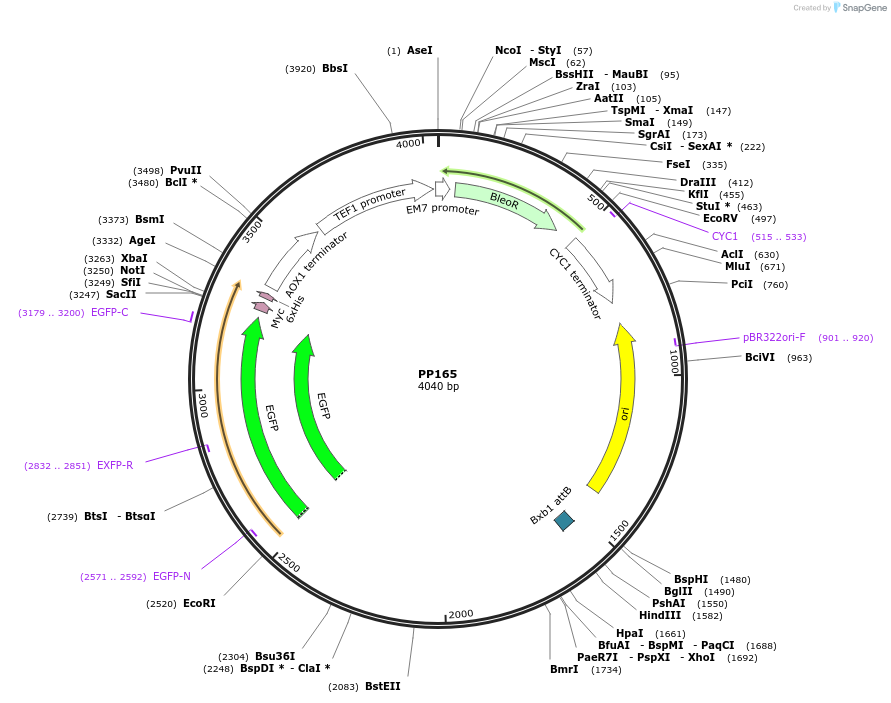
PP165
Plasmid#78985PurposeSupplemental Figure 4. Promoter GCW14. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
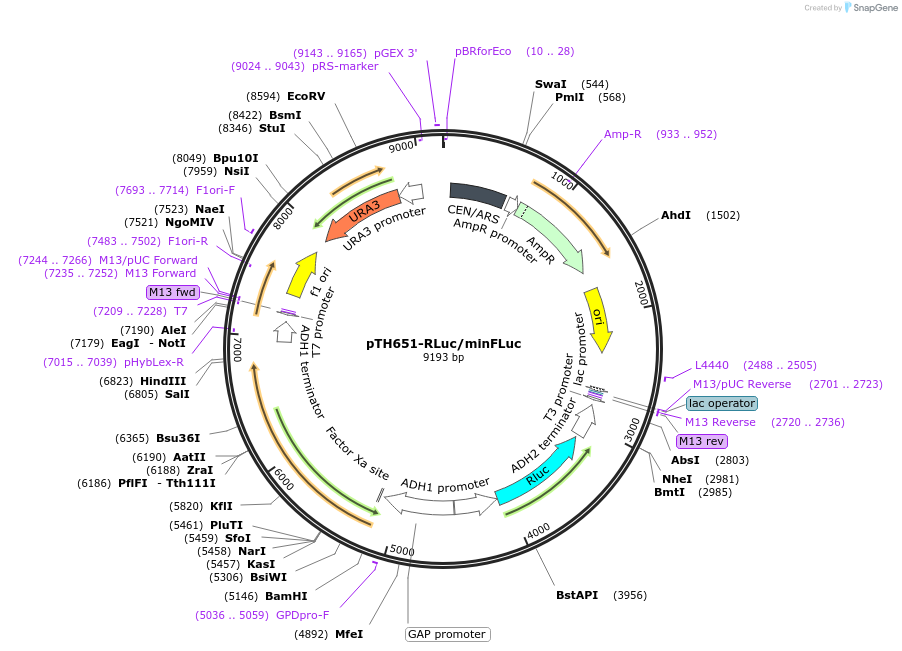
pTH651-RLuc/minFLuc
Plasmid#29697DepositorInsertFirefly Luciferase
ExpressionYeastMutationAll codons of the original FLuc gene were exchang…Available SinceAug. 25, 2011AvailabilityAcademic Institutions and Nonprofits only -
PP275
Plasmid#78968PurposePromoter: GAP, minimal Promoter: GCW14. Figure 4. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceDec. 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
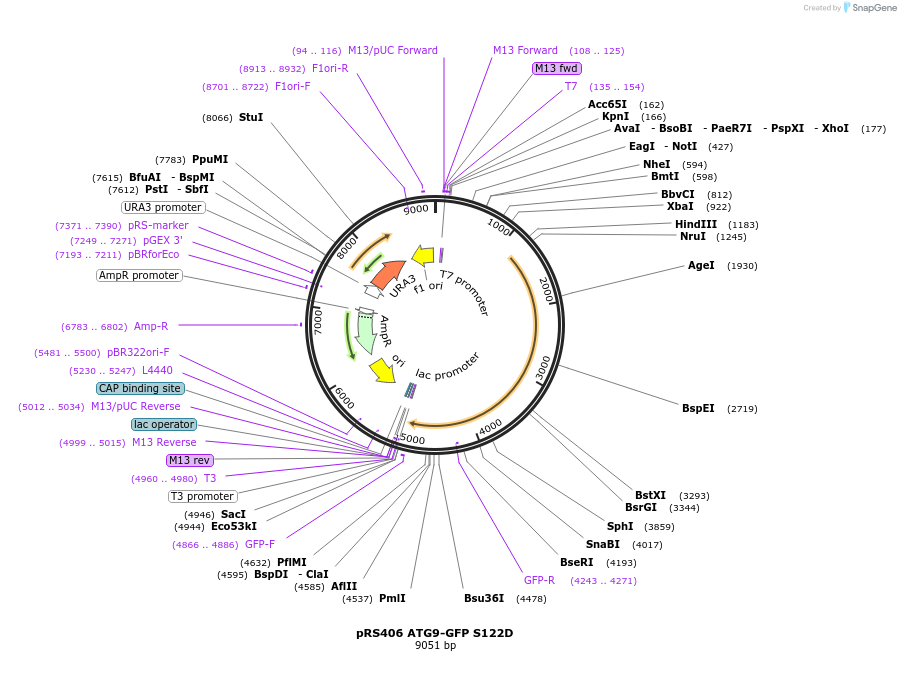
pRS406 ATG9-GFP S122D
Plasmid#114667PurposeExpresses yeast ATG9 (S122D) under the endogenous promoter with a GFP C-terminal tagDepositorInsertATG9
TagsGFPExpressionYeastMutationchanged serine 122 to aspartic acidPromoterATG9Available SinceSept. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
pDR-AmTrac1;2
Plasmid#47765Purposeexpresses AmTrac1;2 in yeastDepositorInsertAmTrac1;2
TagsmcpGFPExpressionYeastPromoterPMAAvailable SinceSept. 16, 2013AvailabilityAcademic Institutions and Nonprofits only -
pDR-AmTrac-LS
Plasmid#47767Purposeexpresses AmTrac-LS in yeastDepositorInsertAmTrac-LS
TagsmcpGFPExpressionYeastPromoterPMAAvailable SinceSept. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
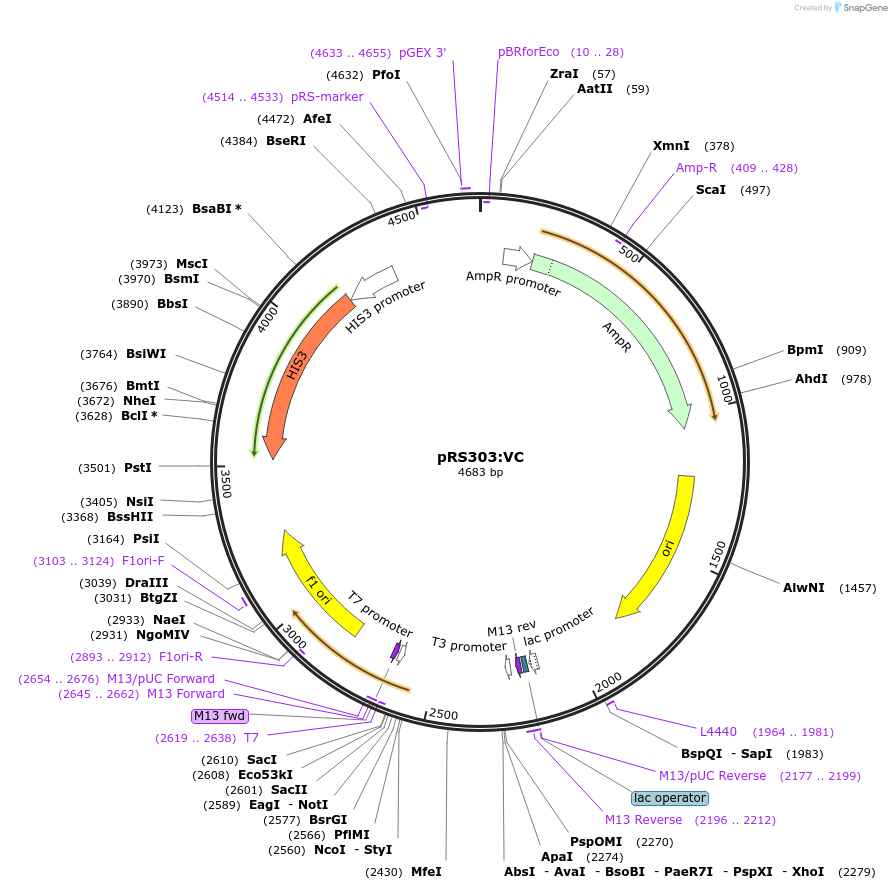
pRS303:VC
Plasmid#37556DepositorInsertVenus C-term (aa 155-238)
ExpressionBacterialAvailable SinceJuly 12, 2012AvailabilityAcademic Institutions and Nonprofits only -
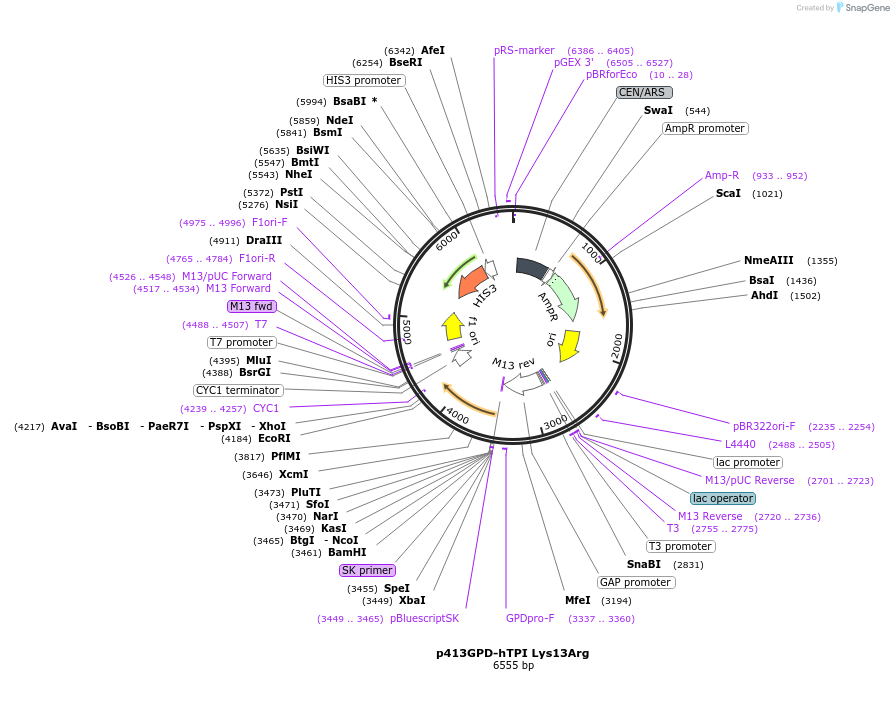
p413GPD-hTPI Lys13Arg
Plasmid#50722Purposeexpression of the human TPI Lys13Arg allele in yeastDepositorInsertTriosephosphate isomerase 1 (TPI1 Human)
ExpressionYeastMutationchanged Lysine 13 to ArgeninePromoterGPDAvailable SinceFeb. 20, 2014AvailabilityAcademic Institutions and Nonprofits only -
PP161
Plasmid#78993PurposeSupplemental Figure 4. Promoter GAP3. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceDec. 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
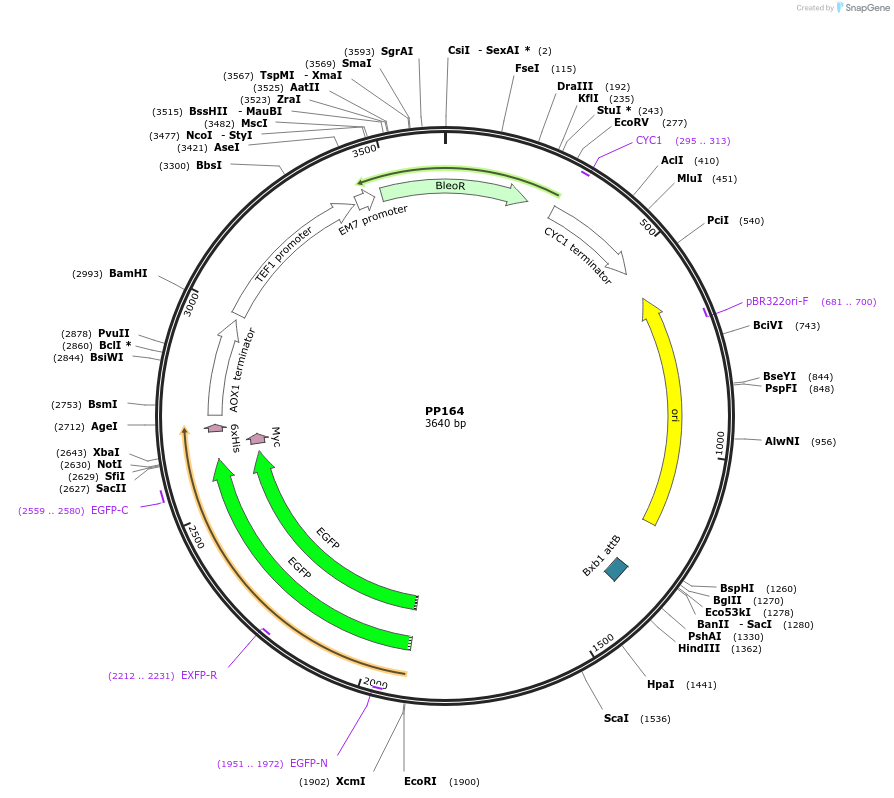
PP164
Plasmid#78988PurposeSupplemental Figure 4. Promoter short ppTEF1. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
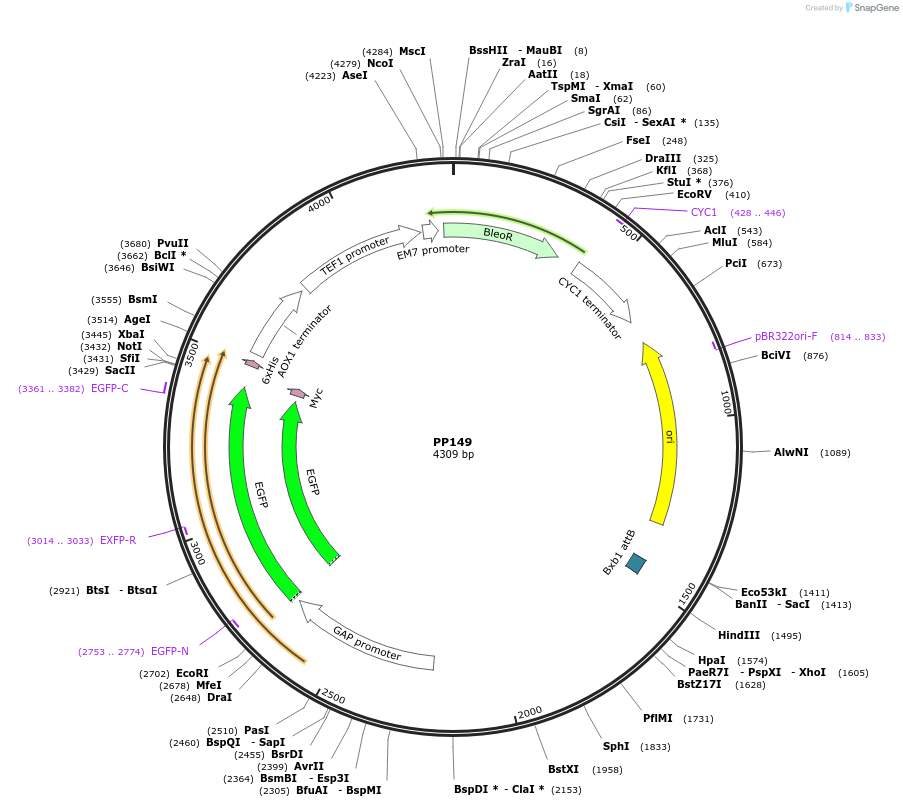
PP149
Plasmid#78989PurposeSupplemental Figure 4. Promoter WT GAP. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
PP154
Plasmid#78991PurposeSupplemental Figure 4. Promoter GAP1. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
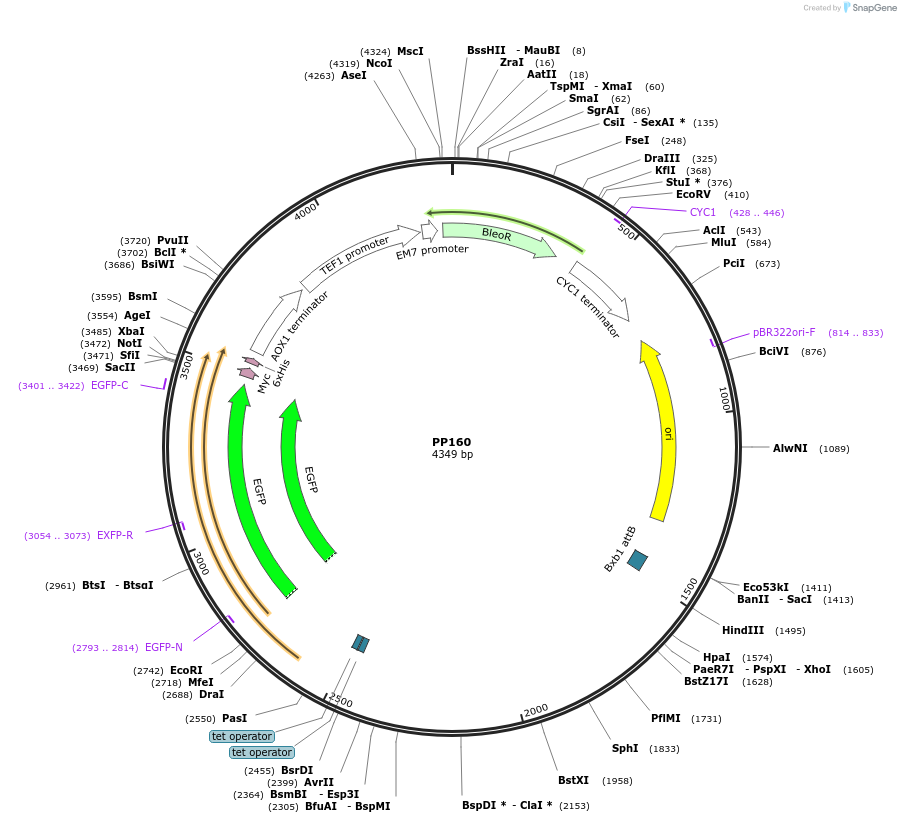
PP160
Plasmid#78992PurposeSupplemental Figure 4. Promoter GAP2. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
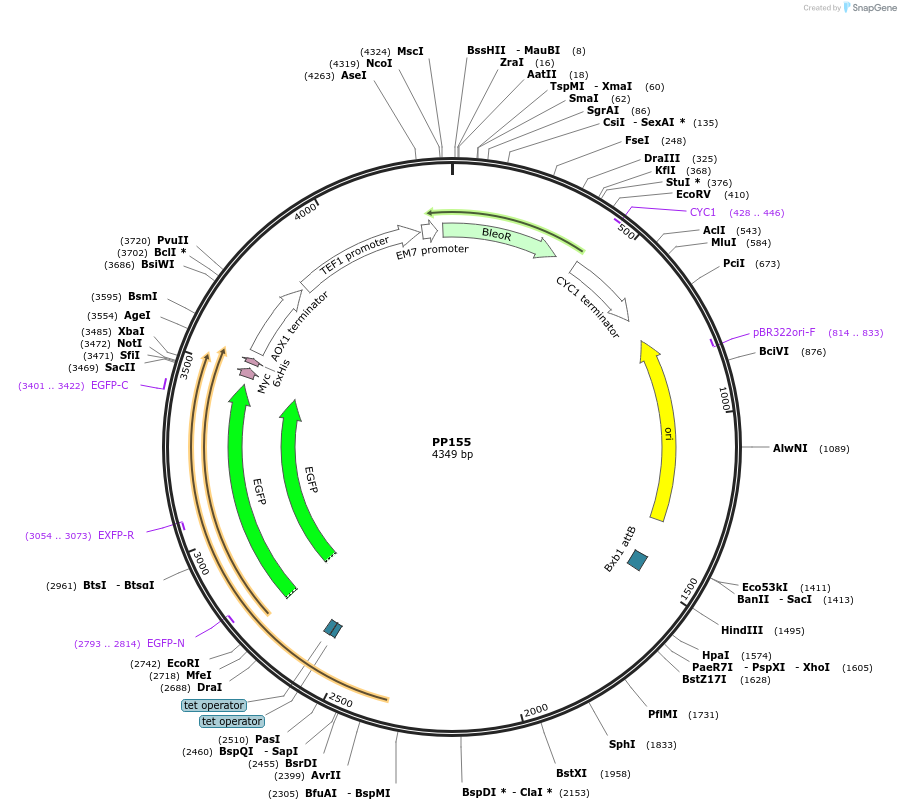
PP155
Plasmid#78994PurposeSupplemental Figure 4. Promoter GAP4. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
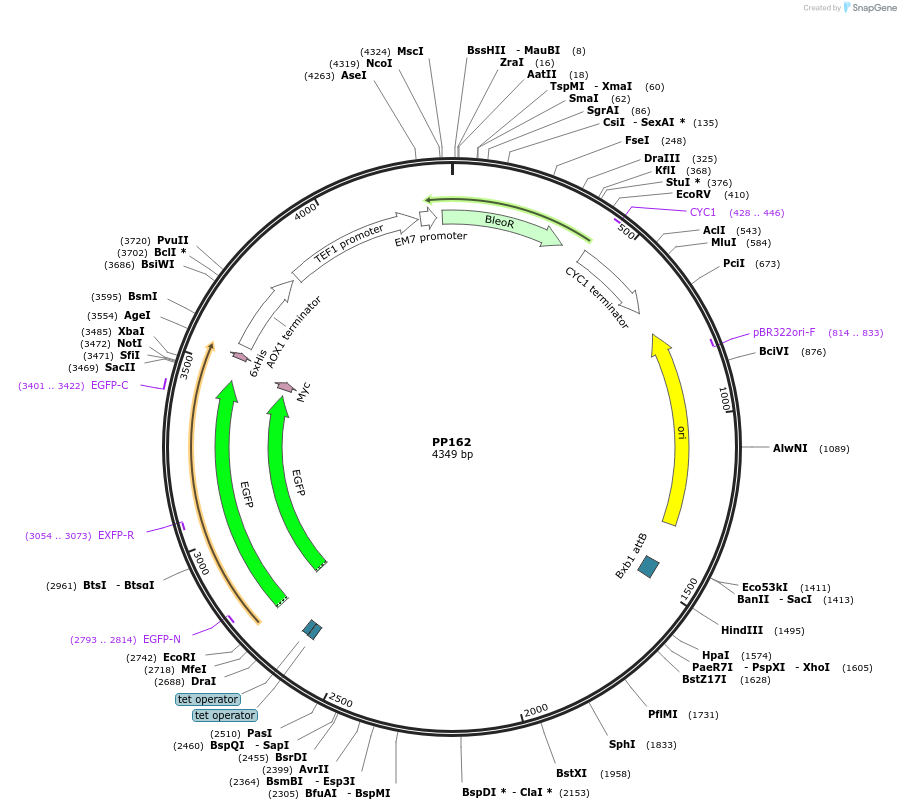
PP162
Plasmid#78995PurposeSupplemental Figure 4. Promoter GAP5. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
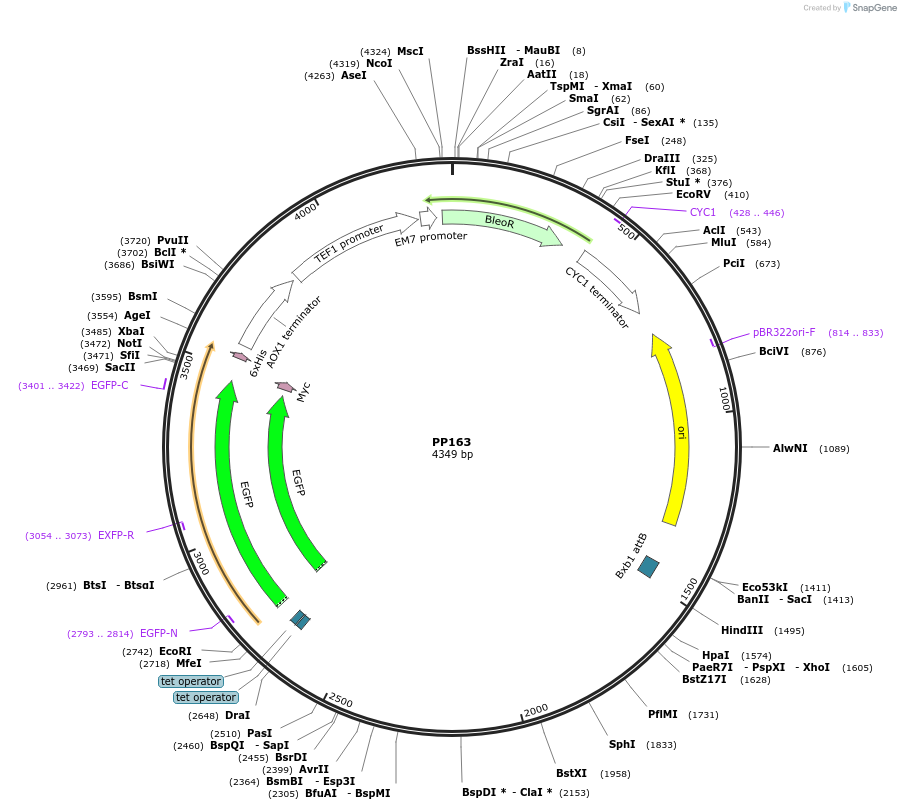
PP163
Plasmid#78996PurposeSupplemental Figure 4. Promoter GAP6. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
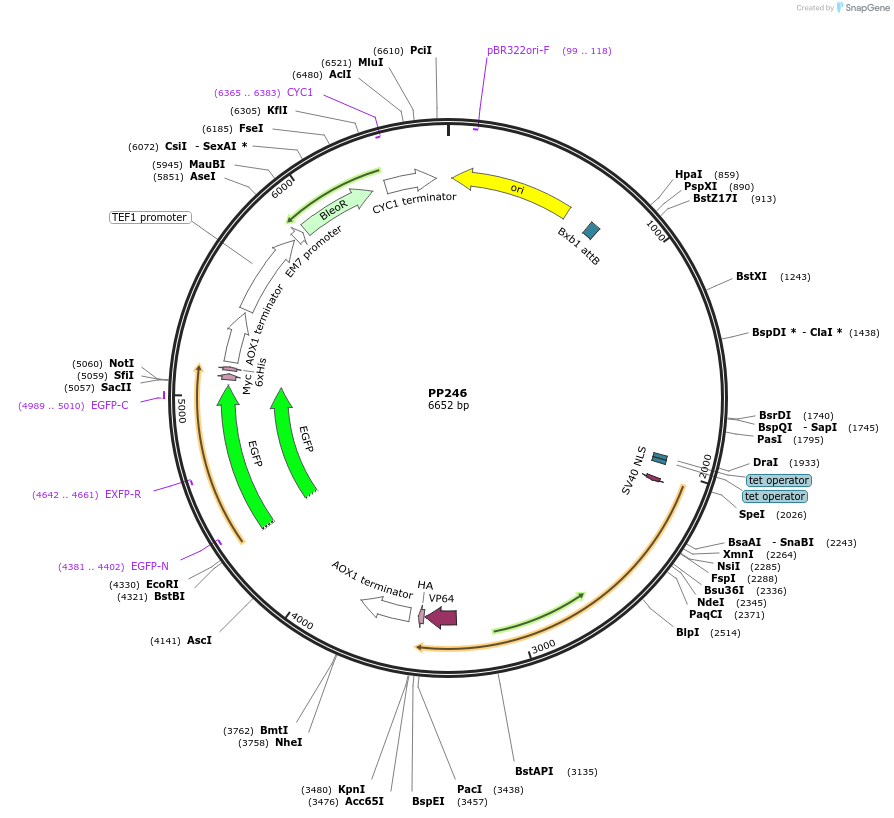
PP246
Plasmid#78966PurposePromoter: GAP6, minimal Promoter: AOX1. Figure 4. Expresses GFP.DepositorInsertGFP
ExpressionYeastAvailable SinceAug. 23, 2016AvailabilityAcademic Institutions and Nonprofits only -
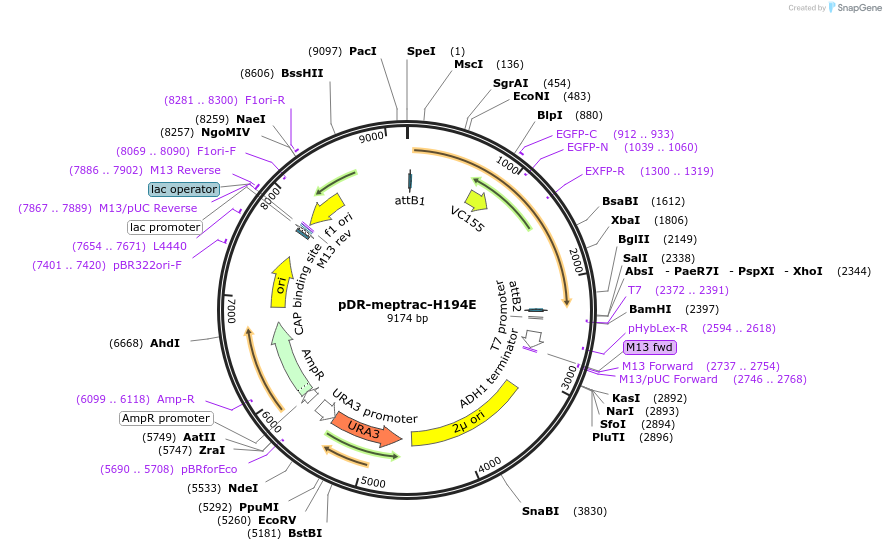
pDR-meptrac-H194E
Plasmid#47764Purposeexpresses MepTrac-H194E in yeastDepositorInsertMepTrac-H194E
TagsmcpGFPExpressionYeastMutationH194EPromoterPMAAvailable SinceSept. 16, 2013AvailabilityAcademic Institutions and Nonprofits only -
pDR-AmTrac-100
Plasmid#47771Purposeexpresses AmTrac-100 in yeastDepositorInsertAmTrac-100
TagsmcpGFPExpressionYeastMutationA141E, T464DPromoterPMAAvailable SinceSept. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
pDR-meptrac
Plasmid#47768Purposeexpresses MepTrac in yeastDepositorInsertmeptrac
TagsmcpGFPExpressionYeastPromoterPMAAvailable SinceSept. 17, 2013AvailabilityAcademic Institutions and Nonprofits only -
pDR-AmTrac-IS
Plasmid#47766Purposeexpresses AmTrac-IS in yeastDepositorInsertAmTrac1;2
TagsmcpGFPExpressionYeastPromoterPMAAvailable SinceSept. 17, 2013AvailabilityAcademic Institutions and Nonprofits only