We narrowed to 35,392 results for: CaS;
-
Plasmid#52140PurposeEncodes module 1 with amino acids 12S-16E for recognition of nt 8GDepositorInsertPumilio homology domain amino acids 25-60 (PUM1 Human)
ExpressionBacterialMutationChanged 16Q to 16E in Module 1Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-2SE
Plasmid#52144PurposeEncodes module 2 with amino acids 12S-16E for recognition of nt 7GDepositorInsertPumilio homology domain amino acids 61-96 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 2Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-3SE
Plasmid#52148PurposeEncodes module 3 with amino acids 12S-16E for recognition of nt 6GDepositorInsertPumilio homology domain amino acids 97-132 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16E in Module 3Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SYR
Plasmid#52158PurposeEncodes module 4 with amino acids 12S-13Y-16R for recognition of nt 5CDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S, 13H to 13Y, and 16Q to 16R in…Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5SR
Plasmid#52162PurposeEncodes module 5 with amino acids 12S-16R for recognition of nt 4CDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16R in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6SE
Plasmid#52164PurposeEncodes module 6 with amino acids 12S-16E for recognition of nt 3GDepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
mCerulean3-Vimentin-7
Plasmid#55452PurposeLocalization: Intermediate Filaments, Excitation: 433, Emission: 475DepositorAvailable SinceDec. 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pZ004 (LseC2c2 locus)
Plasmid#70170PurposeExpresses LseC2c2 locus with one spacer in arrayDepositorInsertLseC2c2 locus with one spacer in array
ExpressionBacterialAvailable SinceMay 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
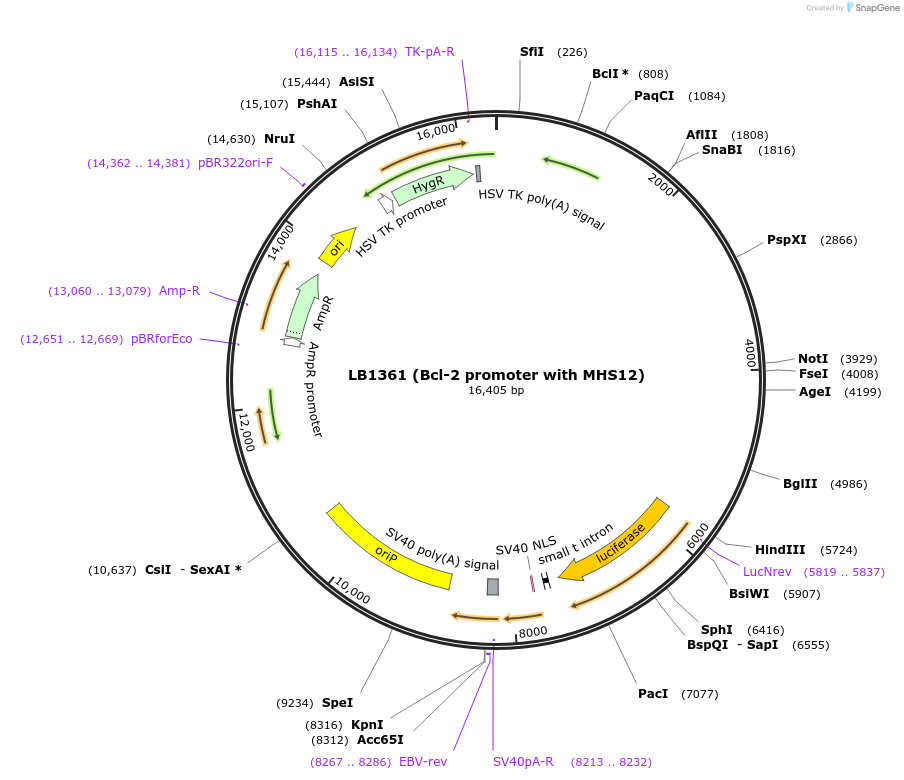
LB1361 (Bcl-2 promoter with MHS12)
Plasmid#15391DepositorInsertB-cell lymphoma/leukemia-2 (BCL2 Human)
UseLuciferaseTagsBcl-2-LuciferaseExpressionBacterial and MammalianMutationBcl-2 full promoter with MHS12Available SinceAug. 1, 2007AvailabilityAcademic Institutions and Nonprofits only -
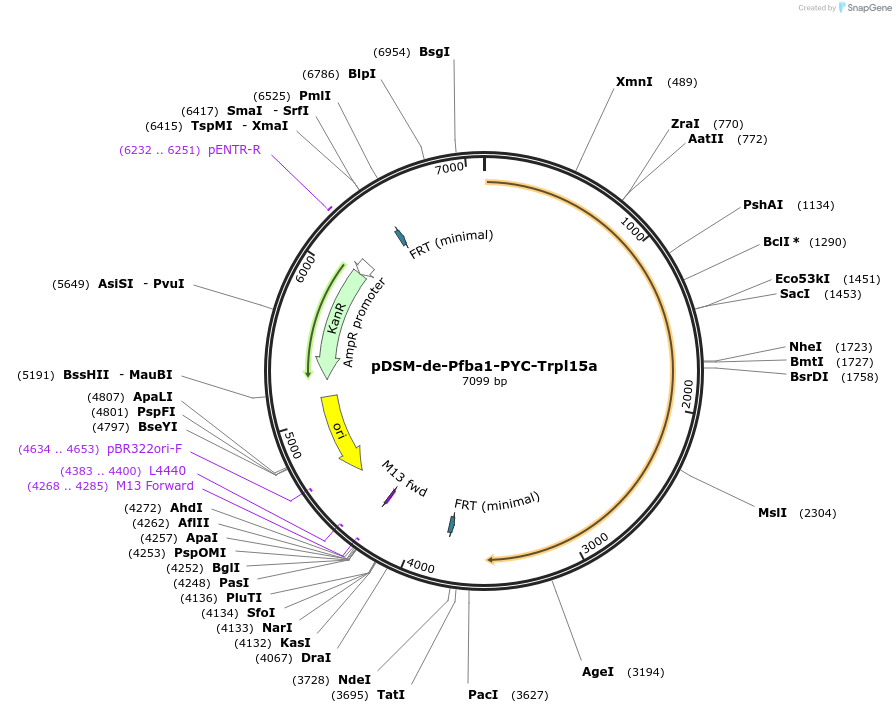
pDSM-de-Pfba1-PYC-Trpl15a
Plasmid#127738PurposeYeast pathway position 5. PYC transcription unit with the FBA1 promoter and RPL15A terminator.DepositorInsertpyruvate carboxylase (PYC1 Synthetic, Budding Yeast)
UseSynthetic BiologyExpressionYeastPromoterPfba1Available SinceJan. 15, 2022AvailabilityAcademic Institutions and Nonprofits only -
pCMV10/3xflag-Clock L57E
Plasmid#47337PurposeMutation used for co-IPsDepositorAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pCMV10/3xflag-Clock L74E
Plasmid#47338PurposeMutation used for co-IPsDepositorAvailable SinceSept. 4, 2013AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SE
Plasmid#52152PurposeEncodes module 4 with amino acids 12S-16E for recognition of nt 5GDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16E in Module 4Available SinceMay 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-8SR
Plasmid#52174PurposeEncodes module 8 with amino acids 12S-16R for recognition of nt 1CDepositorInsertPumilio homology domain amino acids 284-319 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 8Available SinceMay 23, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-1SR
Plasmid#52142PurposeEncodes module 1 with amino acids 12S-16R for recognition of nt 8CDepositorInsertPumilio homology domain amino acids 25-60 (PUM1 Human)
ExpressionBacterialMutationChanged 16Q to 16R in Module 1Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7CQ
Plasmid#52167PurposeEncodes module 7 with amino acids 12C-16Q for recognition of nt 2ADepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12C and 16E to 16Q in Module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7NQ
Plasmid#52169PurposeEncodes module 7 with amino acids 12N-16Q for recognition of nt 2UDepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12N and 16E to 16Q in module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-7SYR
Plasmid#52170PurposeEncodes module 7 with amino acids 12S-13Y-16R for recognition of nt 2CDepositorInsertPumilio homology domain amino acids 241-283 (PUM1 Human)
ExpressionBacterialMutationChanged 13N to 13Y and 16E to 16R in module 7Available SinceMay 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-1NQ
Plasmid#52141PurposeEncodes module 1 with amino acids 12N-16Q for recognition of nt 8UDepositorInsertPumilio homology domain amino acids 25-60 (PUM1 Human)
ExpressionBacterialMutationChanged 12S to 12N in Module 1Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-2CQ
Plasmid#52143PurposeEncodes module 2 with amino acids 12C-16Q for recognition of nt 7ADepositorInsertPumilio homology domain amino acids 61-96 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C in Module 2Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-2SR
Plasmid#52146PurposeEncodes module 2 with amino acids 12S-16R for recognition of nt 7CDepositorInsertPumilio homology domain amino acids 61-96 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 2Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-3NQ
Plasmid#52149PurposeEncodes module 3 with amino acids 12N-16Q for recognition of nt 6UDepositorInsertPumilio homology domain amino acids 97-132 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12N in Module 3Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-3SR
Plasmid#52150PurposeEncodes module 3 with amino acids 12S-16R for recognition of nt 6CDepositorInsertPumilio homology domain amino acids 97-132 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16R in Module 3Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4CQ
Plasmid#52151PurposeEncodes module 4 with amino acids 12C-16Q for recognition of nt 5ADepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SR
Plasmid#52154PurposeEncodes module 4 with amino acids 12S-16R for recognition of nt 5CDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4CYQ
Plasmid#52155PurposeEncodes module 4 with amino acids 12C-13Y-16Q for recognition of nt 5ADepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C and 13H to 13Y in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4SYE
Plasmid#52156PurposeEncodes module 4 with amino acids 12S-13Y-16E for recognition of nt 5GDepositorInsertD43951 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S, 13H to 13Y, and 16Q to 16E in…Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-4NYQ
Plasmid#52157PurposeEncodes module 4 with amino acids 12N-13Y-16Q for recognition of nt 5UDepositorInsertPumilio homology domain amino acids 133-168 (PUM1 Human)
ExpressionBacterialMutationChanged 13H to 13Y in Module 4Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5SE
Plasmid#52160PurposeEncodes module 5 with amino acids 12S-16E for recognition of nt 4GDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12S and 16Q to 16E in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-5NQ
Plasmid#52161PurposeEncodes module 5 with amino acids 12N-16Q for recognition of nt 4UDepositorInsertPumilio homology domain amino acids 169-204 (PUM1 Human)
ExpressionBacterialMutationChanged 12C to 12N in Module 5Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6CQ
Plasmid#52163PurposeEncodes module 6 with amino acids 12C-16Q for recognition of nt 3ADepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12C in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
pChlr-6SR
Plasmid#52166PurposeEncodes module 6 with amino acids 12S-16R for recognition of nt 3CDepositorInsertPumilio homology domain amino acids 205-240 (PUM1 Human)
ExpressionBacterialMutationChanged 12N to 12S and 16Q to 16R in Module 6Available SinceApril 28, 2014AvailabilityAcademic Institutions and Nonprofits only -
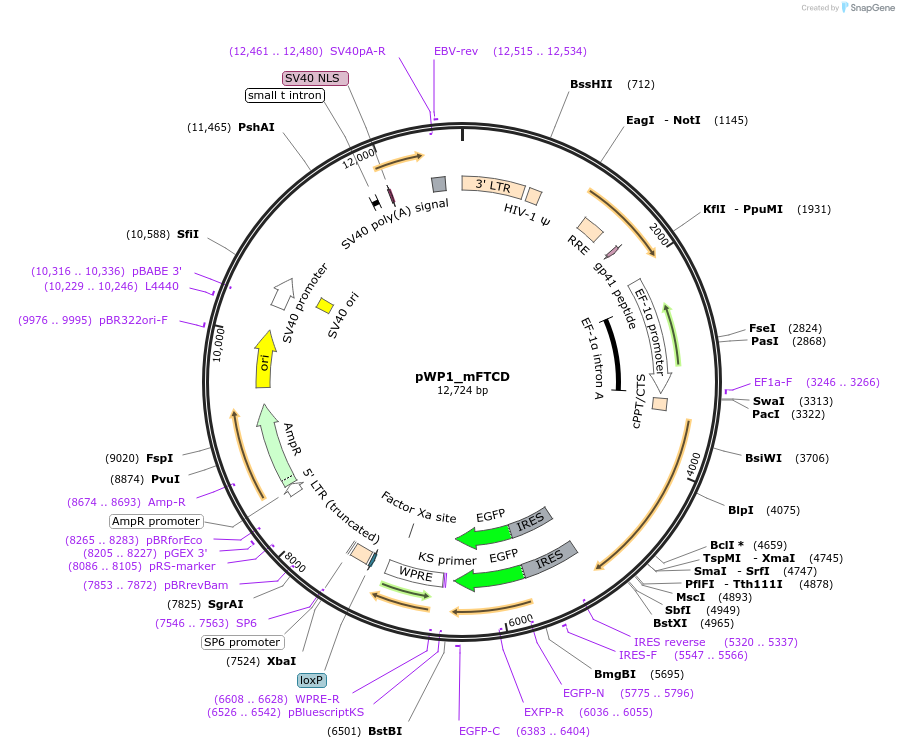
pWP1_mFTCD
Plasmid#102317Purposeexpression of mouse cDNA of FTCD, refractory to the guides in plasmids 1,2DepositorAvailable SinceJuly 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
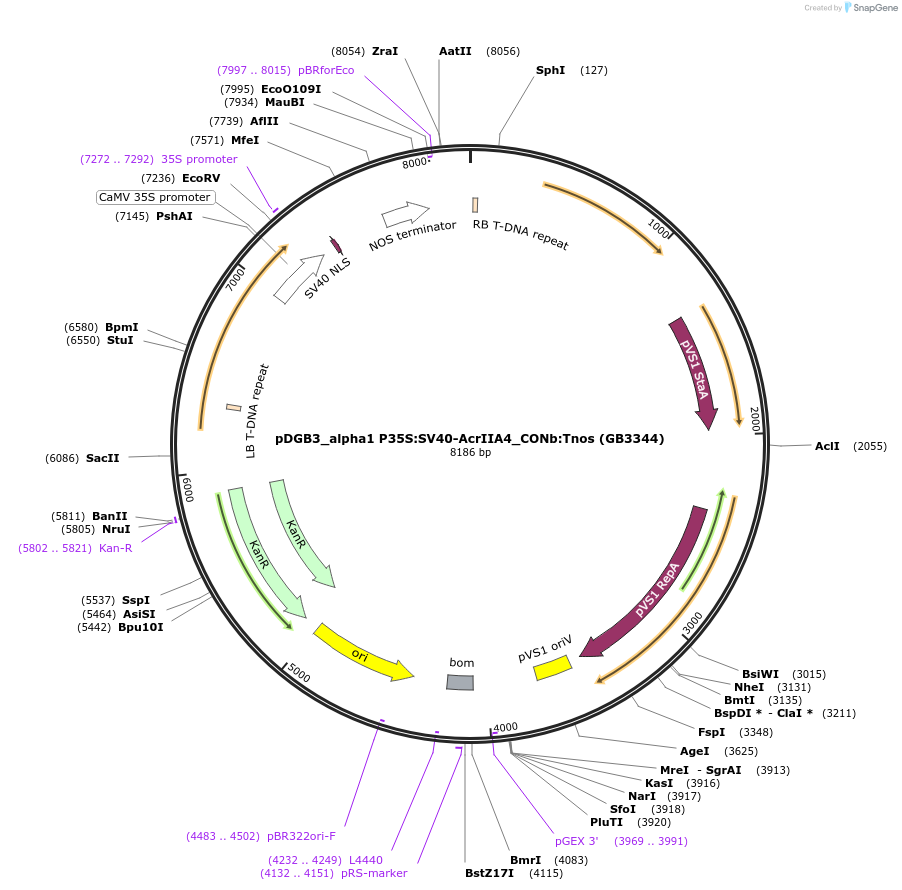
pDGB3_alpha1 P35S:SV40-AcrIIA4_CONb:Tnos (GB3344)
Plasmid#160635PurposeTU for SV40-AcrIIA4 protein Codon optimized for N. benthamiana expression under the regulation of the 35S promoter and Tnos terminatorDepositorInsertAcrIIA4
UseSynthetic BiologyExpressionPlantMutationBsaI and BsmBI sites removedPromoter35SAvailable SinceSept. 20, 2021AvailabilityAcademic Institutions and Nonprofits only -
pCMV10/3xflag-Clock V315R
Plasmid#47342PurposeMutation used for co-IPsDepositorAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
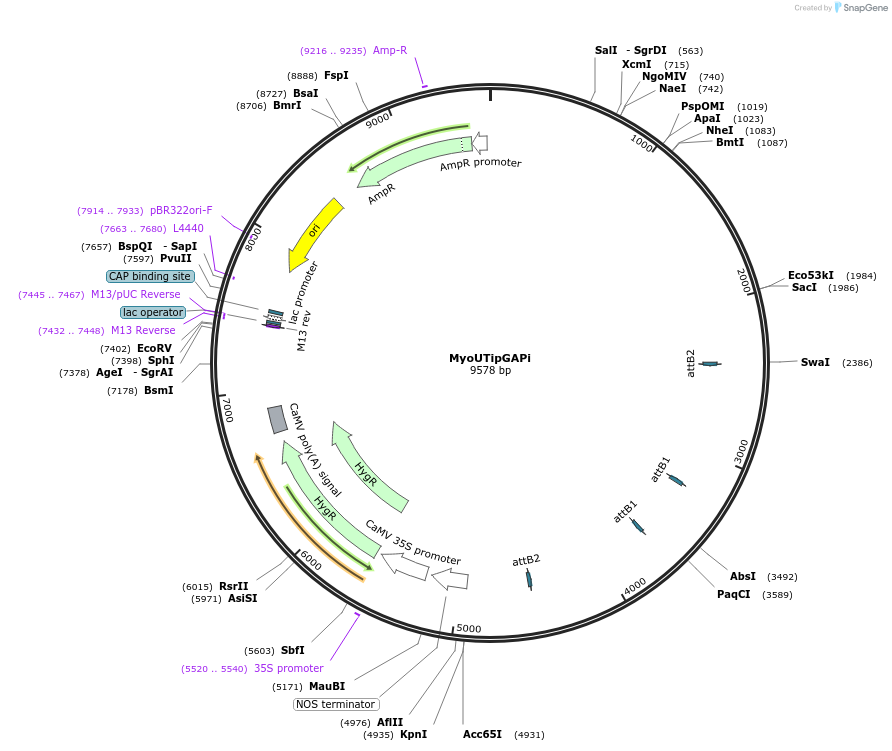
MyoUTipGAPi
Plasmid#127549PurposePositive RNAi control for pGAPi-based RNAi silencing in tandem with APT transcript segmentDepositorInsertMyosin XIa+b 5'UTR and Adenine Phosphorybosyltransferase transcript segment
UseRNAi; Positive controlPromoterUbiquitinAvailable SinceAug. 5, 2020AvailabilityAcademic Institutions and Nonprofits only -
pCMV10/3xflag-Clock W284A
Plasmid#47341PurposeMutation used for co-IPsDepositorAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
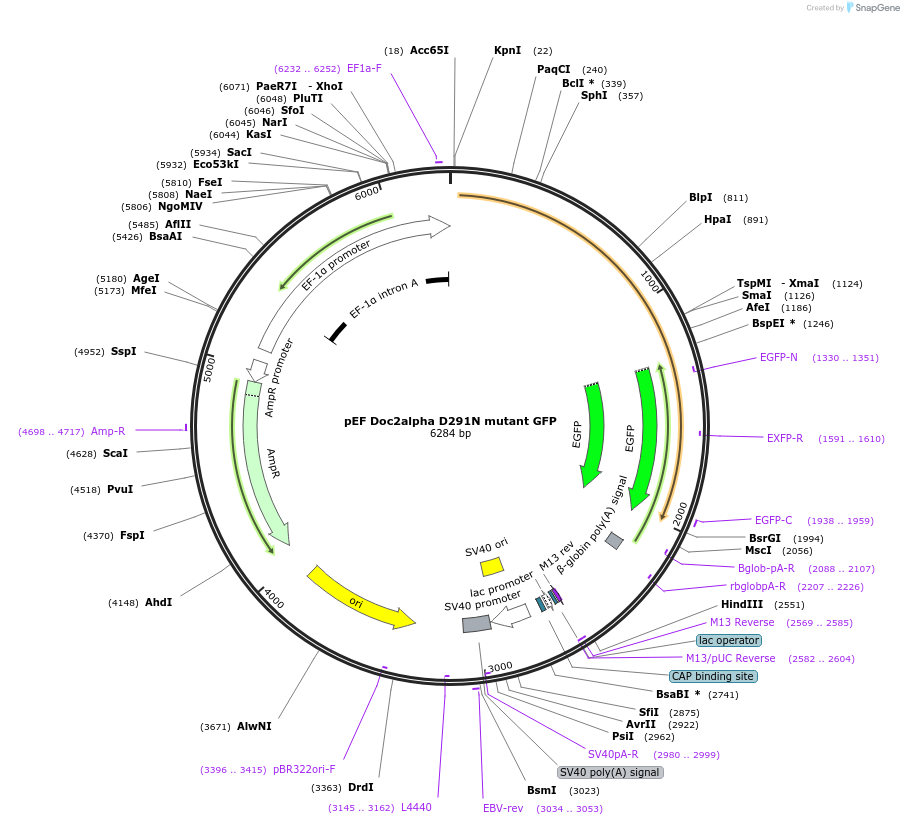
pEF Doc2alpha D291N mutant GFP
Plasmid#128821PurposeTo visualize Doc2alpha D291N mutant in PC12 cellsDepositorAvailable SinceNov. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
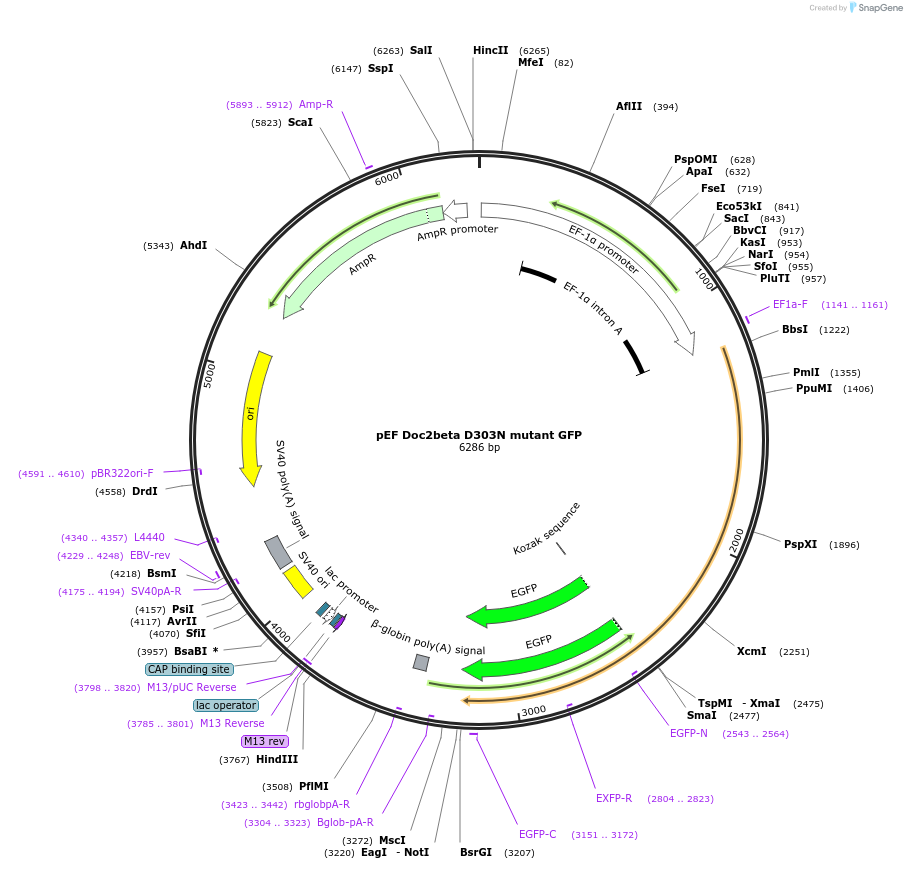
pEF Doc2beta D303N mutant GFP
Plasmid#128822PurposeTo visualize Doc2beta D303N mutant in PC12 cellsDepositorAvailable SinceNov. 9, 2020AvailabilityAcademic Institutions and Nonprofits only -
pET19b-MED29
Plasmid#15415DepositorAvailable SinceAug. 10, 2007AvailabilityAcademic Institutions and Nonprofits only -
mCerulean3-H2B-C-10
Plasmid#55422PurposeLocalization: Nucleus/Histones, Excitation: 433, Emission: 475DepositorInsertH2B (H2BC11 Human)
TagsmCerulean3ExpressionMammalianMutationD26G and V119I in H2BPromoterCMVAvailable SinceDec. 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
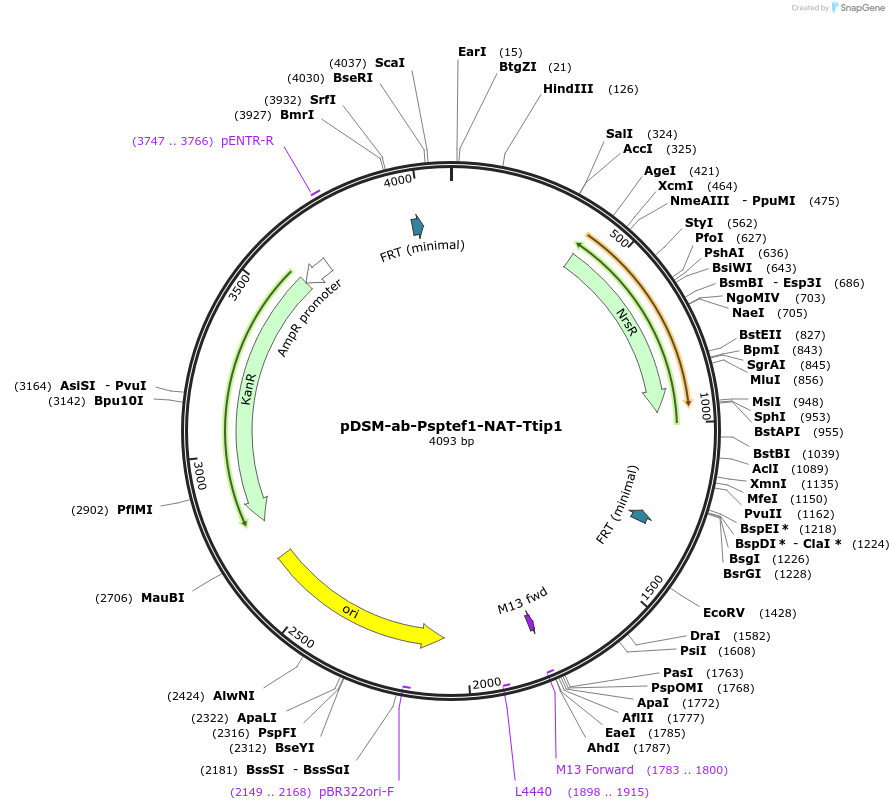
pDSM-ab-Psptef1-NAT-Ttip1
Plasmid#127747PurposeYeast pathway position 2. NatMX transcription unit with the S. paradoxus TEF1 promoter and TIP1 terminator. Nourseothricin resistance.DepositorInsertnourseothricin N-acetyltransferase
UseSynthetic BiologyExpressionYeastPromoterPsptef1Available SinceNov. 13, 2019AvailabilityAcademic Institutions and Nonprofits only -
pET24a_UDP_Xase_E113
Plasmid#73159PurposeExpresses in E. coli, S. pneumoniae TIGR4 UDP-Xase SP_1669 mutant E113ADepositorInsertpET24a
ExpressionBacterialMutationchange glutamate 113 to alanineAvailable SinceMarch 22, 2016AvailabilityAcademic Institutions and Nonprofits only -
XE125 XDsh delta COOH-GFP-CS2+
Plasmid#16784DepositorInsertXdsh-delta-COOH (dvl2.L Frog)
UseXenopus expressionTagsGFPMutationcontains Dsh amino acids 1-588 (deleting 599-736)Available SinceMarch 24, 2008AvailabilityAcademic Institutions and Nonprofits only -
pRKVSV Rac T17N
Plasmid#50537DepositorInsertsmall GTPase, mutated (RAC1 Human)
Tags2X VSVExpressionMammalianMutationT17N (dominant negative)PromoterCMVAvailable SinceJan. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
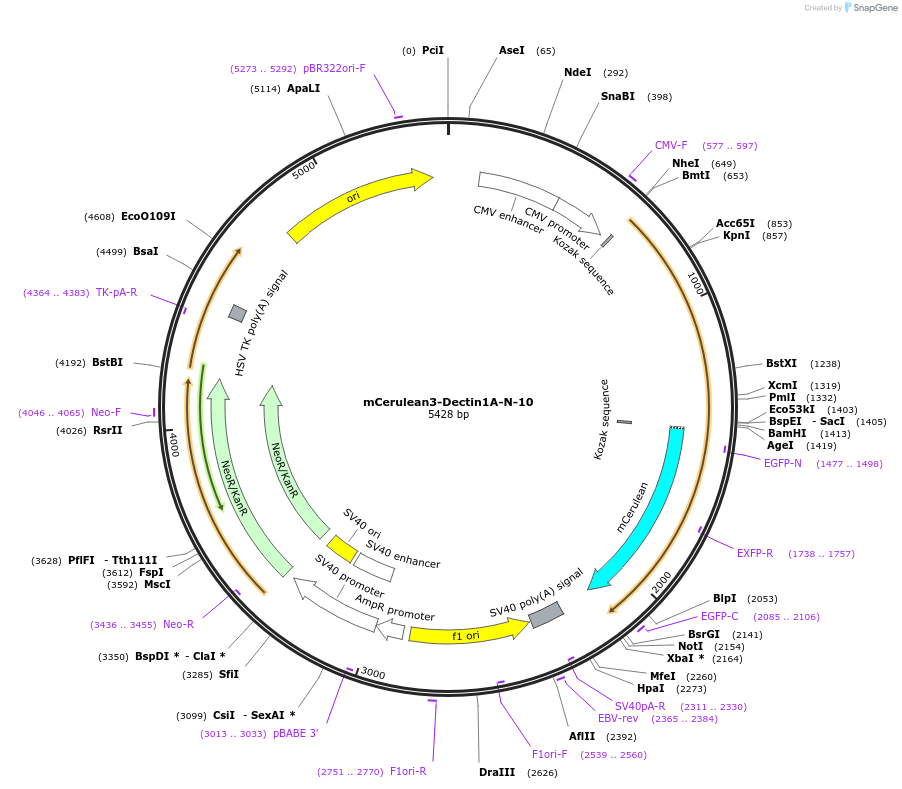
mCerulean3-Dectin1A-N-10
Plasmid#55414PurposeLocalization: Membrane, Excitation: 433, Emission: 475DepositorAvailable SinceOct. 7, 2014AvailabilityAcademic Institutions and Nonprofits only -
mCerulean3-Dectin1A-C-10
Plasmid#55413PurposeLocalization: Membrane, Excitation: 433, Emission: 475DepositorAvailable SinceOct. 10, 2014AvailabilityAcademic Institutions and Nonprofits only -
FLAG-SpEAF 60-251 pBacPAK8
Plasmid#15321DepositorInsertEAF1 (eaf1 Fission Yeast)
UseBaculovirus constructionTagsFLAGMutationN terminal deletion lacking first 59 amino acids …Available SinceAug. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
FLAG-ELL 1-373 pBacPAK8
Plasmid#15328DepositorInserteleven nineteen lysine rich in leukemia (ELL Human)
UseBaculovirus constructionTagsFLAGMutationAmino acids 1-373Available SinceAug. 15, 2007AvailabilityAcademic Institutions and Nonprofits only -
FLAG-ELL 45-621 pBacPAK8
Plasmid#15329DepositorInserteleven nineteen lysine rich in leukemia (ELL Human)
UseBaculovirus constructionTagsFLAGMutationAmino acids 45-621Available SinceAug. 15, 2007AvailabilityAcademic Institutions and Nonprofits only