We narrowed to 8,645 results for: SET
-
Plasmid#105357PurposeSynthetic biologyDepositorInsertpromotor RPS5a (At3g11940; 1700bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pJOG295
Plasmid#105362PurposeSynthetic biologyDepositorInsertpromotor GILT (At4g12960;2100bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pJOG300
Plasmid#105363PurposeSynthetic biologyDepositorInsertpromotor (At1g66850;350bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
pJOG025
Plasmid#105364PurposeSynthetic biologyDepositorInsertpromotor EDS1 (At3g48090)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
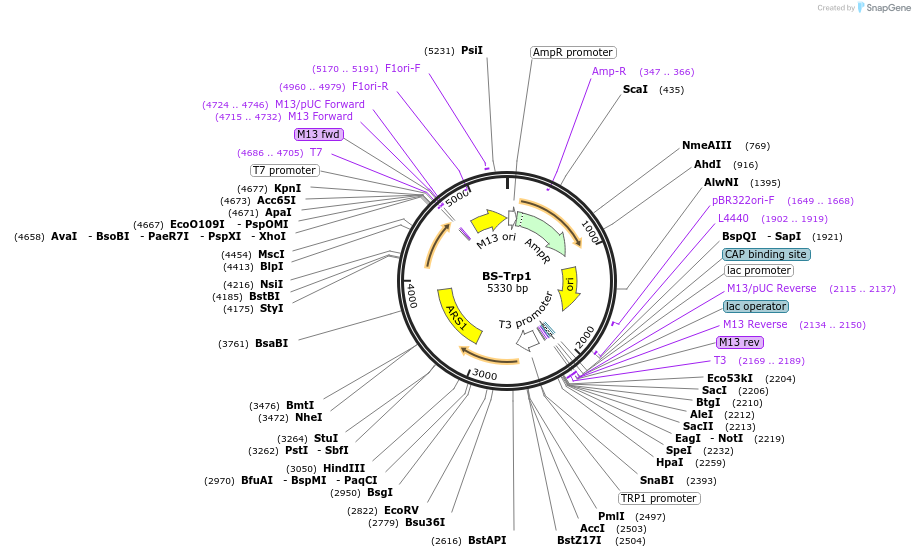
BS-Trp1
Plasmid#69196Purposecomplementation of yeast auxotrophic marker TRP1. Although this is the plasmid described in the paper, the depositor notes that it has a low integration efficiency and Plasmid 69503 should be used.DepositorInsertTRP1
ExpressionYeastAvailable SinceOct. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
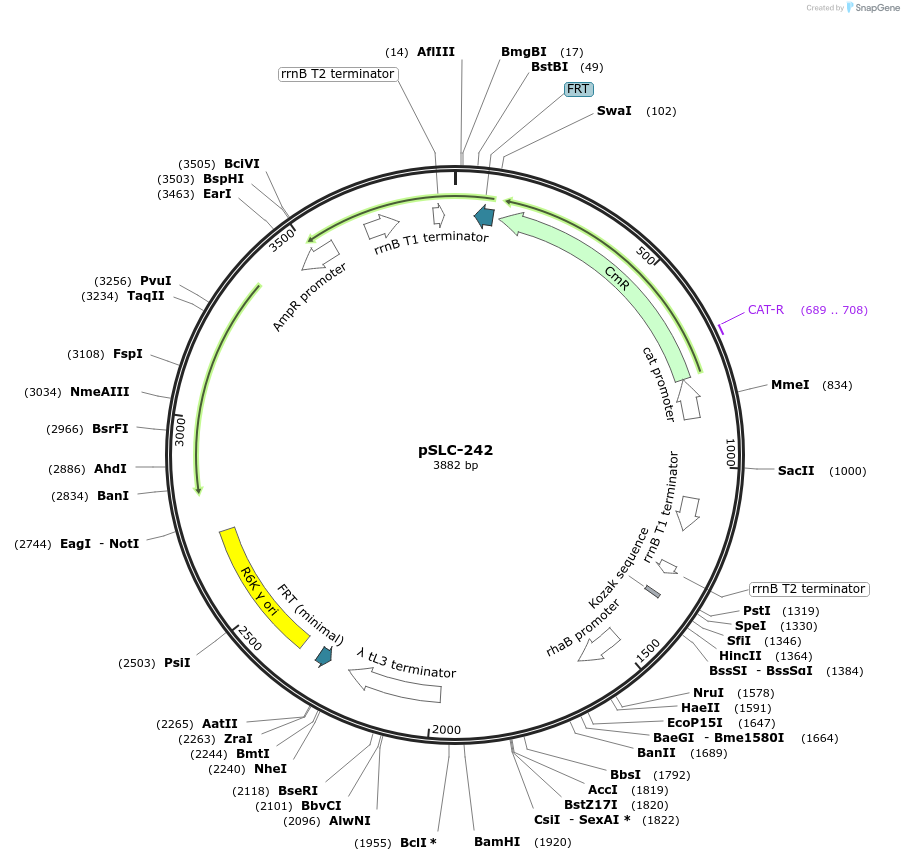
pSLC-242
Plasmid#73189PurposeTemplate plasmid which encodes chloramphenicol resistance gene for positive selection and a toxin gene (relE) under the control of rhamnose induceable promoter (PrhaB) for negative selection.DepositorInsertchloramphenicol (cat Although Ampicillin gene is present, it was inactivated while cloning.)
ExpressionBacterialPromoterchloramphenicol promoterAvailable SinceJuly 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
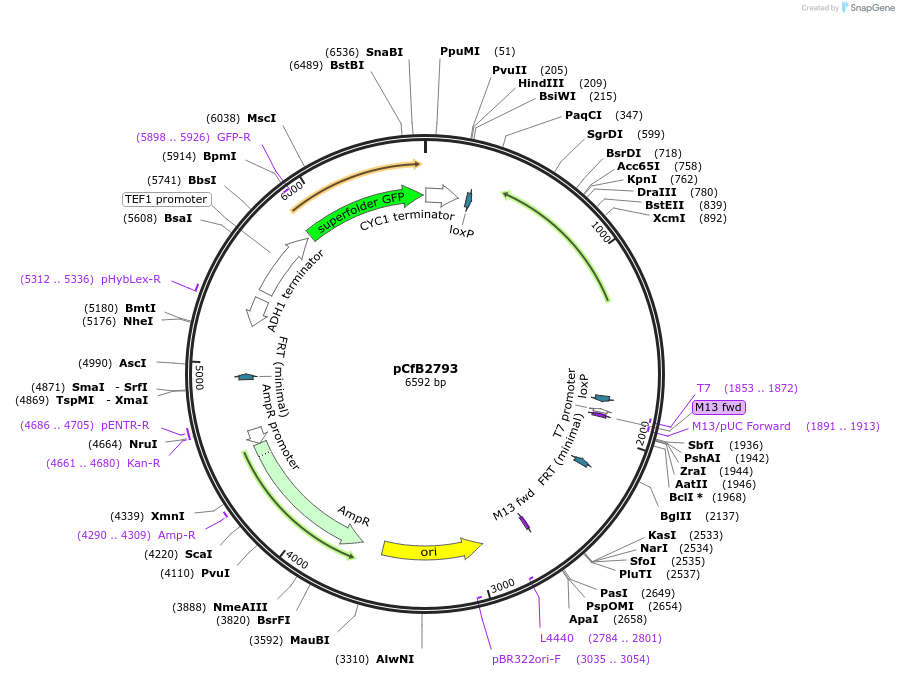
pCfB2793
Plasmid#64043PurposepCfB2793 is a vector for multiple integrations at sites sharing homology with Ty2Cons. The selective marker is Kl.URA3. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoter and noneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
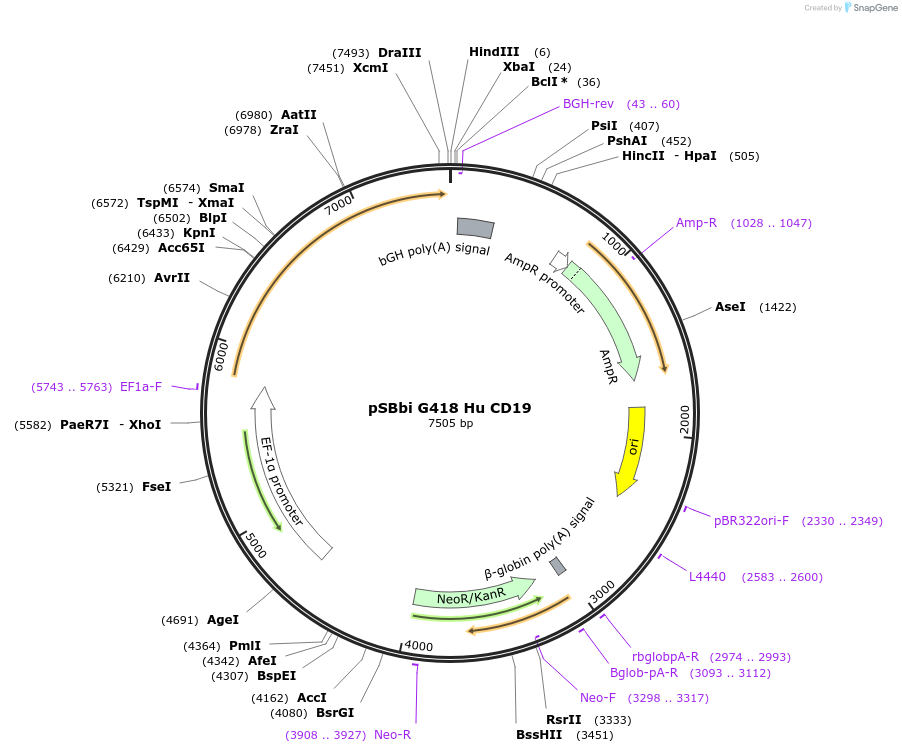
pSBbi G418 Hu CD19
Plasmid#183247PurposeExpression of human CD19 antigen in a Sleeping Beauty transposon vectorDepositorInsertCD19 (CD19 Human)
ExpressionMammalianAvailable SinceOct. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
pSBbi 41BBL CD86 HLA-A201
Plasmid#183248PurposeExpression of human 41BBL, CD86 and HLA-A201 in a Sleeping Beauty transposon vector for use in artificial antigen presenting cellsDepositorInsertsExpressionMammalianAvailable SinceAug. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
ZJOM65
Plasmid#133654PurposeIntegration of VPH1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only -
pCfB2797
Plasmid#63639PurposepCfB2797 is a vector for multiple integrations at sites sharing homology with Ty2Cons. The selective marker is Kl.URA3. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
pCfB2875
Plasmid#63640PurposepCfB2875 is a vector for multiple integrations at sites sharing homology with Ty3Cons. The selective marker is Kl.URA3. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
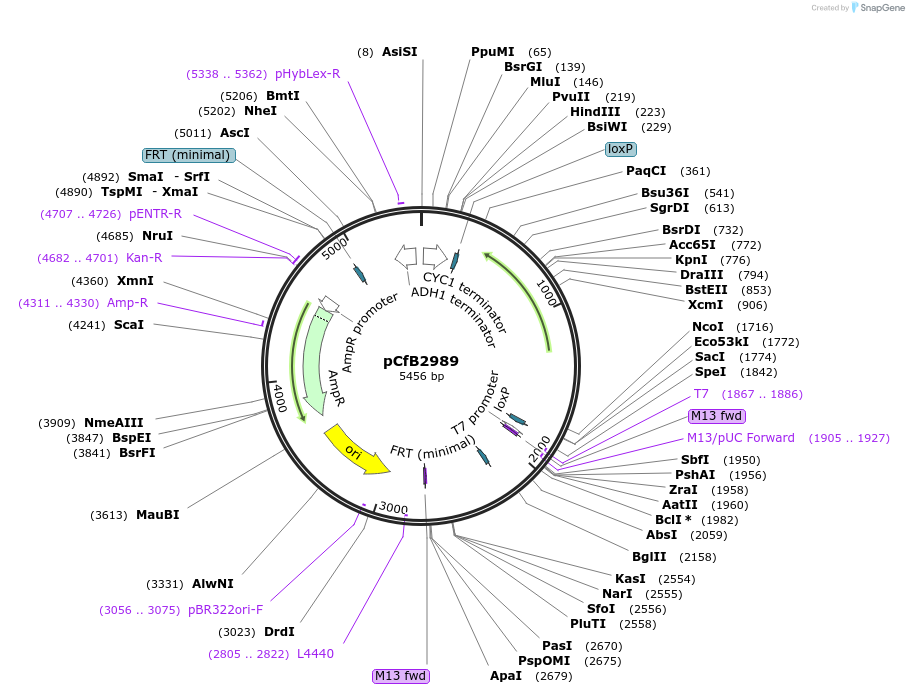
pCfB2989
Plasmid#63636PurposepCfB2989 is a vector for multiple integrations at sites sharing homology with Ty1Cons1. The selective marker is Kl.URA3. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
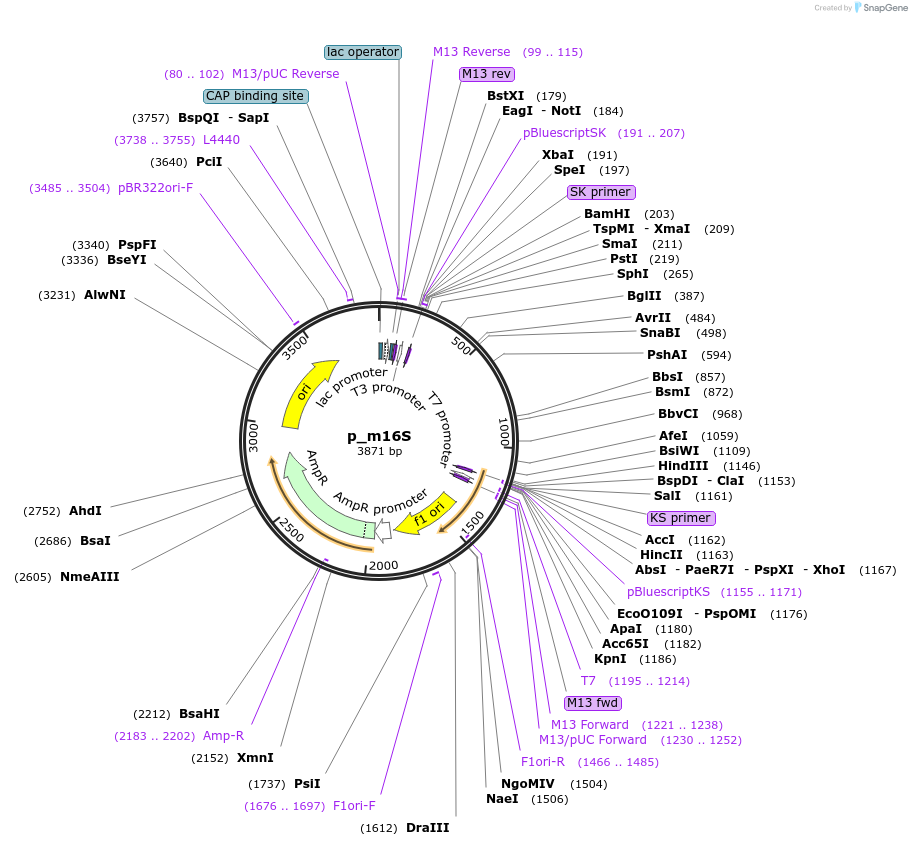
p_m16S
Plasmid#160549PurposeUsed for mycoplasma detection by qPCR. 16S insert truncated internally from the strain Mycoplasma capricolum subsp. capricolum strain California Kid. ATB: CarbenicillineDepositorInsert16S rDNA fragment from M. capricolum subsp. capricolum strain California Kid (gi_83283139)
UseUnspecifiedAvailable SinceDec. 10, 2020AvailabilityAcademic Institutions and Nonprofits only -
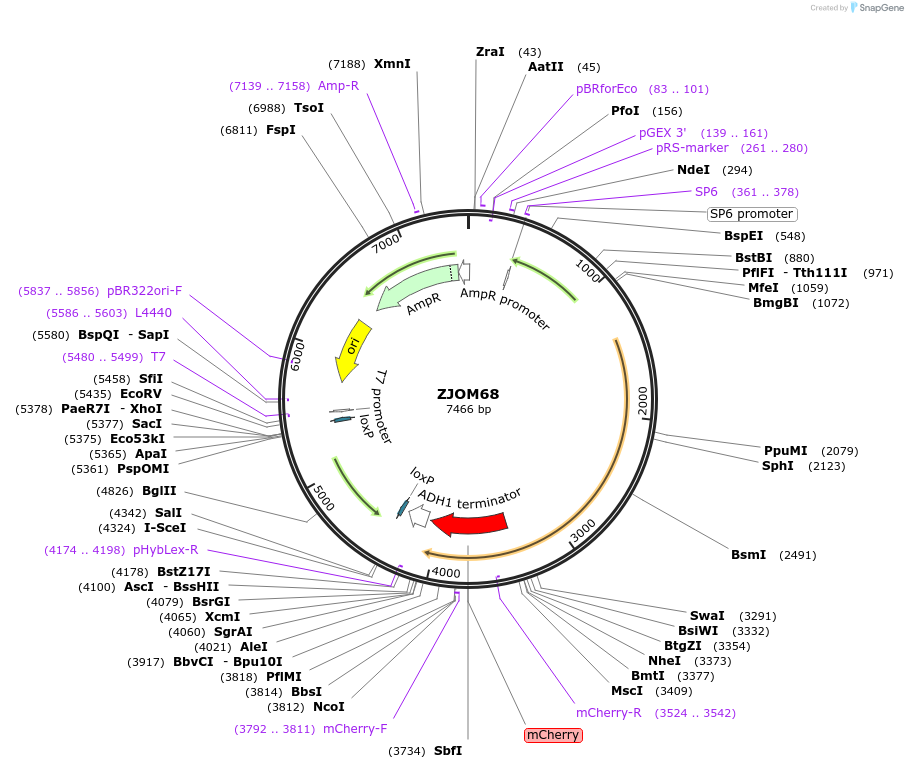
ZJOM68
Plasmid#133657PurposeIntegration of TGL3-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast lipid droplet.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
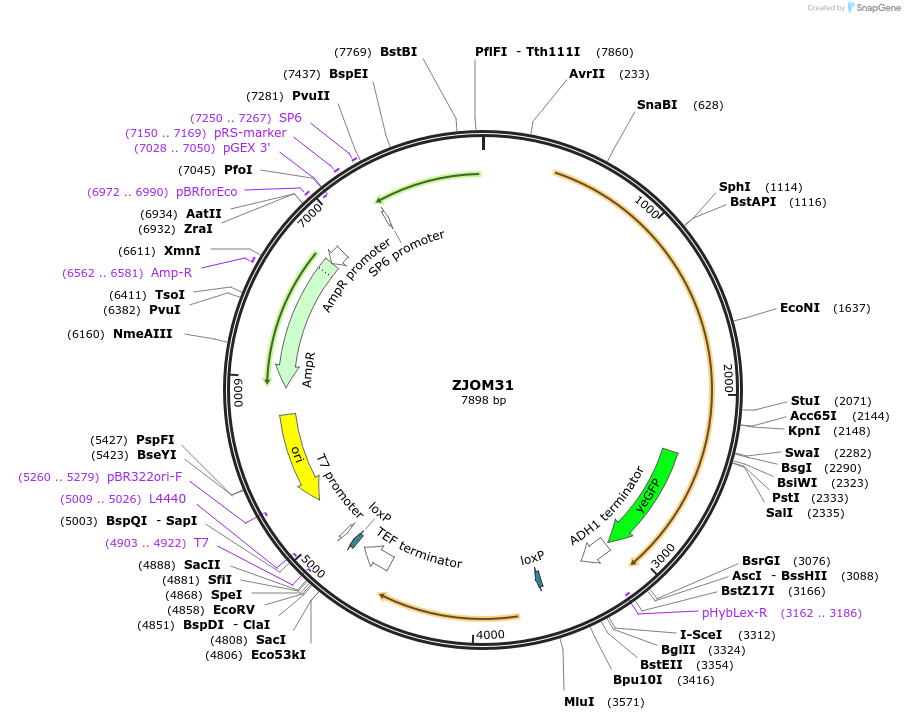
ZJOM31
Plasmid#133645PurposeIntegration of TGL3-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast lipid droplet.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
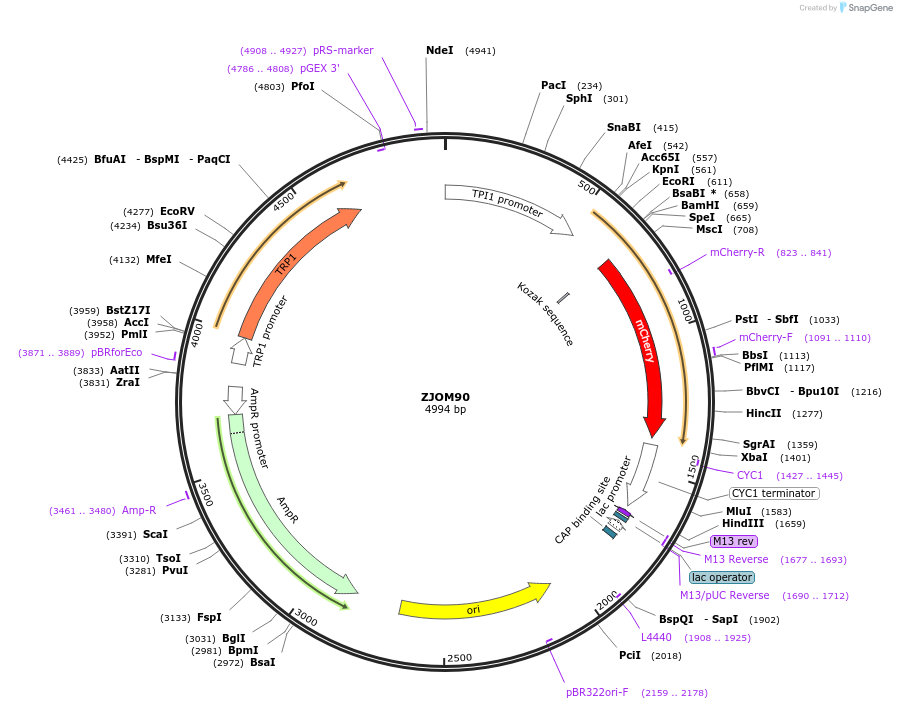
ZJOM90
Plasmid#133647PurposeIntegration of SS-mCherry-HDEL in genome, use auxotrophic marker TRP1(Saccharomyces cerevisiae). Marker for yeast endoplasmic reticulum (ER).DepositorInsertpTPI1-SS-mCherry-HDEL
TagsmCherryExpressionYeastAvailable SinceApril 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
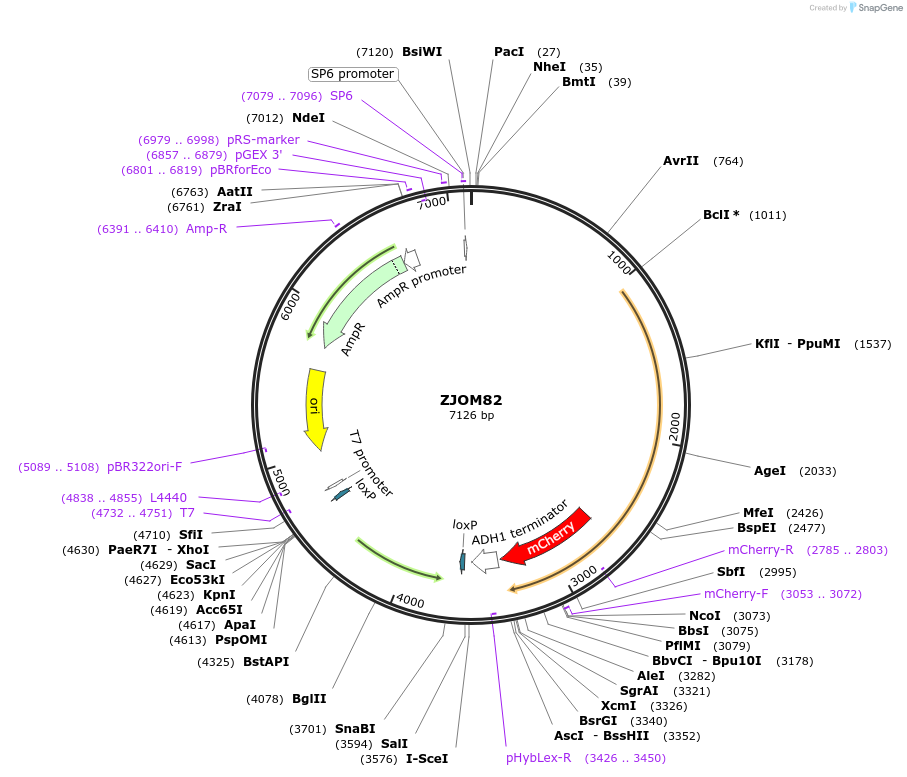
ZJOM82
Plasmid#133648PurposeIntegration of NAB2-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceJan. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
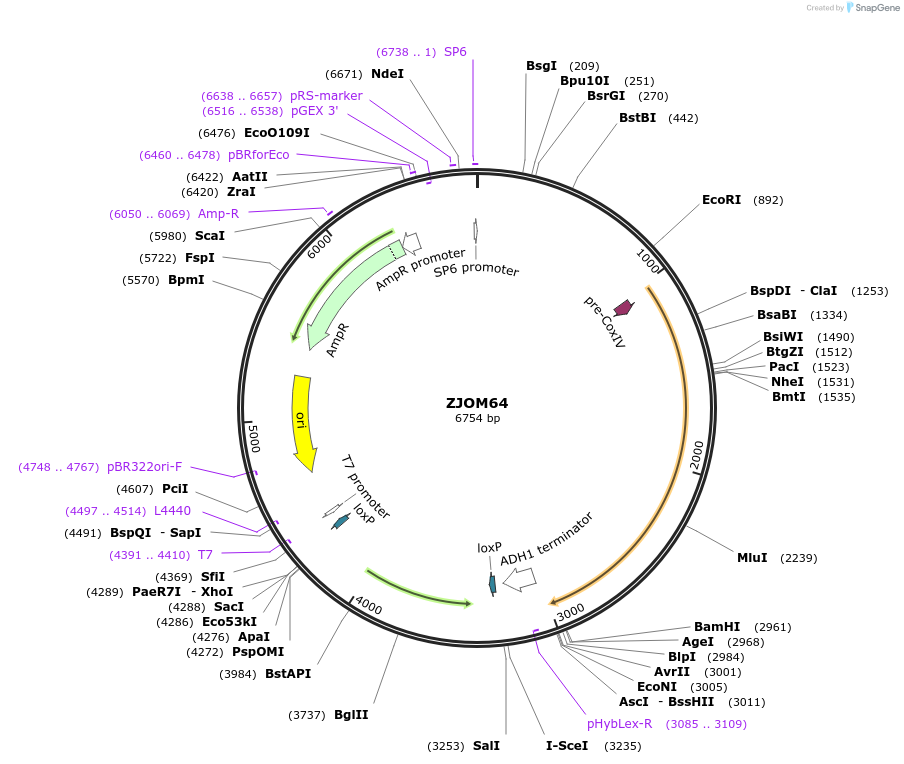
ZJOM64
Plasmid#133655PurposeIntegration of COX4-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
ZJOM135
Plasmid#133659PurposeIntegration of SEC63-2mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
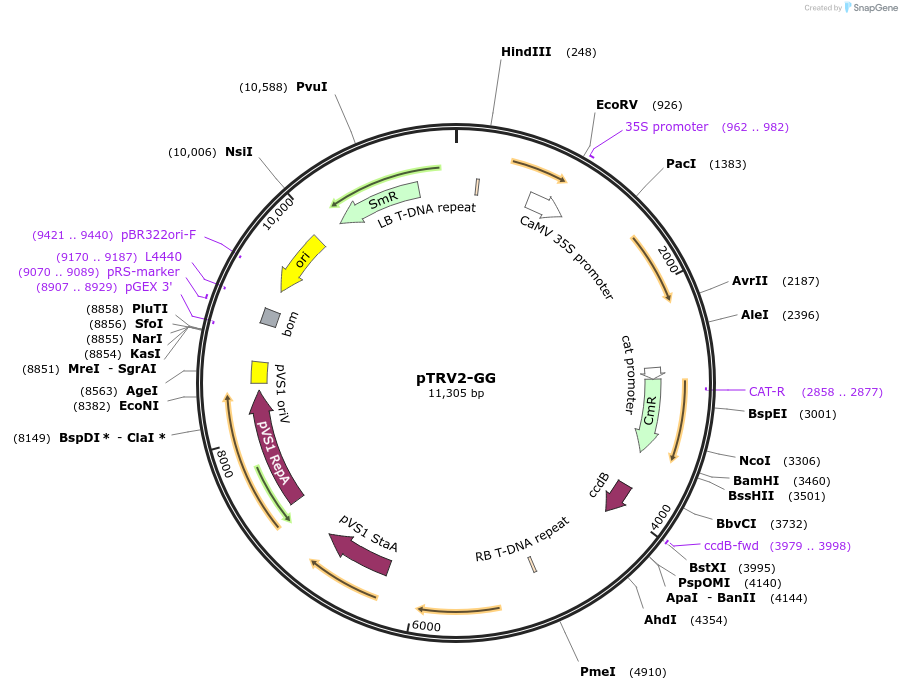
pTRV2-GG
Plasmid#105349Purposevirus-induced gene silencingDepositorInsertTRV-RNA2(5')
UseIn planta gene silencing; viralMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
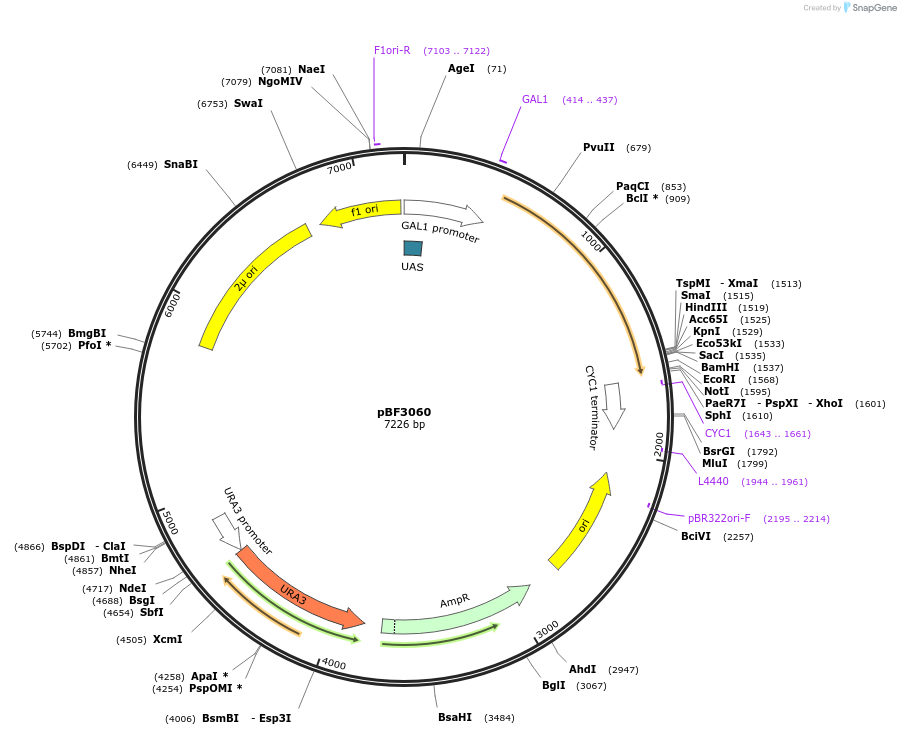
pBF3060
Plasmid#26853PurposeExpression of yeast codon-optimized CreA recombinase from the GAL1 promoterDepositorInsertCreA recombinase
UseCre/LoxExpressionYeastMutationN/APromoterGAL1Available SinceDec. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
ZJOM143
Plasmid#133667PurposeIntegration of VPH1-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
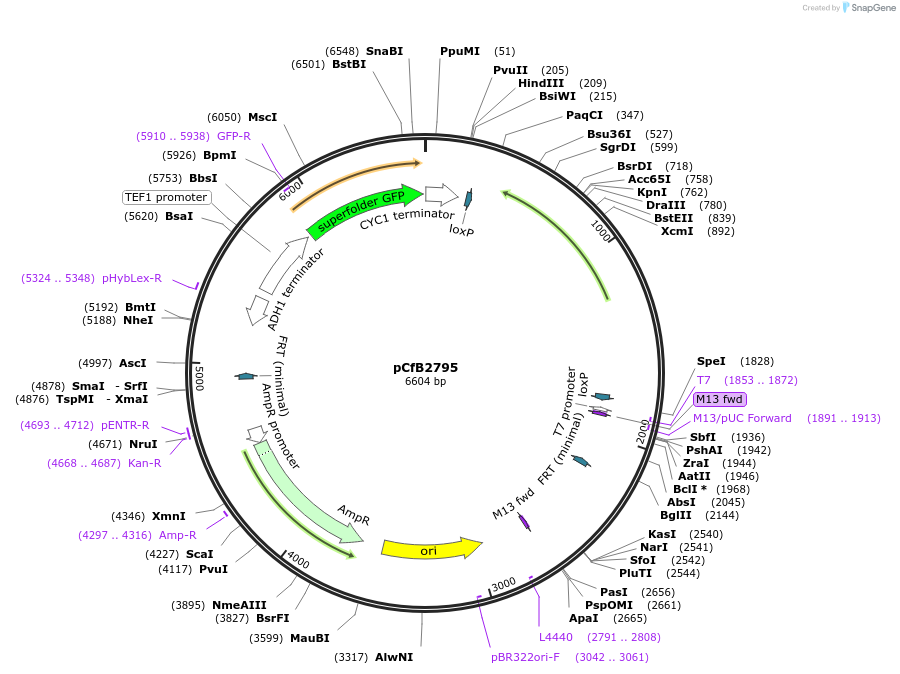
pCfB2795
Plasmid#63650PurposepCfB2795 is a vector for multiple integrations at sites sharing homology with Ty1Cons1. The selective marker is Kl.URA3. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoterAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
ZJOM119
Plasmid#133661PurposeIntegration of NAB2-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast nucleus.DepositorAvailable SinceDec. 4, 2019AvailabilityAcademic Institutions and Nonprofits only -
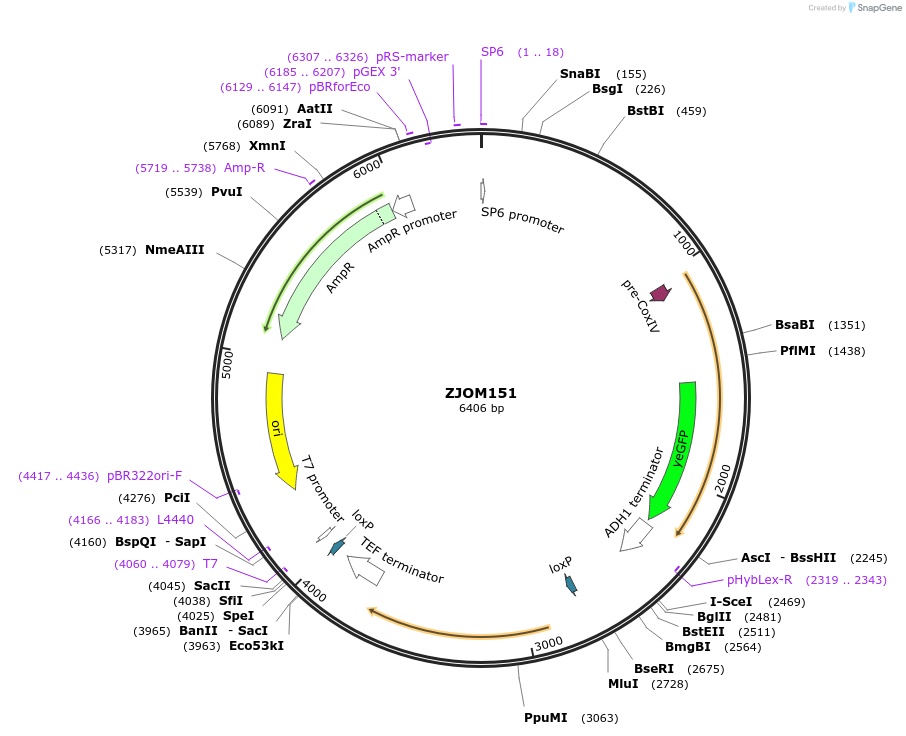
ZJOM151
Plasmid#133643PurposeIntegration of COX4-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast mitochondria.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
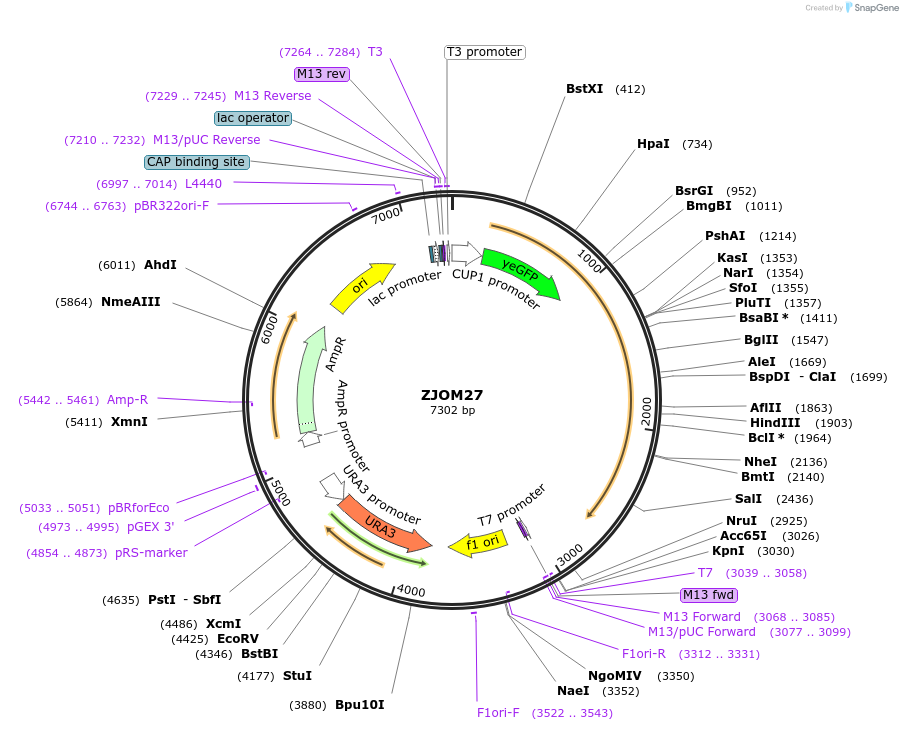
ZJOM27
Plasmid#133641PurposeIntegration of GFP-PHO8 as an additional copy in genome, use auxotrophic marker URA3(Saccharomyces cerevisiae). Marker for yeast vacuole.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
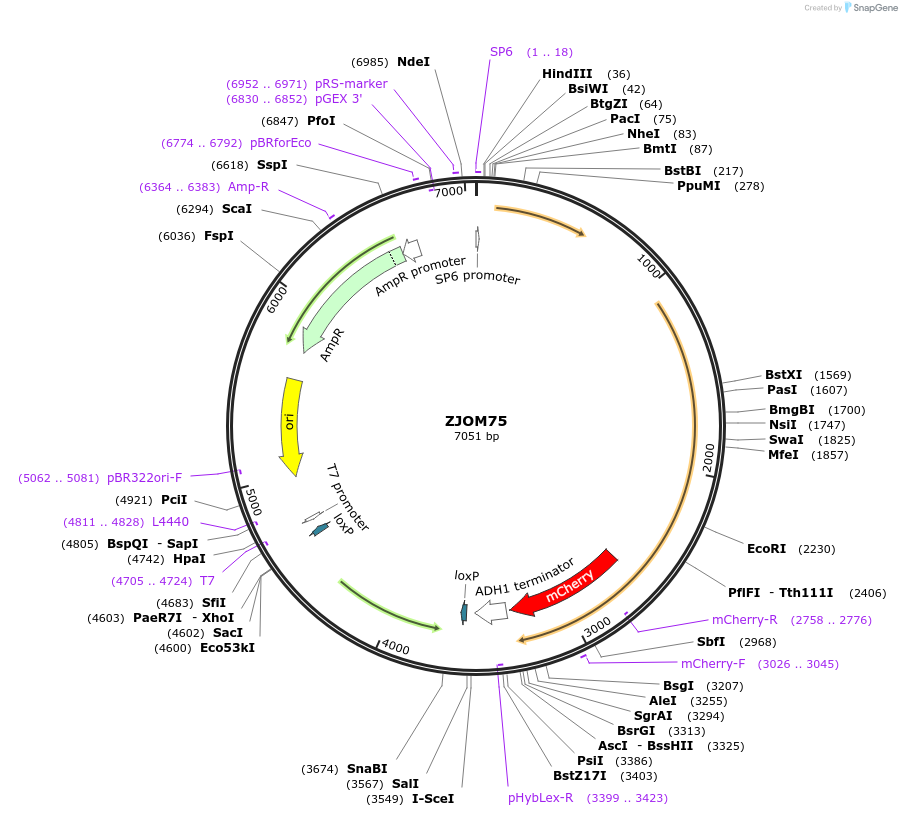
ZJOM75
Plasmid#133649PurposeIntegration of ANP1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast early golgi.DepositorAvailable SinceJan. 2, 2020AvailabilityAcademic Institutions and Nonprofits only -
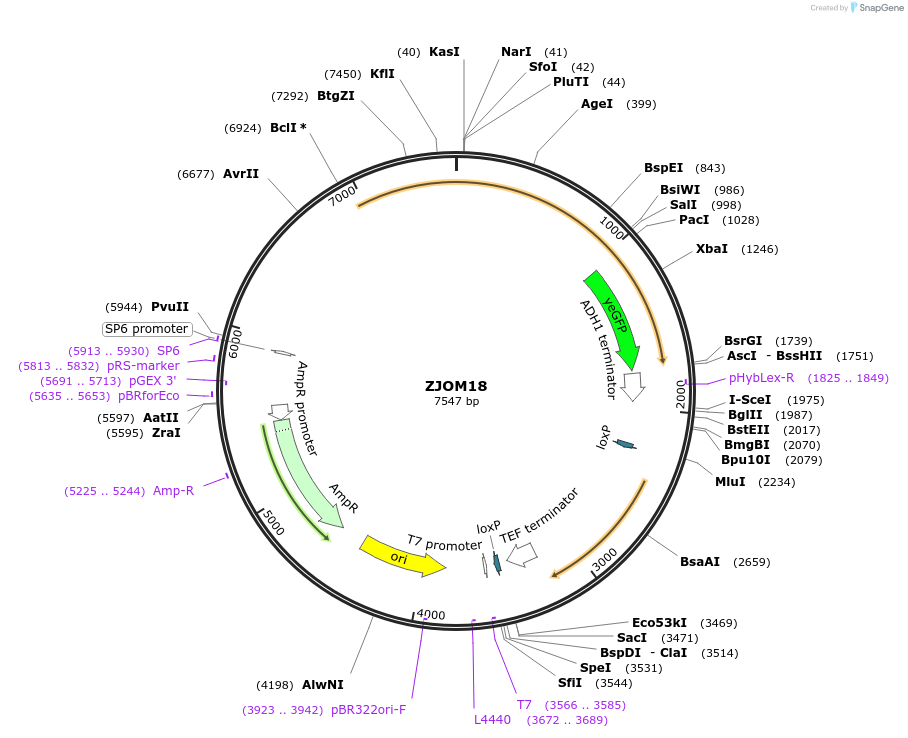
ZJOM18
Plasmid#133631PurposeIntegration of NAB2-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast nucleus.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
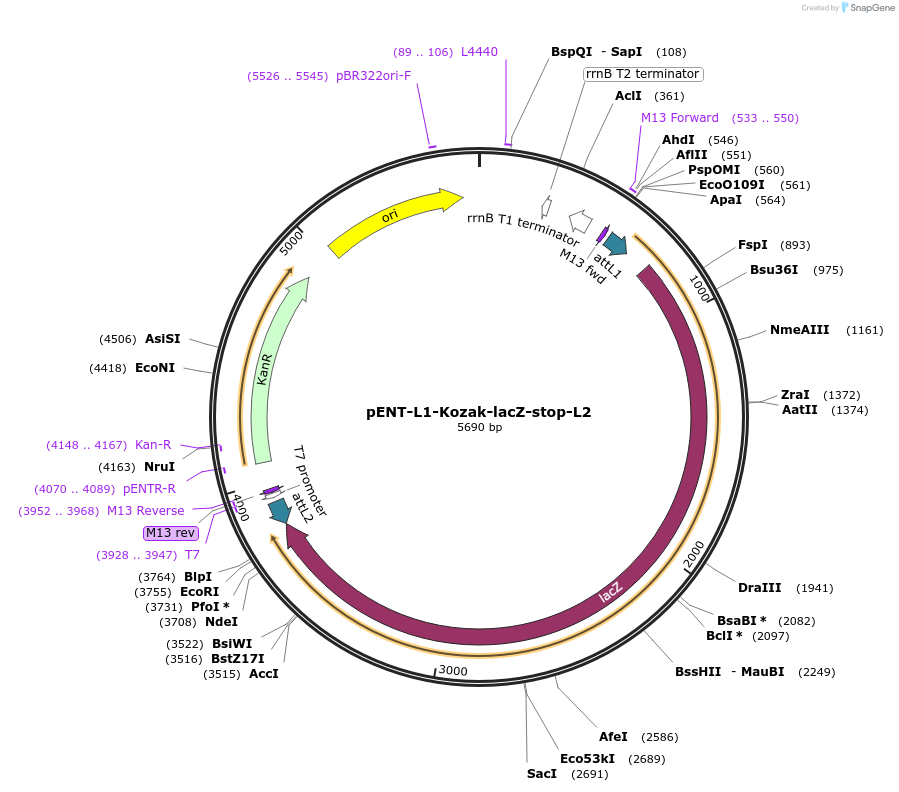
pENT-L1-Kozak-lacZ-stop-L2
Plasmid#186359PurposeEntry clone with ORF encoding lacZ flanked by Gateway recombination sequences.DepositorInsertlacZ (lacZ E. coli, Synthetic)
UseExpression of lacz reporterAvailable SinceSept. 28, 2022AvailabilityAcademic Institutions and Nonprofits only -
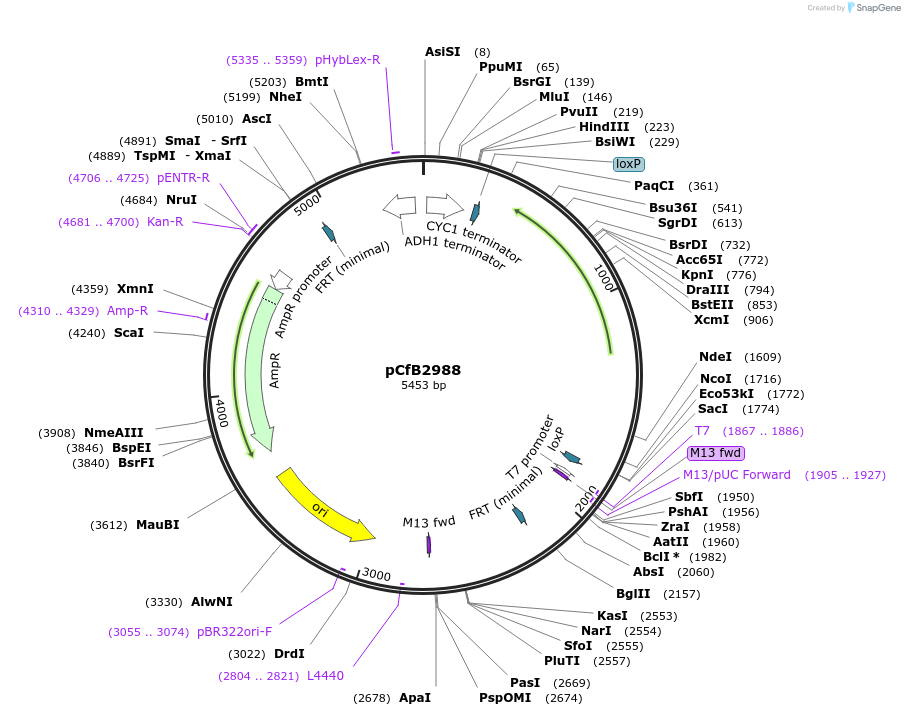
pCfB2988
Plasmid#63638PurposepCfB2988 is a vector for multiple integrations at sites sharing homology with Ty1Cons2. The selective marker is Kl.URA3. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
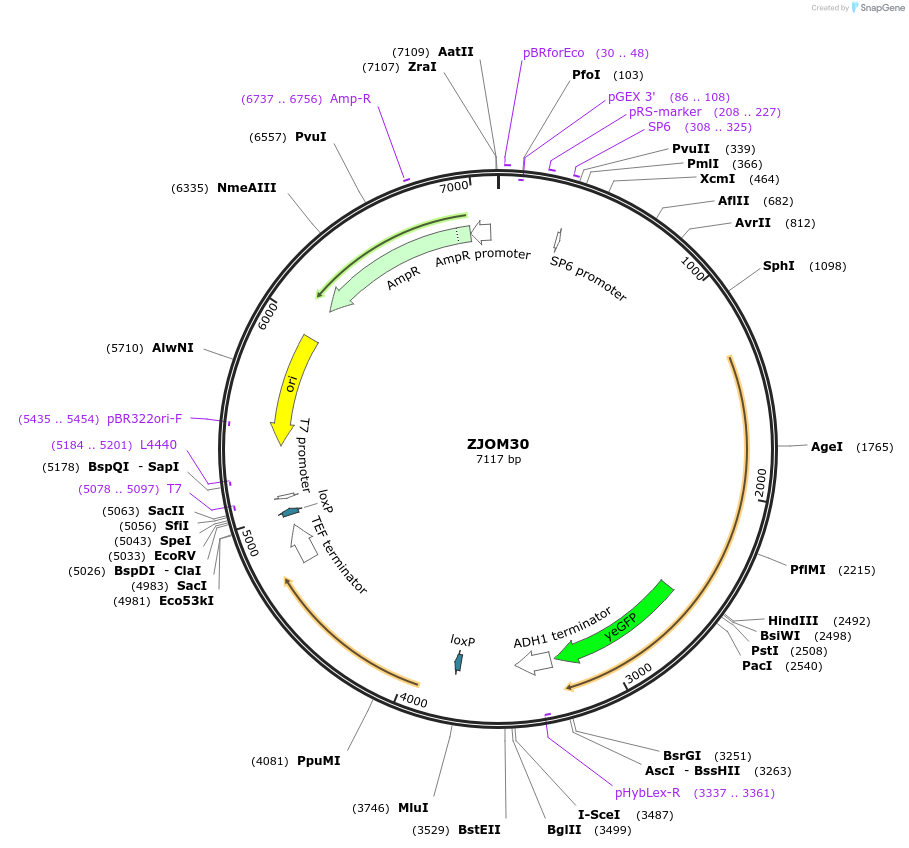
ZJOM30
Plasmid#133673PurposeIntegration of ERG6-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Present on ER and lipid droplets.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
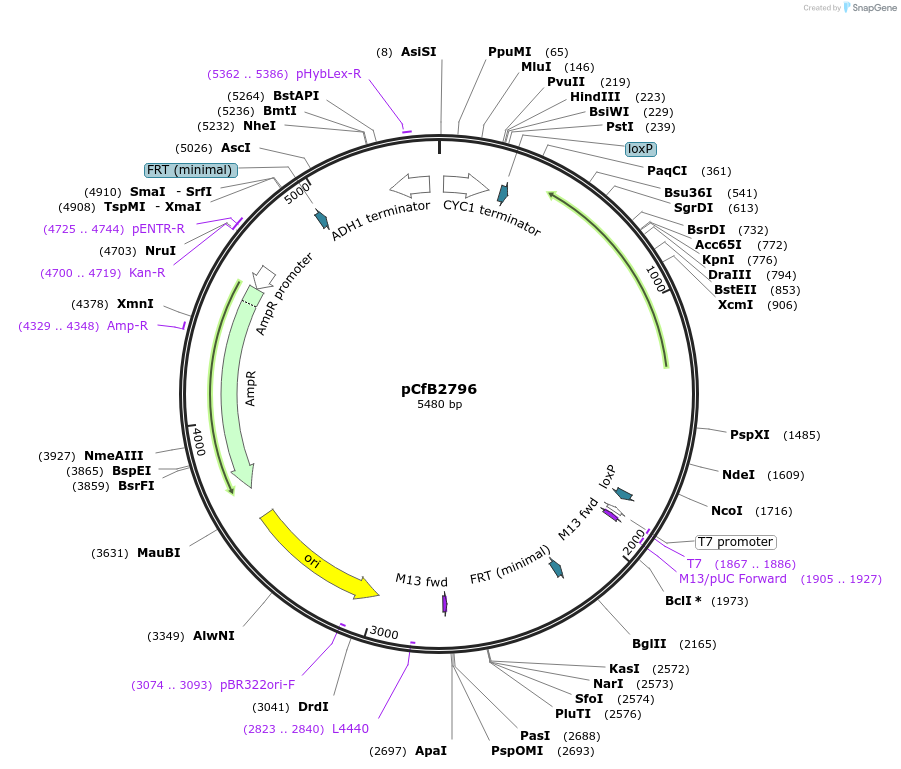
pCfB2796
Plasmid#63641PurposepCfB2796 is a vector for multiple integrations at sites sharing homology with Ty4Cons. The selective marker is Kl.URA3. Additionally it contains a USER cassette.DepositorTypeEmpty backboneUseCre/LoxExpressionYeastPromoternoneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
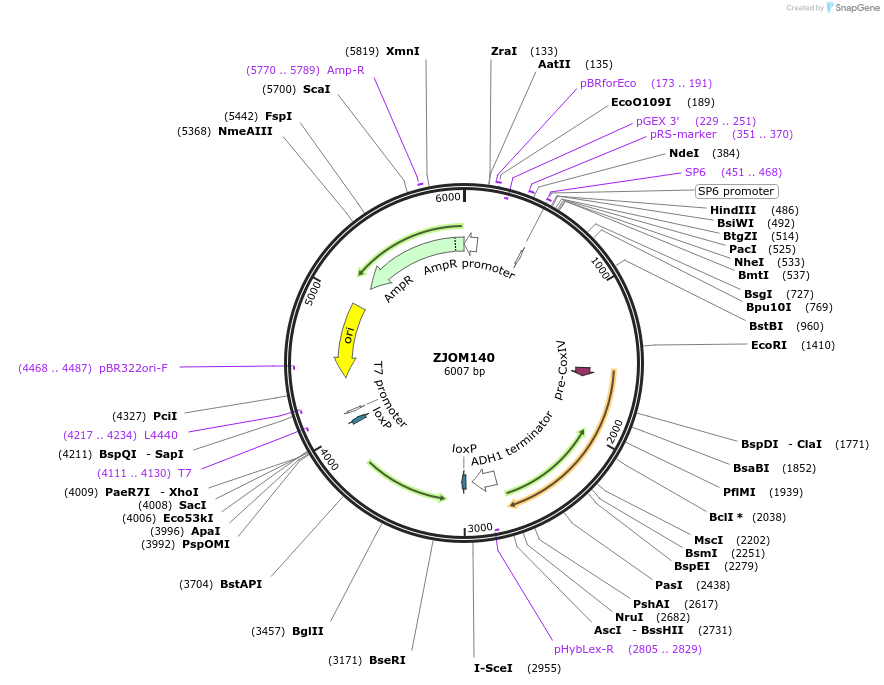
ZJOM140
Plasmid#133668PurposeIntegration of COX4-mTagBFP2 as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast mitochondria.DepositorAvailable SinceFeb. 11, 2020AvailabilityAcademic Institutions and Nonprofits only -
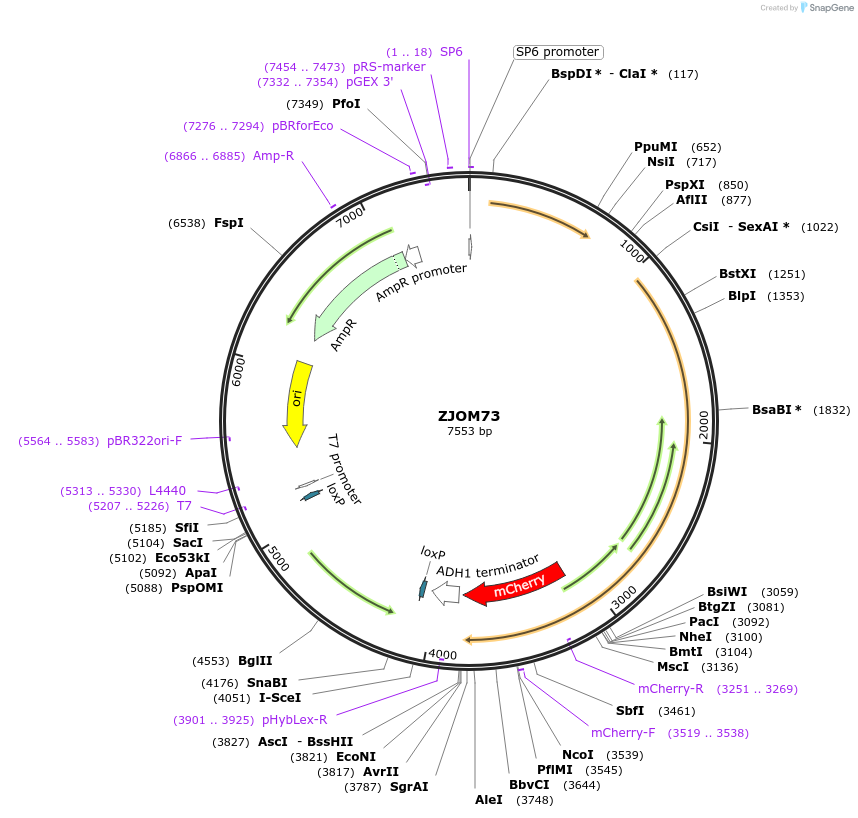
ZJOM73
Plasmid#133651PurposeIntegration of CHS5-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
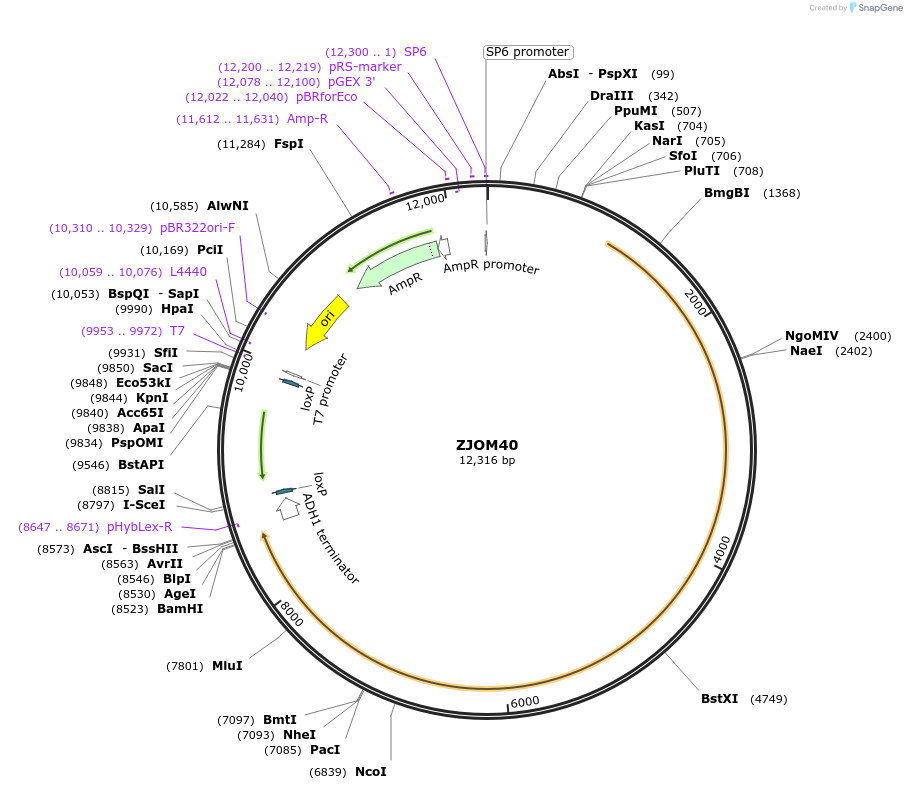
ZJOM40
Plasmid#133650PurposeIntegration of SEC7-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast late golgi/early endosome.DepositorAvailable SinceApril 23, 2020AvailabilityAcademic Institutions and Nonprofits only -
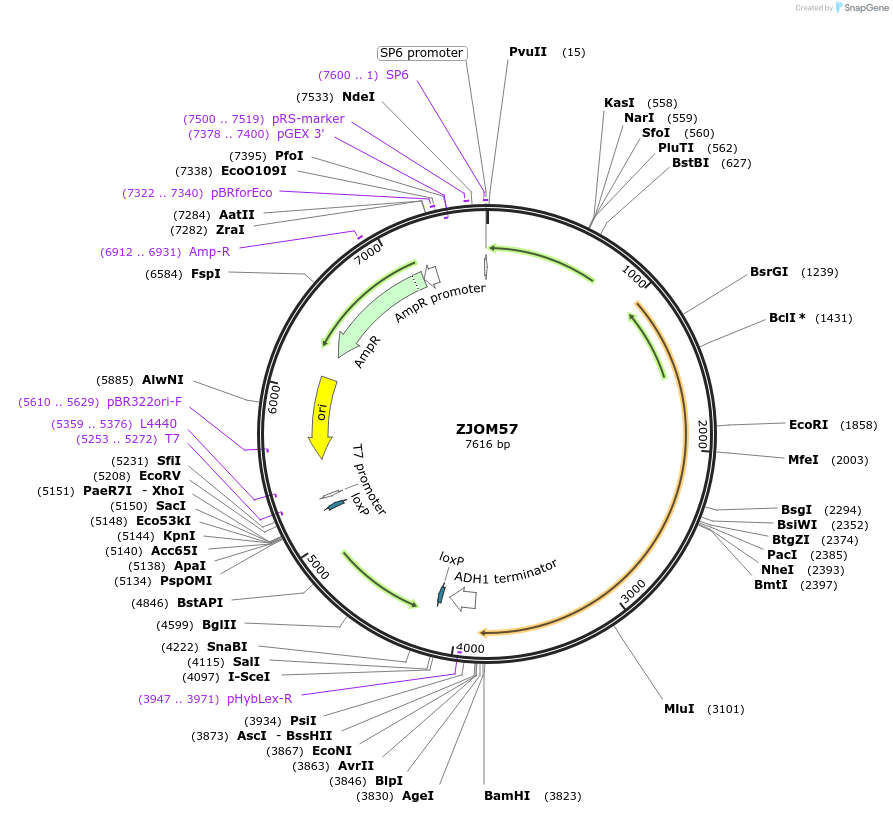
ZJOM57
Plasmid#133656PurposeIntegration of PEX3-DuDre as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast peroxisome.DepositorAvailable SinceNov. 25, 2020AvailabilityAcademic Institutions and Nonprofits only -
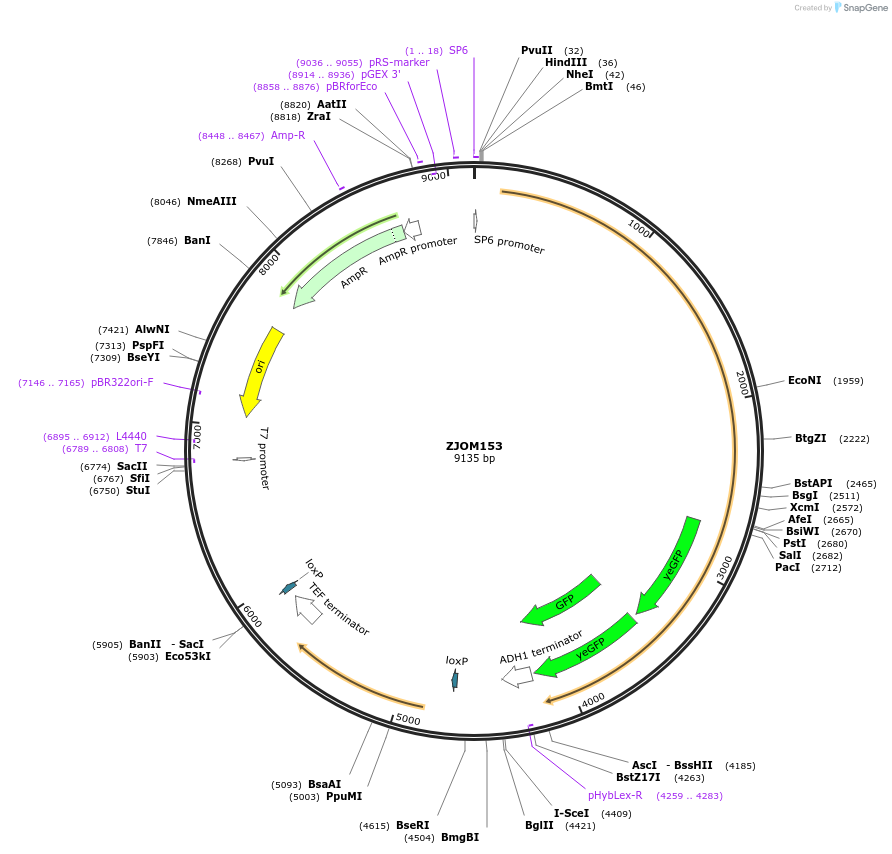
ZJOM153
Plasmid#133642PurposeIntegration of 2xGFP tag at VPH1 C terminus, use auxotrophic marker URA3(Candida albicans). Marker for yeast vacuole.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
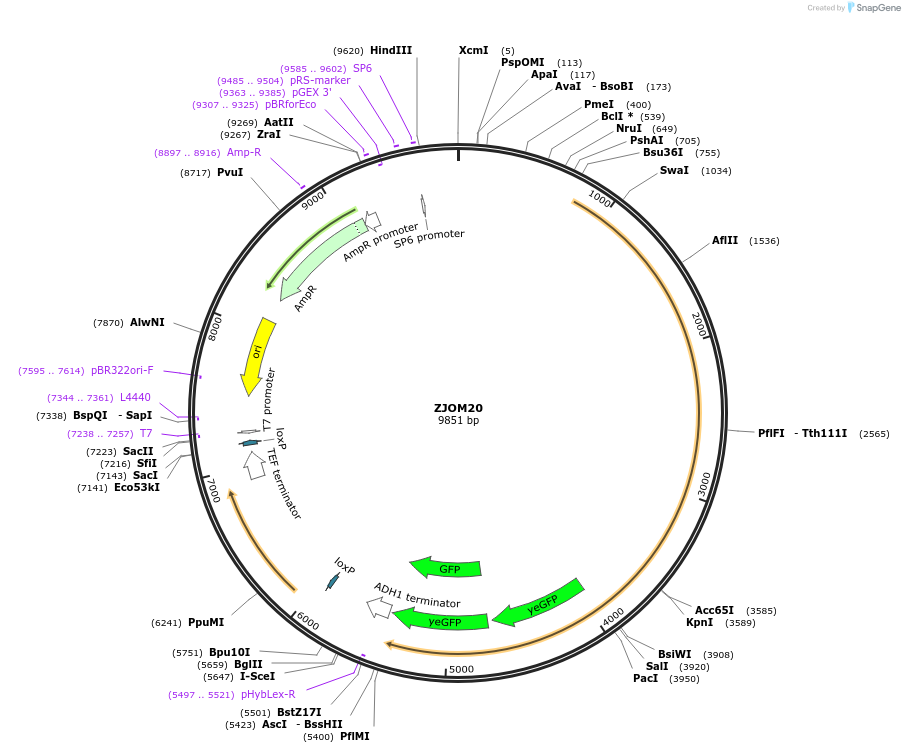
ZJOM20
Plasmid#133644PurposeIntegration of PEX1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast peroxisome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
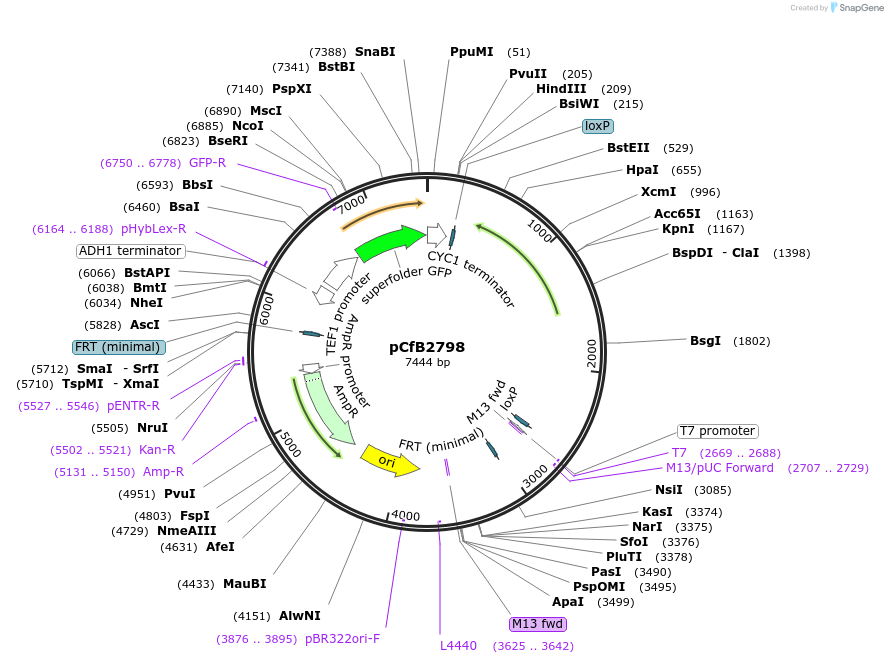
pCfB2798
Plasmid#63659PurposepCfB2798 is a vector for multiple integrations at sites sharing homology with Ty4Cons. The selective marker is Kl.LEU2. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoterAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
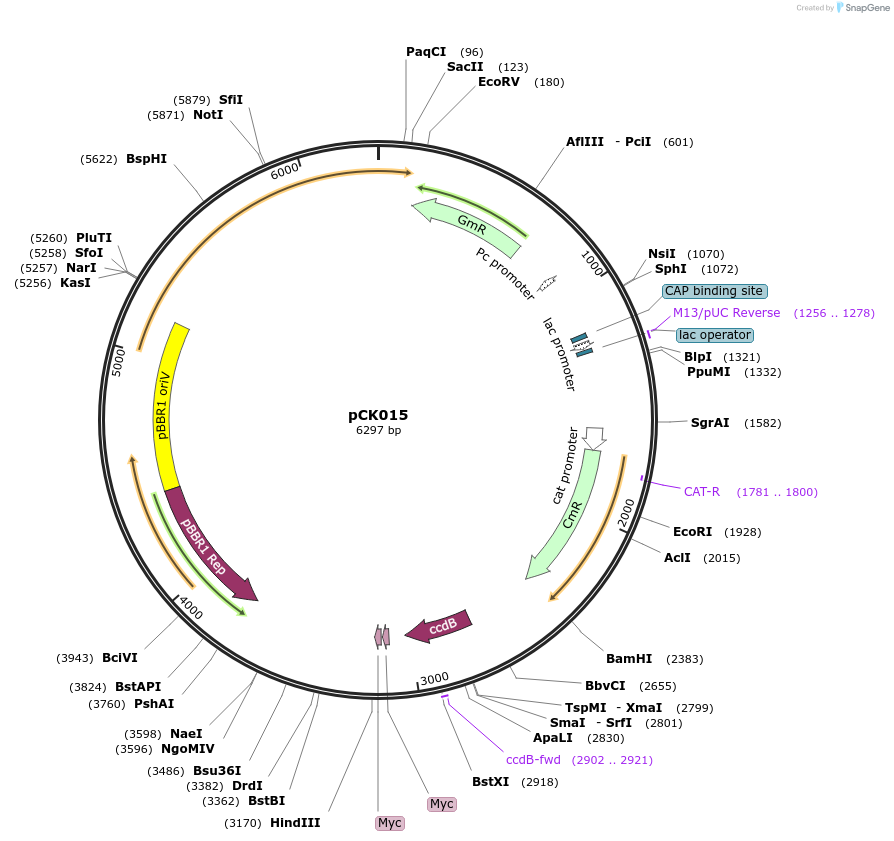
pCK015
Plasmid#105347PurposeBacterial translocation fo proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
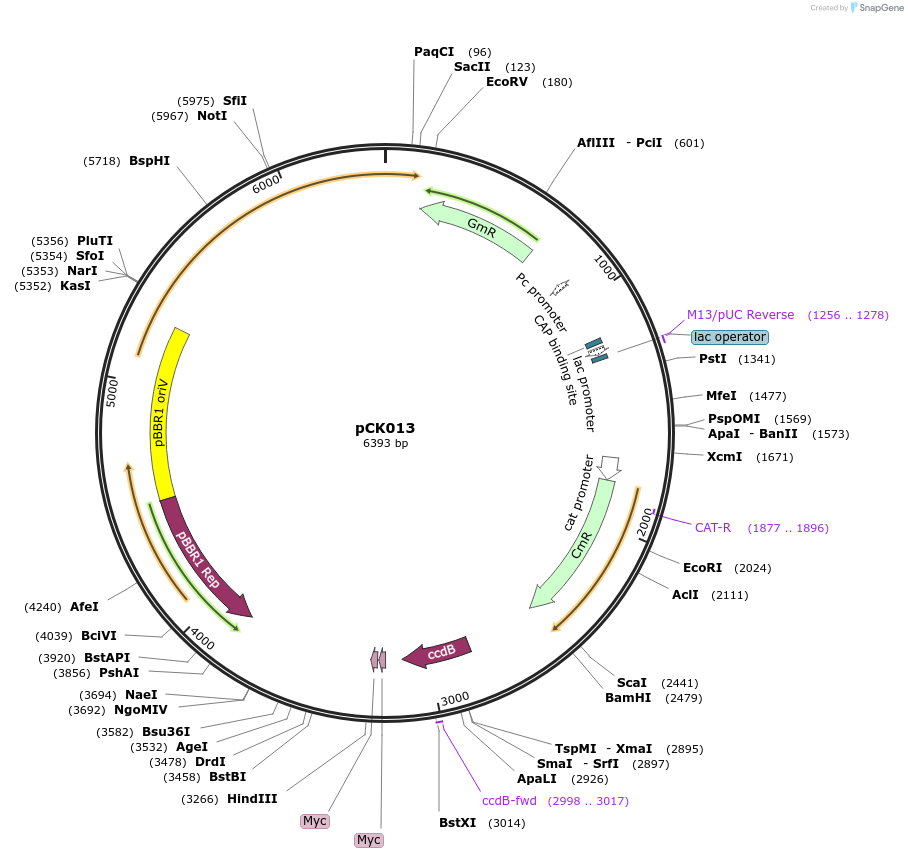
pCK013
Plasmid#105345PurposeBacterial translocation fo proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
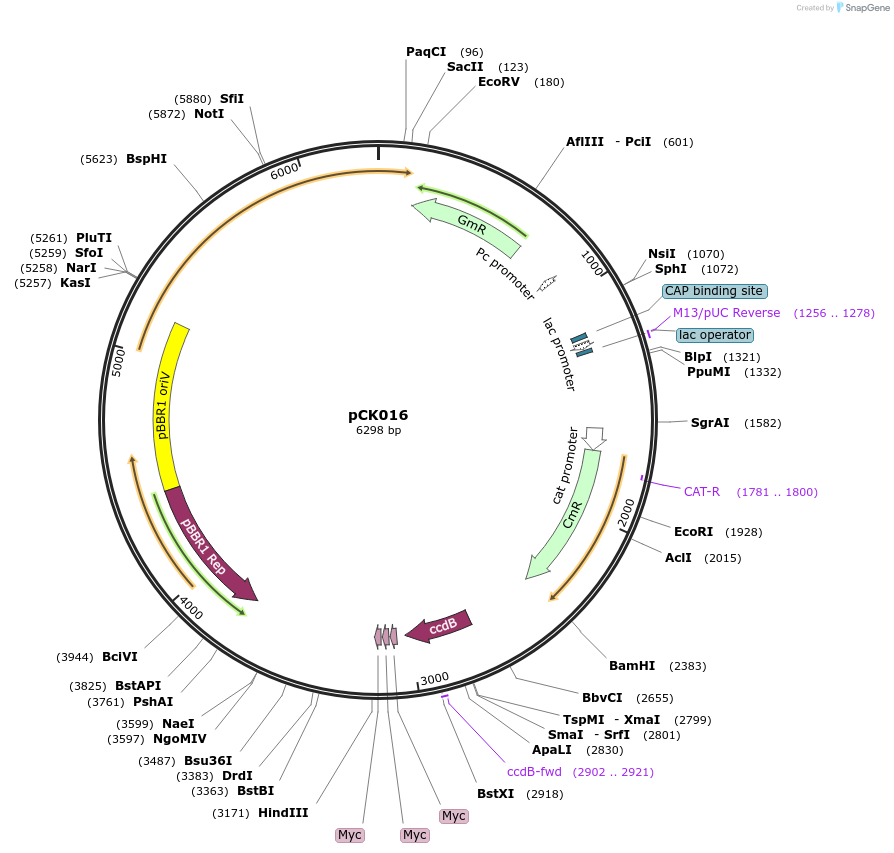
pCK016
Plasmid#105348PurposeBacterial translocation for proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
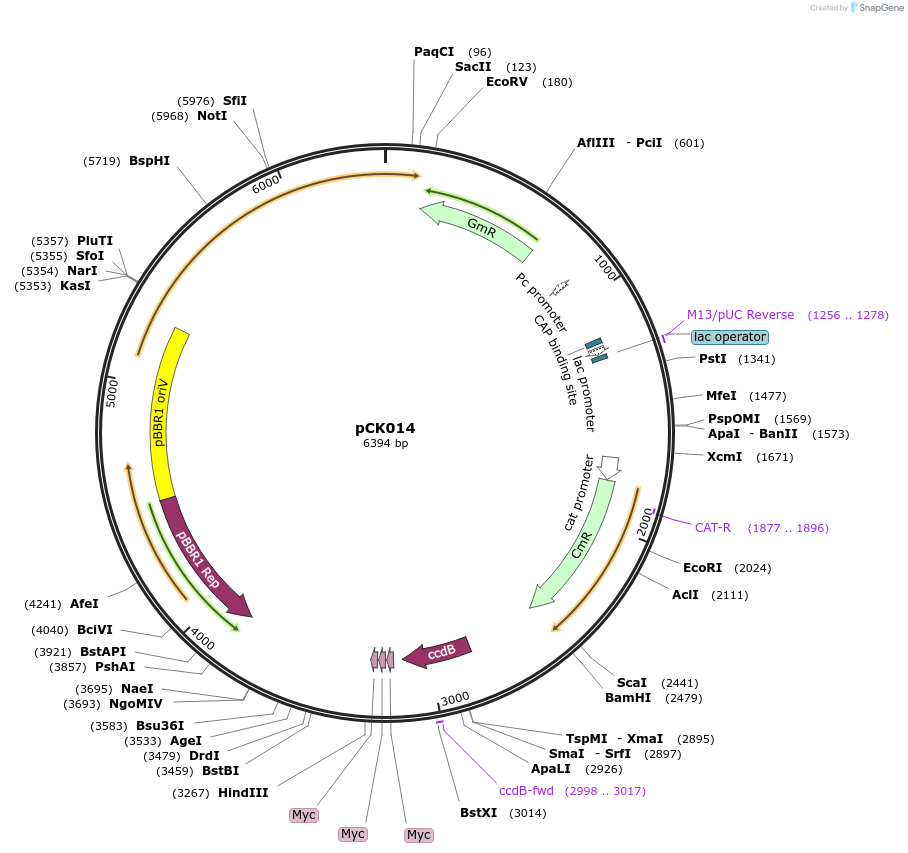
pCK014
Plasmid#105346PurposeBacterial translocation fo proteins into plant cellsDepositorInsertsecretion signal, ccdB cassette
UseBacterial secretionMutationBsaI restriction sites removedAvailable SinceMay 10, 2018AvailabilityAcademic Institutions and Nonprofits only -
ZJOM5
Plasmid#133635PurposeIntegration of CHS5-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
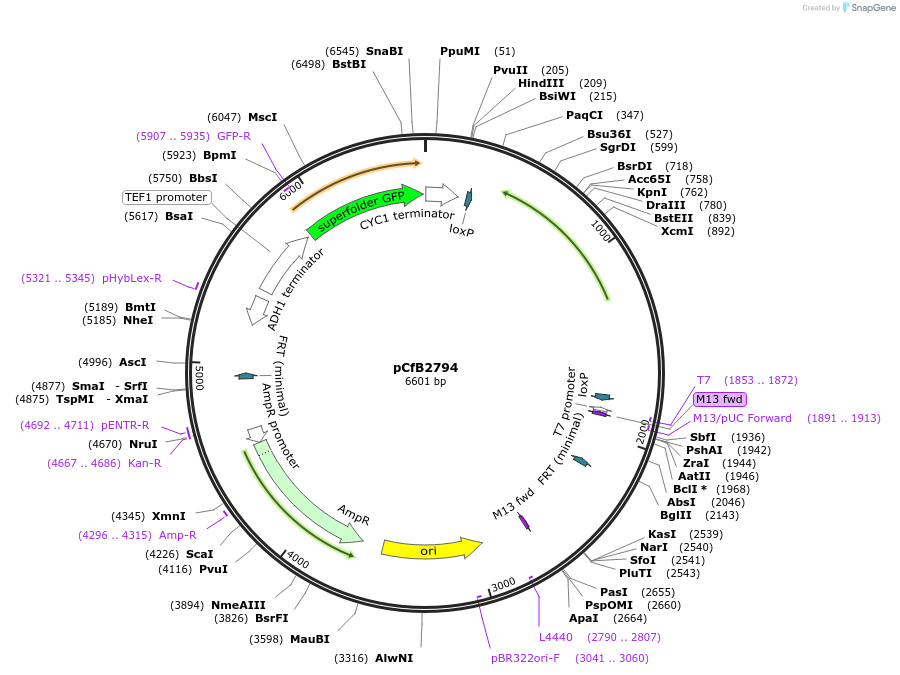
pCfB2794
Plasmid#63651PurposepCfB2794 is a vector for multiple integrations at sites sharing homology with Ty1Cons2. The selective marker is Kl.URA3. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoterAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
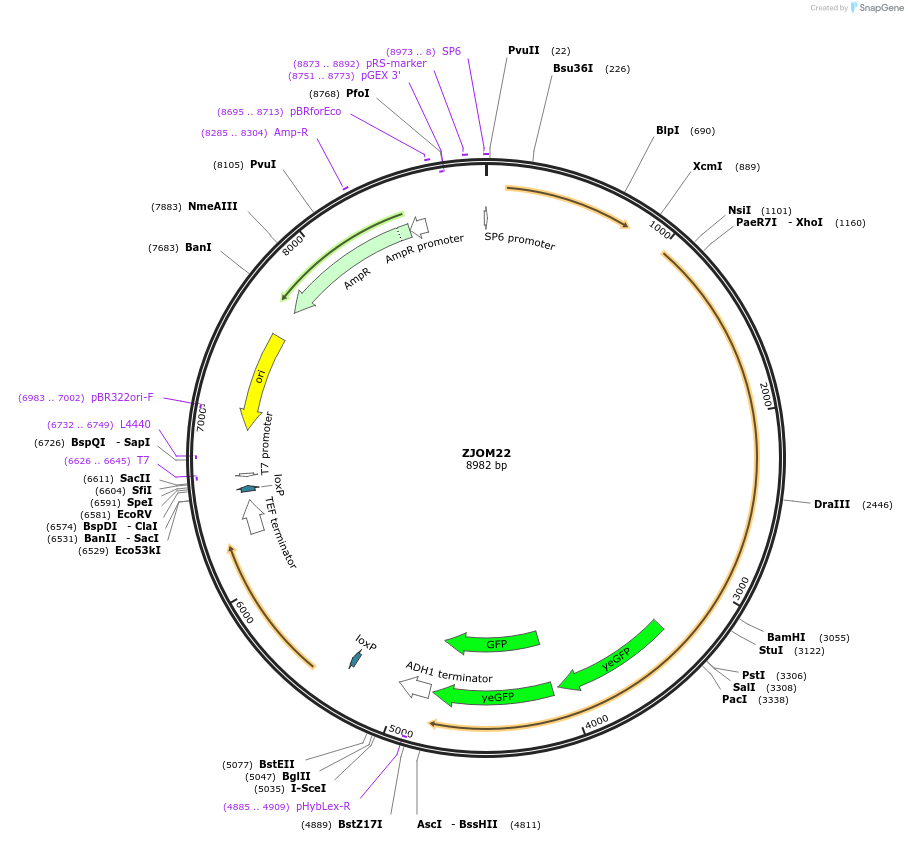
ZJOM22
Plasmid#133629PurposeIntegration of EMC1-2GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast endoplasmic reticulum (ER).DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only -
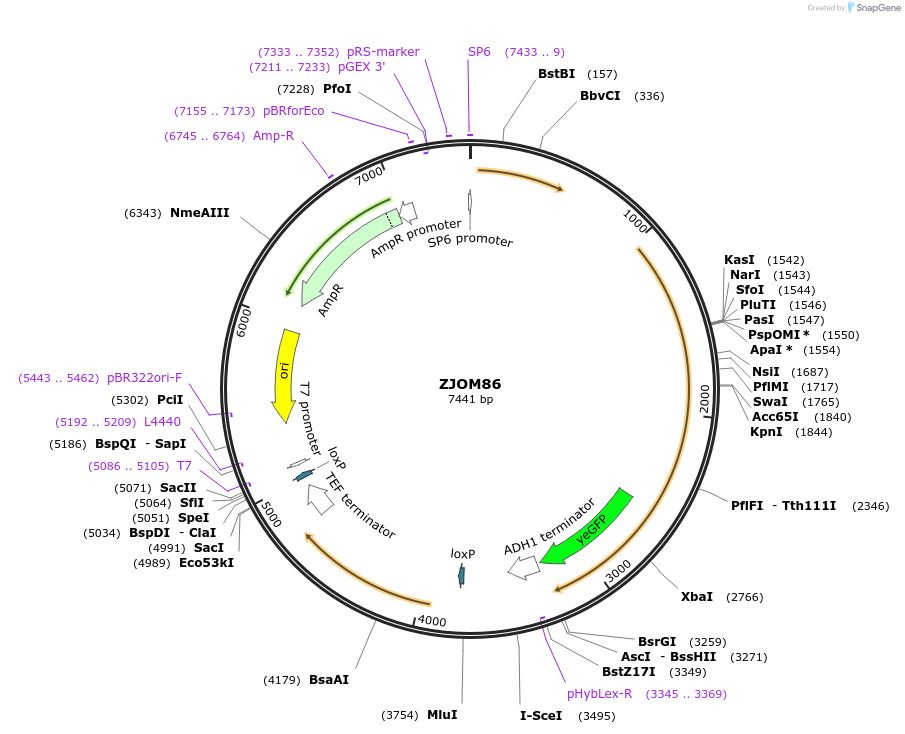
ZJOM86
Plasmid#133634PurposeIntegration of ANP1-GFP as an additional copy in genome, use auxotrophic marker URA3(Candida albicans). Marker for yeast early golgi.DepositorAvailable SinceDec. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
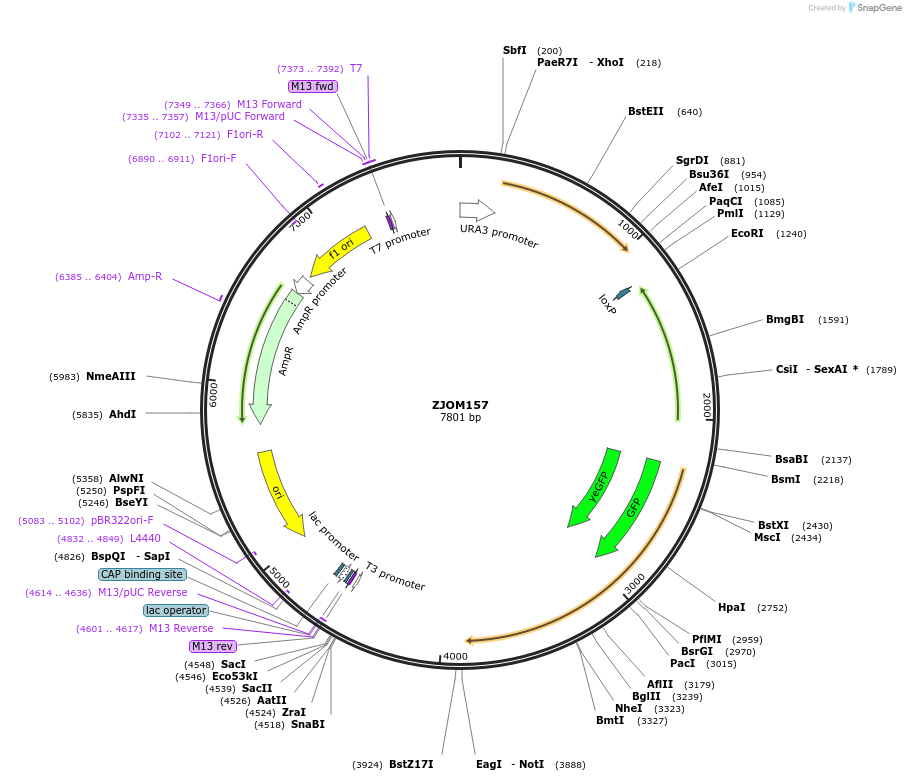
ZJOM157
Plasmid#133640PurposeIntegration of GFP-PEP12 as an additional copy in genome, use auxotrophic marker URA3(Kluyveromyces lactis).Marker for yeast late endosome.DepositorAvailable SinceFeb. 19, 2020AvailabilityAcademic Institutions and Nonprofits only -
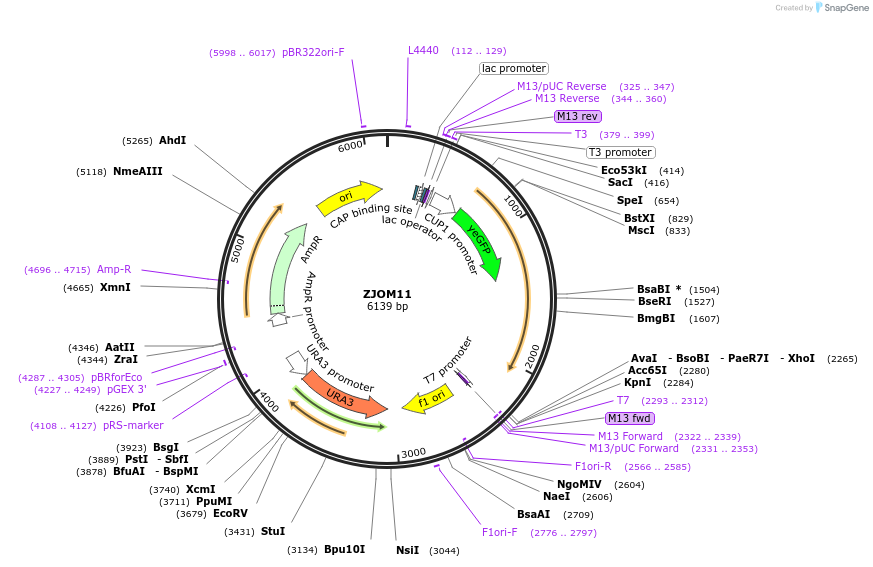
ZJOM11
Plasmid#133637PurposeIntegration of GFP-TLG1 as an additional copy in genome, use auxotrophic marker URA3(Saccharomyces cerevisiae). Marker for yeast late golgi/early endosome.DepositorAvailable SinceFeb. 12, 2020AvailabilityAcademic Institutions and Nonprofits only