We narrowed to 6,763 results for: poly
-
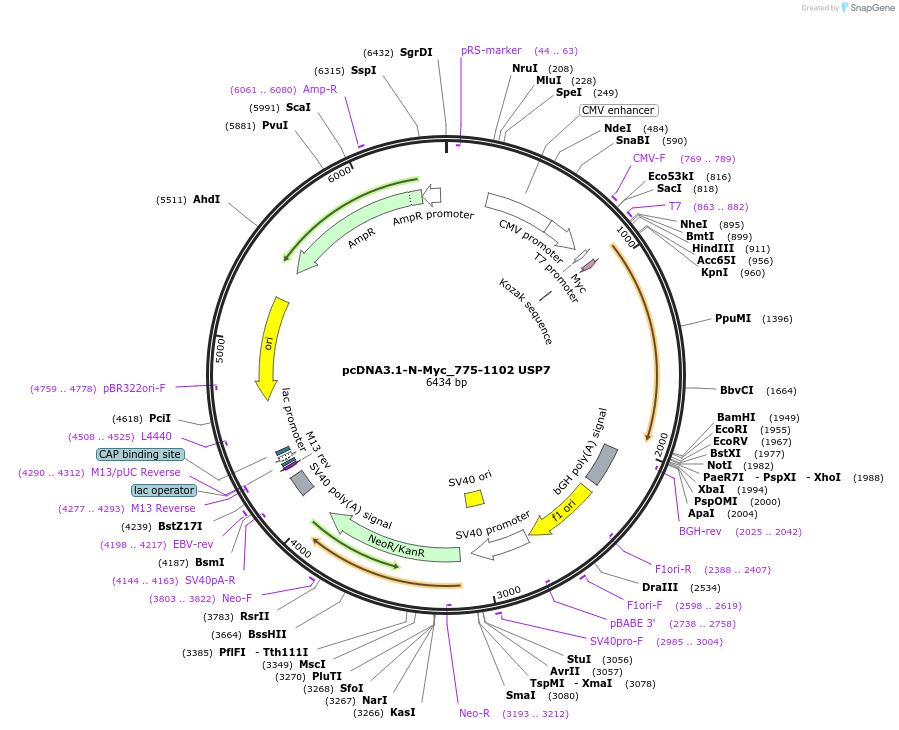
Plasmid#131250PurposeMammalian expression of the USP7 UBL3-5 domains with an N-terminal Myc tagDepositorInsertUbiquitin carboxyl-terminal hydrolase 7 (USP7 Human)
TagsMycExpressionMammalianMutationEncodes USP7 UBL domains 3-5 (USP7 amino acids 77…Available SinceDec. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
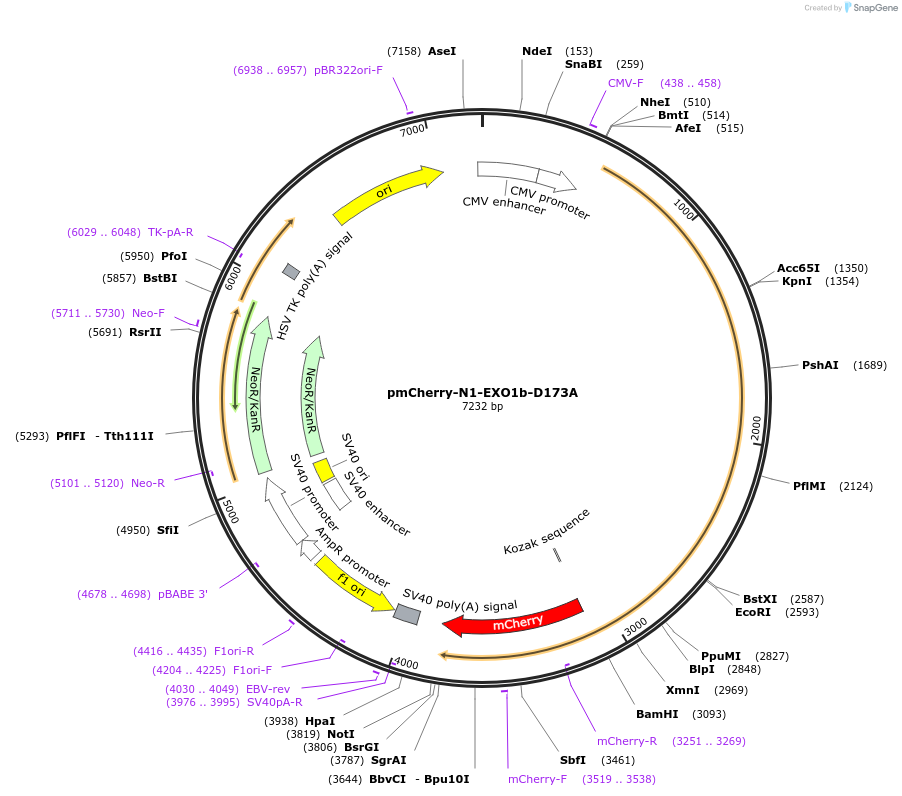
pmCherry-N1-EXO1b-D173A
Plasmid#111622PurposeExpression of human catalitic dead Exonuclease 1 (EXO1) mCherry-tagged in mammalian cellsDepositorAvailable SinceJune 26, 2018AvailabilityAcademic Institutions and Nonprofits only -
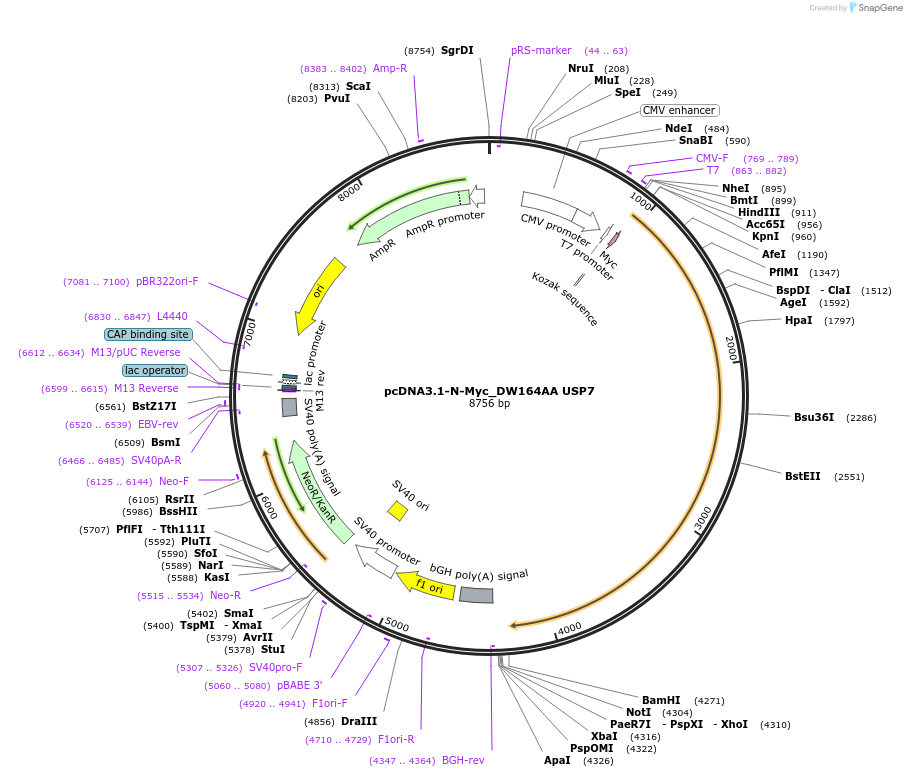
pcDNA3.1-N-Myc_DW164AA USP7
Plasmid#131252PurposeMammalian expression of N-terminally Myc-tagged DW164AA USP7DepositorInsertUbiquitin carboxyl-terminal hydrolase 7 (USP7 Human)
TagsMycExpressionMammalianMutationContains a point mutation that converts USP7 aspa…Available SinceDec. 17, 2020AvailabilityAcademic Institutions and Nonprofits only -
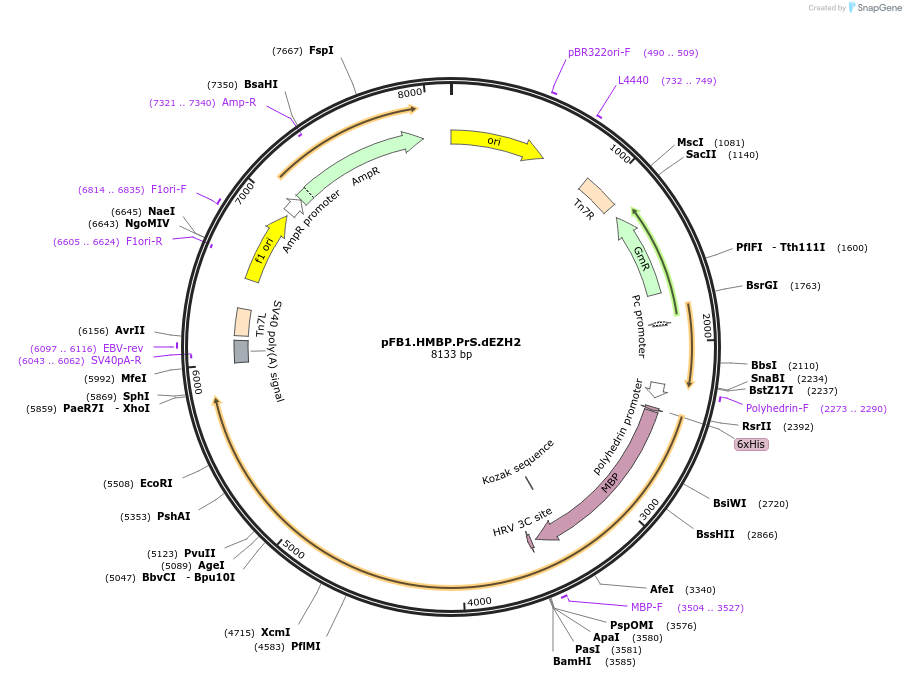
pFB1.HMBP.PrS.dEZH2
Plasmid#220241PurposeExpression of an EZH2 catalytic site mutant in insect cells.DepositorAvailable SinceJune 12, 2024AvailabilityAcademic Institutions and Nonprofits only -
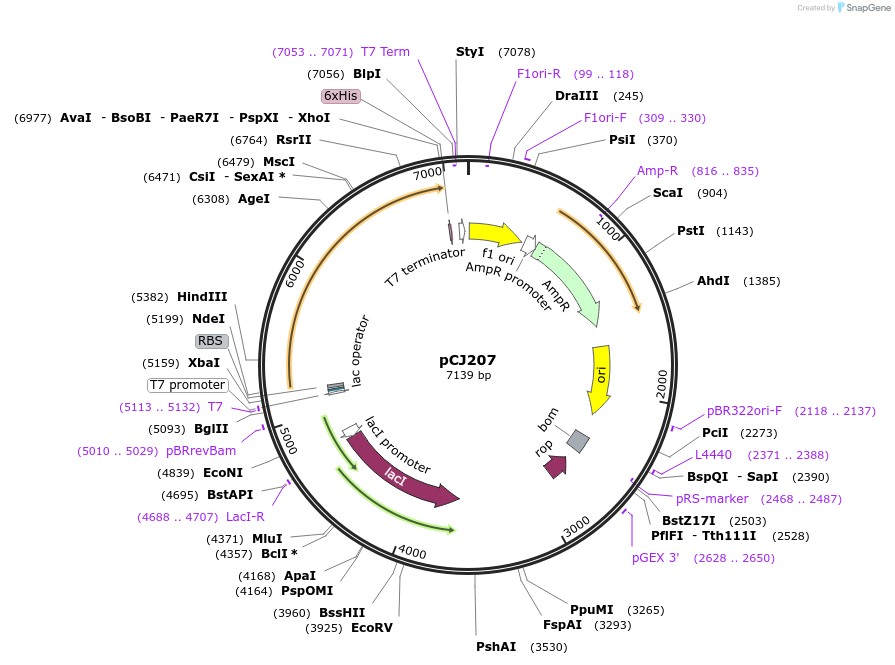
pCJ207
Plasmid#162680PurposepET-21b(+) based plasmid for expression of the putative MHETase from Hydrogenophaga sp. PML113 (Genbank WP_083293388.1) with C-terminal His tag, codon optimized for expression in E. coli K12.DepositorInsertPutative MHETase from Hydrogenophaga sp. PML113 (Genbank WP_083293388.1) with signal peptide
TagsHisExpressionBacterialMutationcodon optimized for expression in E. coli K12PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
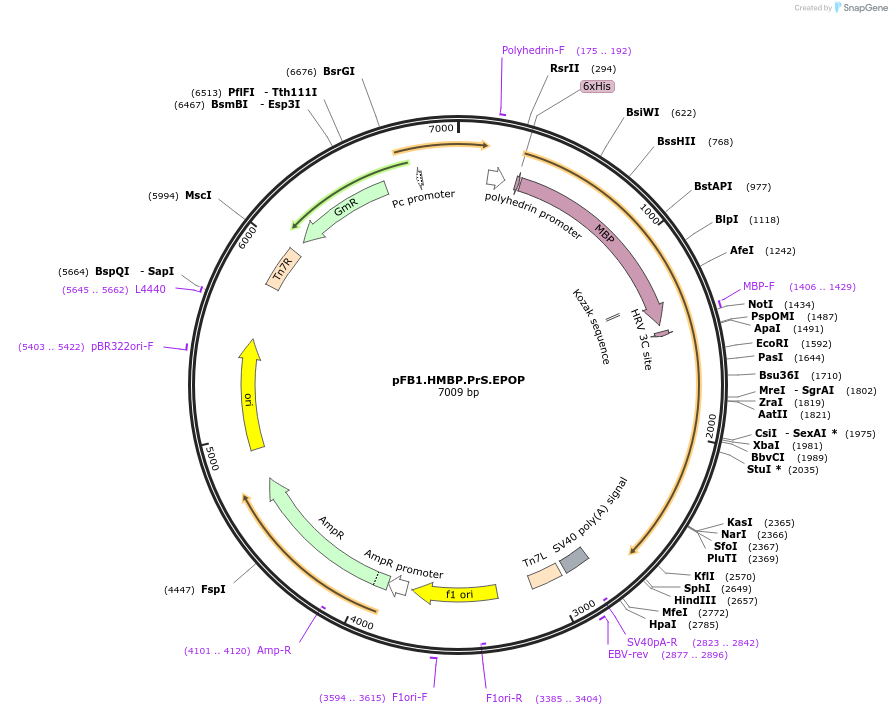
pFB1.HMBP.PrS.EPOP
Plasmid#125170PurposeExpresses human EPOP in insect cells, under a C3-cleavable (PreScission Protease) N-terminal hexahistidine-MBP tag.DepositorAvailable SinceMay 7, 2020AvailabilityAcademic Institutions and Nonprofits only -
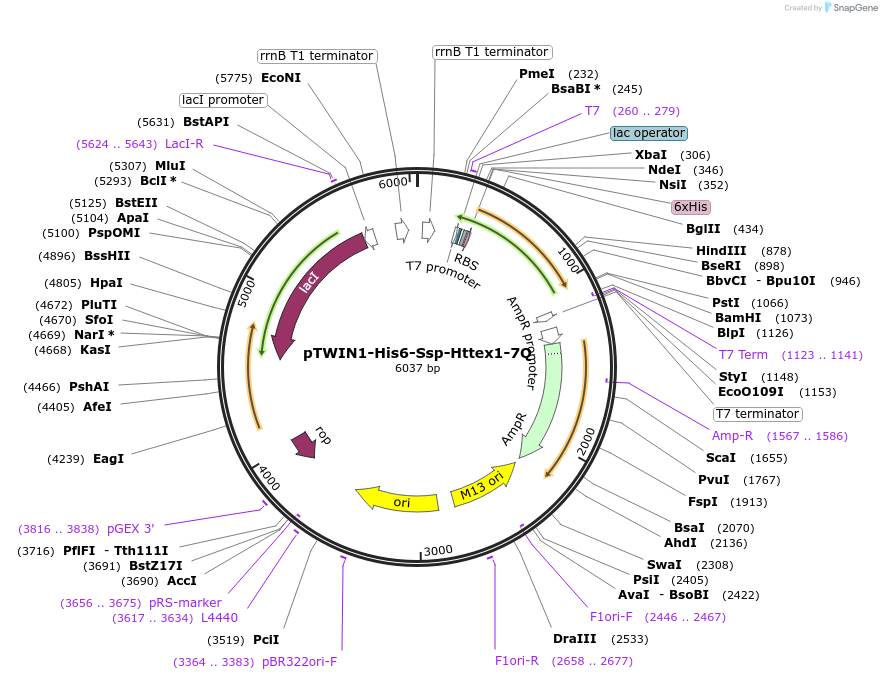
pTWIN1-His6-Ssp-Httex1-7Q
Plasmid#84347PurposeExpression of the human Huntingtin Exon1 protein containing 7Q in E.coliDepositorInsertHuntingtin Exon 1 (HTT Human)
TagsN-terminal His6-tagged intein SspExpressionBacterialPromoterT7Available SinceDec. 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
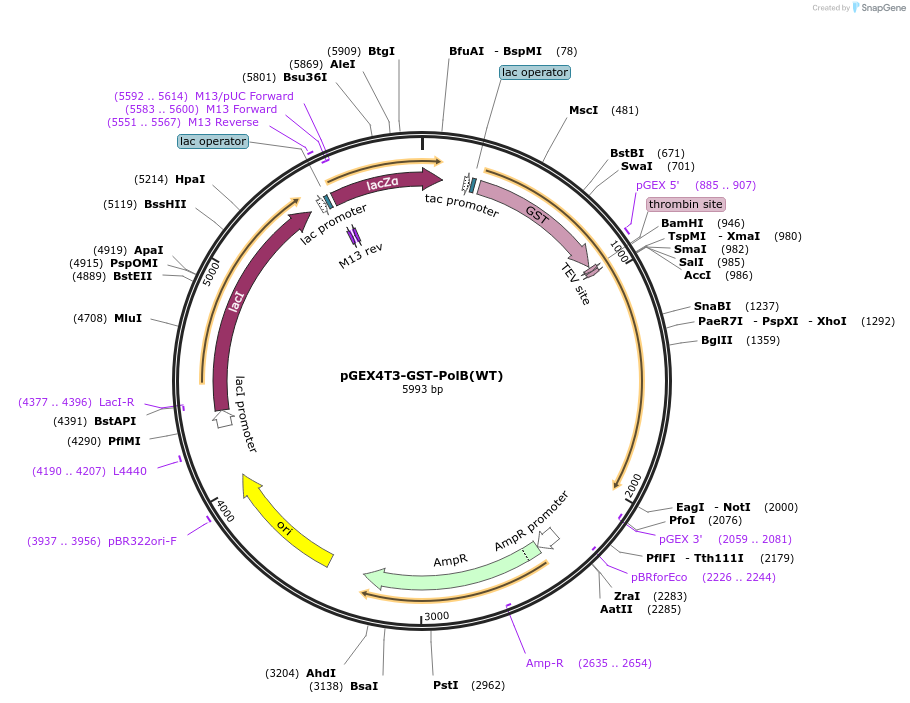
pGEX4T3-GST-PolB(WT)
Plasmid#177131PurposeBacterial vector for expression of an N-terminal GST fusion of PolB(WT) with a TEV protease site located between the GST tag and PolB(WT)DepositorAvailable SinceDec. 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
pET21a-Med25-VBD-6His
Plasmid#64773PurposeBacterial expression of His-tagged Med25-VBDDepositorAvailable SinceJune 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
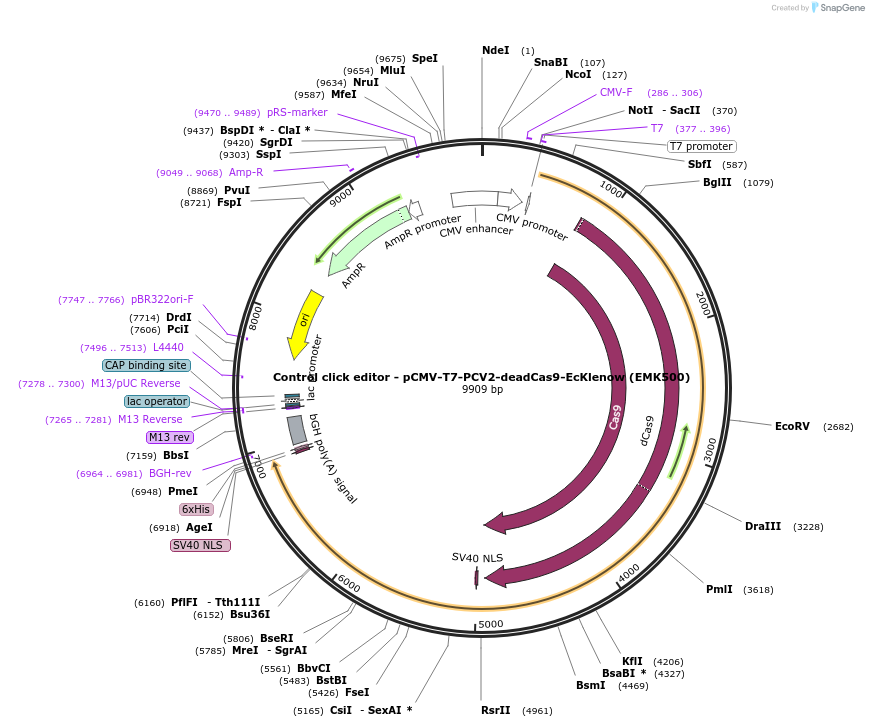
Control click editor - pCMV-T7-PCV2-deadCas9-EcKlenow (EMK500)
Plasmid#208944PurposeA control CE1 construct with catalytically inactivated SpCas9, expressed from CMV or T7 promoters.DepositorInsertPCV2-XTEN-dSpCas9-BPNLS-EcKlenow(-exo)-BPNLS
UseCRISPR; In vitro transcription; t7 promoterTagsBPNLSExpressionMammalianMutationdSpCas9(D10A/H840A);EcKlenow(-exo;D355A/E357A)PromoterCMV and T7Available SinceNov. 22, 2023AvailabilityAcademic Institutions and Nonprofits only -
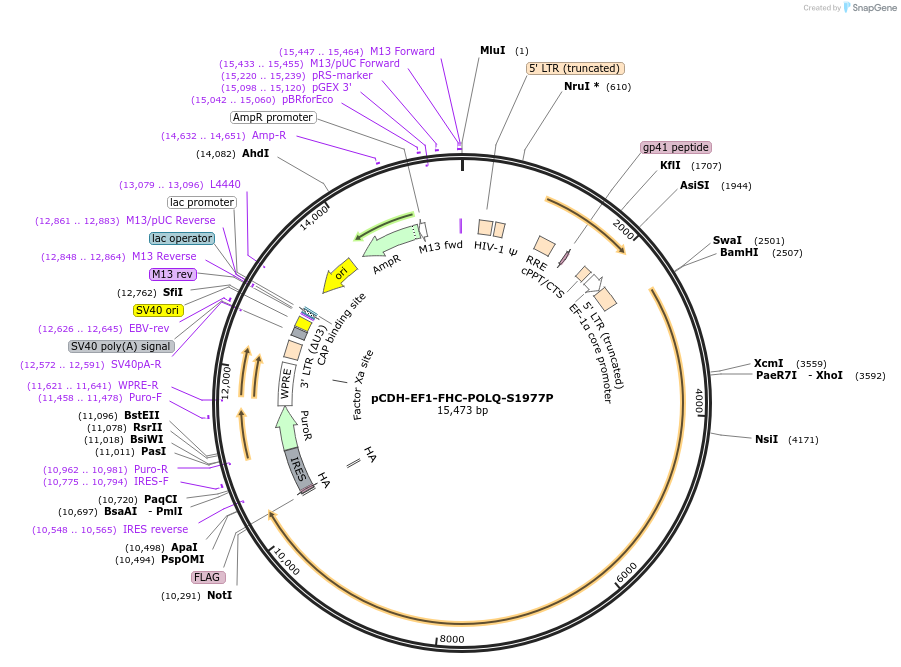
pCDH-EF1-FHC-POLQ-S1977P
Plasmid#64876Purposelentiviral expression of human POLQ S1977P mutant (mimicks the chaos1 mutation)DepositorInsertPOLQ (POLQ Human)
UseLentiviralTagsFLAG and HAExpressionMammalianMutationS1977P, mimicking the chaos1 mutationPromoterEF1alphaAvailable SinceJuly 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
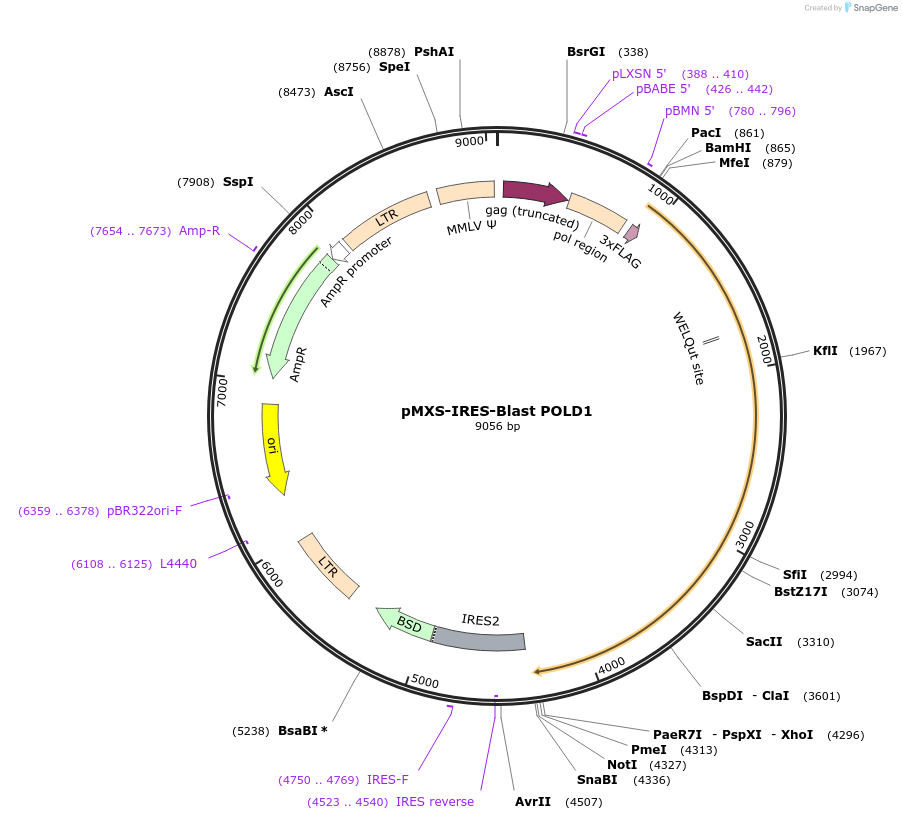
pMXS-IRES-Blast POLD1
Plasmid#160805PurposeExpress POLD1DepositorAvailable SinceJan. 26, 2021AvailabilityAcademic Institutions and Nonprofits only -
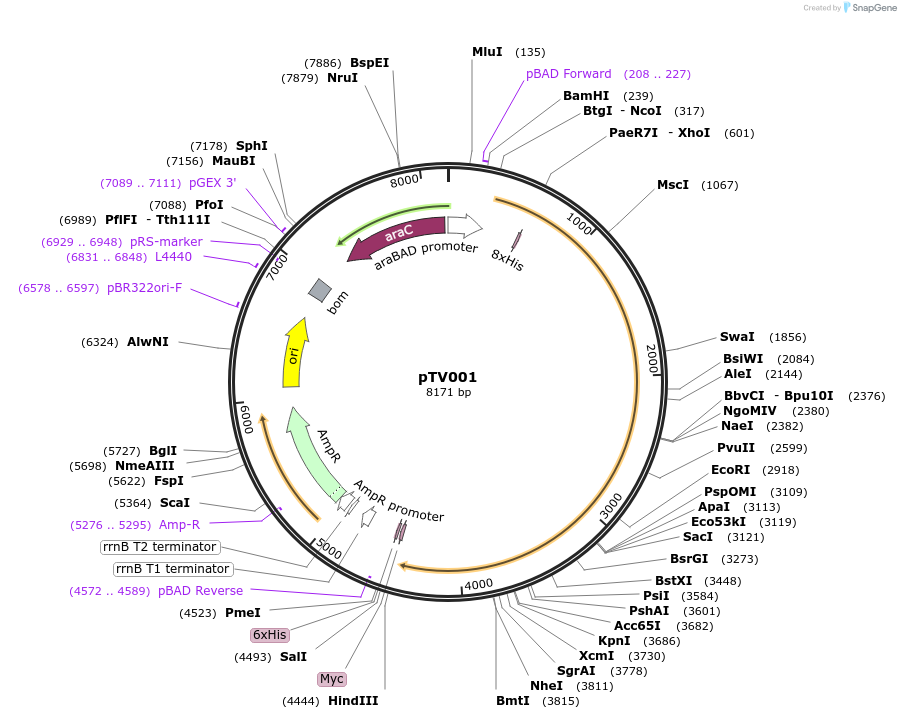
pTV001
Plasmid#130265PurposeEatA expression plasmid containing 8His encoding region in permissive site of eatA geneDepositorInserteatA::His8
Tagspolyhistidine (internal)ExpressionBacterialMutation8His polyhistidine encoding region and XhoI site …PromoteraraBADAvailable SinceNov. 18, 2021AvailabilityAcademic Institutions and Nonprofits only -
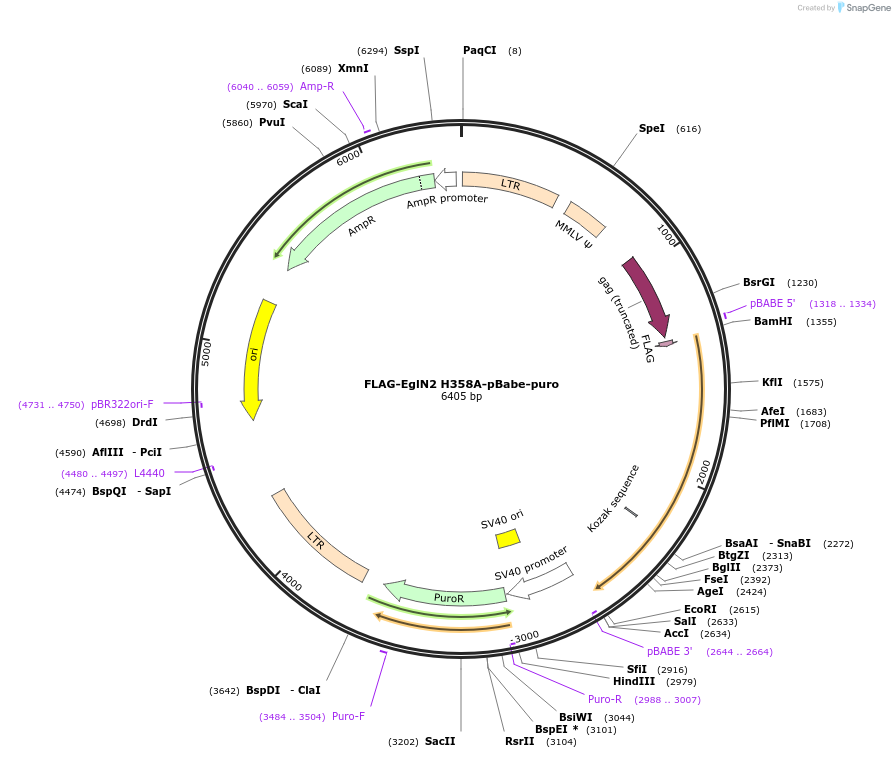
FLAG-EglN2 H358A-pBabe-puro
Plasmid#22705DepositorInsertEgg laying Nine - 2 (EglN2) (EGLN2 Human)
UseRetroviralTagsFLAGExpressionMammalianMutationmutated cDNA: point mutation in codon 358 changi…Available SinceDec. 29, 2009AvailabilityAcademic Institutions and Nonprofits only -
pCAG-Flag-Avi-ires-CHD5
Plasmid#68878PurposeTransient mammalian expression of CHD5 for BirA taggingDepositorAvailable SinceSept. 17, 2015AvailabilityAcademic Institutions and Nonprofits only -
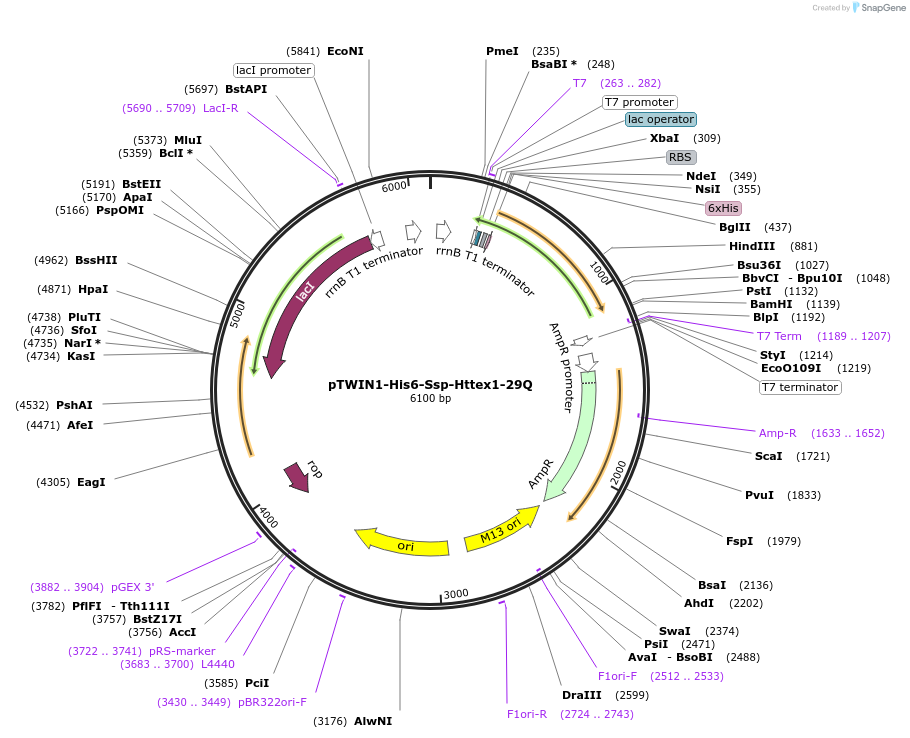
pTWIN1-His6-Ssp-Httex1-29Q
Plasmid#84350PurposeExpression of the human Huntingtin Exon1 protein containing 29Q in E.coliDepositorInsertHuntingtin Exon 1 (HTT Human)
TagsN-terminal Ssp intein (His-tagged)ExpressionBacterialPromoterT7Available SinceJan. 18, 2018AvailabilityAcademic Institutions and Nonprofits only -
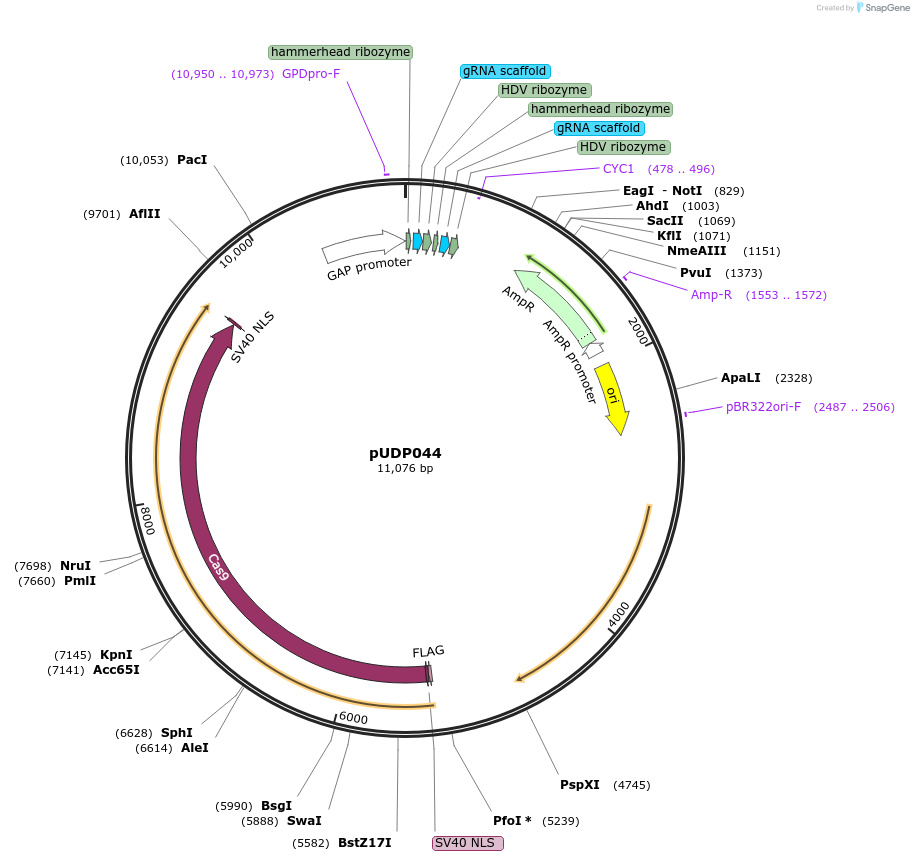
pUDP044
Plasmid#101168PurposepUDP004 expressing a polycistronic g-RNA array for Cas9 editing targeting the genes SeATF1 and SeATF2 and Spcas9D147Y P411T in S. pastorianus (HH-gRNASeATF1-HDV-linker-HH-gRNASeATF2-HDV)DepositorInsertpolycistronic HH-gRNA-HDV-HH-gRNA-HDV array targetting SeATF1 and 2 in S. pastorianus
UseCRISPRExpressionYeastPromoterScTDH3Available SinceDec. 4, 2017AvailabilityAcademic Institutions and Nonprofits only -
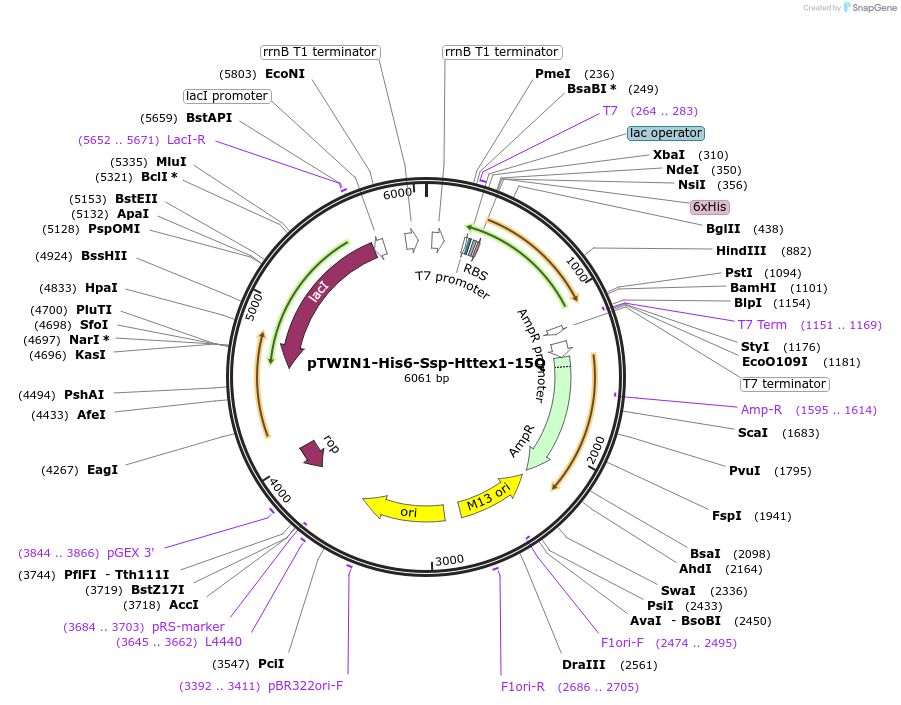
pTWIN1-His6-Ssp-Httex1-15Q
Plasmid#84348PurposeExpression of the human Huntingtin Exon1 protein containing 15Q in E.coliDepositorInsertHuntingtin Exon1 (HTT Human)
TagsN-terminal Ssp intein (His-tagged)ExpressionBacterialPromoterT7Available SinceDec. 19, 2017AvailabilityAcademic Institutions and Nonprofits only -
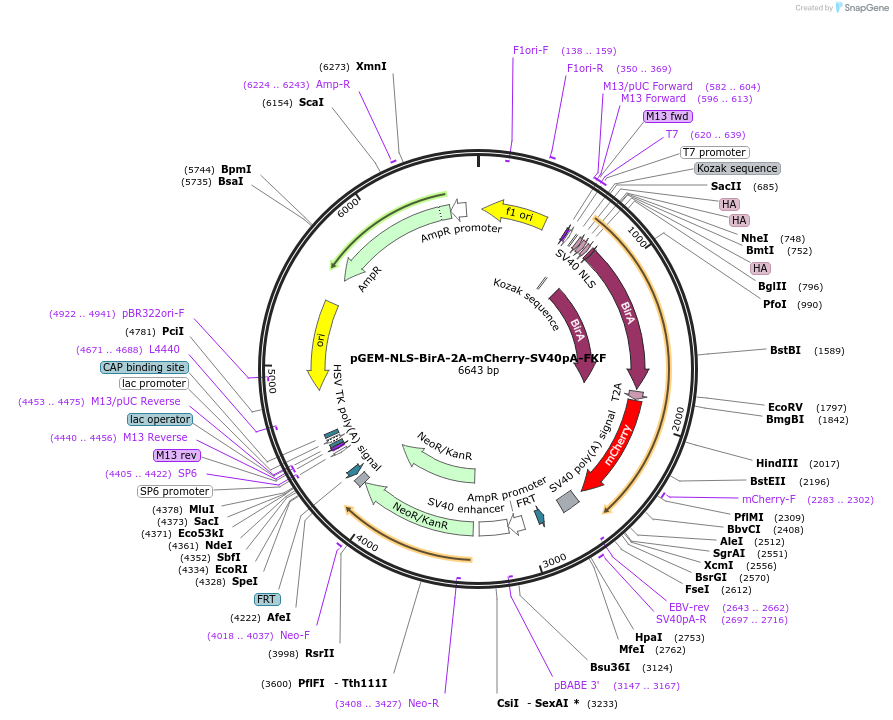
pGEM-NLS-BirA-2A-mCherry-SV40pA-FKF
Plasmid#79889PurposeDonor cassette containing HA-tagged biotin ligase (BirA) with NLS, a Thosea asigna virus peptide (2A), mCherry protein and SV40 polyA tail, followed by FRT site-flanked Kanamycin selection geneDepositorInsertHA-tagged NLS-BirA, 2A, mCherry protein and SV40 polyA, followed by FRT site-flanked Kanamycin selection gene
ExpressionBacterialAvailable SinceOct. 11, 2016AvailabilityAcademic Institutions and Nonprofits only -
pET41s-GST-ATF6a-AD
Plasmid#64774Purposebacterial expression of GST-tagged ATF6 (1-150)DepositorAvailable SinceJune 8, 2015AvailabilityAcademic Institutions and Nonprofits only