We narrowed to 6,701 results for: proc
-
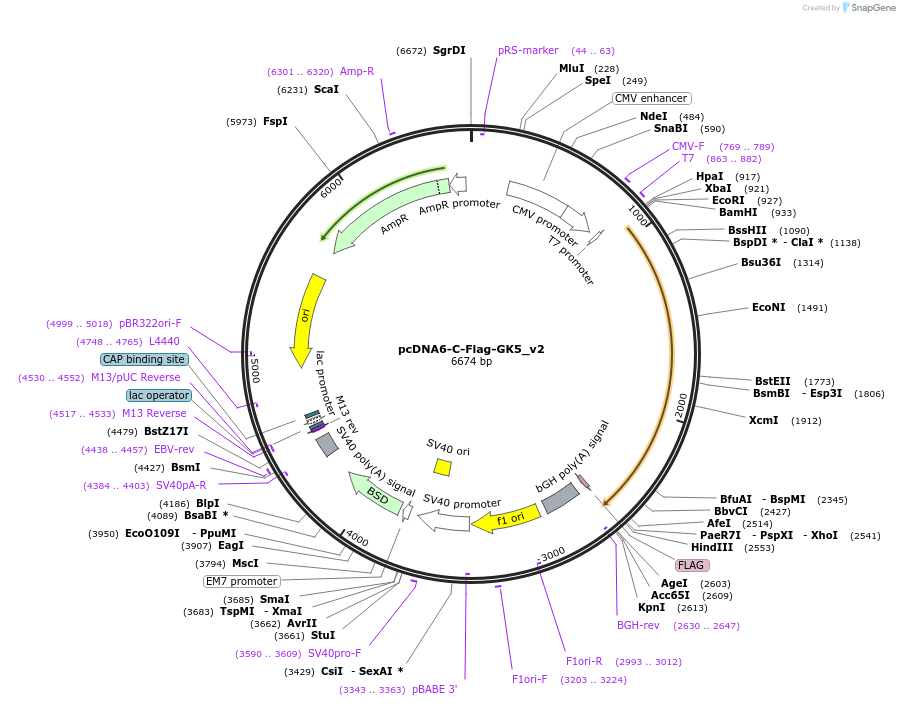
Plasmid#134282PurposeExpresses Flag-tagged GK5 (isoform 2) in mammalian cellsDepositorInsertGk5 (Gk5 Mouse)
TagsFlagExpressionMammalianMutationmouse Gk5 isoform 2; 534 amino acidsPromoterCMVAvailable SinceMarch 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
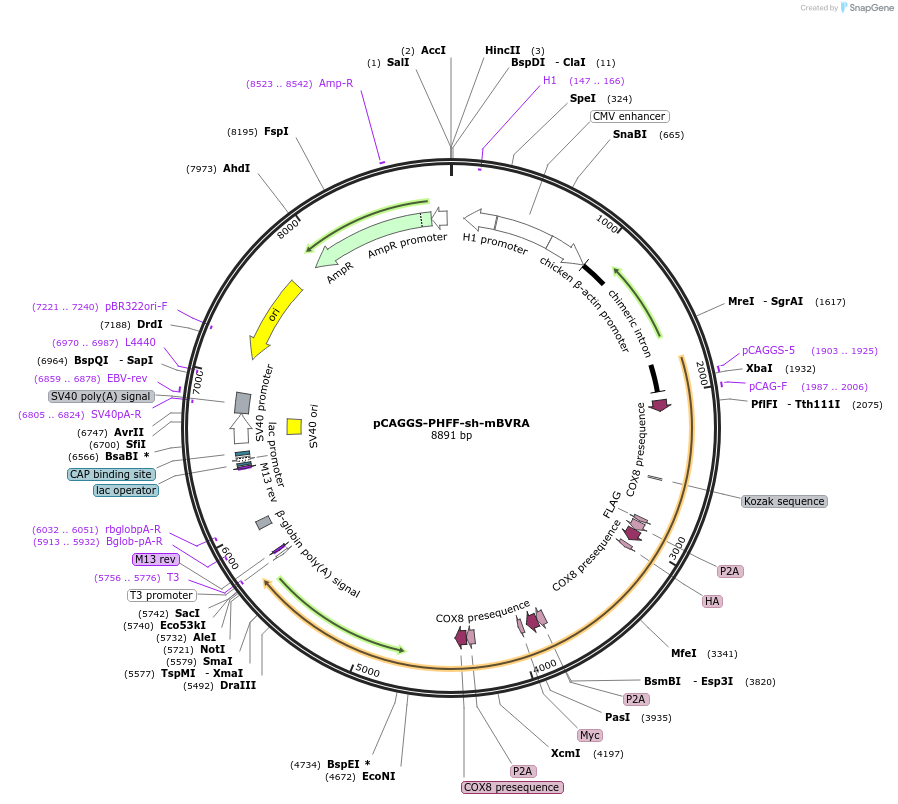
pCAGGS-PHFF-sh-mBVRA
Plasmid#100286PurposeExpression vector for phycocyanobilin (PCB) synthesis and shRNA for mouse BVRA gene in mammalian cellsDepositorInsertsMTS-PcyA-FLAG-P2A-MTS-HA-HO1-P2A-MTS-Myc-Fd-P2A-MTS-Fnr-T7 (synthetic genes)
shRNA for mouse BVRA
TagsFLAG, T7 and Mitochondria-targeting sequence (MTS…ExpressionMammalianMutationAll genes are codon-optimized for expression in h…PromoterCAG promoter and H1 promoterAvailable SinceSept. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
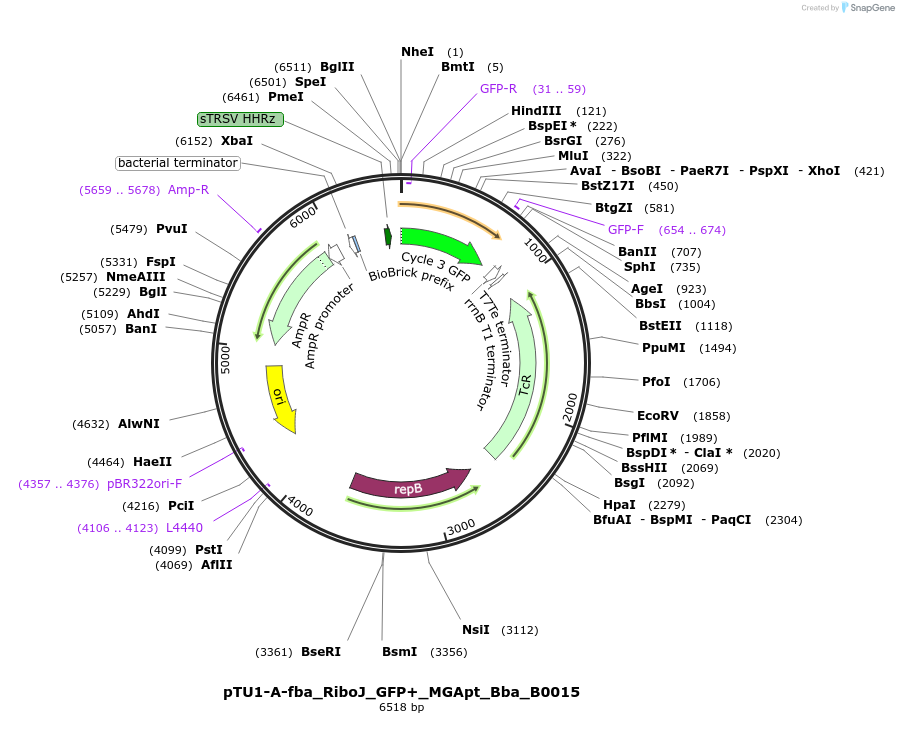
pTU1-A-fba_RiboJ_GFP+_MGApt_Bba_B0015
Plasmid#107582PurposeB. megaterium DSM319 fba promoter, GFP+ with malachite green mRNA aptamer for Bacillus cell-free transcription-translation. Bacillus shuttle vector backbone, colE1/AmpR (E. coli), RebB/TetA (Bacillus)DepositorInsertGFP+
UseSynthetic Biology; E. coli and bacillus shuttle v…TagsB. megaterium DSM319 xylA leader sequence (MTSSKI…ExpressionBacterialMutationF64L/ S65T/ Q80R/ F99S/ M153T/ V163APromoterfba promoter Bacillus megaterium DSM319Available SinceApril 30, 2018AvailabilityAcademic Institutions and Nonprofits only -
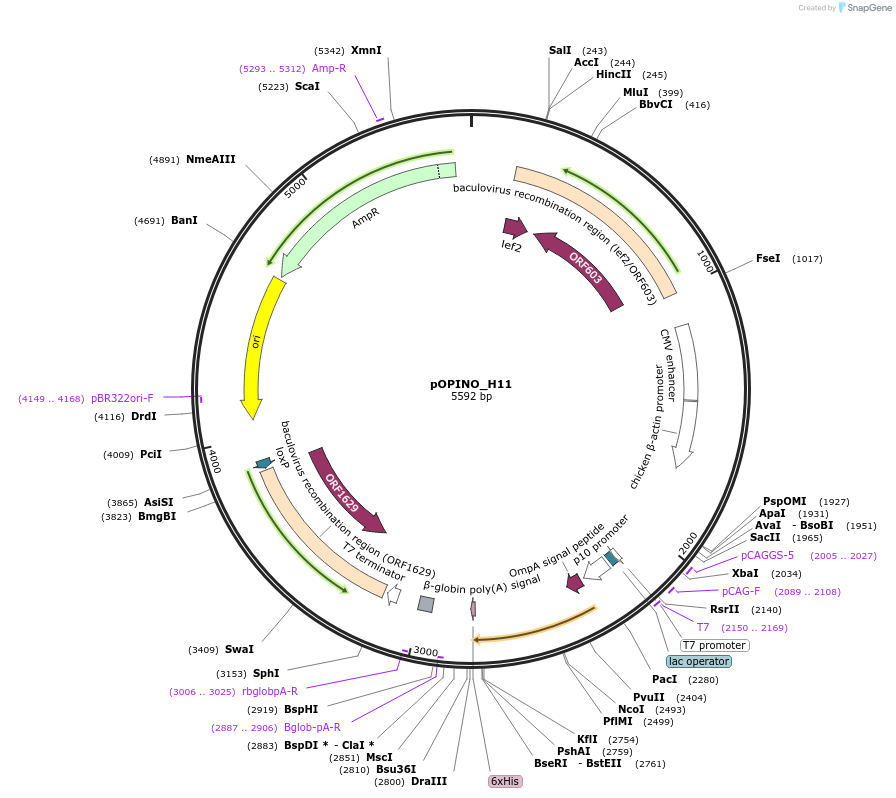
pOPINO_H11
Plasmid#184277PurposeExpresses nanobody H11 in E. coliDepositorInsertNanobody H11
TagsHis6ExpressionBacterial, Insect, and Mamm…PromoterT7Available SinceFeb. 27, 2024AvailabilityAcademic Institutions and Nonprofits only -
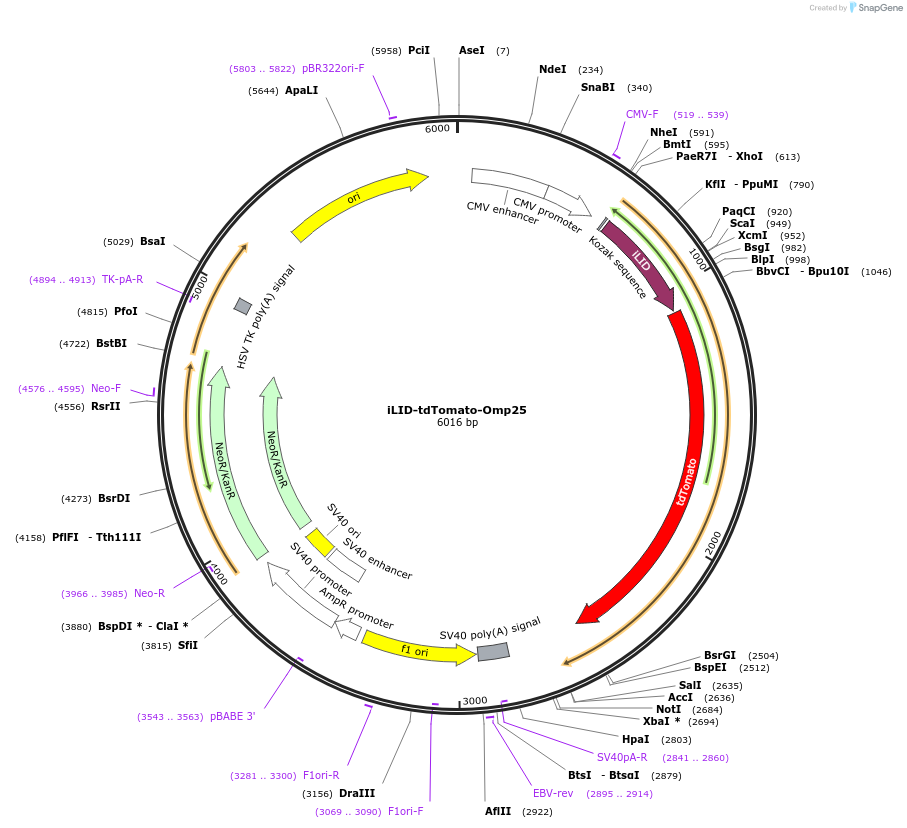
iLID-tdTomato-Omp25
Plasmid#214400PurposeExpresses fusion of iLID with tdTomato and transmembrane domain of Omp25DepositorInsertiLID-tdTomato-Omp25 (Synj2bp Synthetic, Rat)
TagsiLID-TdTomato-Omp25ExpressionMammalianPromoterCMVAvailable SinceMarch 20, 2024AvailabilityAcademic Institutions and Nonprofits only -
pInducer10-SAE1_sh1
Plasmid#168984PurposeExpresses RFP cDNA and SAE1 shRNA 1 in mammalian cellsDepositorAvailable SinceJune 1, 2021AvailabilityAcademic Institutions and Nonprofits only -
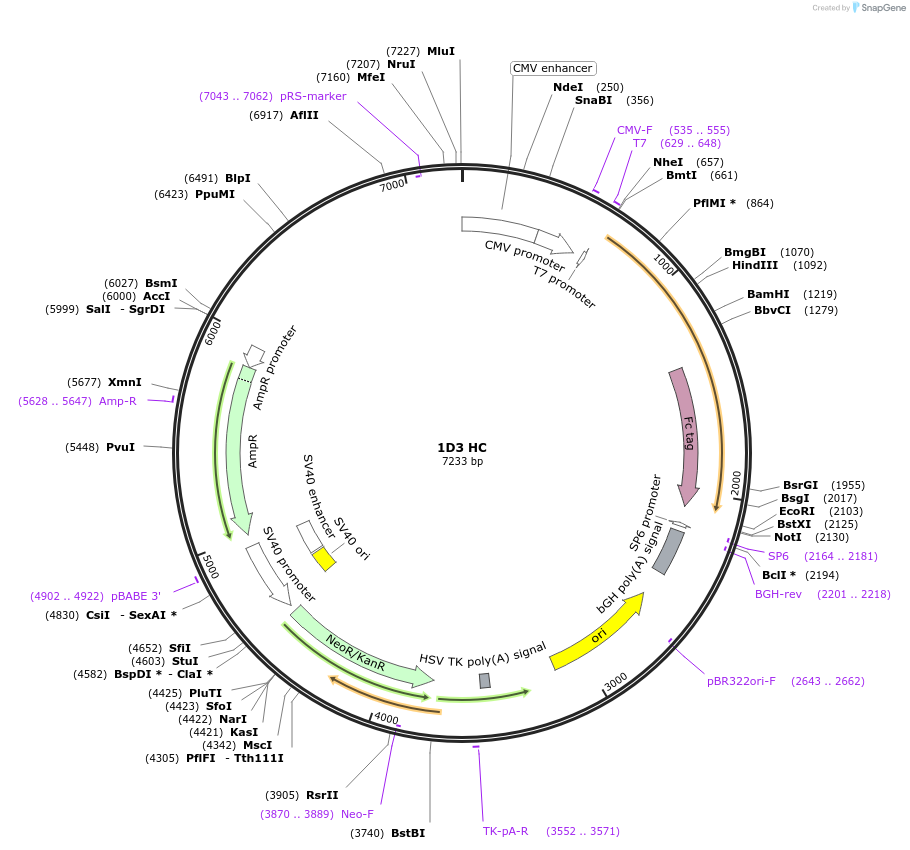
1D3 HC
Plasmid#170670PurposeMammalian Expression Plasmid of Anti-cytochrome c, 1D3 HC (Human)DepositorInsertHC of anti-cytochrome c, 1D3 (human) recombinant mouse monoclonal antibody
ExpressionMammalianAvailable SinceJuly 12, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
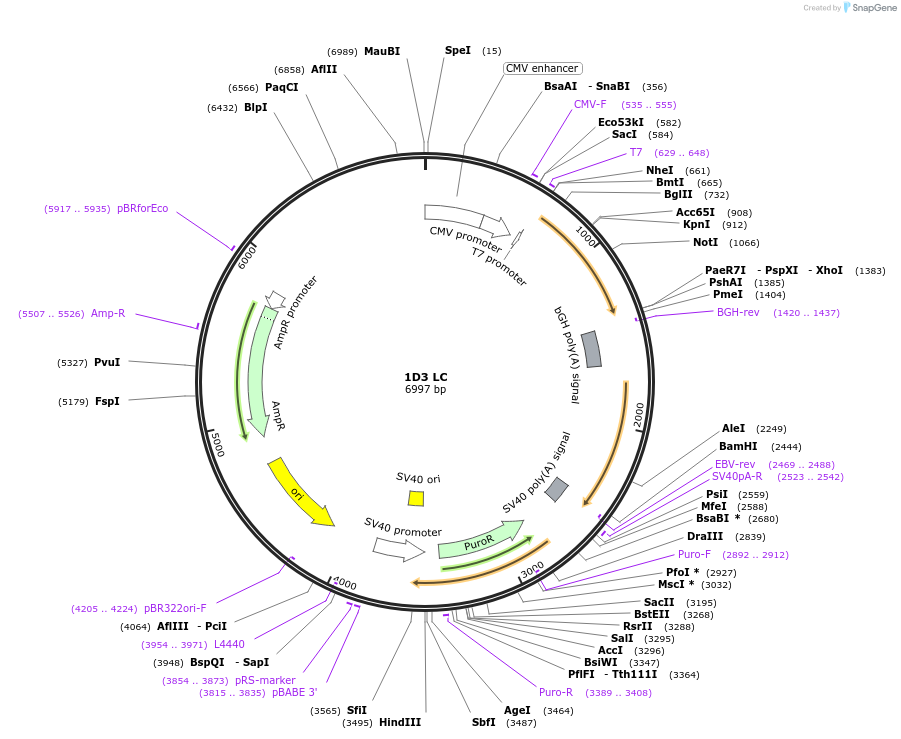
1D3 LC
Plasmid#170669PurposeMammalian Expression Plasmid of Anti-cytochrome c, 1D3 LC (Human)DepositorInsertLC of anti-cytochrome c, 1D3 (human) recombinant mouse monoclonal antibody
ExpressionMammalianAvailable SinceJune 10, 2021AvailabilityIndustry, Academic Institutions, and Nonprofits -
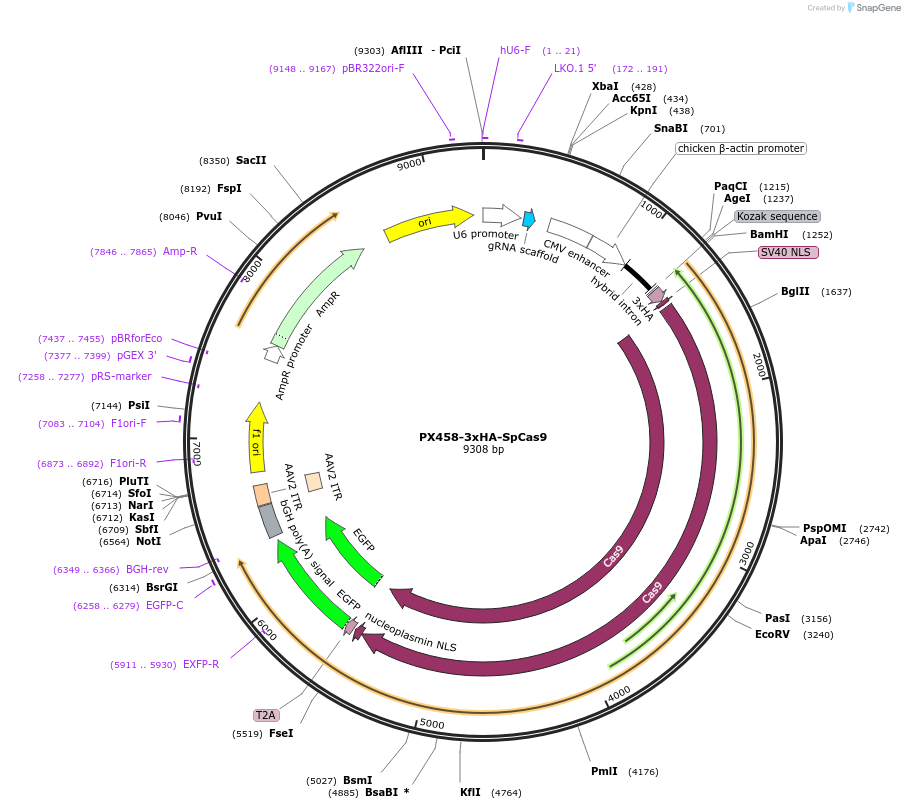
PX458-3xHA-SpCas9
Plasmid#130968Purpose3xHA tagged Cas9 from S. pyogenes with T2A-EGFP and cloning backbone for sgRNADepositorInsert3xHA-NLS-SpCas9
UseCRISPRTags3x HA, EGFP, and NLSExpressionMammalianPromoterCbhAvailable SinceNov. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
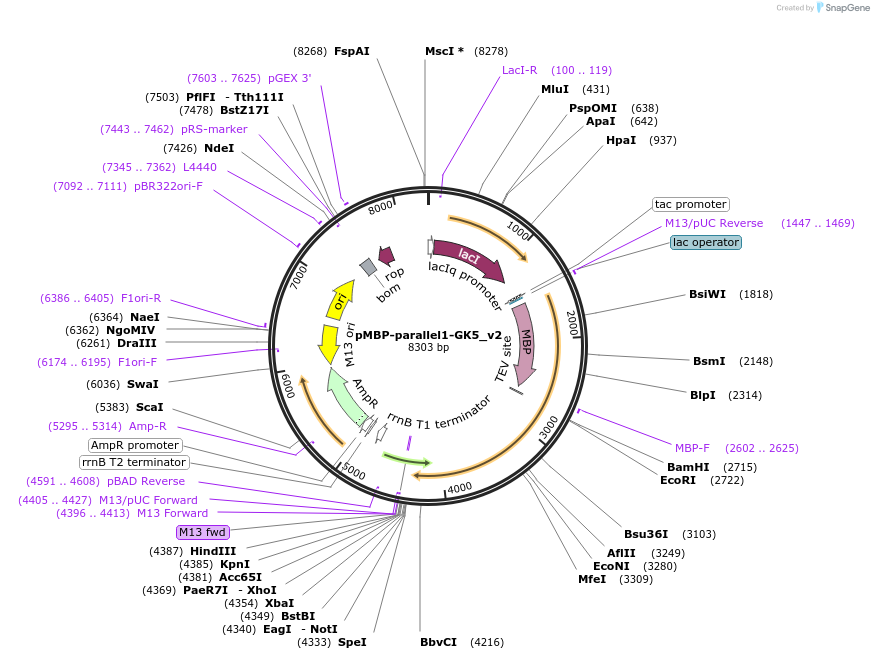
pMBP-parallel1-GK5_v2
Plasmid#134288PurposeExpresses GK5 (isoform 2) in bacteriaDepositorInsertGk5 (Gk5 Mouse)
TagsMBPExpressionBacterialMutationmouse Gk5 isoform 2; 534 amino acidsPromoterT7Available SinceMarch 20, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
-
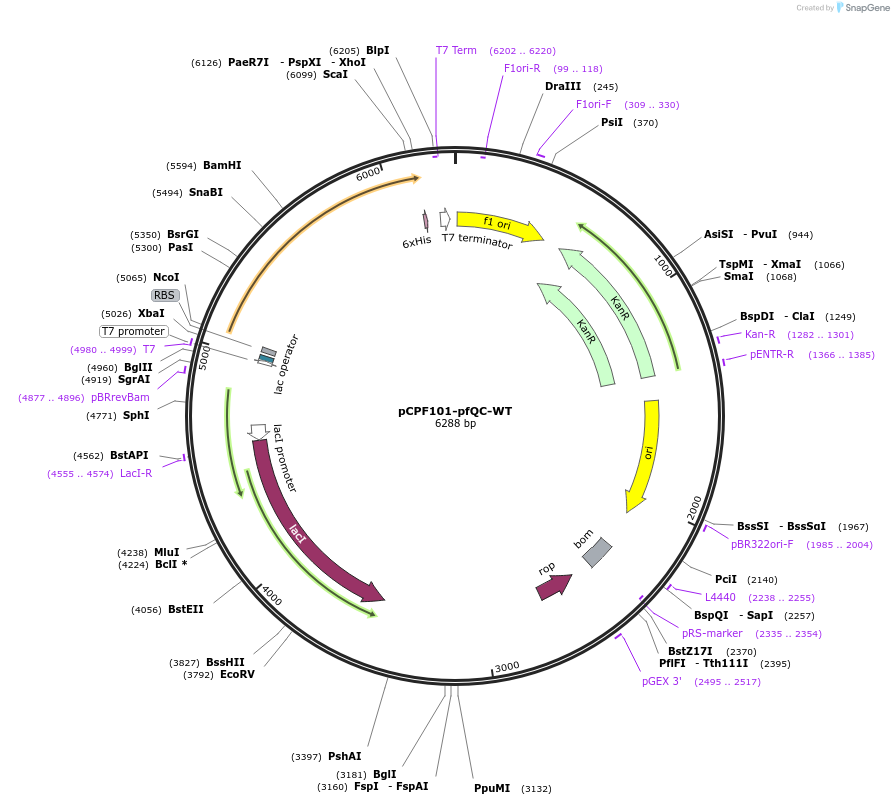
pCPF101-pfQC-WT
Plasmid#184877Purposeexpresses soluble plasmodium falciparum QC protein with a histagDepositorInsertpfQC-wt
TagsHis tagExpressionBacterialAvailable SinceJuly 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
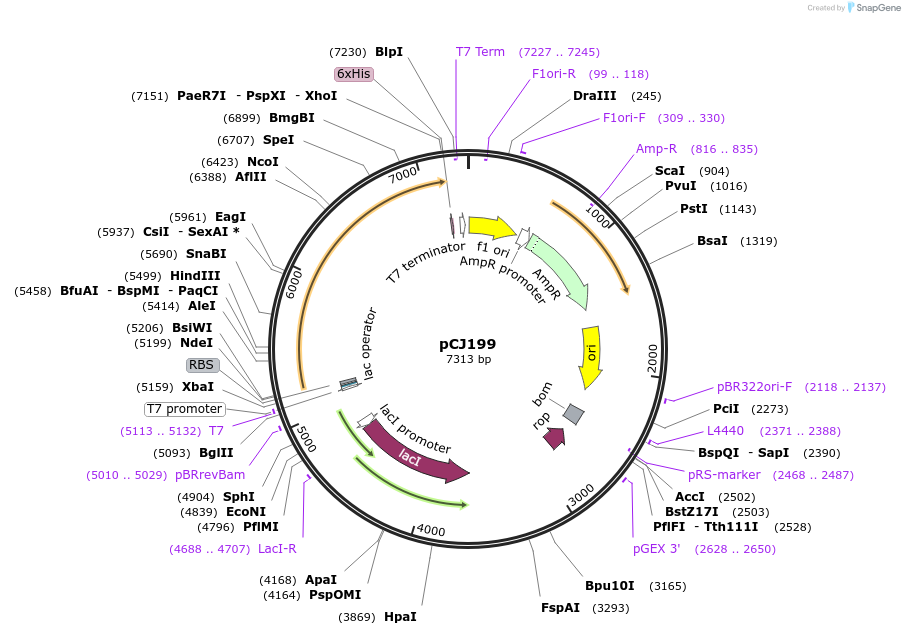
pCJ199
Plasmid#162672PurposepET-21b(+) based plasmid for expression of the putative MHET hydrolase from Comamonas thiooxydans (Genbank WP_080747404.1) with C-terminal His tag, codon optimized for expression in E. coli K12.DepositorInsertPutative MHET hydrolase from Comamonas thiooxydans (Genbank WP_080747404.1) with signal peptide
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
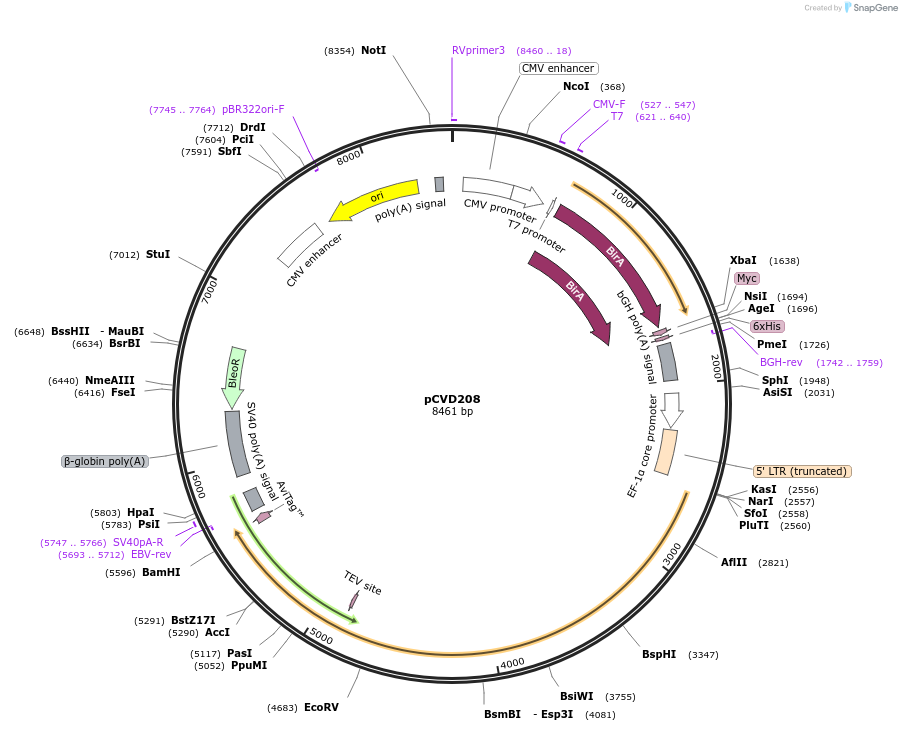
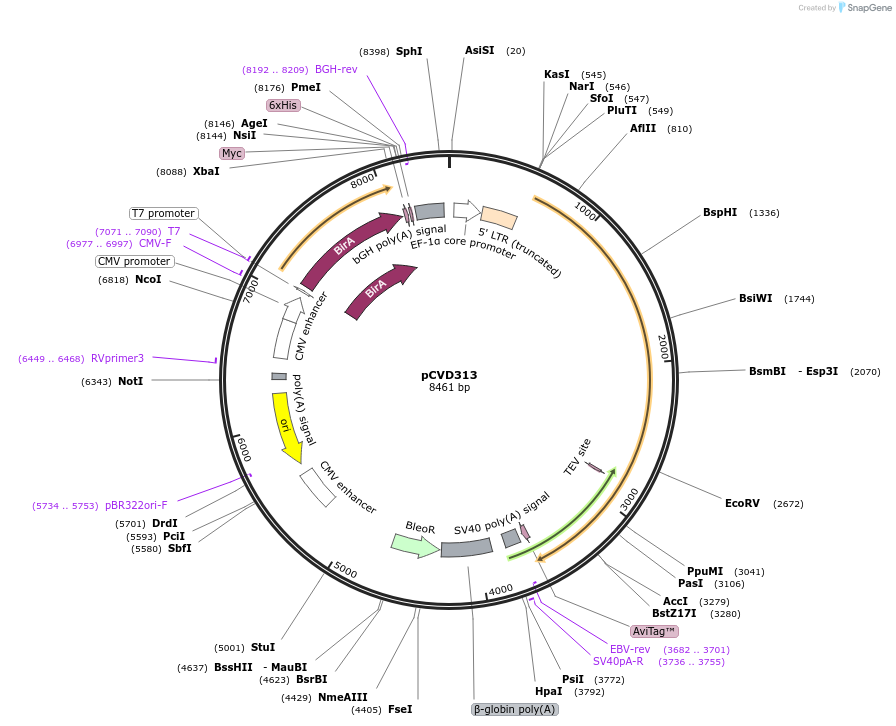
pCVD313
Plasmid#159771PurposeExpression vector for mutated Fc-fused ACE2(740)DepositorAvailable SinceFeb. 12, 2021AvailabilityAcademic Institutions and Nonprofits only -
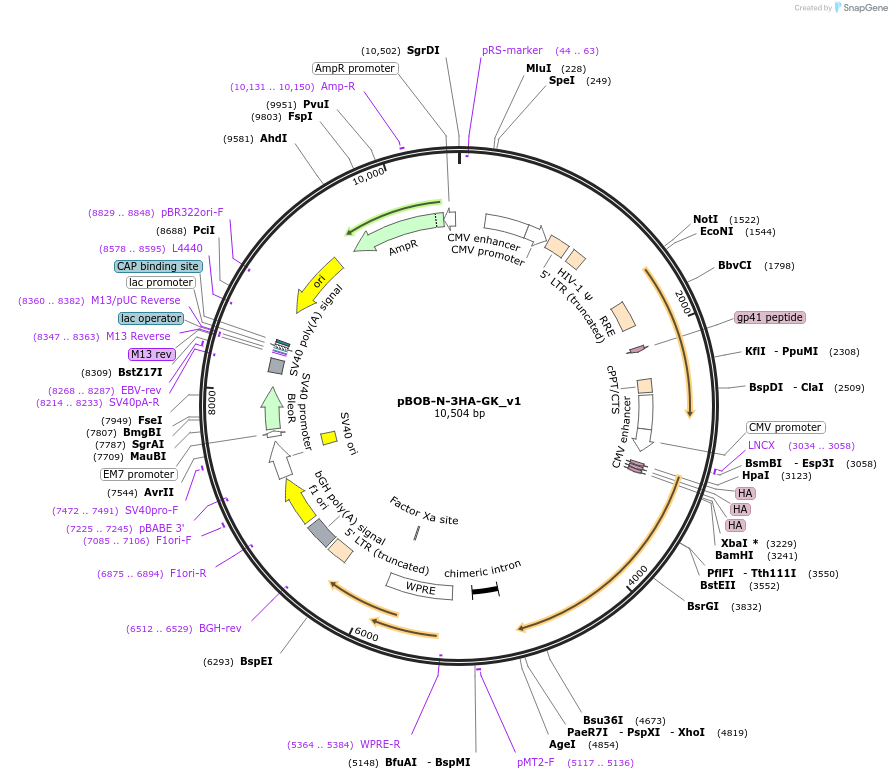
pBOB-N-3HA-GK_v1
Plasmid#134275PurposeLentivector encoding 3XHA-tagged GK (isoform 1)DepositorInsertGk (Gk Mouse)
UseLentiviralTags3X HAExpressionMammalianMutation524 amino acids, isoform 1PromoterCMVAvailable SinceMay 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
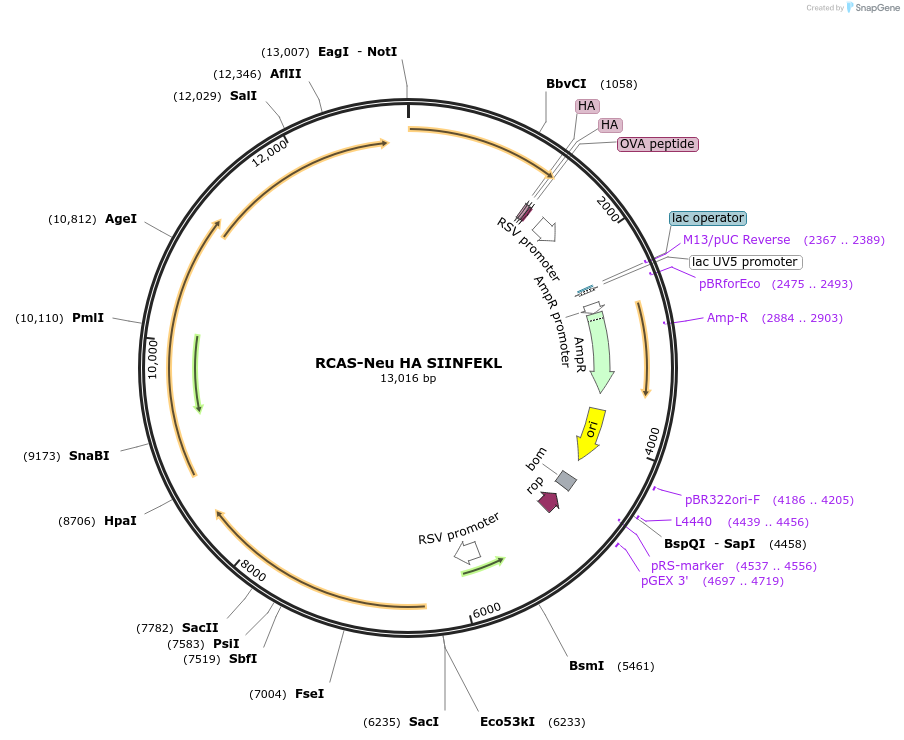
RCAS-Neu HA SIINFEKL
Plasmid#125832PurposeRetroviral expression of rat Neu cDNADepositorInsertNeu (Erbb2 Rat)
UseRetroviralTags2x HA and SIINFEKLExpressionMammalianMutationVal to Glu point mutation of codon 664 and trunca…Available SinceJuly 23, 2019AvailabilityAcademic Institutions and Nonprofits only -
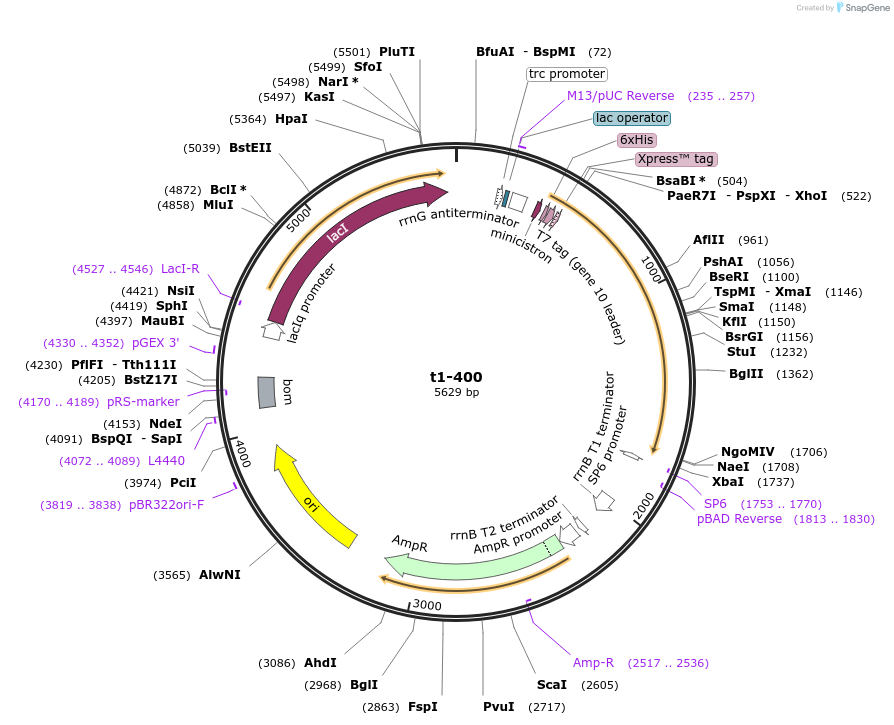
t1-400
Plasmid#166577PurposeFor expression of human talin head (residues 1-400) in E. coli. N-terminal His6-tag, Xpress-epitope (DLYDDDDK) and enterokinase cleavage site for tag removal.DepositorInsertTln1 Head (1-400) (TLN1 Human)
TagsHis6-tag, Xpress-epitope (DLYDDDDK) and enterokin…ExpressionBacterialMutationresidues 1-400 onlyAvailable SinceApril 13, 2021AvailabilityAcademic Institutions and Nonprofits only -
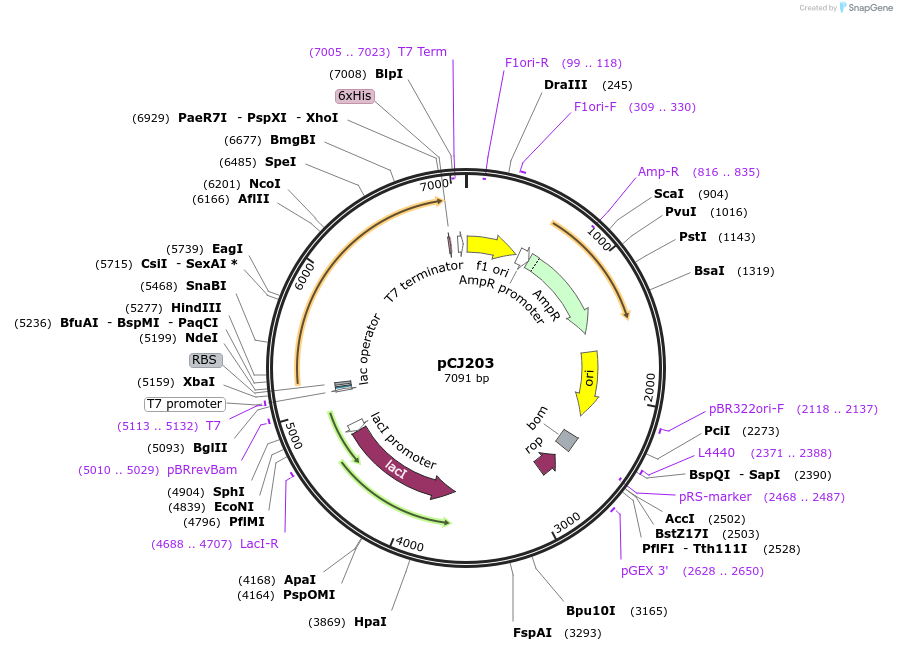
pCJ203
Plasmid#162678PurposepET-21b(+) based plasmid for expression of the putative MHET hydrolase from Comomonas thiooxydans (Genbank WP_080747404.1) with deletion of the predicted signal peptide (Phe2:Ala75)DepositorInsertPutative MHETase from Comomonas thiooxydans (Genbank WP_080747404.1) without 75 residue signal peptide
TagsHisExpressionBacterialMutationdeltaF2-A75; codon optimized for expression in E.…PromoterT7/lacAvailable SinceFeb. 3, 2021AvailabilityAcademic Institutions and Nonprofits only -
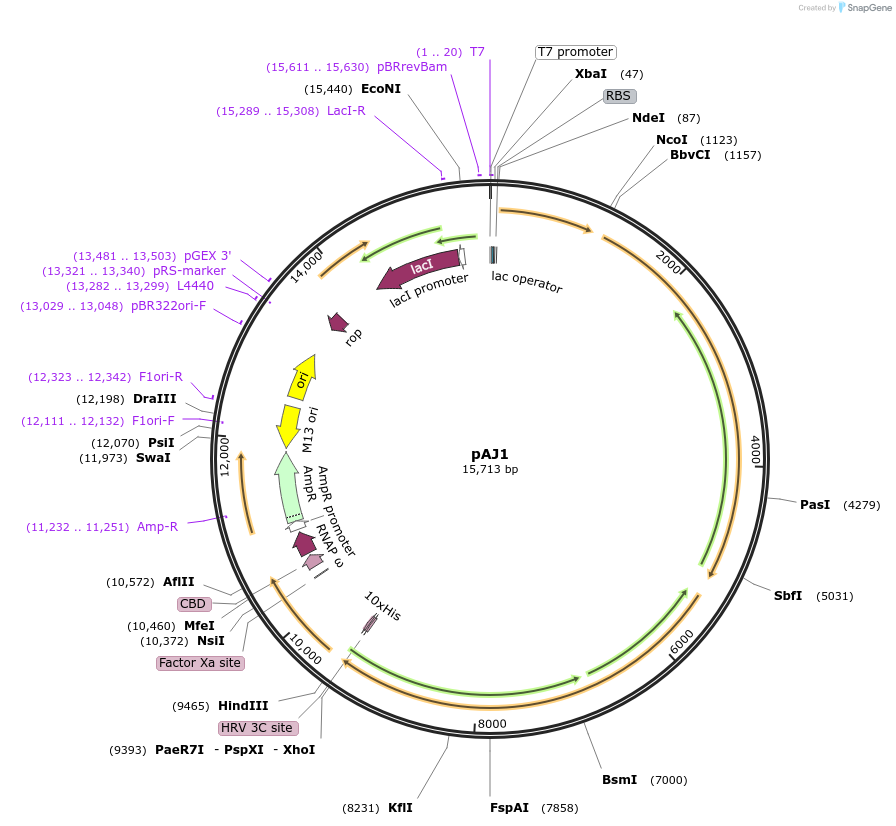
pAJ1
Plasmid#138379PurposepEcRNAP6 derivative encoding Escherichia coli core RNA polymerase subunits: RpoA, RpoB, RpoC(His)10, RpoZDepositorTagsHis10ExpressionBacterialMutationClamp toe deletion: Δ(Tyr144-Lys179) ::(G144-S145…PromoterT7Available SinceApril 1, 2020AvailabilityAcademic Institutions and Nonprofits only