We narrowed to 1,740 results for: plasmid for e coli
-
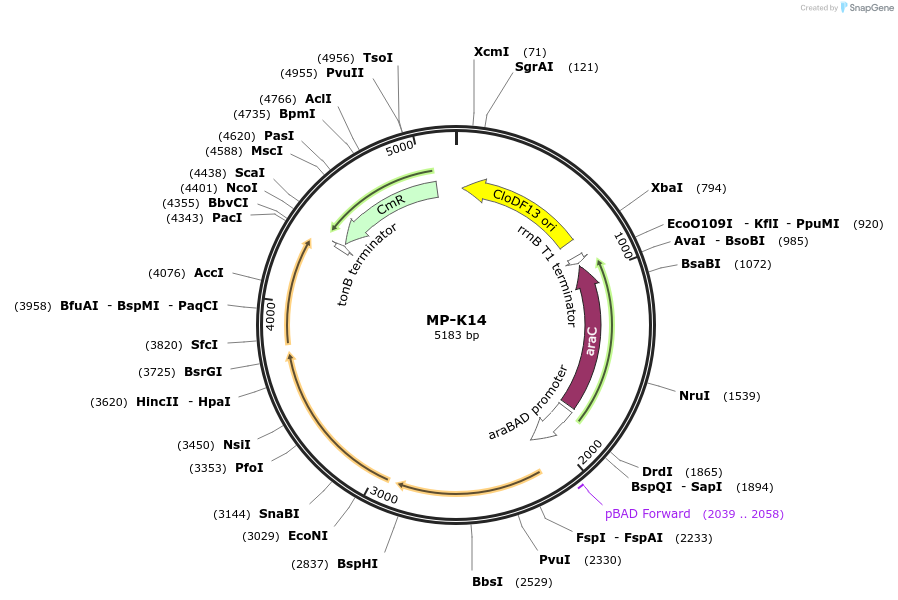
Plasmid#69658PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA(RBS+)
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
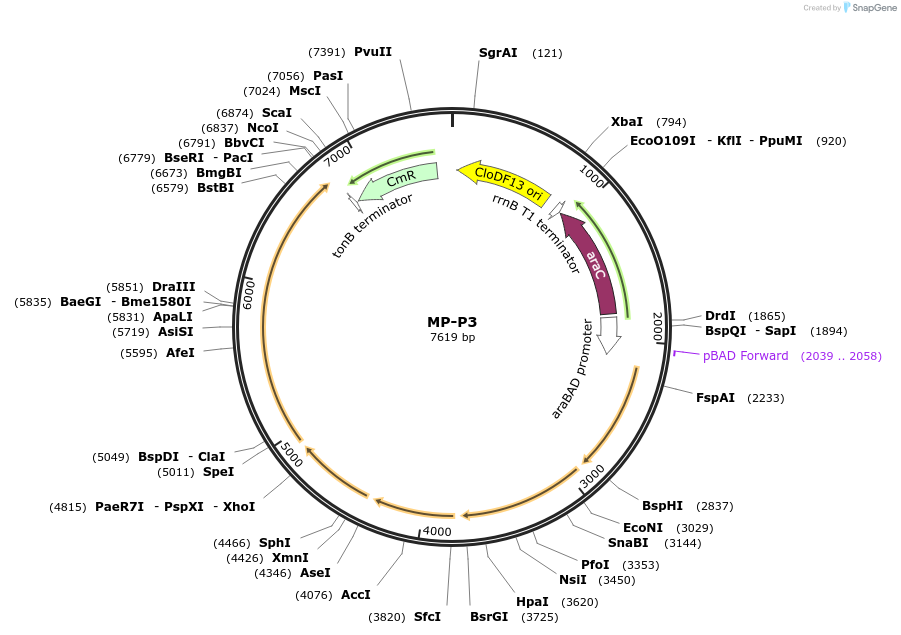
MP-P3
Plasmid#69662PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA emrR mutL713
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
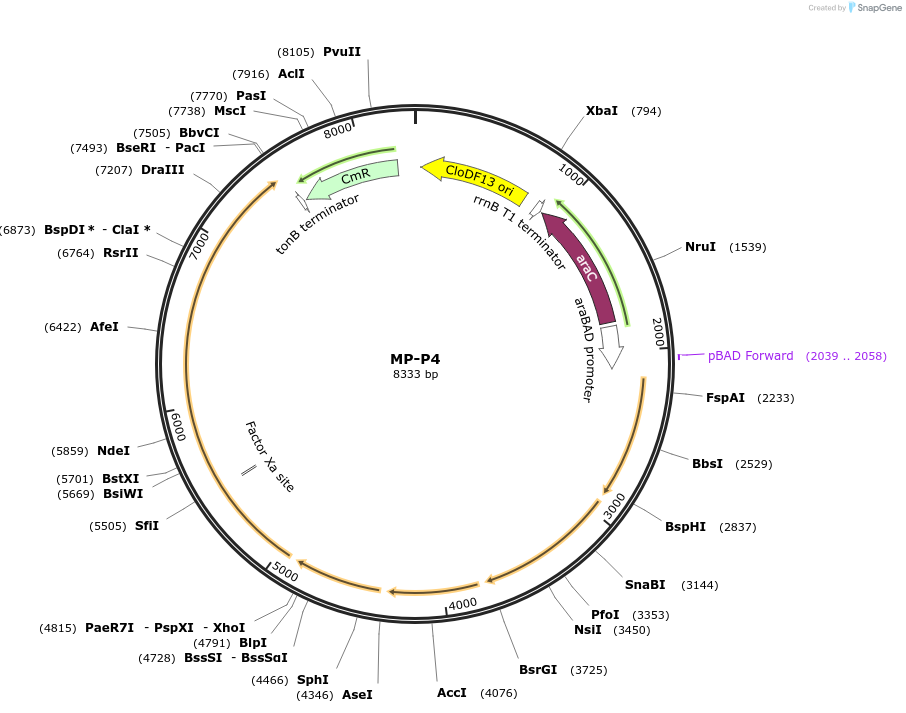
MP-P4
Plasmid#69663PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA emrR mutS503'
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
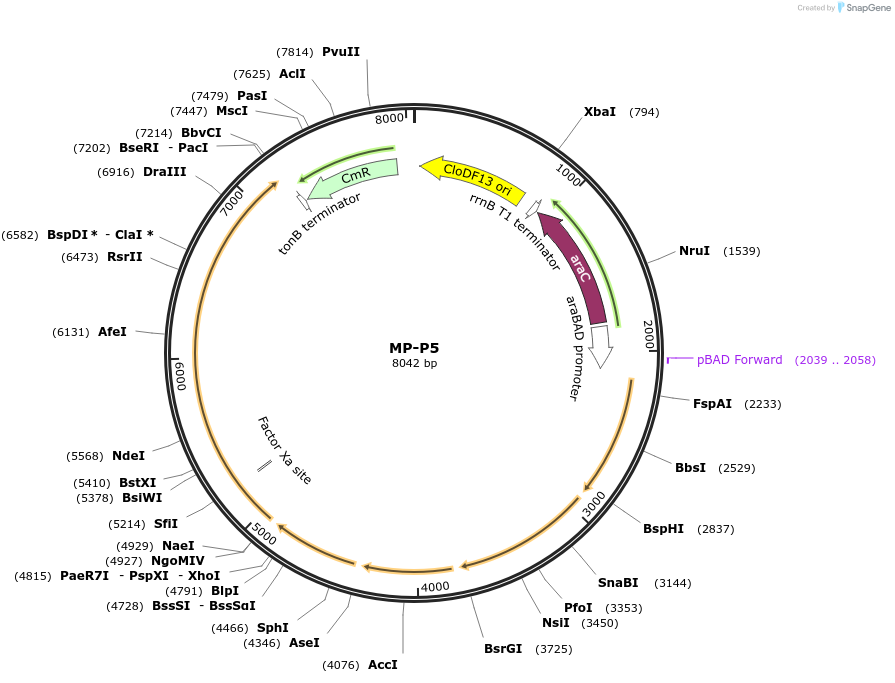
MP-P5
Plasmid#69664PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA emrR mutSdN
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
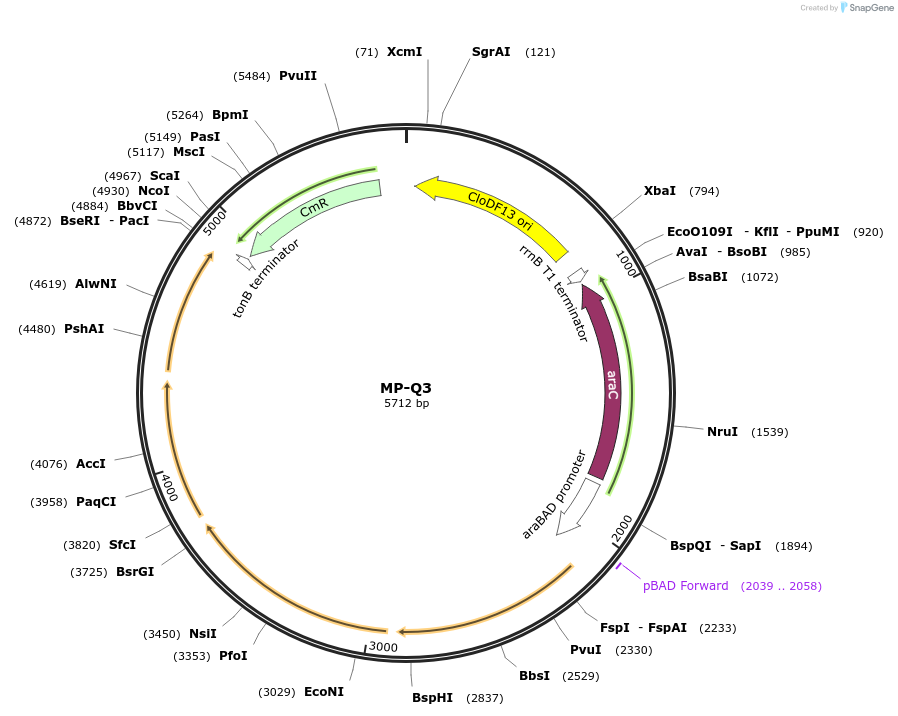
MP-Q3
Plasmid#69673PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA yffI
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
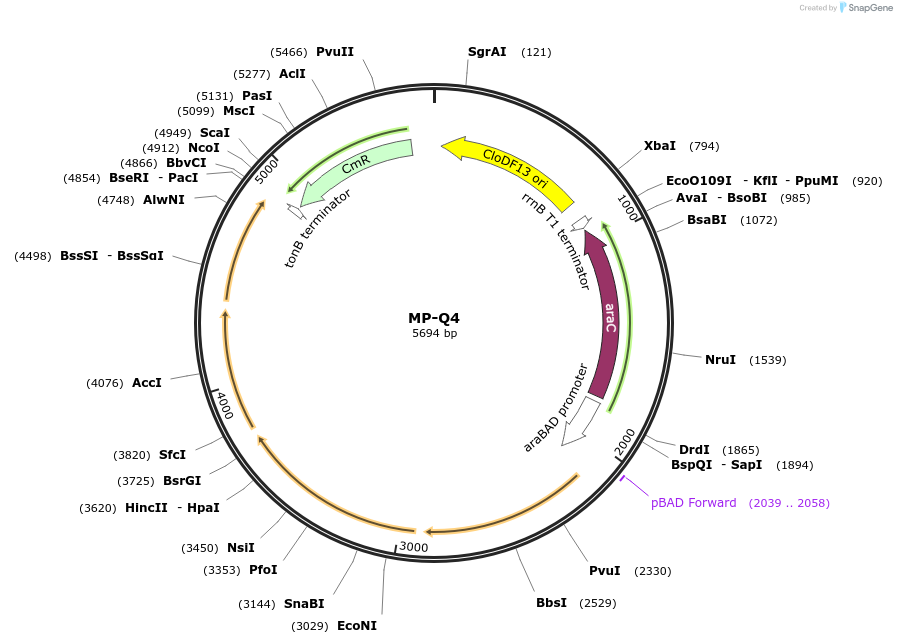
MP-Q4
Plasmid#69674PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA yfjY
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
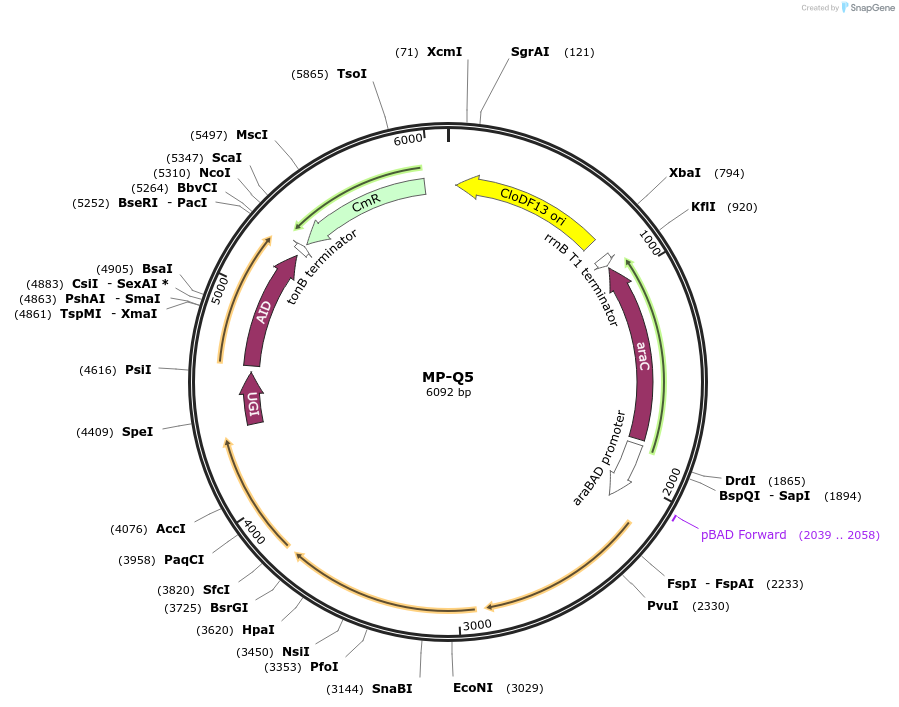
MP-Q5
Plasmid#69675PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA ugi AID
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
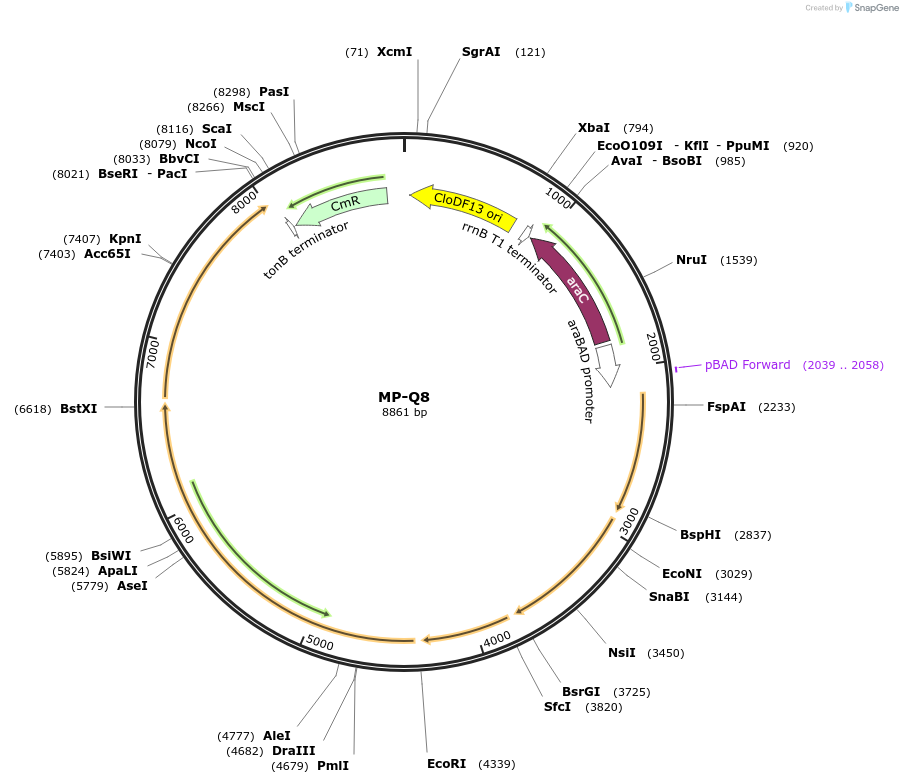
MP-Q8
Plasmid#69678PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA nrdAB
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
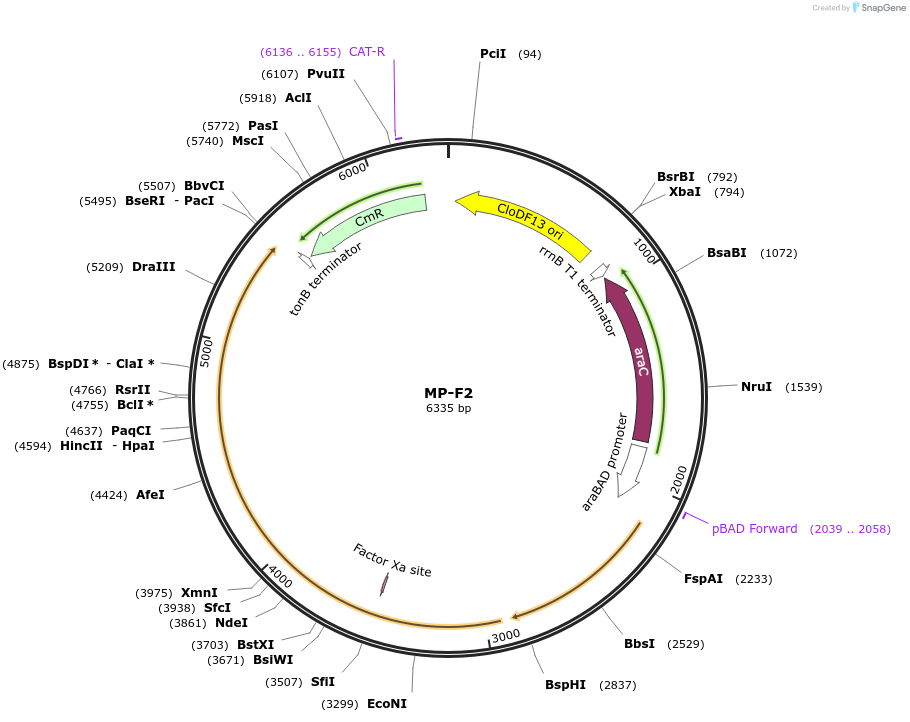
MP-F2
Plasmid#69638PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutS538'
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
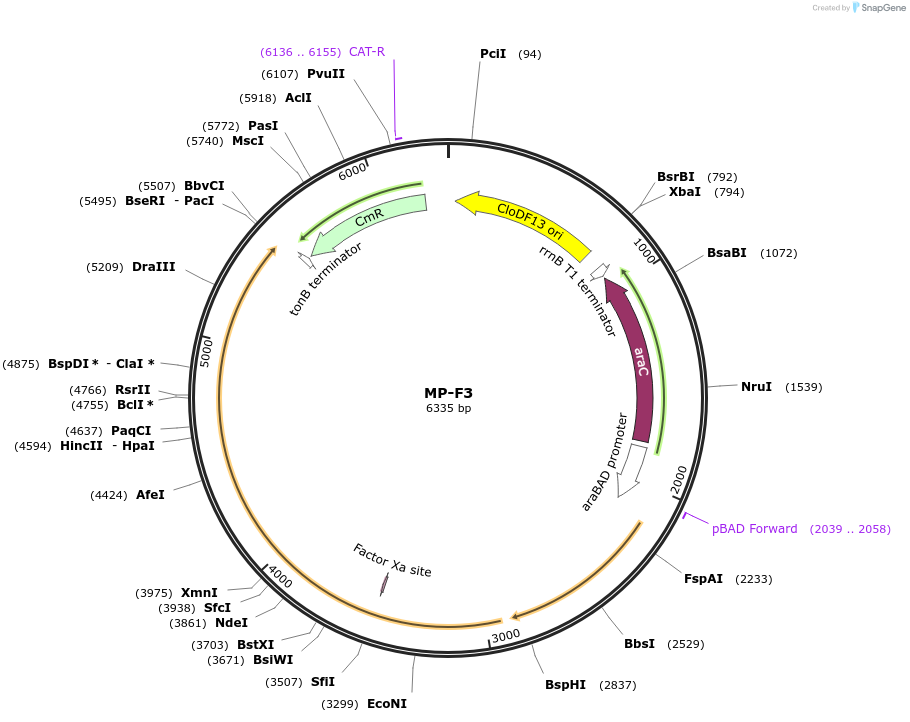
MP-F3
Plasmid#69639PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutS503'
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
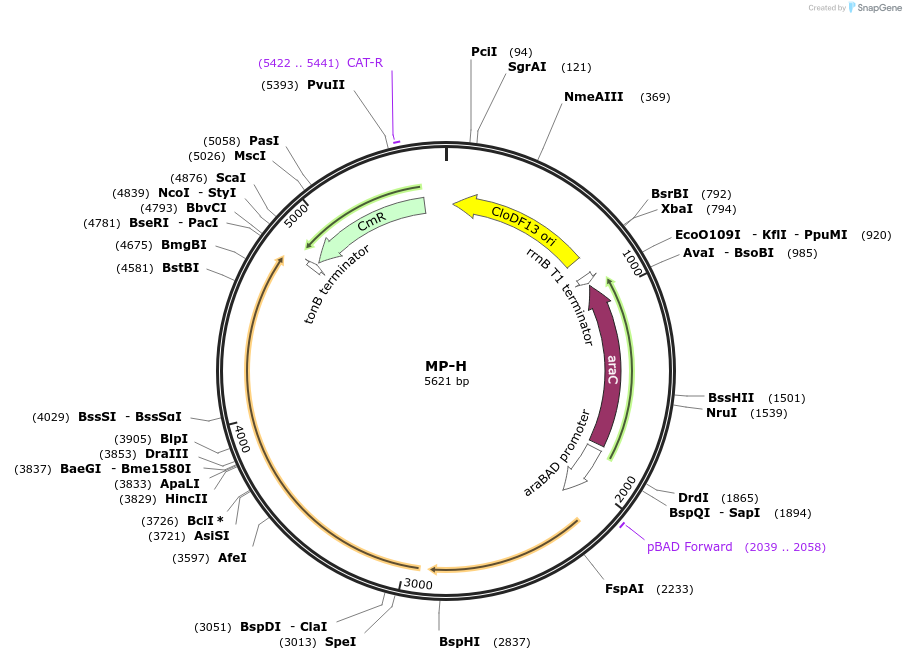
MP-H
Plasmid#69640PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutL705
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
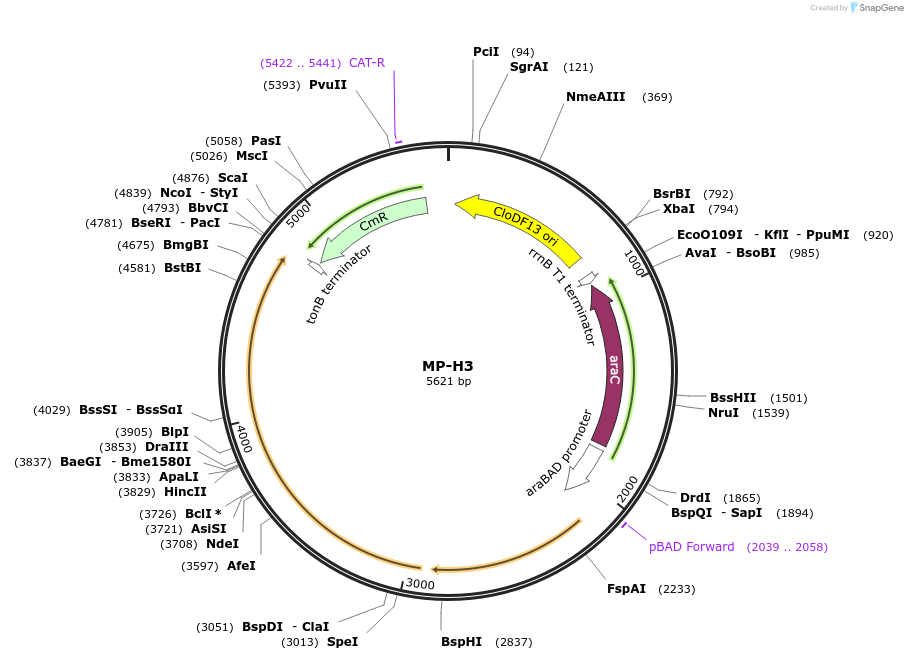
MP-H3
Plasmid#69642PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutL(R261H)
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
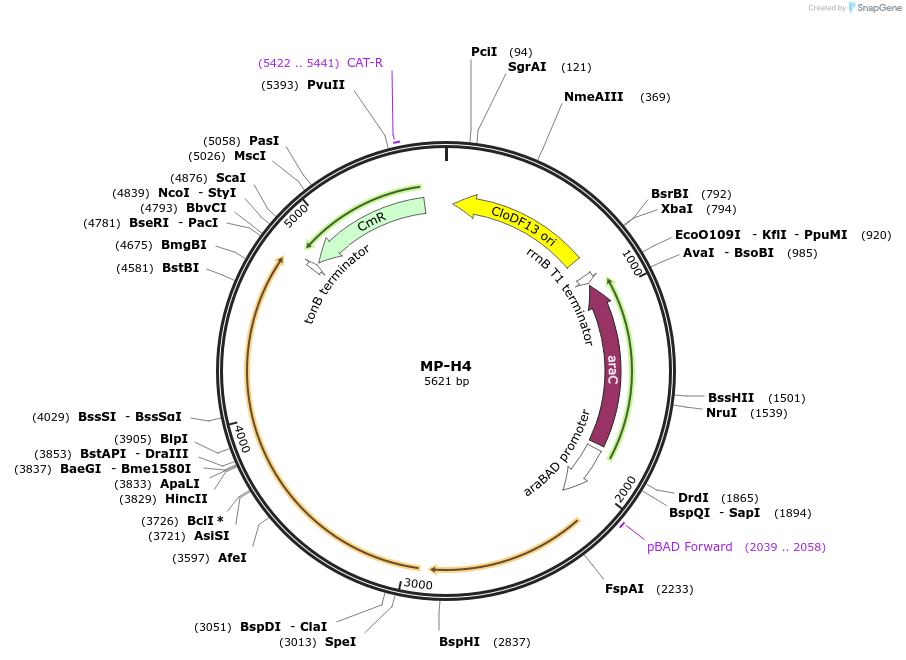
MP-H4
Plasmid#69643PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutL(K307A)
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
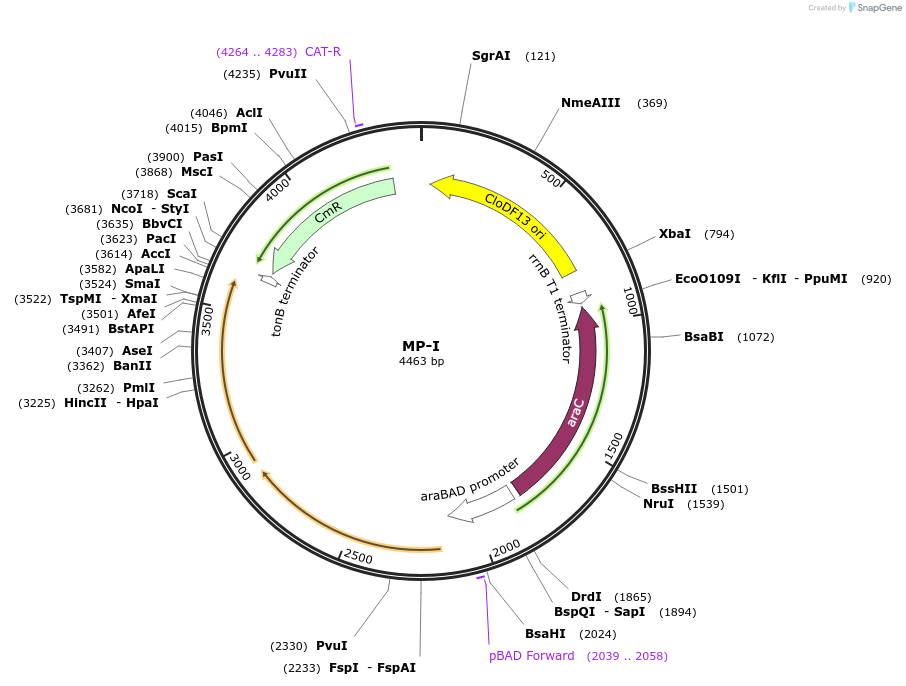
MP-I
Plasmid#69644PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutH(E56A)
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
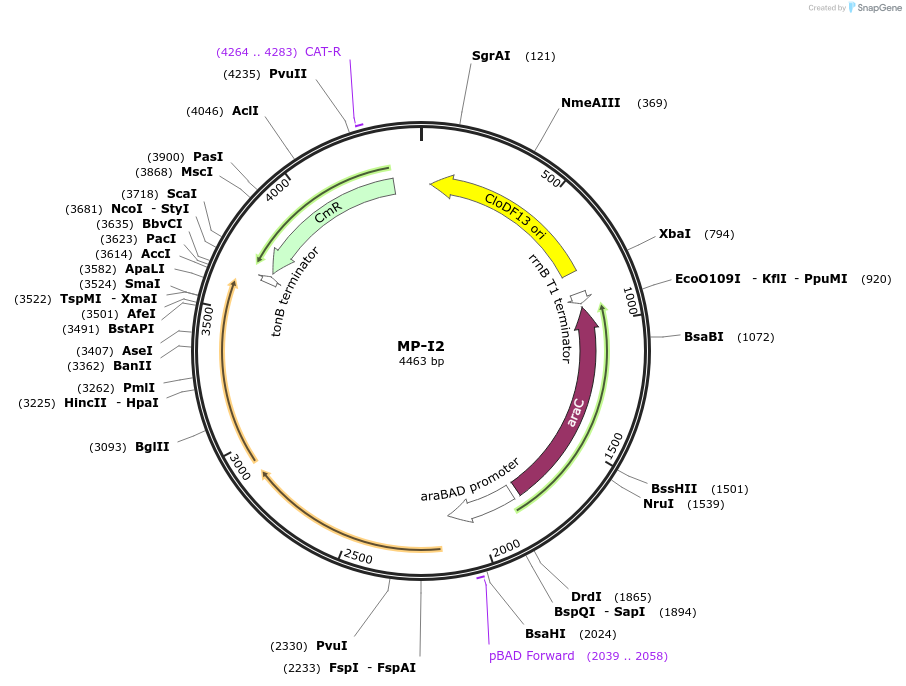
MP-I2
Plasmid#69645PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutH(K79E)
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
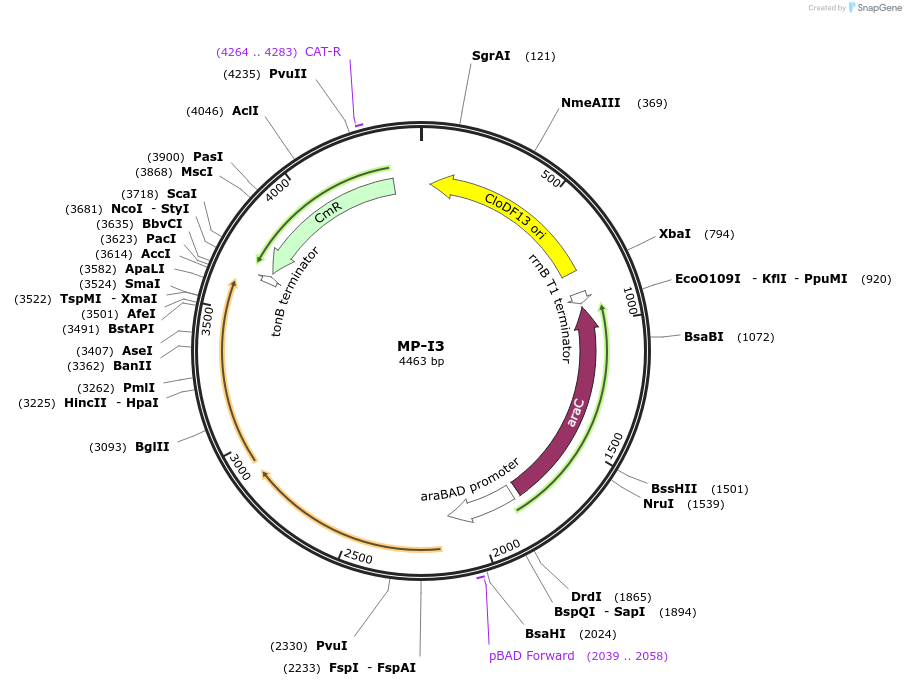
MP-I3
Plasmid#69646PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 mutH(K116E)
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
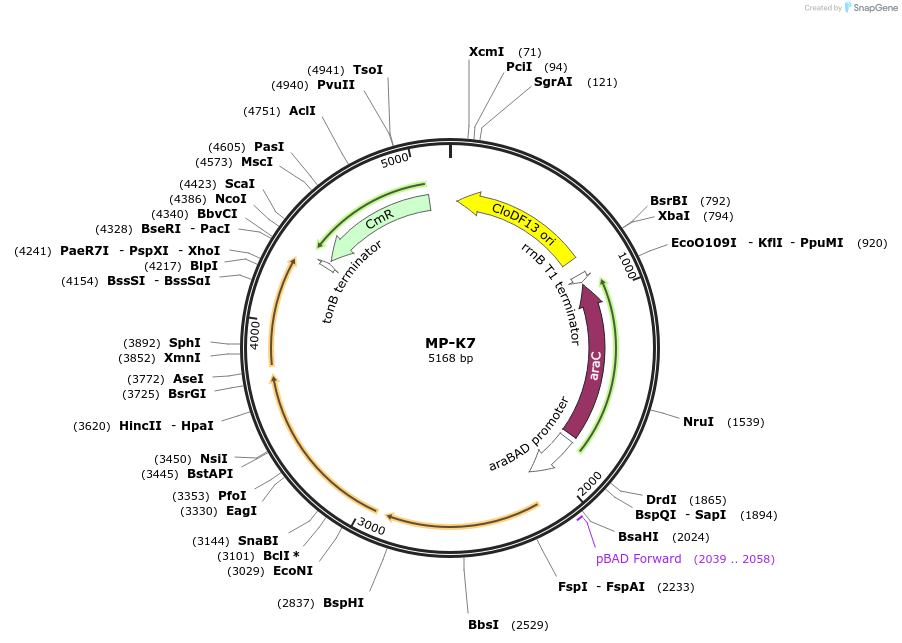
MP-K7
Plasmid#69651PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam emrR
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
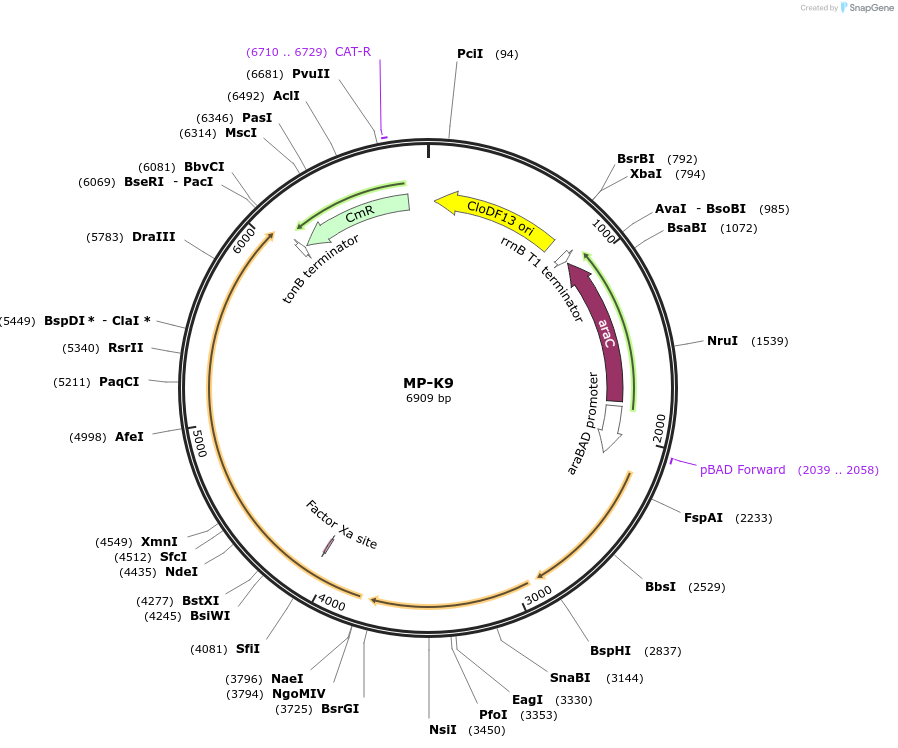
MP-K9
Plasmid#69653PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam mutSdN
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
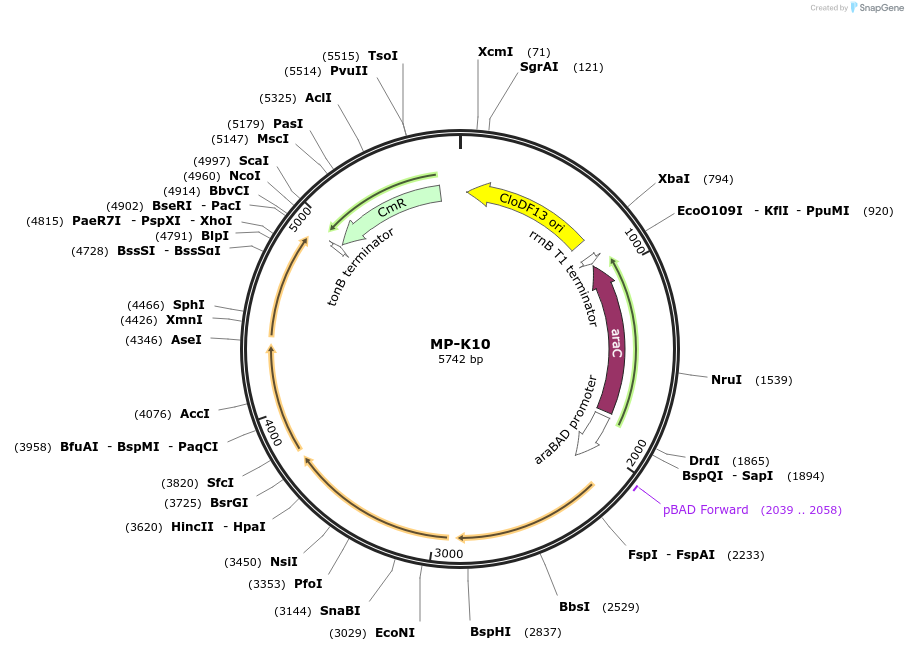
MP-K10
Plasmid#69654PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA emrR
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
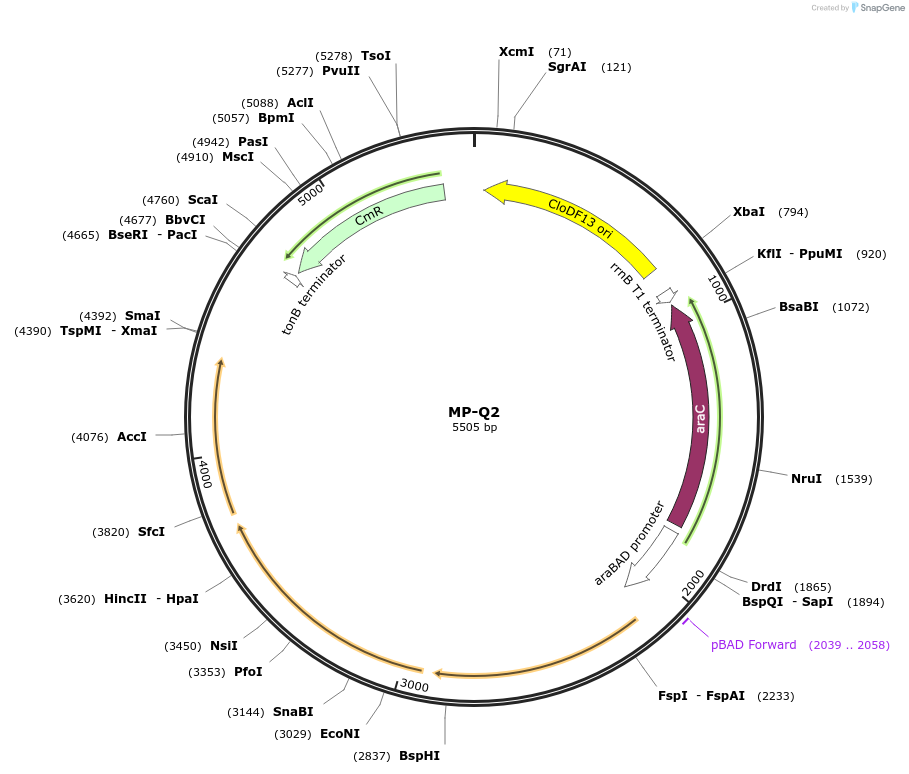
MP-Q2
Plasmid#69672PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dam seqA cchA
ExpressionBacterialPromoterE. coli PBADAvailable SinceOct. 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
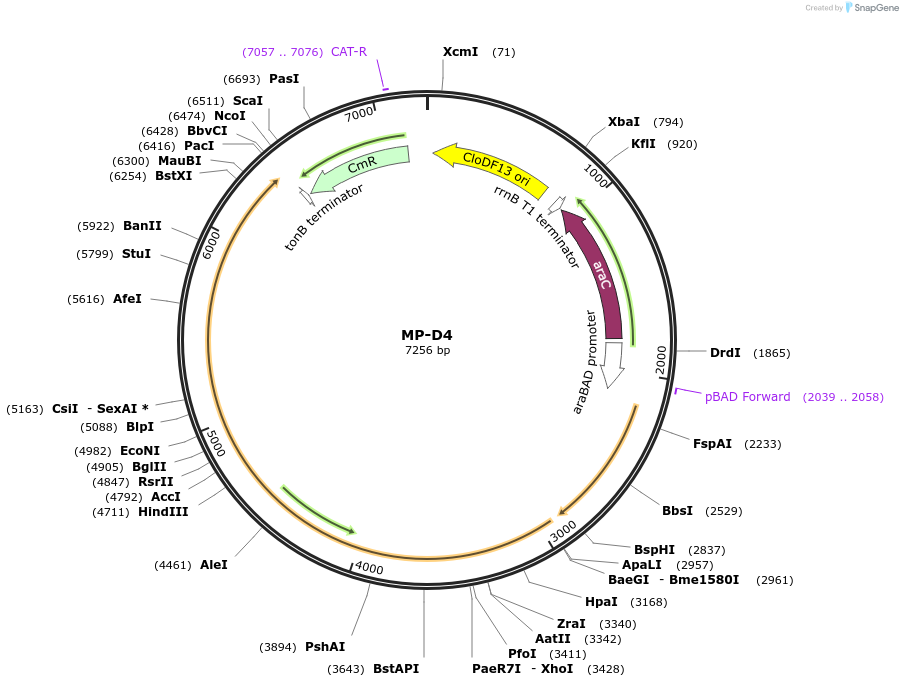
MP-D4
Plasmid#69635PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dnaE1026
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
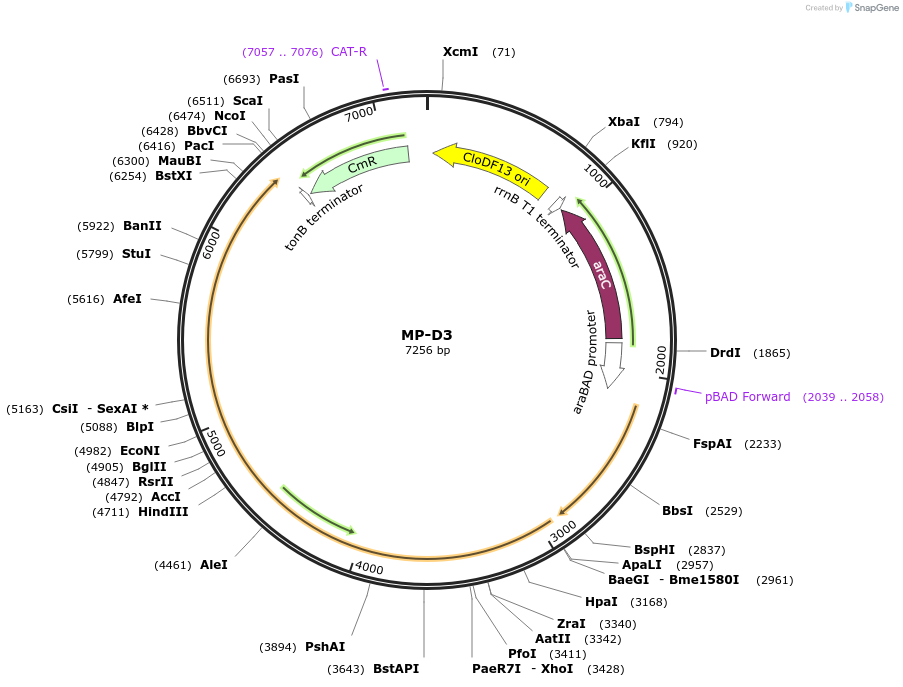
MP-D3
Plasmid#69634PurposeUntargetted Bacterial MutagenesisDepositorInsertdnaQ926 dnaE486
ExpressionBacterialPromoterE. coli PBADAvailable SinceSept. 21, 2015AvailabilityAcademic Institutions and Nonprofits only -
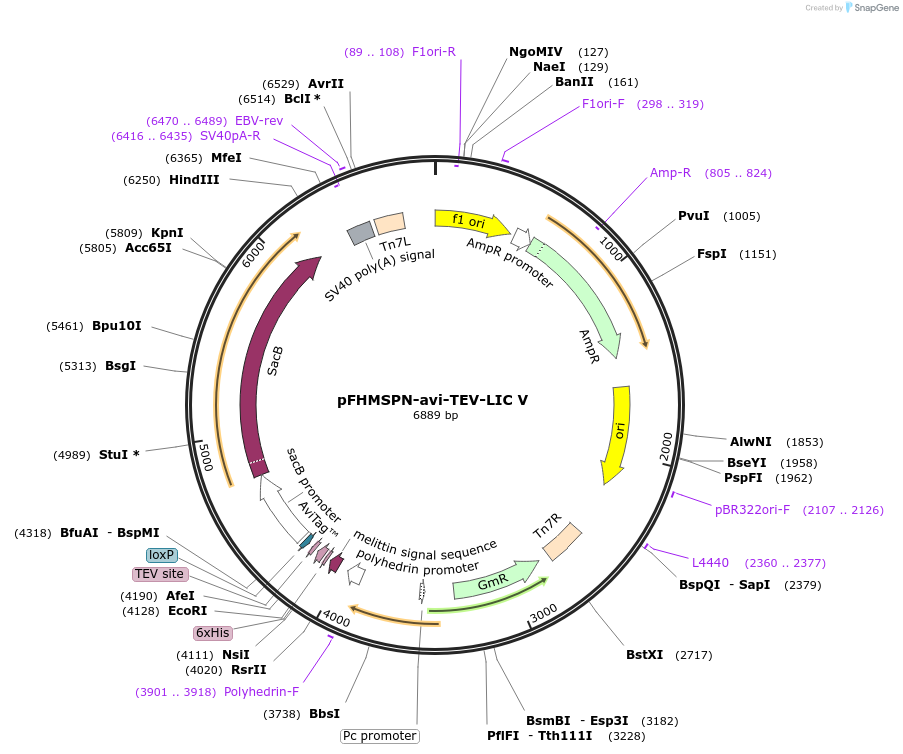
pFHMSPN-avi-TEV-LIC V
Plasmid#220885PurposeDonor vector to generate recombinant baculovirus by site-specific transposition in an E. coli host.DepositorTypeEmpty backboneUseBaculovirus expresssionTagsHoneybee melittin signal followed by His, avi and…Available SinceJune 20, 2024AvailabilityIndustry, Academic Institutions, and Nonprofits -
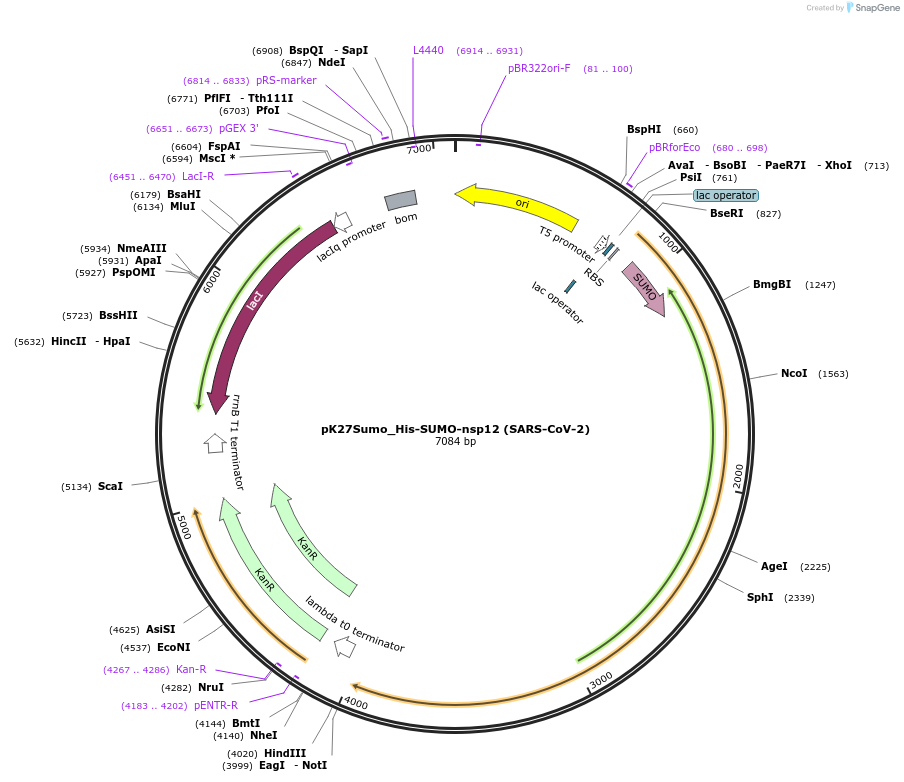
pK27Sumo_His-SUMO-nsp12 (SARS-CoV-2)
Plasmid#169188PurposeTo express His-SUMO-nsp12 (SARS-CoV-2) in E. coliDepositorInsert14His-SUMO-nsp12 (ORF1ab Synthetic, SARS-CoV-2)
Tags14His-SUMO (Ulp1-cleavable)ExpressionBacterialMutationCodon optimised for E. coliPromoterT5Available SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
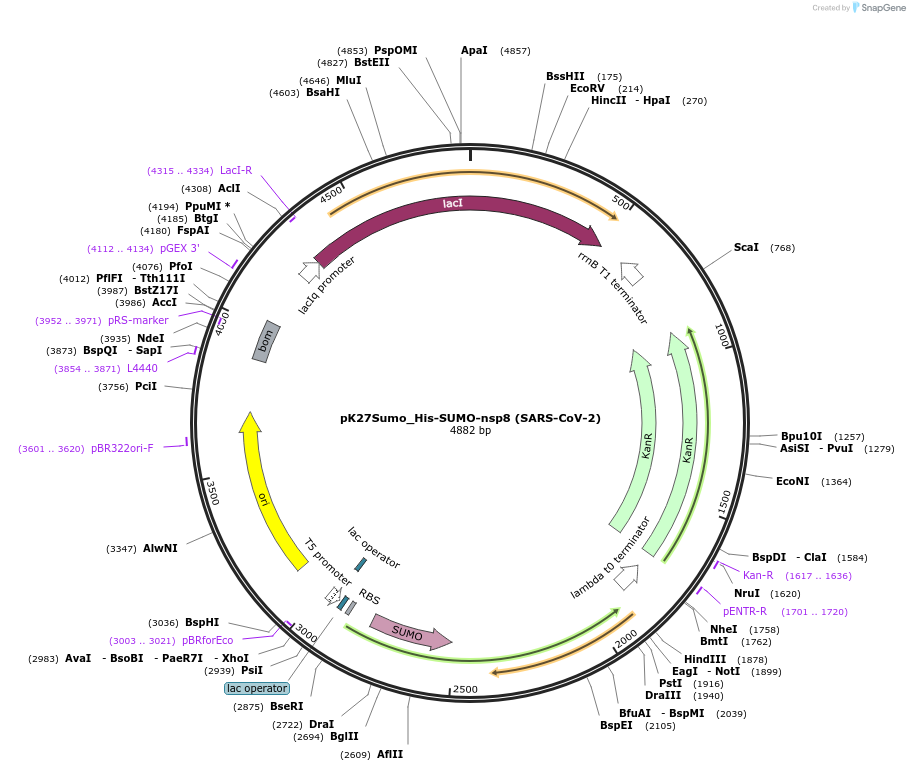
pK27Sumo_His-SUMO-nsp8 (SARS-CoV-2)
Plasmid#169187PurposeTo express His-SUMO-nsp8 (SARS-CoV-2) in E. coliDepositorInsert14His-SUMO-nsp8 (ORF1ab Synthetic, SARS-CoV-2)
Tags14His-SUMO (Ulp1-cleavable)ExpressionBacterialMutationCodon optimised for E. coliPromoterT5Available SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
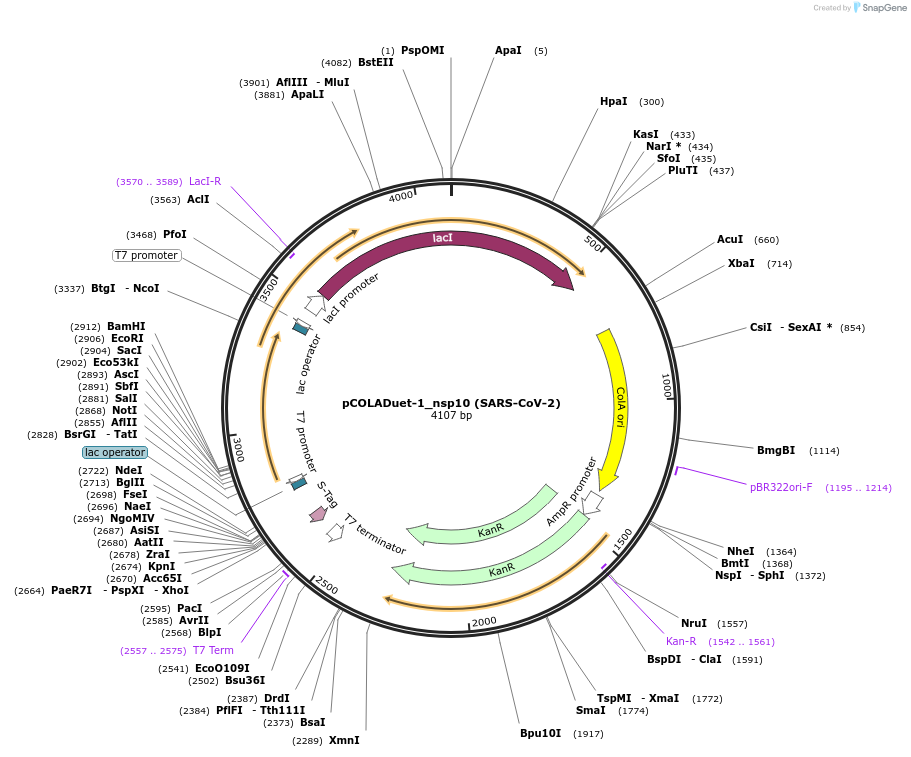
pCOLADuet-1_nsp10 (SARS-CoV-2)
Plasmid#169158PurposeTo express SARS-CoV-2 nsp10 in E. coliDepositorInsertnsp10 (ORF1ab SARS-CoV-2)
TagsMethionineExpressionBacterialMutationCodon optimised for E. coliPromoterT7Available SinceJuly 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
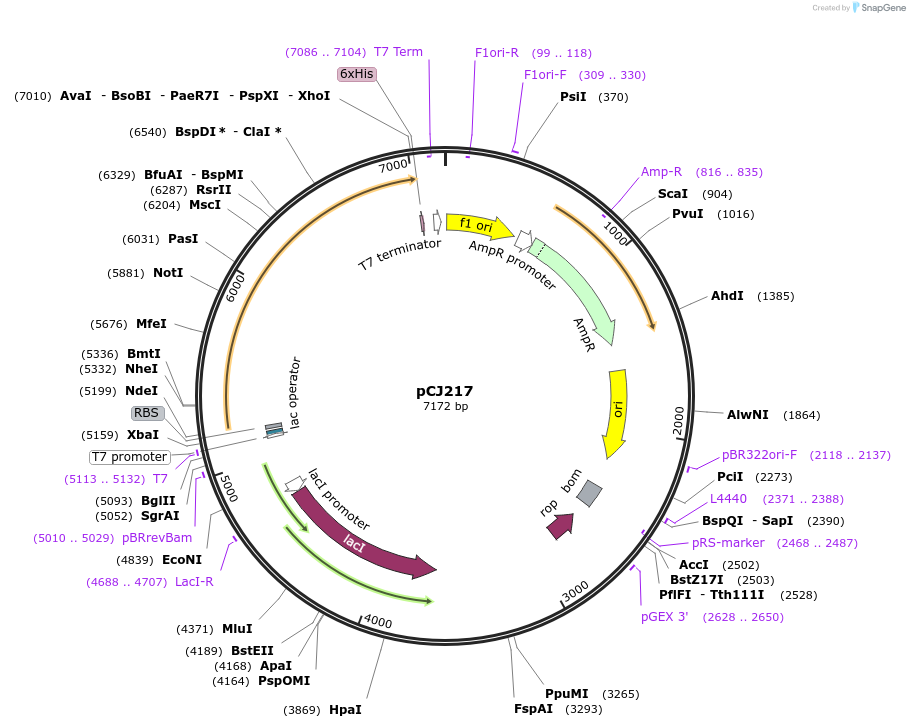
pCJ217
Plasmid#162685PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating two point mutations, G489C and S530C, to introduce a 6th disulfide bond (from PETase).DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with mutations to introduce a 6th disulfide bond from PETase
TagsHisExpressionBacterialMutationG489C, S530C; codon optimized for E. coli K12PromoterT7/lacAvailable SinceJan. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
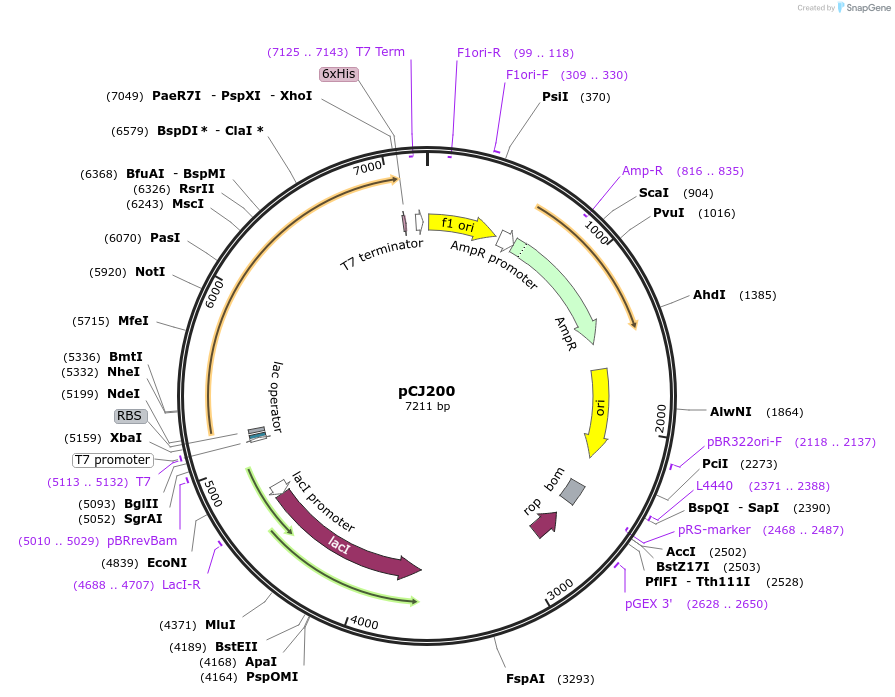
pCJ200
Plasmid#162673PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C and a sequence Tyr75:Phe104 to introduce a 6th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C and a sequence Y75-F104 to introduce a 6th…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
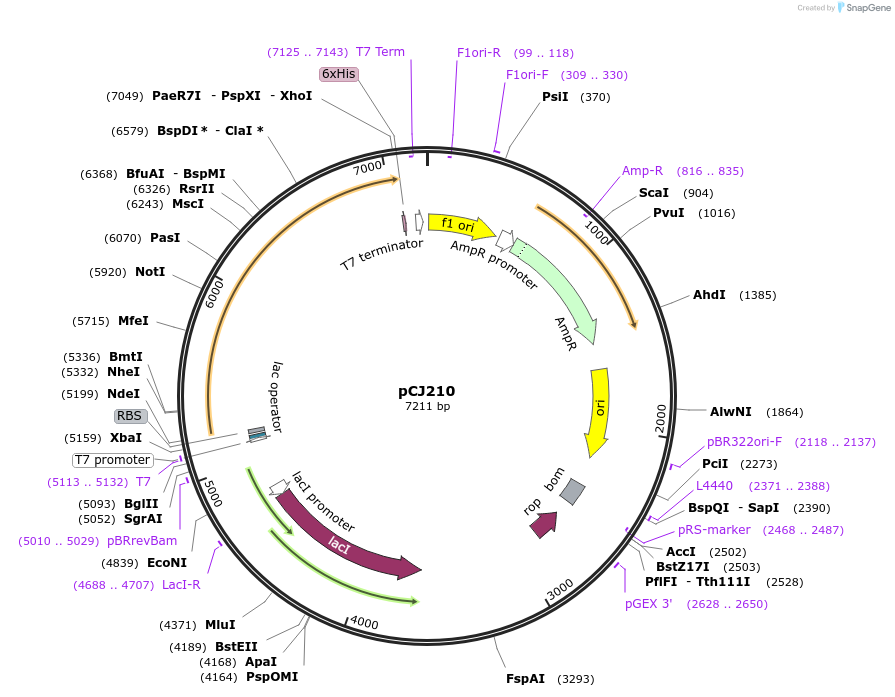
pCJ210
Plasmid#162683PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating S136C, G489C, S530C, and a sequence Tyr75:Phe104 to introduce a 6th and 7th disulfide bond as in AoFaeB-2.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1)
TagsHisExpressionBacterialMutationS136C, G489C, S530C, and a sequence Y75-F104 to i…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
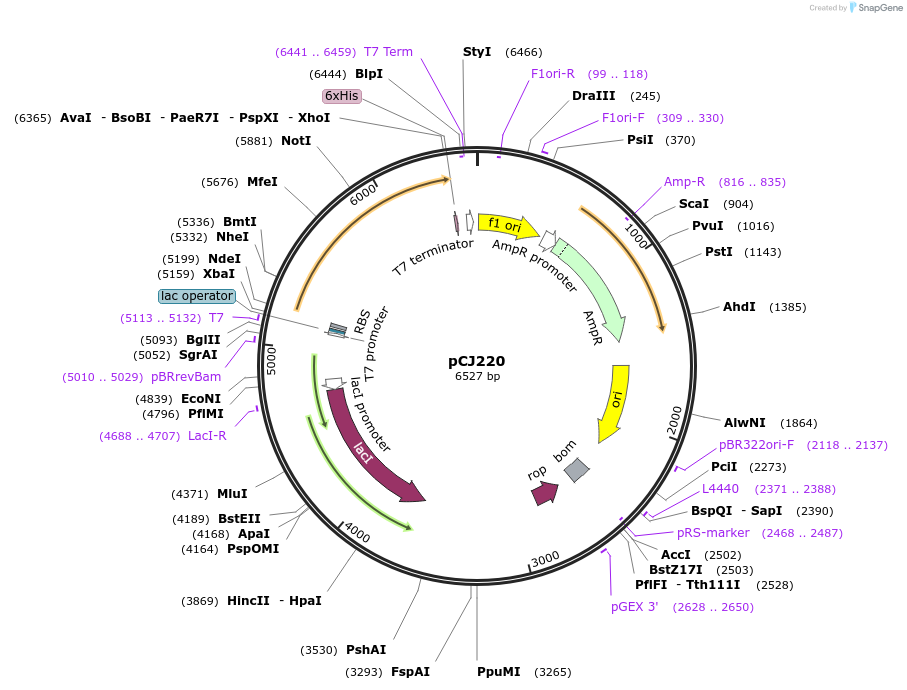
pCJ220
Plasmid#162686PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224W and C529S mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1) with lid deletion and C224W and C529S mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224W, C529S; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
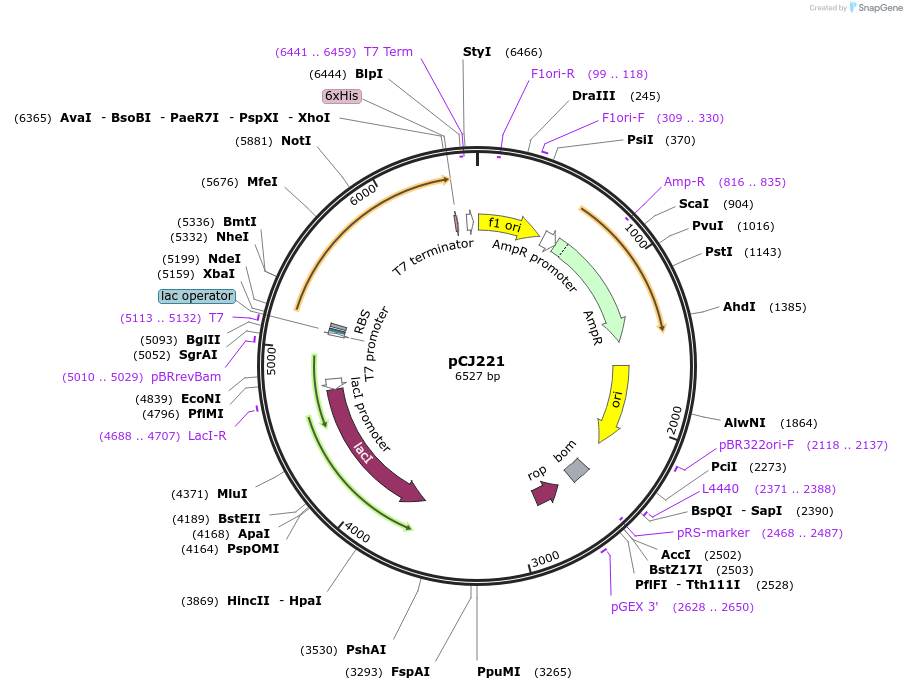
pCJ221
Plasmid#162687PurposepET-21b(+) based plasmid for expression of MHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), codon optimized for expression in E. coli K12, with C-terminal His tag, incorporating lid deletion (Gly251:Thr472) and C224H and C529F mutations from PETase active site.DepositorInsertMHETase from Ideonella sakaiensis 201-F6 (Genbank GAP38911.1), with lid deletion and C224H and C529F mutations
TagsHisExpressionBacterialMutationdeltaG251-T472, C224H, C529F; codon optimized for…PromoterT7/lacAvailable SinceJan. 7, 2021AvailabilityAcademic Institutions and Nonprofits only -
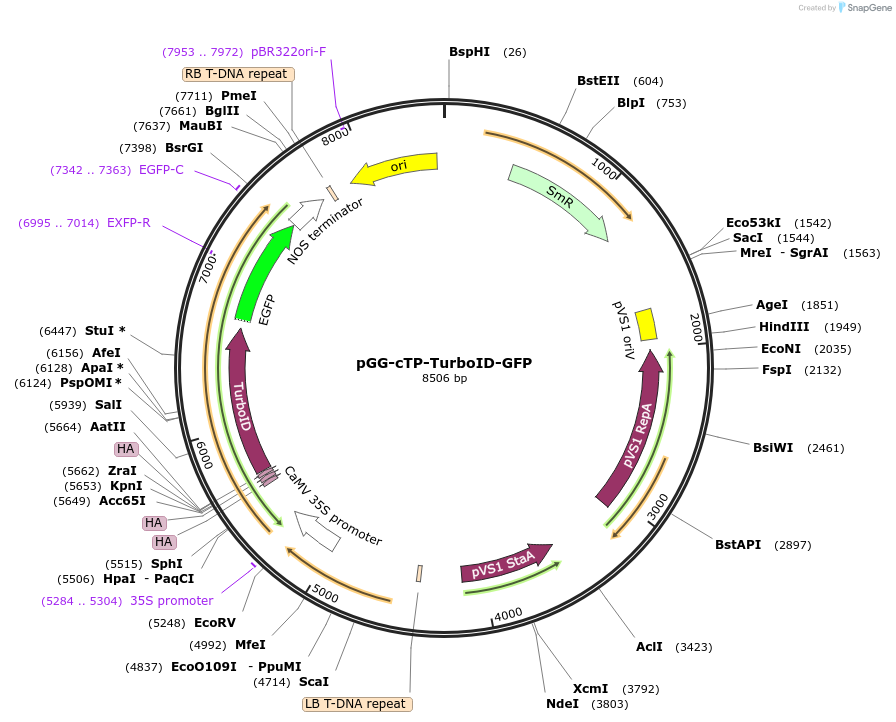
pGG-cTP-TurboID-GFP
Plasmid#209403PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a chloroplast transit peptide (cTP) in plantaDepositorInsertPM-3XHA-TurbOID
TagsGFPExpressionPlantPromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
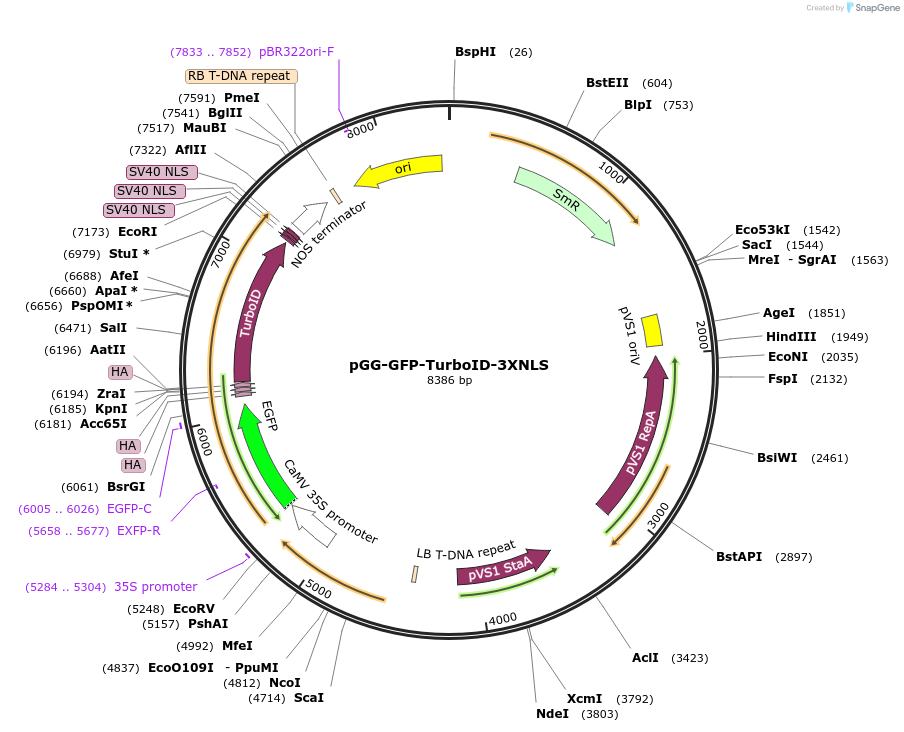
pGG-GFP-TurboID-3XNLS
Plasmid#209400PurposeTransiently expressing and visualizating the subcellular localization of TurboID fused with 3X nuclear localization signal (NLS) in plantaDepositorInsert3XHA-TurboID-3XNLS
TagsGFPExpressionPlantPromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
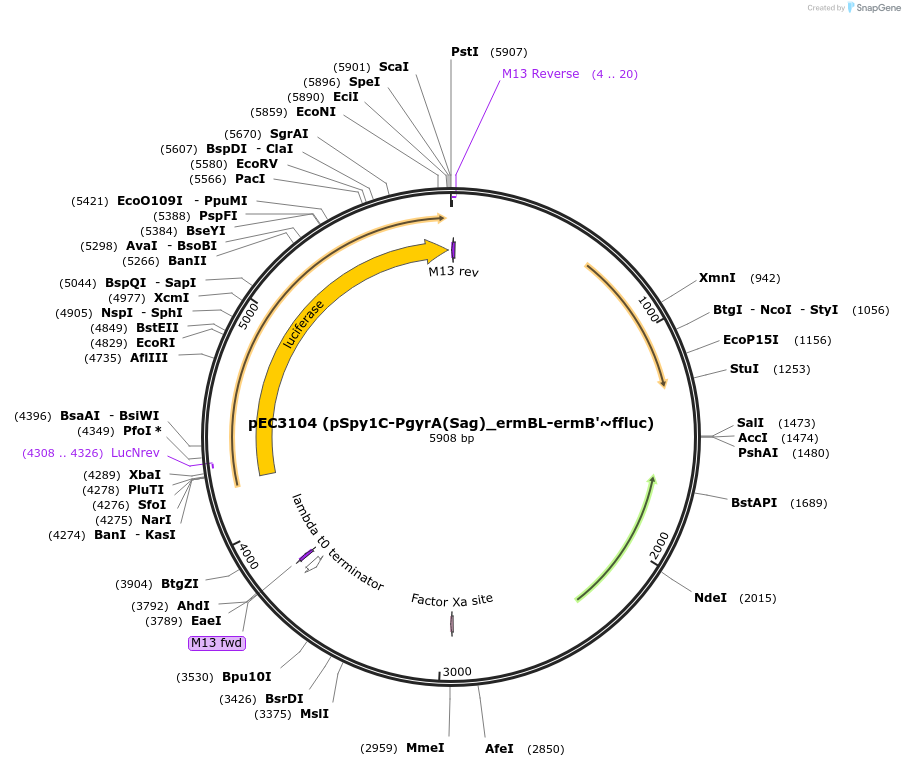
pEC3104 (pSpy1C-PgyrA(Sag)_ermBL-ermB'~ffluc)
Plasmid#218513Purposereplicative E. coli-S. pyogenes shuttle plasmid for erythromycin-inducible expression of the Firefly luciferaseDepositorInsertffluc
UseLuciferaseExpressionBacterialMutationdeletion of the Plac_mrfp cassettePromoterPgyrA_ermBL-ermB'Available SinceSept. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
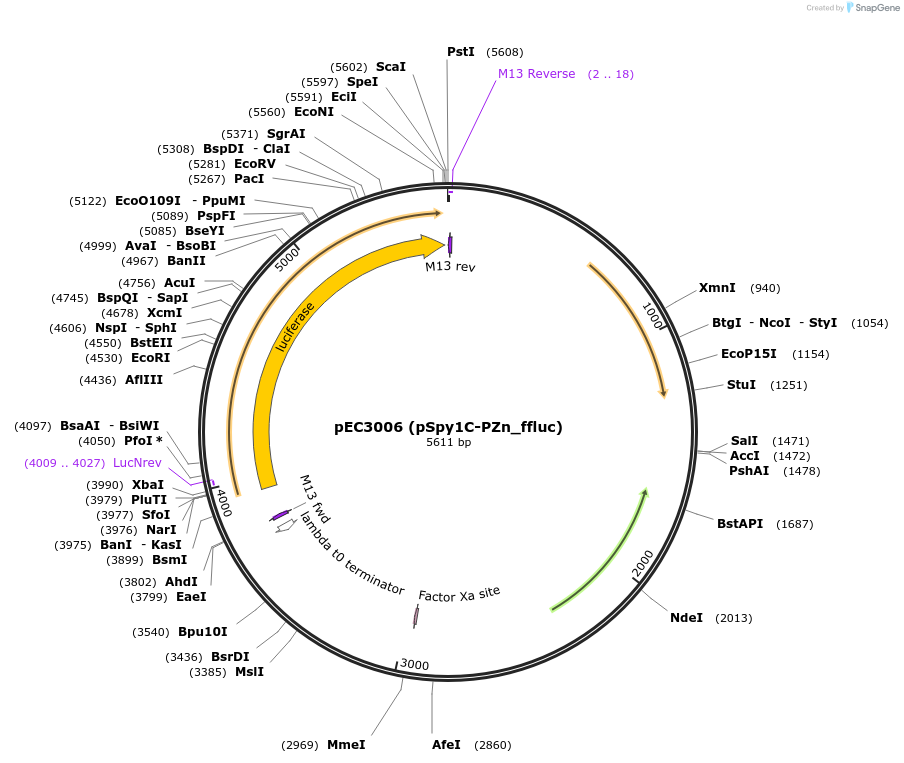
pEC3006 (pSpy1C-PZn_ffluc)
Plasmid#218515Purposereplicative E. coli-S. pyogenes shuttle plasmid for zinc-inducible expression of the Firefly luciferase in S. pyogenesDepositorInsertffluc
UseLuciferaseExpressionBacterialMutationdeletion of Plac_mrfp cassettePromoterPZn (PczcD)Available SinceSept. 3, 2024AvailabilityAcademic Institutions and Nonprofits only -
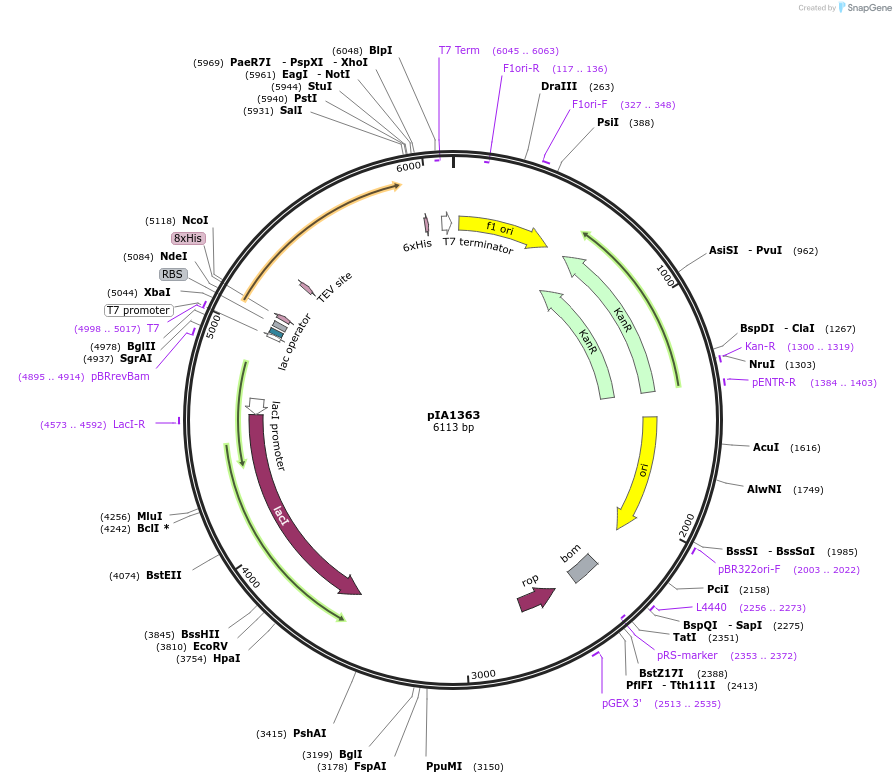
pIA1363
Plasmid#166861PurposeExpression of codon optimized SARS-COV-2 Nsp8 in E. coliDepositorAvailable SinceMarch 23, 2021AvailabilityAcademic Institutions and Nonprofits only -
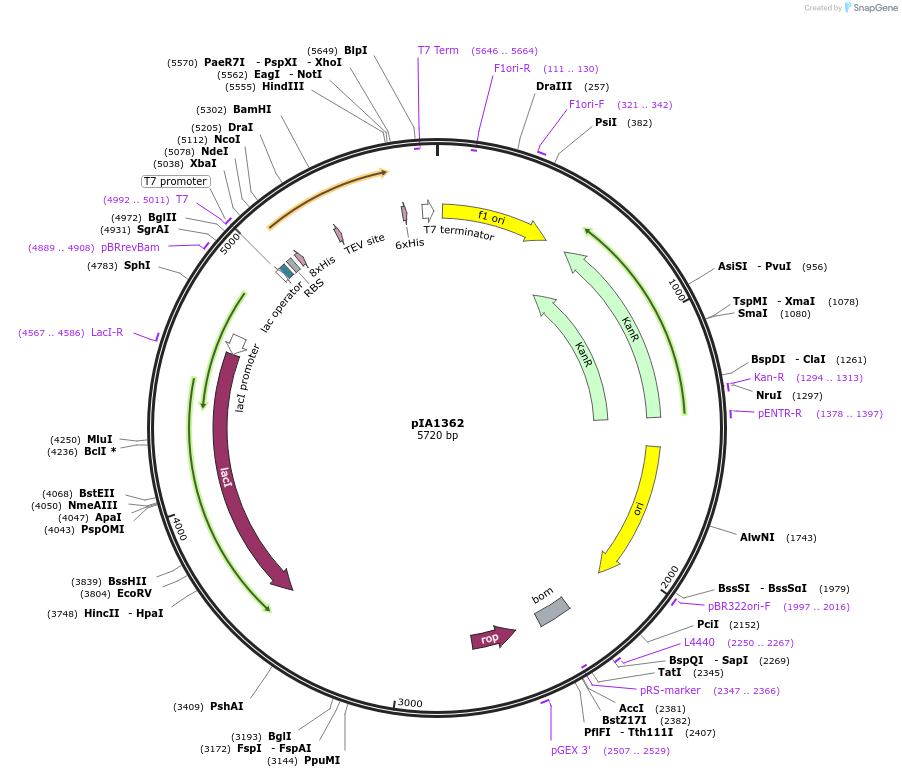
pIA1362
Plasmid#166860PurposeExpression of codon optimized SARS-COV-2 Nsp7 in E. coliDepositorAvailable SinceMarch 16, 2021AvailabilityAcademic Institutions and Nonprofits only -
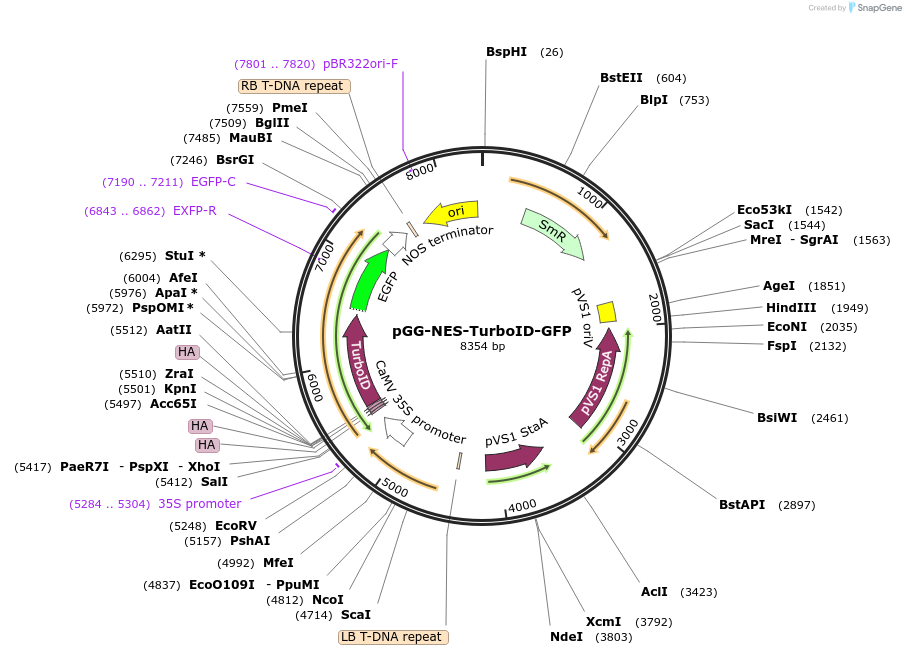
pGG-NES-TurboID-GFP
Plasmid#209401PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a nuclear export signal (NES) in plantaDepositorInsertNES-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
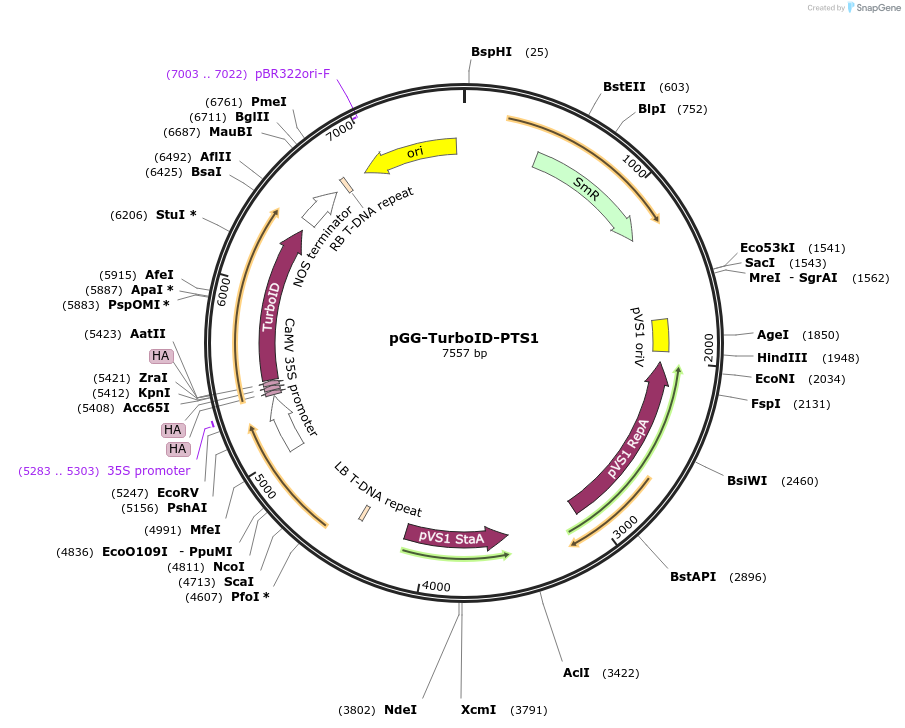
pGG-TurboID-PTS1
Plasmid#209398PurposeTransiently expressing TurboID fused with a peroxisomal targeting signal type I (PTS1) in plantaDepositorInsert3XHA-TurboID-PTS1
ExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
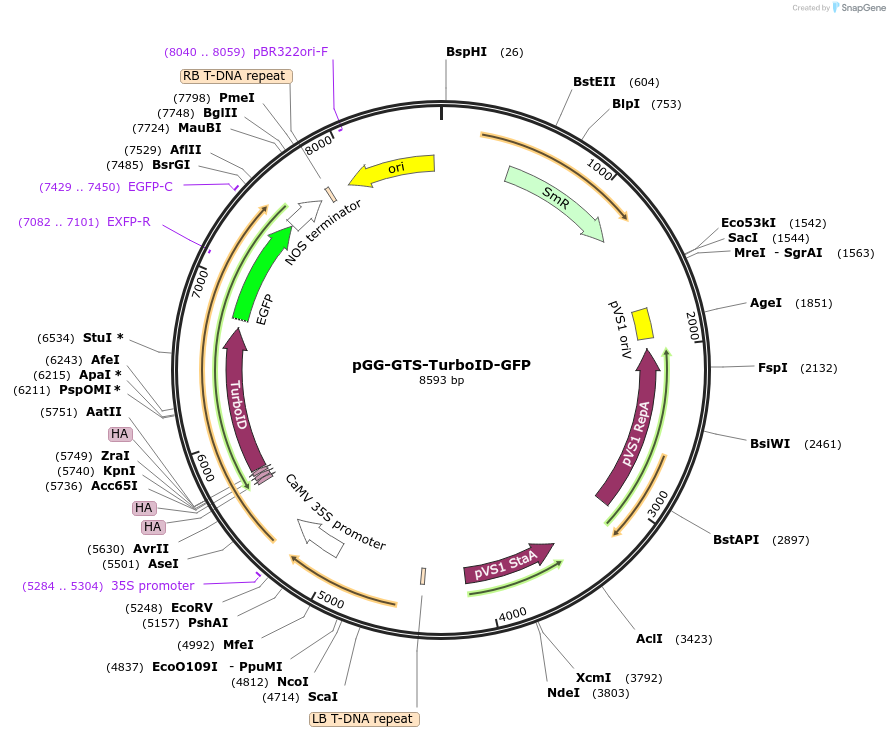
pGG-GTS-TurboID-GFP
Plasmid#209405PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a Golgi targeting sequence (GTS) in plantaDepositorInsertGTS-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
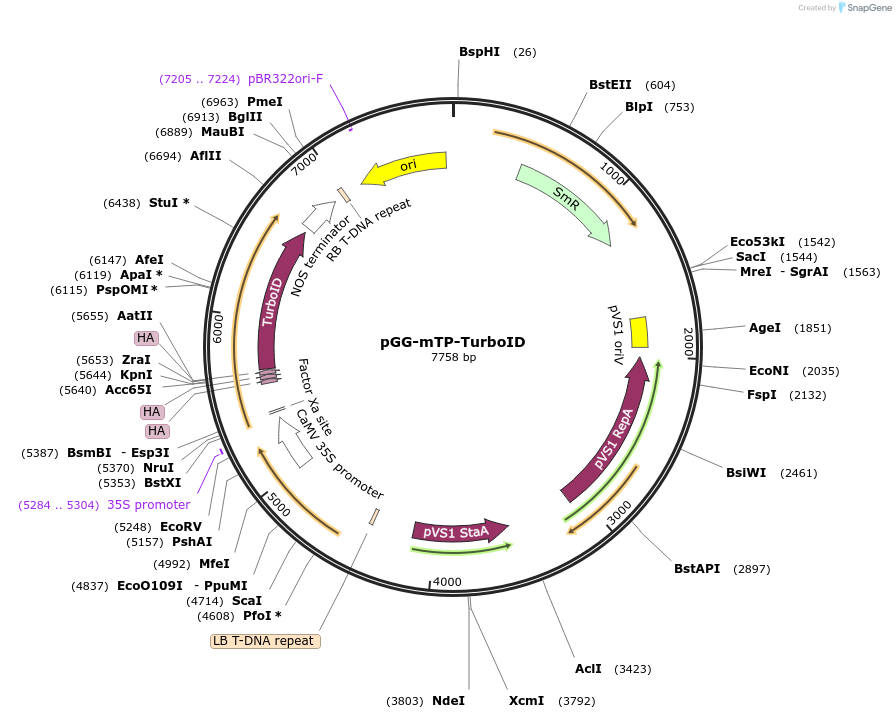
pGG-mTP-TurboID
Plasmid#209396PurposeTransiently expressing TurboID fused with a Mitochondrial transit peptide (mTP) in plantaDepositorInsertmTP-3XHA-TurboID
ExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
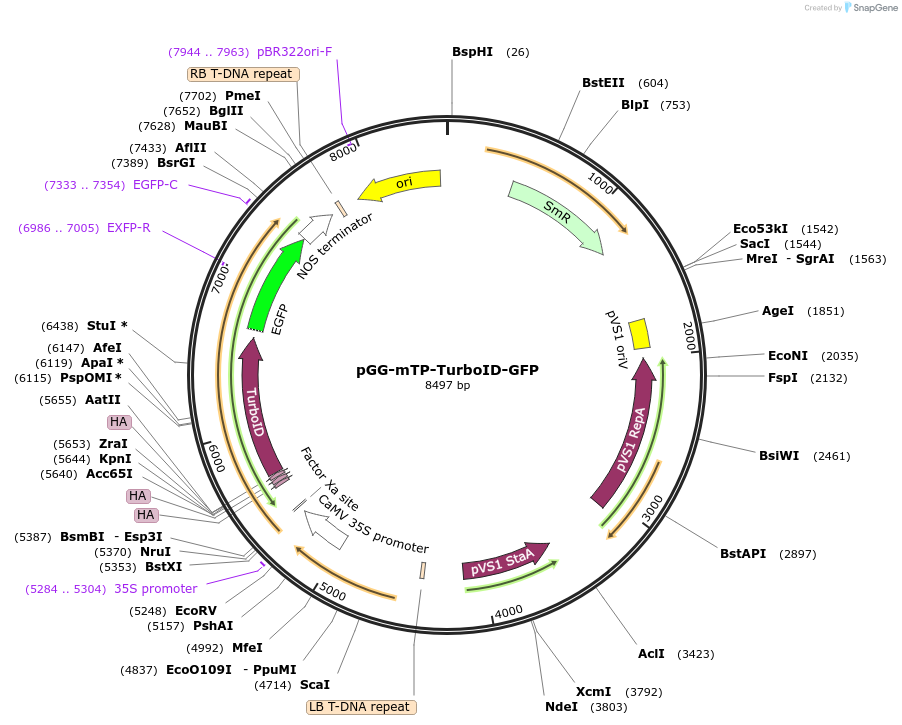
pGG-mTP-TurboID-GFP
Plasmid#209404PurposeTransiently expressing and visualizing the subcellular localization of TurboID fused with a Mitochondrial transit peptide (mTP) in plantaDepositorInsertmTP-3XHA-TurboID
TagsGFPExpressionPlantMutationTurboID gene sequence C249APromoterCauliflower mosaic virus (CaMV) 35S promoterAvailable SinceJan. 5, 2024AvailabilityAcademic Institutions and Nonprofits only -
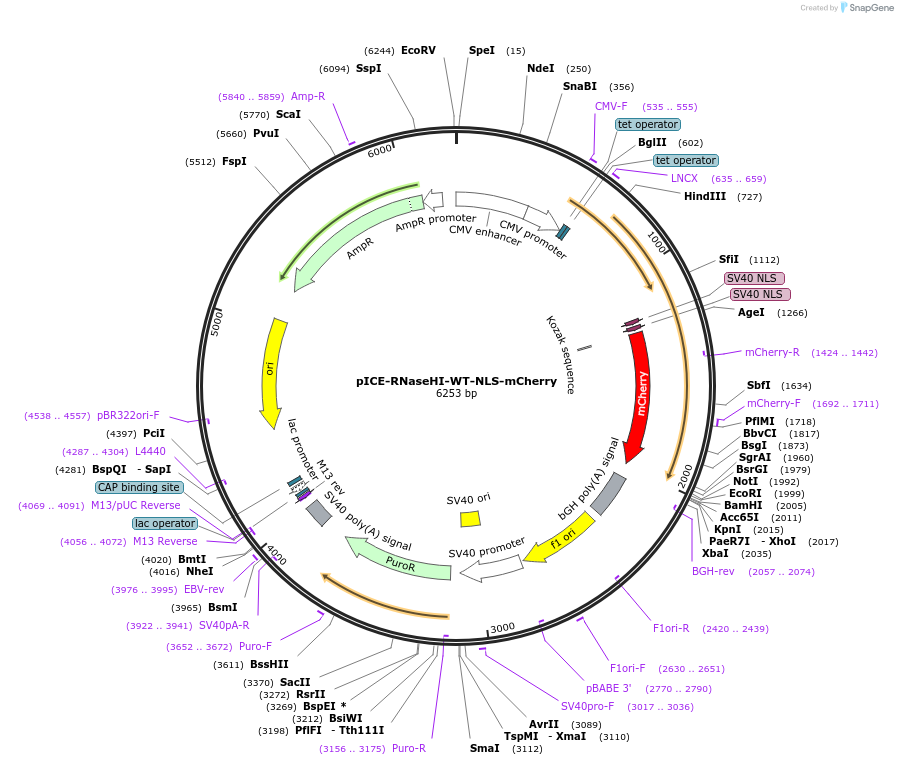
pICE-RNaseHI-WT-NLS-mCherry
Plasmid#60365PurposePlasmid for expression of bacterial RNase HI tagged with NLS-mCherry that can be used to degrade R-loops. Confers resistance to Puromycin. Expression is inducible in T-REx cells.DepositorInsertRNase HI
TagsNLS from SV40 T antigen and mCherryExpressionMammalianPromoterCMV-tetAvailable SinceJan. 8, 2015AvailabilityAcademic Institutions and Nonprofits only -
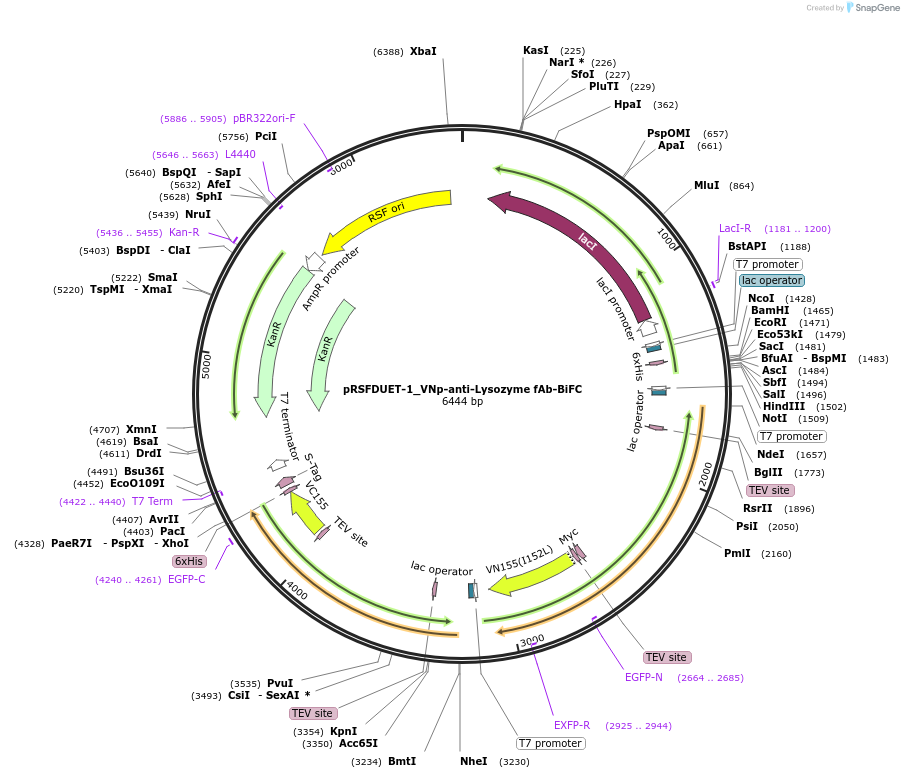
pRSFDUET-1_VNp-anti-Lysozyme fAb-BiFC
Plasmid#214950PurposeBacterial expression of anti-chicken lysozyme Fab light and heavy chain VNp fusions. Each chain is fused to separate halves of a BiFC protein -Venus fluorescence signifies Hc/Lc heterodimer formation.DepositorInsertsVNp-anti-Lysozyme Fab heavy chain amino-BiFC fusion
VNp-anti-Lysozyme Fab light chain amino-BiFC fusion
Tags6xHis, Amino half of venus BIFC fragment, Carboxy…ExpressionBacterialPromoterT7Available SinceJan. 24, 2025AvailabilityAcademic Institutions and Nonprofits only -
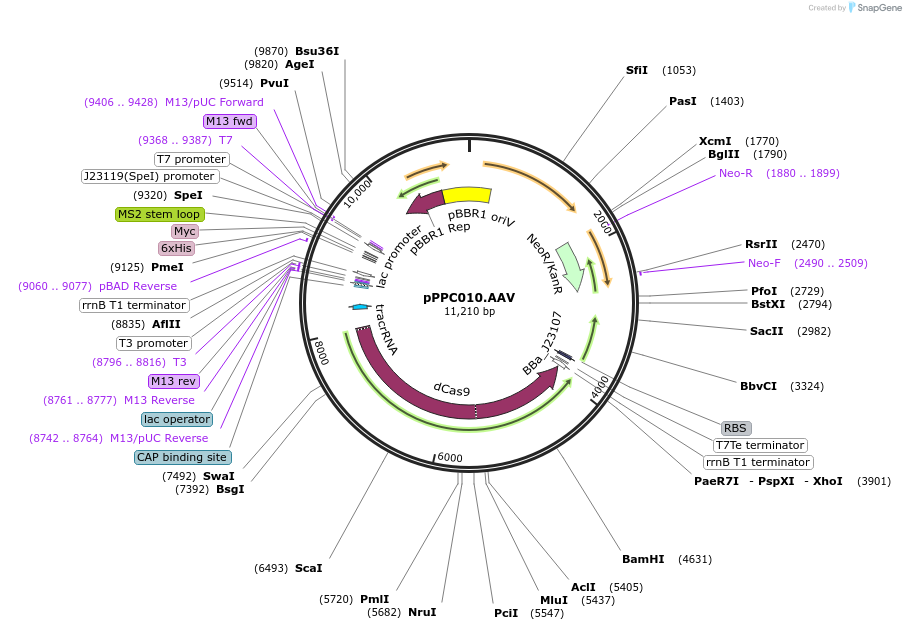
pPPC010.AAV
Plasmid#171175PurposeExpression of Sp.pCas9-dCas9, BBa_J23107-MCP-SoxS(R93A/S101A), and hAAVS1 scRNA on pBBR1-KmR plasmidDepositorInsertshAAVS1 scRNA
Sp.pCas9-dCas9_BBa_J23107-MCP-SoxS(R93A, S101A)
UseCRISPRExpressionBacterialMutationSoxS has R93A and S101A mutationsPromoterBBa_J23119 and Sp.pCas9, BBa_J23107Available SinceOct. 6, 2021AvailabilityAcademic Institutions and Nonprofits only -
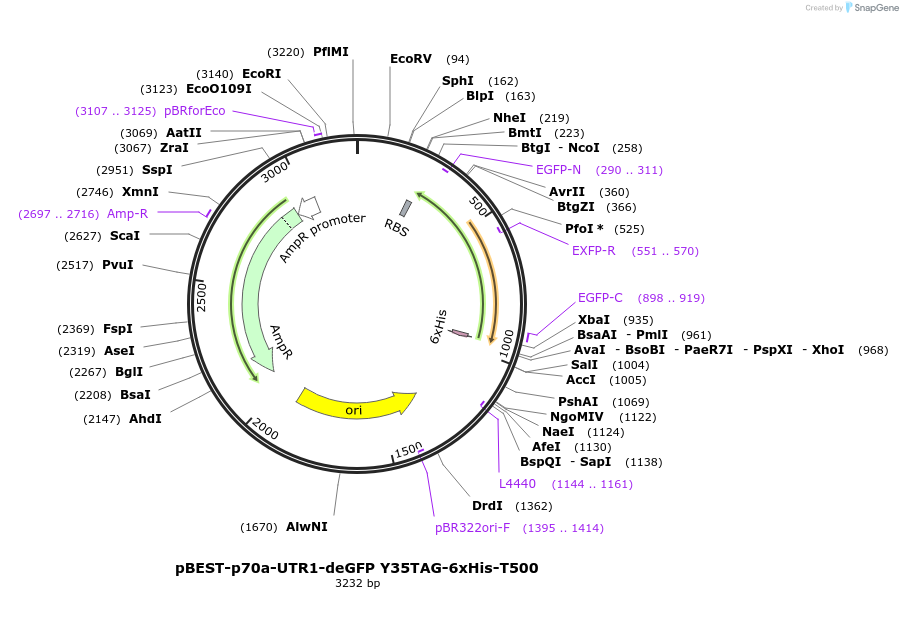
pBEST-p70a-UTR1-deGFP Y35TAG-6xHis-T500
Plasmid#92225PurposeExpression of deGFP mutant Y35TAG for ncAA incoporation using the p70a promoter and UTR1DepositorInsertdeGFP Y35TAG
UseSynthetic Biology; Cell free protein synthesis (c…Tags6xHisExpressionBacterialMutationResidue Y35 mutated into TAG (Amber) stop codon f…Promoterp70aAvailable SinceOct. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
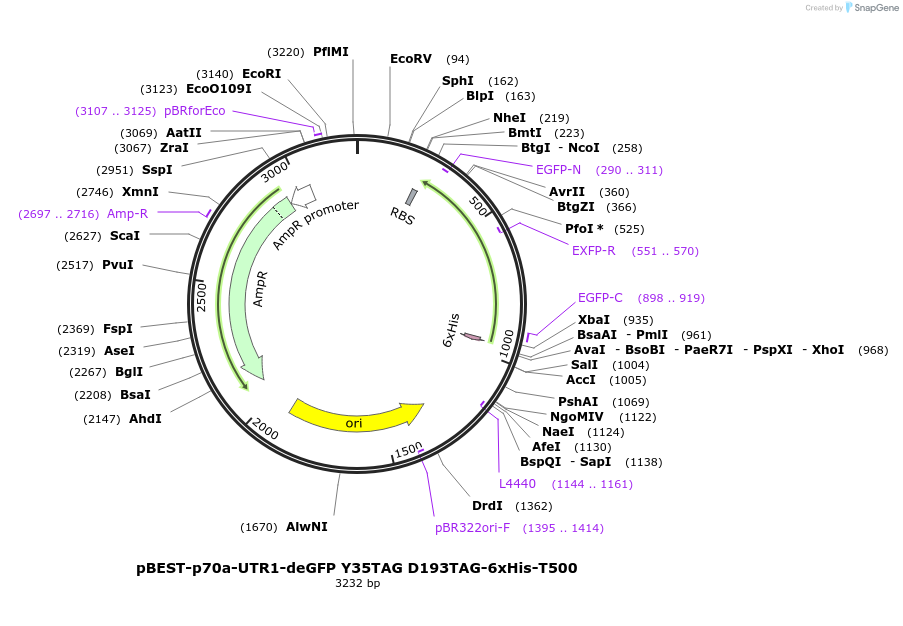
pBEST-p70a-UTR1-deGFP Y35TAG D193TAG-6xHis-T500
Plasmid#92227PurposeExpression of deGFP mutant Y35TAG D193TAG for ncAA incoporation using the p70a promoter and UTR1DepositorInsertdeGFP Y35TAG D193TAG
UseSynthetic Biology; Cell free protein synthesis (c…Tags6xHisExpressionBacterialMutationResidues Y35 D193 were mutated into TAG (Amber) s…Promoterp70aAvailable SinceOct. 23, 2017AvailabilityAcademic Institutions and Nonprofits only -
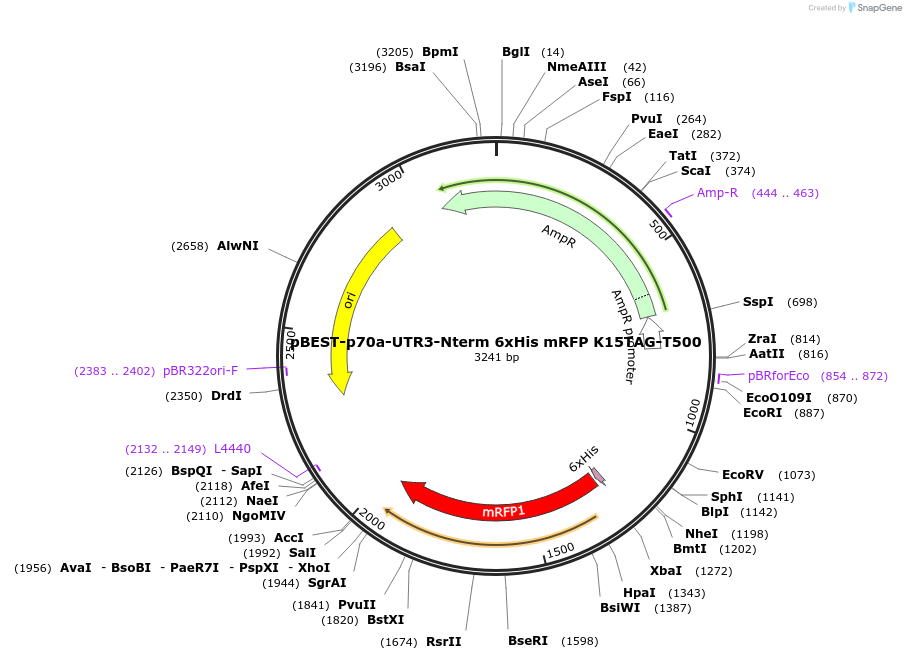
pBEST-p70a-UTR3-Nterm 6xHis mRFP K15TAG-T500
Plasmid#92319PurposeExpression of NTerm 6xHis tagged mRFP mutant K15TAG for ncAA incoporation using the p70a promoter and UTR3DepositorInsertOptimized NTerm 6xHis tagged monomeric RFP (mRFP)
UseSynthetic Biology; Cell free protein synthesis (c…Tags6xHisExpressionBacterialPromoterp70aAvailable SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
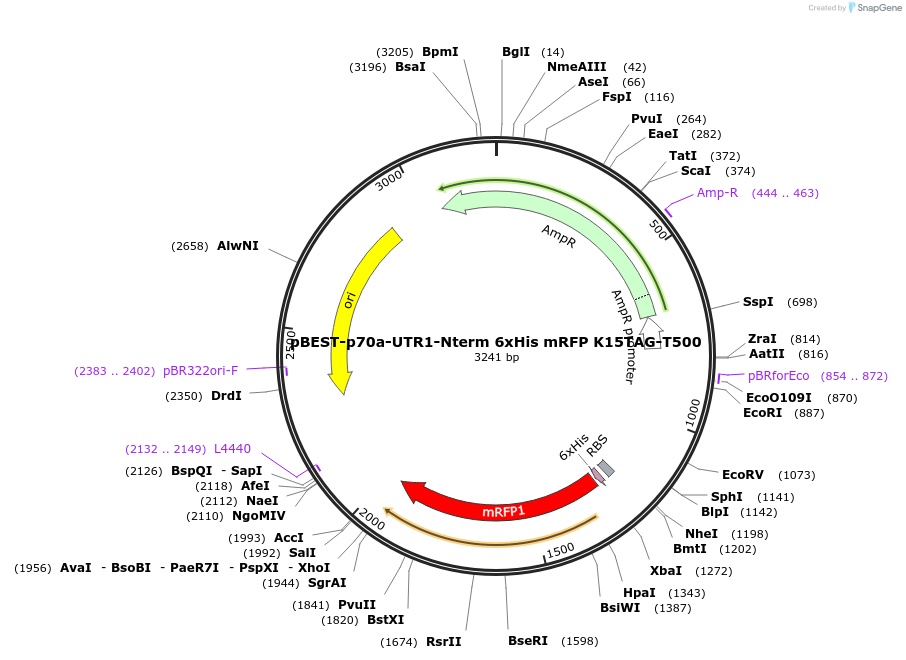
pBEST-p70a-UTR1-Nterm 6xHis mRFP K15TAG-T500
Plasmid#92318PurposeExpression of NTerm 6xHis tagged mRFP mutant K15TAG for ncAA incoporation using the p70a promoter and UTR1DepositorInsertOptimized NTerm 6xHis tagged monomeric RFP (mRFP)
UseSynthetic Biology; Cell free protein synthesis (c…Tags6xHisExpressionBacterialPromoterp70aAvailable SinceJuly 5, 2017AvailabilityAcademic Institutions and Nonprofits only -
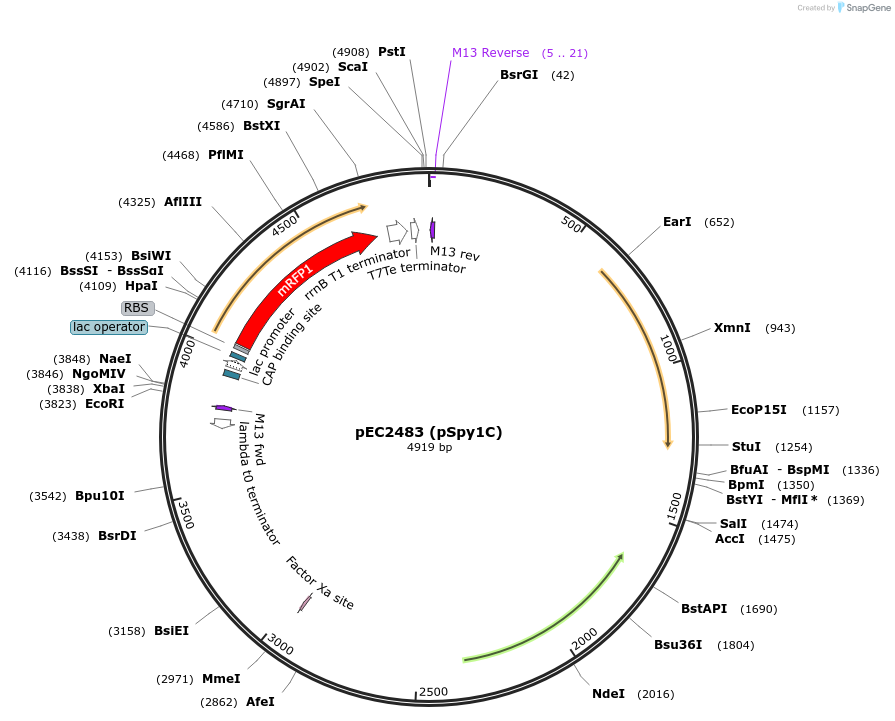
pEC2483 (pSpy1C)
Plasmid#220248Purposereplicative E. coli-S. pyogenes shuttle plasmid for heterologous gene expressionDepositorTypeEmpty backboneExpressionBacterialAvailable SinceSept. 3, 2024AvailabilityAcademic Institutions and Nonprofits only