We narrowed to 3,273 results for: RFP
-
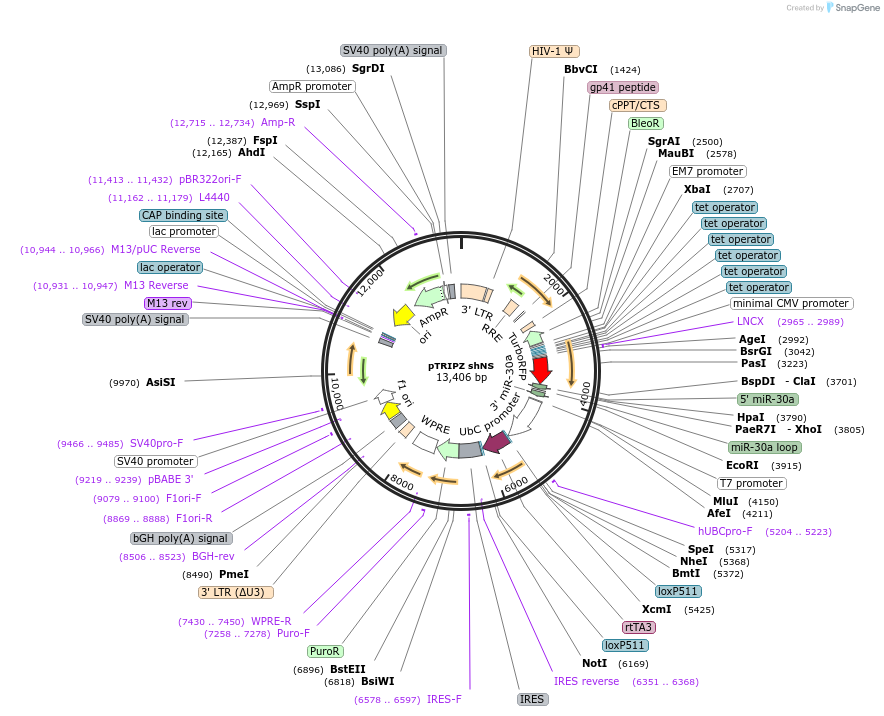
Plasmid#127696PurposeDoxycyclin inducible pTRIPZ lentivirus vector containing a non-silencing sequence for both human and mouseDepositorInsertshNS
UseLentiviralExpressionMammalianAvailable SinceNov. 1, 2019AvailabilityAcademic Institutions and Nonprofits only -
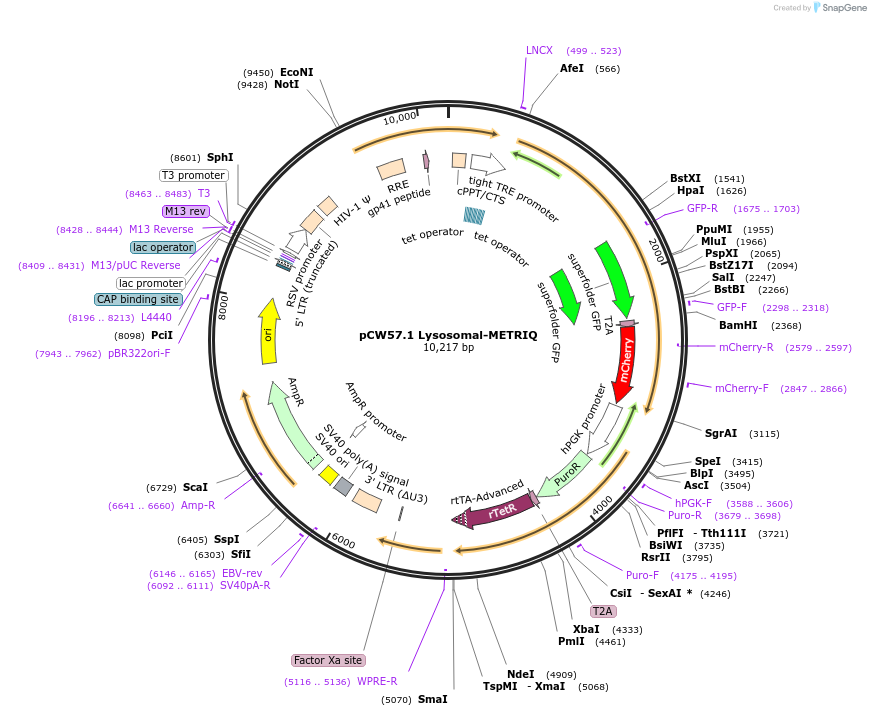
pCW57.1 Lysosomal-METRIQ
Plasmid#135401PurposeExpresses lysosomal-METRIQ (DNAse II-sfGFP together with cytosolic RFP) to measure lysosomal activityDepositorInsertDNAse II alpha (DNASE2 Human)
UseLentiviralTagsT2A, mCherry, and sfGFPExpressionMammalianMutationSingle-nucleotide polymorphism C432TAvailable SinceFeb. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
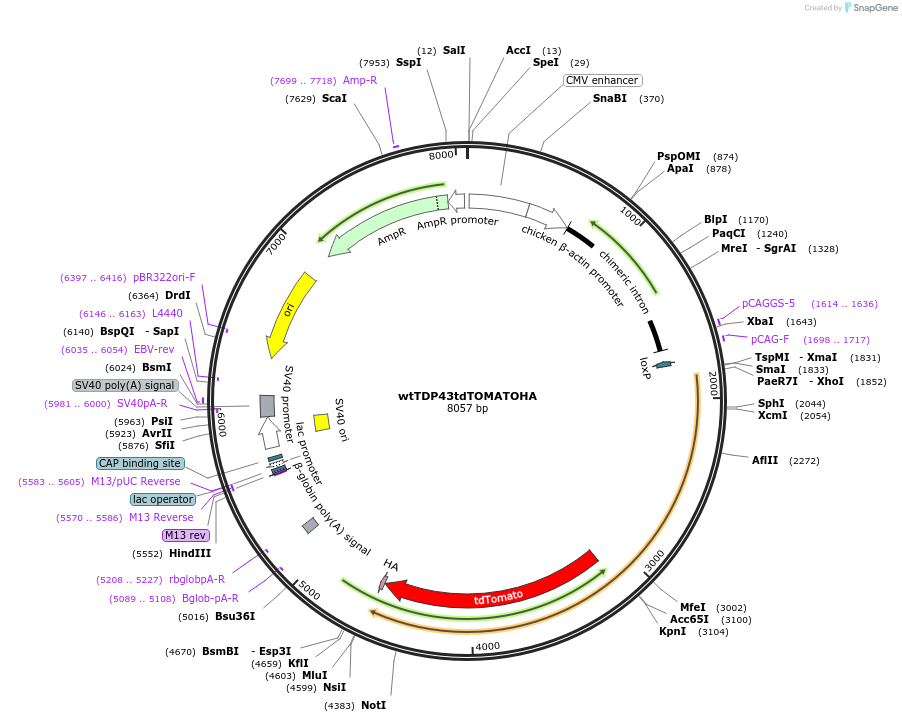
wtTDP43tdTOMATOHA
Plasmid#28205DepositorAvailable SinceJune 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
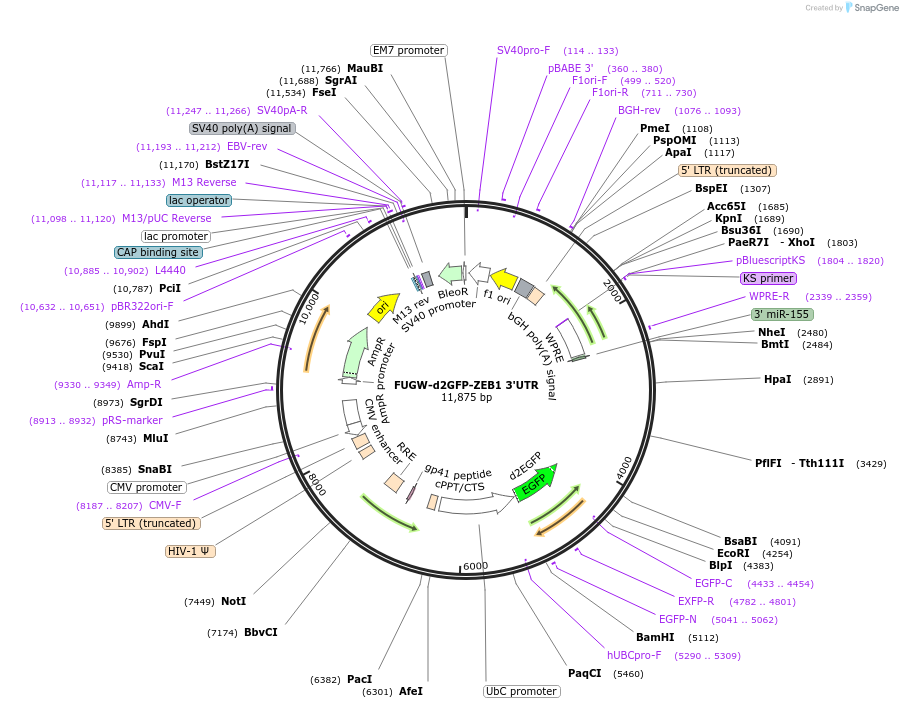
FUGW-d2GFP-ZEB1 3'UTR
Plasmid#79601PurposeFUGW lentiviral vector with constitutively expressed destabilized GFP (d2GFP) regulated by the human ZEB1 3'UTR. When used in combination with Ecad-RFP, comprises the Z-cad dual sensor.DepositorInsertd2GFP-Zeb1 3'UTR
UseLentiviralAvailable SinceJuly 13, 2016AvailabilityAcademic Institutions and Nonprofits only -
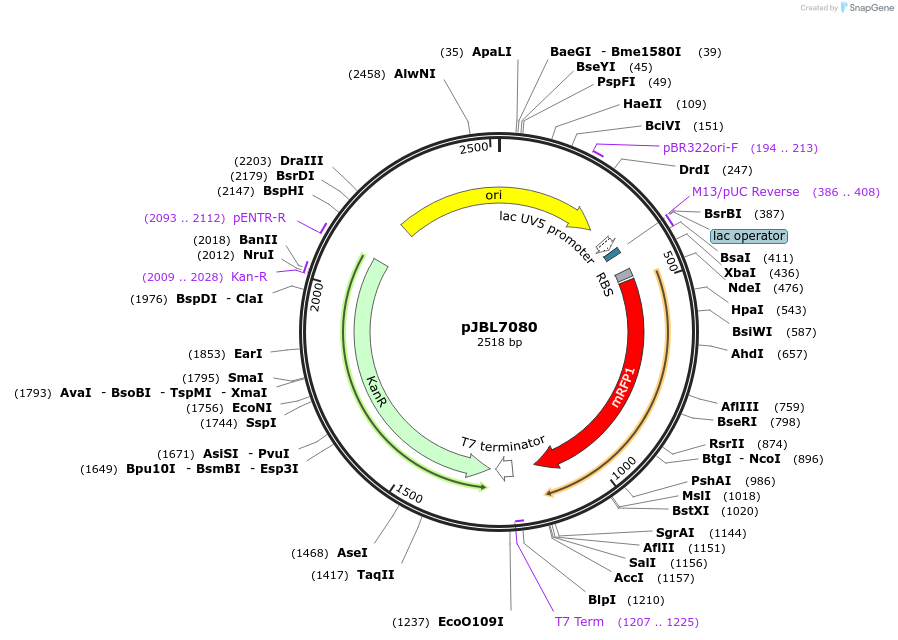
pJBL7080
Plasmid#167245Purposecell-free lac sensor with RFP outputDepositorInsertpLac-mRFP1
UseSynthetic BiologyAvailable SinceJuly 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
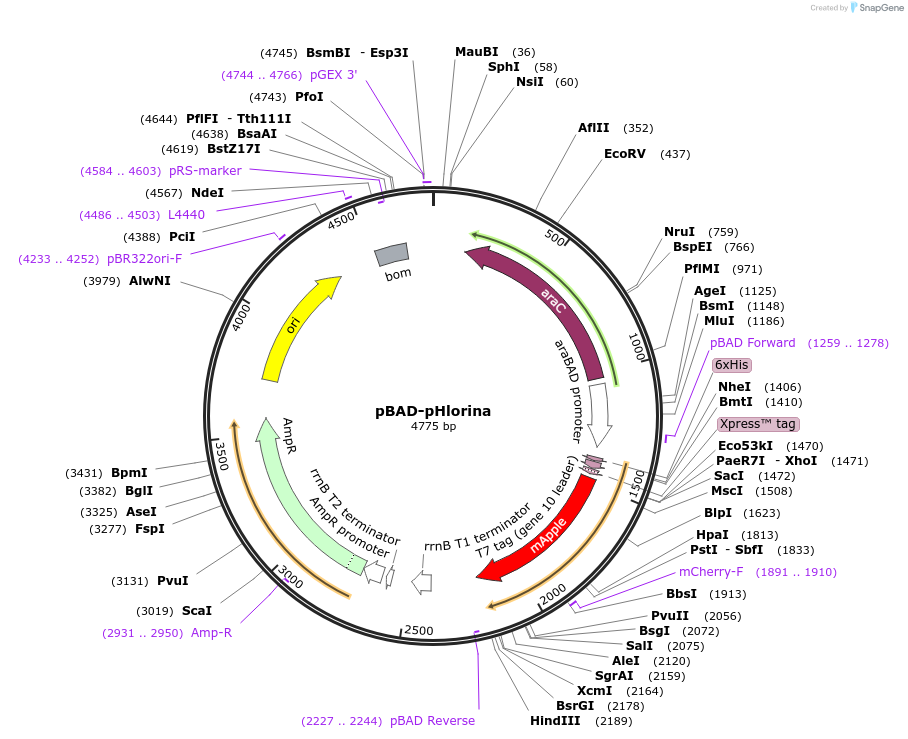
pBAD-pHlorina
Plasmid#182763PurposepH-sensitive RFP pHlorina for detection of efferocytosisDepositorInsertpHlorina
ExpressionBacterialAvailable SinceSept. 27, 2022AvailabilityAcademic Institutions and Nonprofits only -
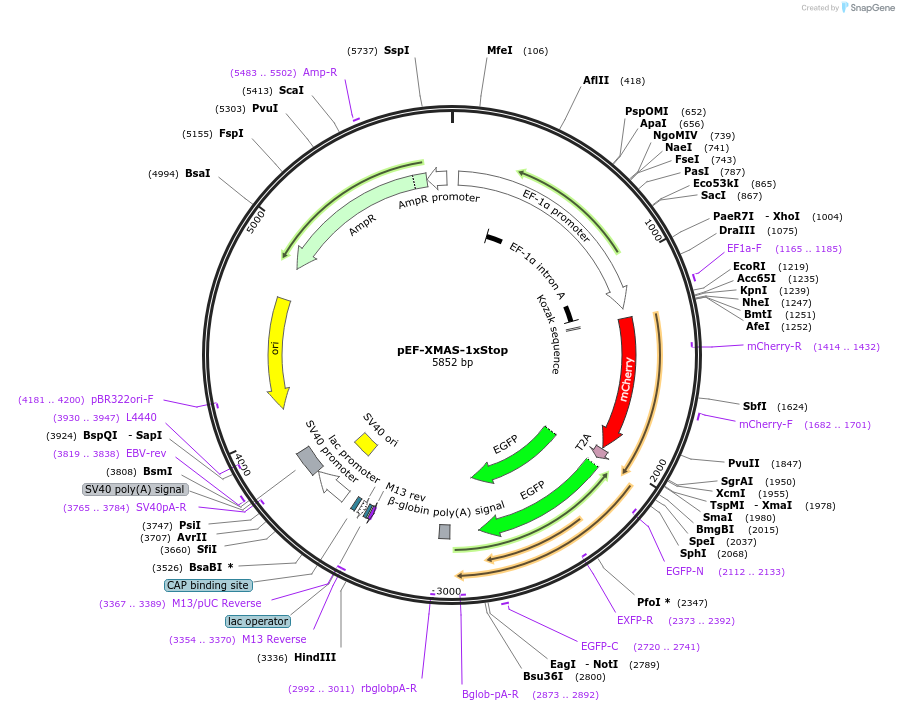
pEF-XMAS-1xStop
Plasmid#164411PurposeTransient Reporter for Editing Enrichment (TREE) Plasmid. GFP turns on with adenosine base editing (ABE). 1 stop codon.DepositorInsertmCherry
UseCRISPR and Synthetic BiologyTagsT2A-GFPExpressionMammalianPromoterEF1-alphaAvailable SinceAug. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
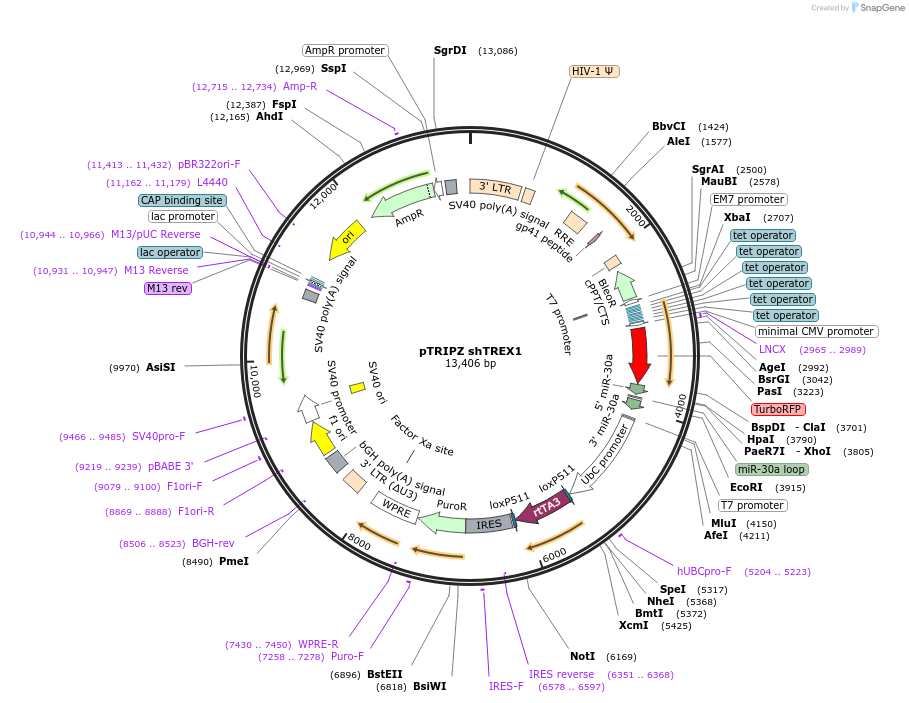
pTRIPZ shTREX1
Plasmid#127700PurposeDoxycyclin inducible shRNA knockdown of both human and mouse TREX1 geneDepositorAvailable SinceFeb. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
TDP43 NOTAG1
Plasmid#28206DepositorAvailable SinceJuly 1, 2011AvailabilityAcademic Institutions and Nonprofits only -
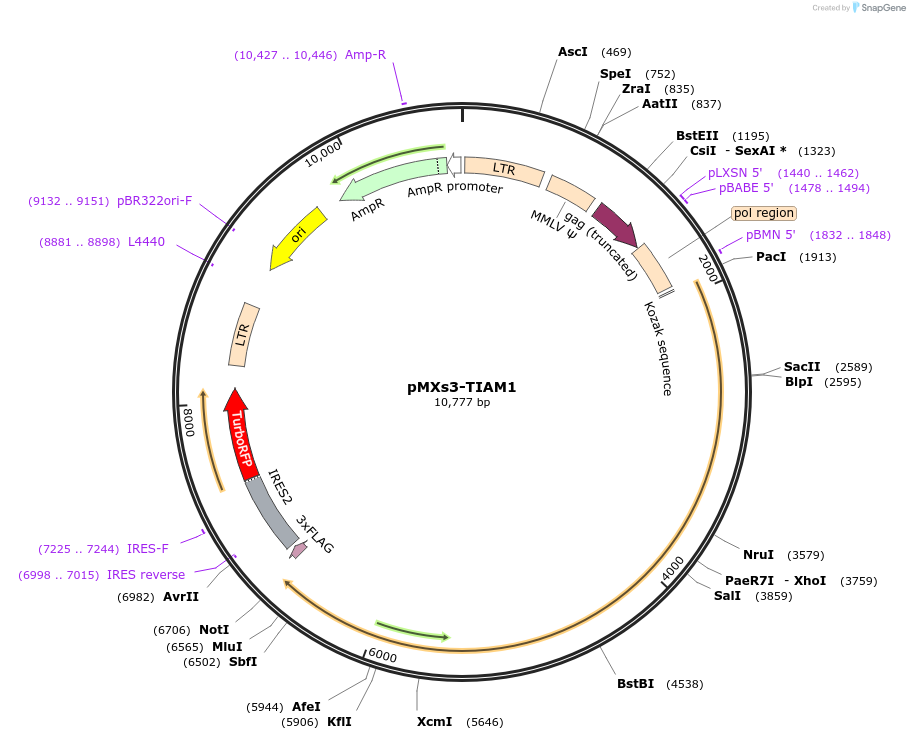
pMXs3-TIAM1
Plasmid#86143PurposeRetroviral vector expressing TIAM1-IRES-RFPDepositorAvailable SinceFeb. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
TDP43 NOTAG2
Plasmid#28207DepositorAvailable SinceAug. 17, 2011AvailabilityAcademic Institutions and Nonprofits only -
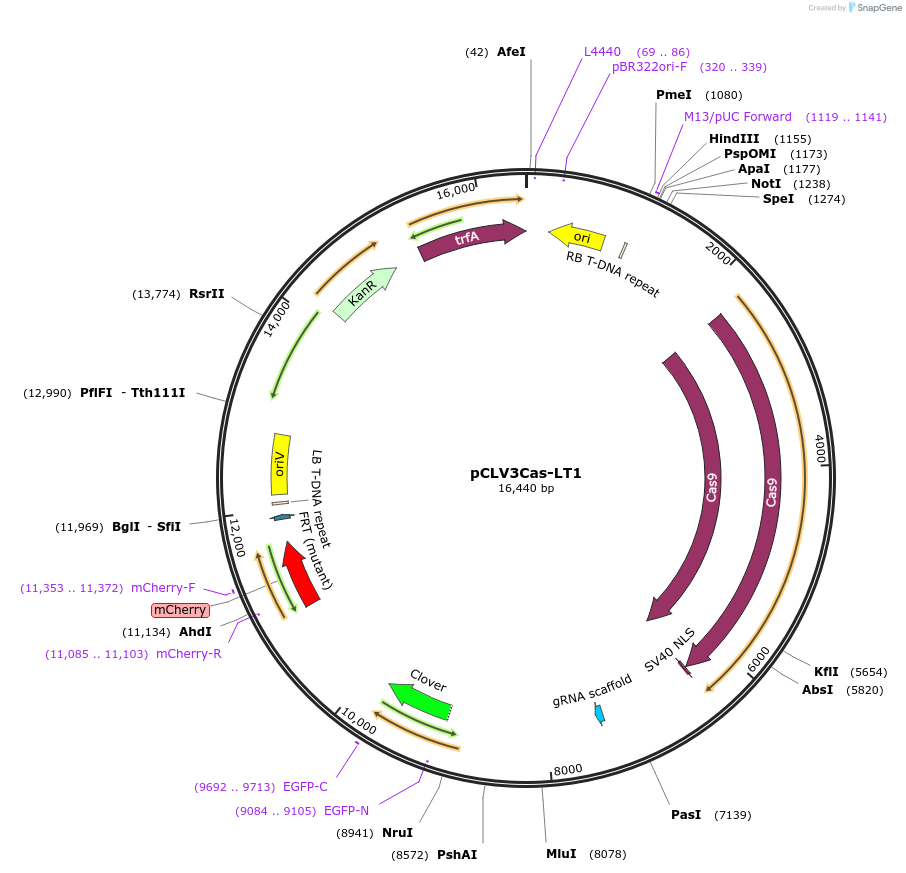
pCLV3Cas-LT1
Plasmid#210549PurposeBinary vector expressing the Lineage Tracing system. Cas9 expression driven by CLV3 promoter. LT1 reporterDepositorInsertCLV3p::spCas9::PEA3At::U6p::AAVS1gRNA::pHTR5::LT1::mCherrySSM
ExpressionPlantMutation2bp insertion after AAVS1 target sequence that di…Available SinceMarch 25, 2024AvailabilityAcademic Institutions and Nonprofits only -
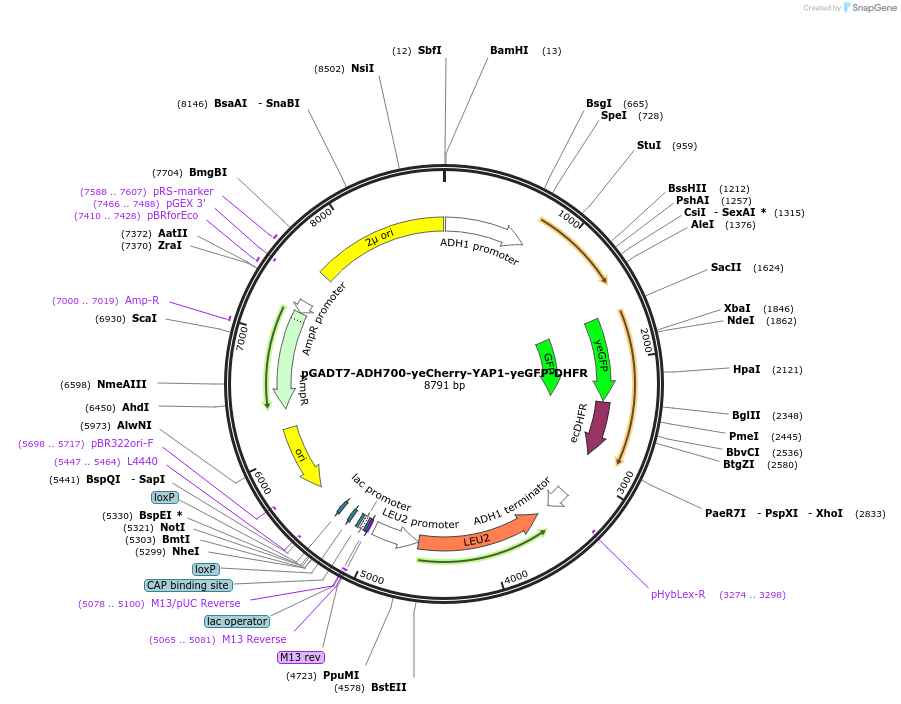
pGADT7-ADH700-yeCherry-YAP1-yeGFP-DHFR
Plasmid#24549DepositorInsertpADH1 yeCherry::YAP1 yeGFP-DHFR (YAP1 Budding Yeast)
ExpressionYeastMutation1. Truncated version of ADH700 promoter (SbfI/Spe…Available SinceMay 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
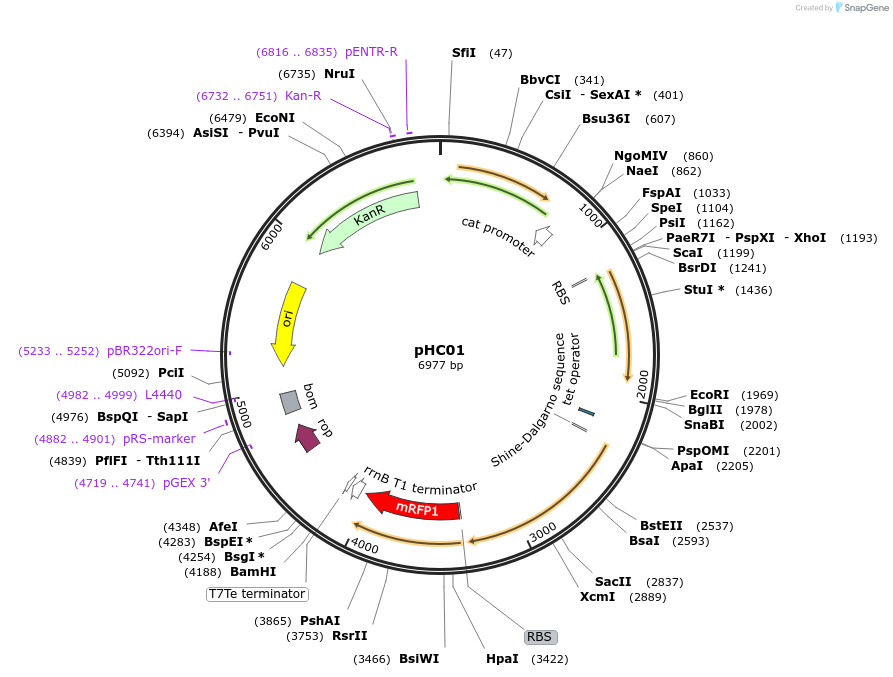
pHC01
Plasmid#106900PurposeRatiometric gas biosensor plasmid for 3-oxo-C12-HSL sensingDepositorInsertsLasR
Methyl Halide Transferase
Ethylene Forming Enzyme
Red Fluorescent Protein
ExpressionBacterialAvailable SinceMarch 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
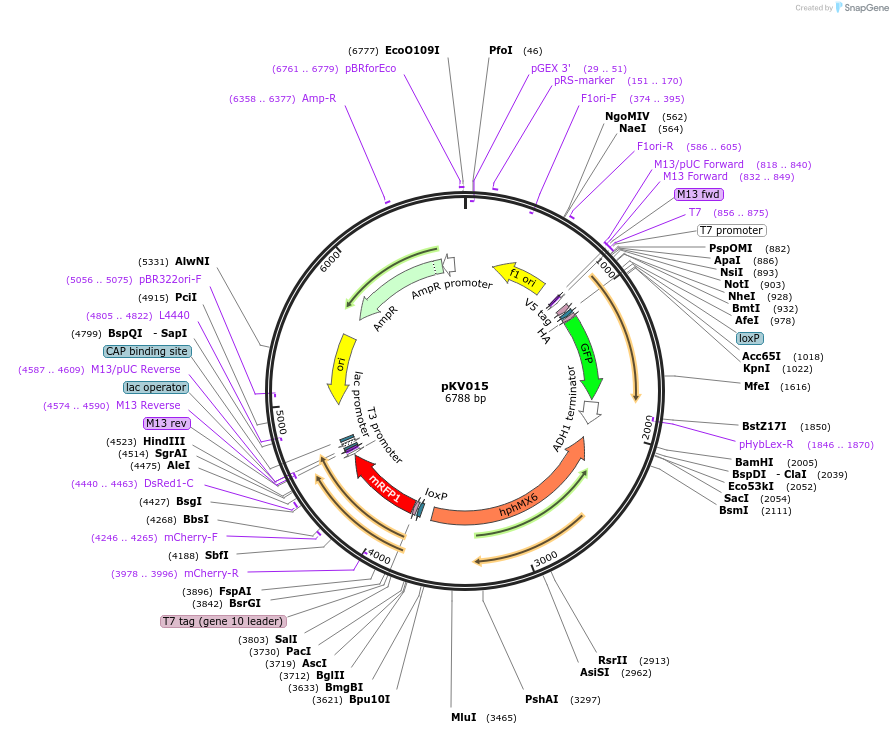
pKV015
Plasmid#64766PurposeRecombination-Induced Tag Exchange (RITE) Cassette, HA-GFP to a T7-RFP tag, invariable V5 tag, Hygro selectionDepositorInsertRITE cassette
UseRite cassettePromoternoneAvailable SinceJuly 6, 2015AvailabilityAcademic Institutions and Nonprofits only -
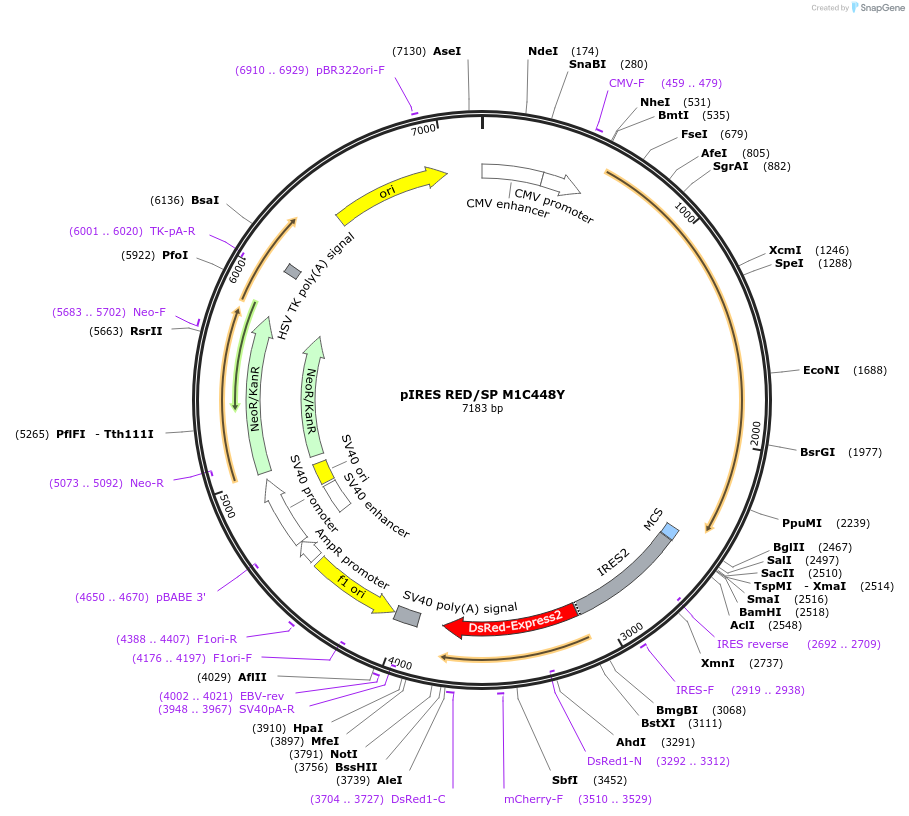
pIRES RED/SP M1C448Y
Plasmid#89323PurposeExpression of RFP and M1C448Y spastin in mammalian cellsDepositorAvailable SinceApril 11, 2017AvailabilityAcademic Institutions and Nonprofits only -
TDP43 NOTAG6
Plasmid#28209DepositorAvailable SinceSept. 8, 2011AvailabilityAcademic Institutions and Nonprofits only -
TDP43 NOTAG3
Plasmid#28208DepositorAvailable SinceSept. 8, 2011AvailabilityAcademic Institutions and Nonprofits only -
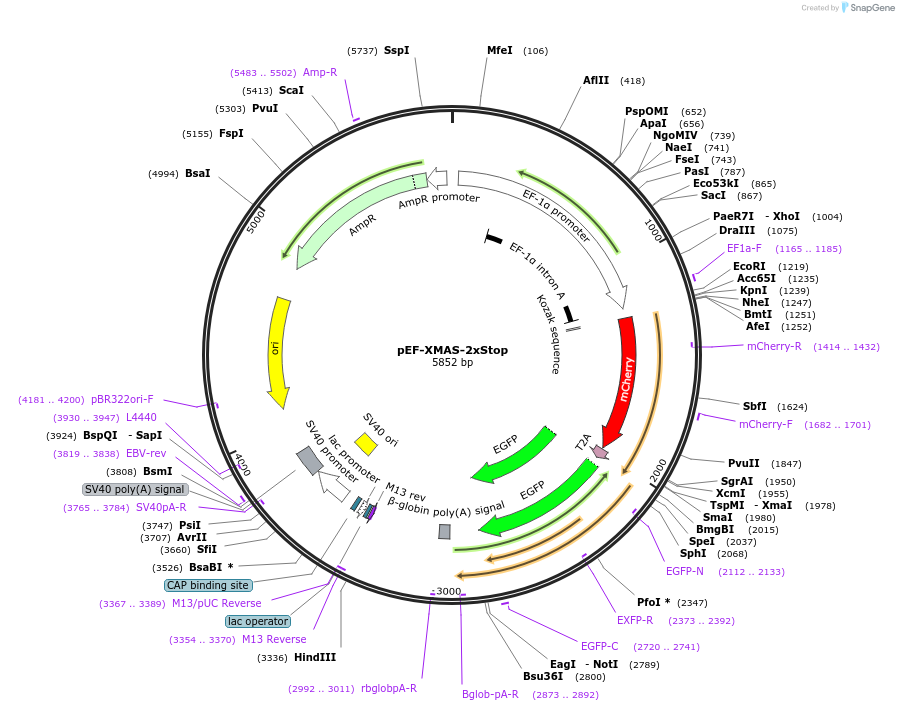
pEF-XMAS-2xStop
Plasmid#164412PurposeTransient Reporter for Editing Enrichment (TREE) Plasmid. GFP turns on with adenosine base editing (ABE). 2 stop codons.DepositorInsertmCherry
UseCRISPR and Synthetic BiologyTagsT2A-GFPExpressionMammalianPromoterEF1-alphaAvailable SinceAug. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
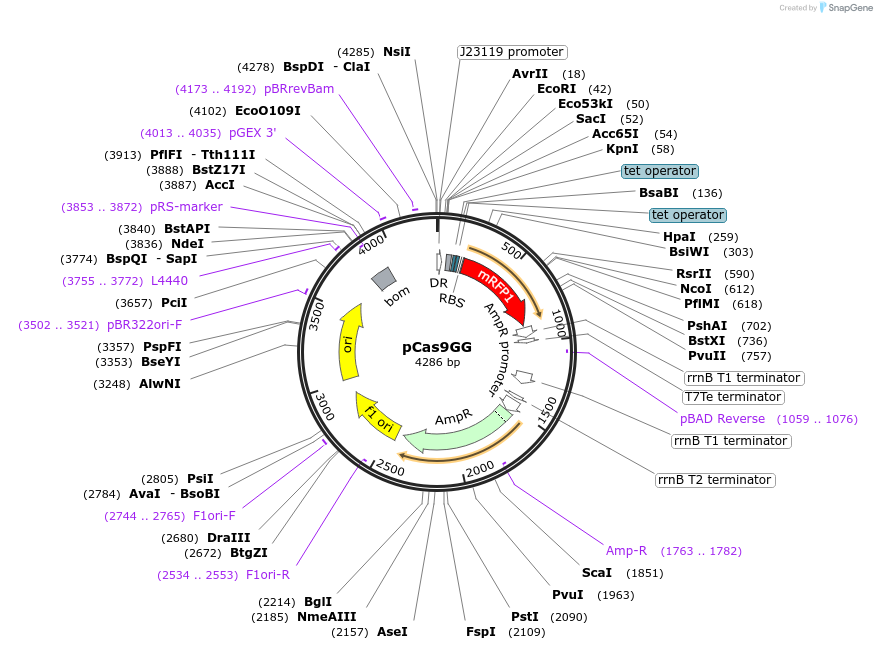
pCas9GG
Plasmid#131009PurposeBackbone plasmid for generating CRISPR arrays for SpCas9 using CRATES. Contains a direct repeat and a RFP-dropout cassette.DepositorInsertsdirect repeat of SpCas9
promoter PJ23119
mRFP expression cassette
UseCRISPRExpressionBacterialPromoterBba_R0040 TetRAvailable SinceSept. 18, 2019AvailabilityAcademic Institutions and Nonprofits only