We narrowed to 22,416 results for: Tes
-
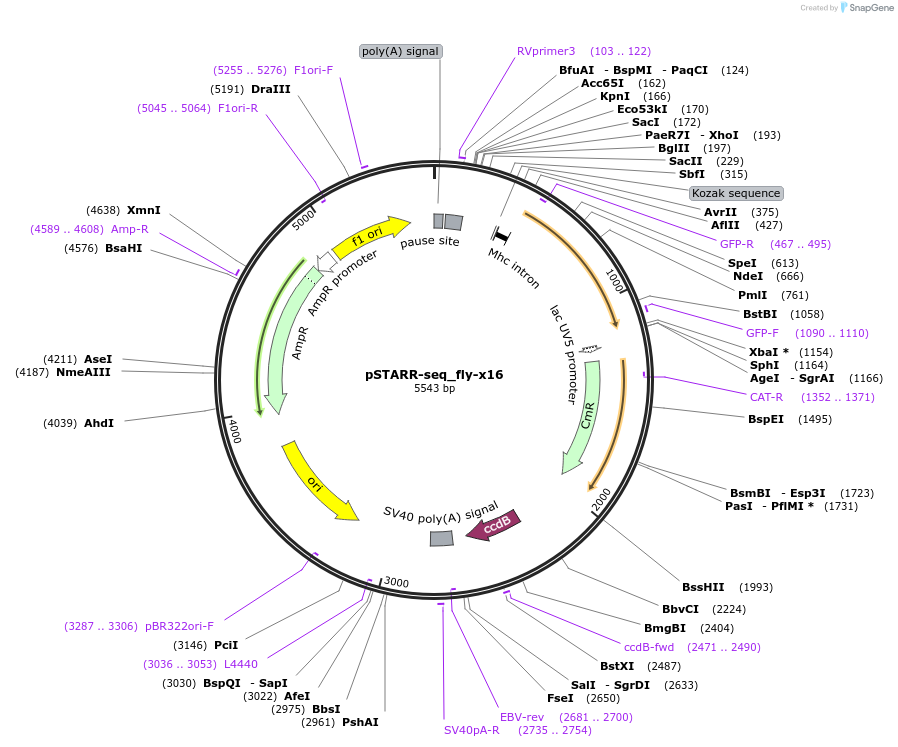
Plasmid#71505PurposeflySTARR-seq screening vector with x16 core promoterDepositorTypeEmpty backboneUseFlystarr-seq screening vectorTagssgGFP (SuperGlo, Qbiogene, Inc)ExpressionInsectAvailable SinceFeb. 5, 2016AvailabilityAcademic Institutions and Nonprofits only
-
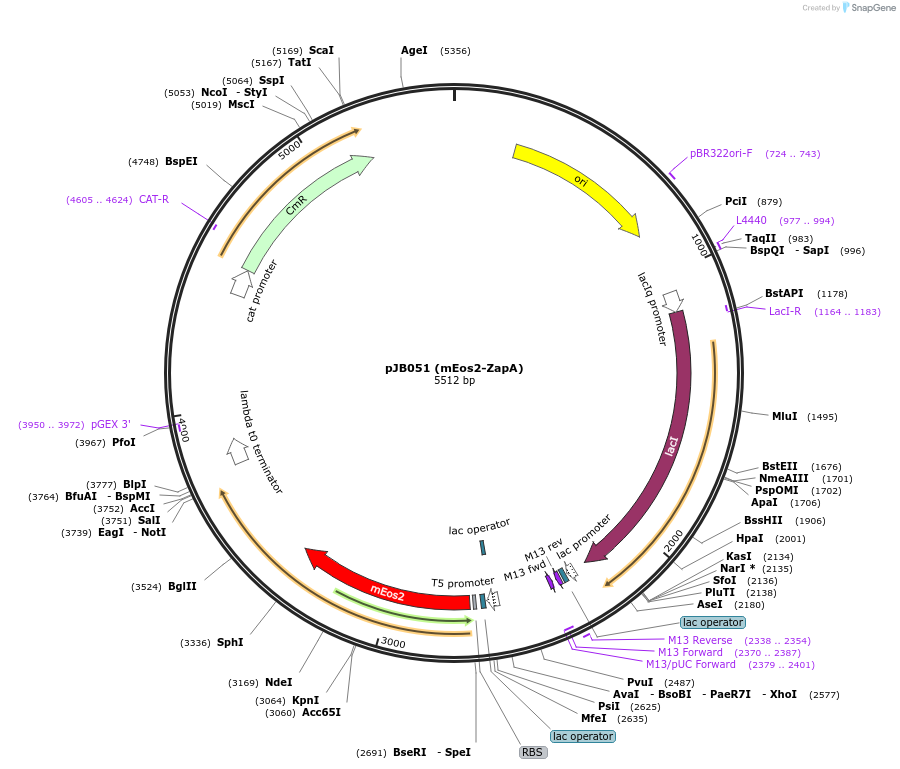
pJB051 (mEos2-ZapA)
Plasmid#72650PurposeInducible expression of mEos2-ZapA in bacteriaDepositorInsertmEos2-ZapA
TagsmEos2ExpressionBacterialPromoterT5-lacAvailable SinceFeb. 1, 2016AvailabilityAcademic Institutions and Nonprofits only -
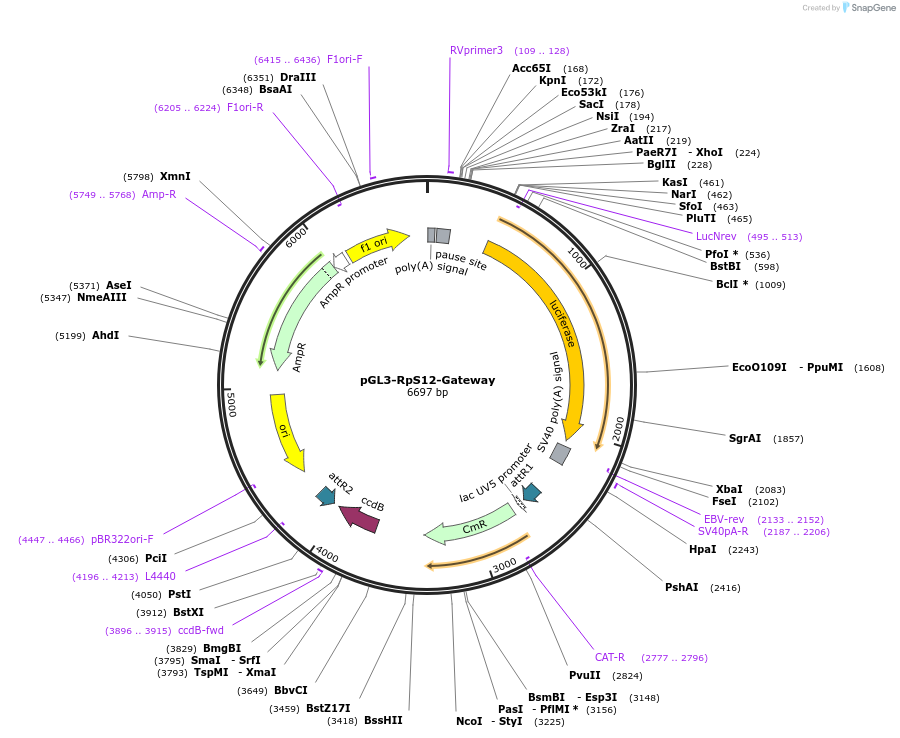
pGL3-RpS12-Gateway
Plasmid#71508PurposeflySTARR-seq validation vector with Gateway cassette downstream of the firefly luciferase gene and the RpS12 core promoterDepositorTypeEmpty backboneUseFlystarr-seq validation vectorTagsluciferaseExpressionInsectAvailable SinceDec. 29, 2015AvailabilityAcademic Institutions and Nonprofits only -
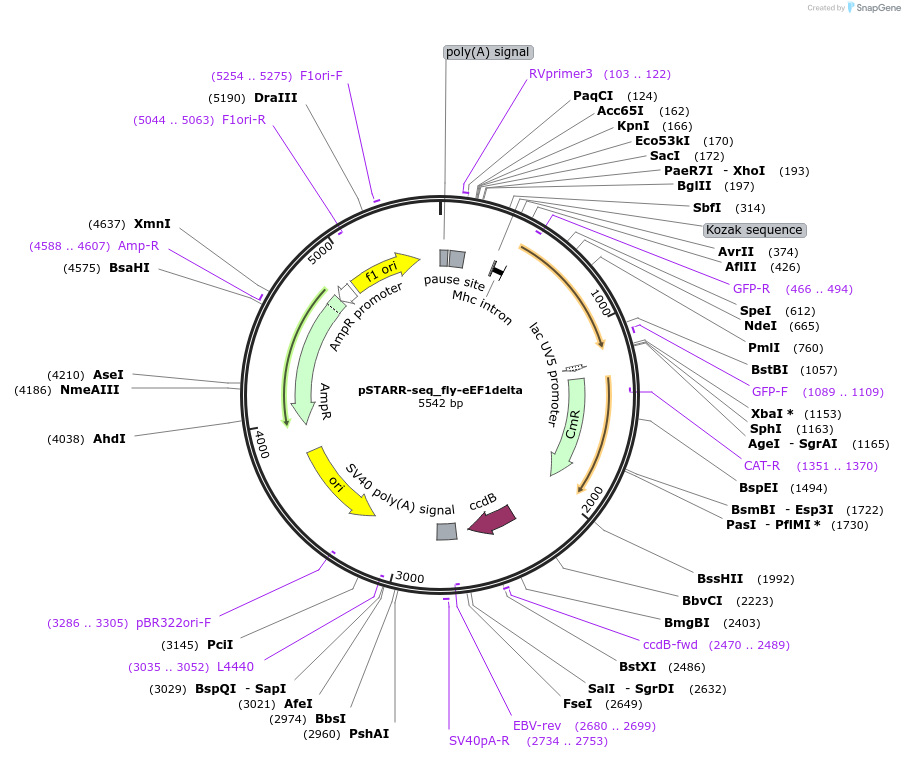
pSTARR-seq_fly-eEF1delta
Plasmid#71501PurposeflySTARR-seq screening vector with eEF1delta core promoterDepositorTypeEmpty backboneUseFlystarr-seq screening vectorTagssgGFP (SuperGlo, Qbiogene, Inc)ExpressionInsectAvailable SinceDec. 10, 2015AvailabilityAcademic Institutions and Nonprofits only -
pBGSA FPR1 R123A
Plasmid#66721PurposeConstitutive expression of FPR1 R123A in mammalian cells.DepositorAvailable SinceJune 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
pBGSA FPR1 D71A
Plasmid#66720PurposeConstitutive expression of FPR1 D71A in mammalian cells.DepositorAvailable SinceJune 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
pOGGUSPlus
Plasmid#64400PurposeOat globulin Glo1 promoter driving expression of GUSPlusDepositorInsertOat globulin Glo1 promoter
TagsGUSplus (modified Staphylococcus sp. b-glucuronid…ExpressionPlantPromoterOat globulin Glo1Available SinceJune 9, 2015AvailabilityAcademic Institutions and Nonprofits only -
pOGGFPR
Plasmid#64398PurposeOat globulin Glo1 promoter driving expression of GFPDepositorInsertOat globulin Glo1 promoter
TagsGFPExpressionPlantPromoterOat globulin Glo1Available SinceJune 2, 2015AvailabilityAcademic Institutions and Nonprofits only -
pIhh4.5k-Luc
Plasmid#53603PurposeReporter construct containing a 4.5 kb promoter region of the Ihh gene in front of a luciferase gene for in vitro DNA transfection assaysDepositorInsertIhh promoter
UseLuciferase; ReporterAvailable SinceJune 1, 2015AvailabilityAcademic Institutions and Nonprofits only -
psiCHECK-2-Tob1-3'UTR
Plasmid#61774PurposeLuciferase reporter containing the 3'UTR of mouse Tob1DepositorInsertmouse Tob1 3'UTR
UseLuciferaseAvailable SinceMay 14, 2015AvailabilityAcademic Institutions and Nonprofits only -
psiCHECK-2-Skp2-3'UTR
Plasmid#61784PurposeLuciferase reporter containing the 3'UTR of mouse Skp2DepositorInsertmouse Skp2 3'UTR
UseLuciferaseAvailable SinceMay 14, 2015AvailabilityAcademic Institutions and Nonprofits only -
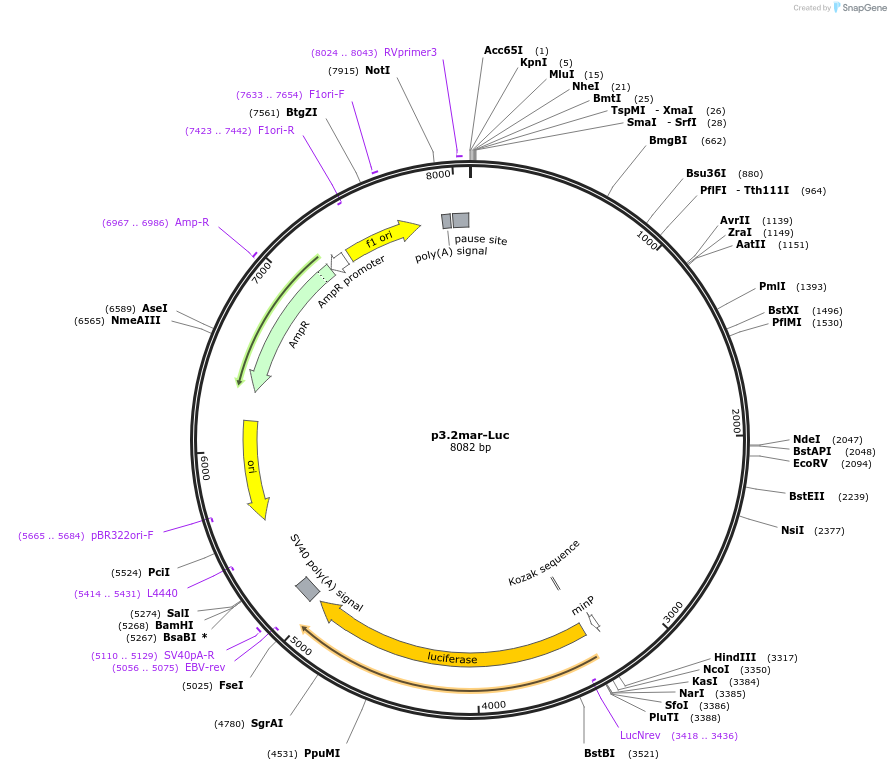
p3.2mar-Luc
Plasmid#63696Purpose3.2 kb marine armor plate enhancer driving Luciferase expressionDepositorInsert3.2 kb marine stickleback armor plate enhancer
TagsLuciferaseExpressionMammalianPromoterTATA boxAvailable SinceApril 7, 2015AvailabilityAcademic Institutions and Nonprofits only -
pAD-g6
Plasmid#61497PurposeFor expression of C. briggsae gld-1, used for rescue of C. elegans q485 homozygotesDepositorAvailable SinceFeb. 25, 2015AvailabilityAcademic Institutions and Nonprofits only -
tdEos-Supervillin-45
Plasmid#57671PurposeLocalization: Membrane/Actin, Excitation: 505 / 569, Emission: 516 / 581DepositorInsertSupervillin
TagstdEosExpressionMammalianPromoterCMVAvailable SinceJan. 16, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
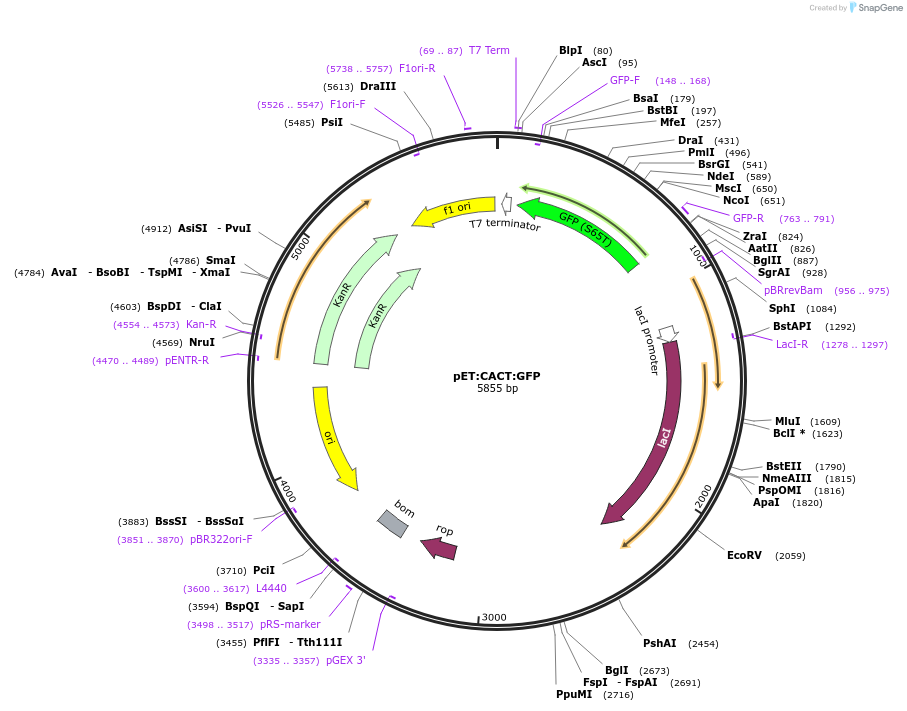
pET:CACT:GFP
Plasmid#60737PurposeA modified pET28 driving expression of GFP, biobrick part Bba_E0040. No LacO site downstream of PT7. T7 promoter has base pairs 'CACT' between -11 and -8.DepositorInsertGFP
UseSynthetic BiologyExpressionBacterialAvailable SinceNov. 5, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCMV-HA-mIFITM1-C103A
Plasmid#58419PurposeExpresses murine IFITM1-C103A with an N-terminal HA tag in mammalian cells.DepositorInsertIFITM1 (Ifitm1 Mouse)
TagsHAExpressionMammalianMutationCysteine 103 to AlaninePromoterCMV IEAvailable SinceSept. 9, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCMV-HA-mIFITM1-C83A
Plasmid#58418PurposeExpresses murine IFITM1-C83A with an N-terminal HA tag in mammalian cells.DepositorInsertIFITM1 (Ifitm1 Mouse)
TagsHAExpressionMammalianMutationCysteine 83 to AlaninePromoterCMV IEAvailable SinceSept. 9, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCMV-HA-mIFITM1-C50A
Plasmid#58417PurposeExpresses murine IFITM1-C50A with an N-terminal HA tag in mammalian cells.DepositorInsertIFITM1 (Ifitm1 Mouse)
TagsHAExpressionMammalianMutationCysteine 50 to AlaninePromoterCMV IEAvailable SinceSept. 9, 2014AvailabilityAcademic Institutions and Nonprofits only -
pCMV-HA-mIFITM1-C49A
Plasmid#58416PurposeExpresses murine IFITM1-C49A with an N-terminal HA tag in mammalian cellsDepositorInsertIFITM1 (Ifitm1 Mouse)
TagsHAExpressionMammalianMutationCysteine 49 to AlaninePromoterCMV IEAvailable SinceSept. 9, 2014AvailabilityAcademic Institutions and Nonprofits only -
pGL3-PIM2mut2
Plasmid#51850Purposepromoter region of PIM2 lacking the forkhead binding site #1DepositorInsertPIM2 (PIM2 Human)
UseLuciferase; Reporter vectorAvailable SinceJuly 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMiR-HPV16-E1-m5
Plasmid#53702PurposeLuciferase reporter assay for HPV16 E7 with point mutation on miR-375 binding siteDepositorInsertHPV16-E1(partial)
UseLuciferaseMutationMutation in miR-375 binding site m5 (refer to cit…Available SinceJune 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMiR-HPV16-E1-m4
Plasmid#53701PurposeLuciferase reporter assay for HPV16 E7 with point mutation on miR-375 binding siteDepositorInsertHPV16-E1(partial)
UseLuciferaseMutationMutation in miR-375 binding site m4 (refer to cit…Available SinceJune 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMiR-HPV16-E1-m3
Plasmid#53700PurposeLuciferase reporter assay for HPV16 E7 with point mutation on miR-375 binding siteDepositorInsertHPV16-E1(partial)
UseLuciferaseMutationMutation in miR-375 binding site m3 (refer to cit…Available SinceJune 26, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMiR-HPV16-E7-m2
Plasmid#53698PurposeLuciferase reporter assay for HPV16 E7 with point mutation on miR-375 binding siteDepositorInsertHPV16-E7
UseLuciferaseMutationMutation in miR-375 binding site m2 (refer to cit…Available SinceJune 19, 2014AvailabilityAcademic Institutions and Nonprofits only -
pMiR-HPV16-E7-m1
Plasmid#53697PurposeLuciferase reporter assay for HPV16 E7 with point mutation on miR-375 binding siteDepositorInsertHPV16-E7
UseLuciferaseMutationMutation in miR-375 binding site m1 (refer to cit…Available SinceJune 19, 2014AvailabilityAcademic Institutions and Nonprofits only -
CS2-HA-dASUN
Plasmid#47041PurposeExpression of tagged-dASUNDepositorAvailable SinceSept. 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
TV3-mCHY-dASUN
Plasmid#47034PurposeDrosophila testes specific expression of fluorescent dASUNDepositorAvailable SinceSept. 10, 2013AvailabilityAcademic Institutions and Nonprofits only -
TV3-eGFP-dASUN
Plasmid#47033PurposeDrosophila testes specific expression of fluorescent dASUNDepositorAvailable SinceSept. 10, 2013AvailabilityAcademic Institutions and Nonprofits only -
pFA5
Plasmid#46791PurposeTo express a version of Fun30 without CUE domain in budding yeastDepositorAvailable SinceAug. 8, 2013AvailabilityAcademic Institutions and Nonprofits only -
pFA12
Plasmid#46792PurposeTo express a version of Fun30 with a mutated CUE domain in budding yeastDepositorAvailable SinceAug. 8, 2013AvailabilityAcademic Institutions and Nonprofits only