We narrowed to 7,692 results for: Lif;
-
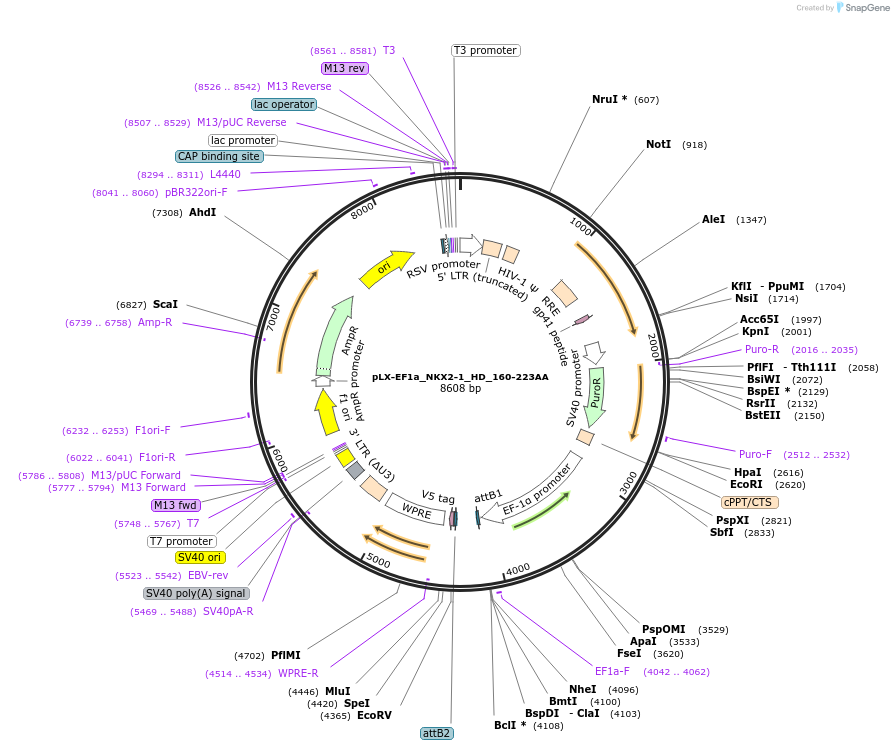
Plasmid#221489PurposeNKX2-1 N-terminal and C-terminal truncation (homeodomain only) mutation lentiviral overexpressionDepositorInsertNKX2-1 (NKX2-1 Human)
UseLentiviralTagsV5ExpressionMammalianMutationDeletion of 1-159AA N-terminal and 224-371AA C-te…PromoterEF1aAvailable SinceJuly 1, 2024AvailabilityAcademic Institutions and Nonprofits only -
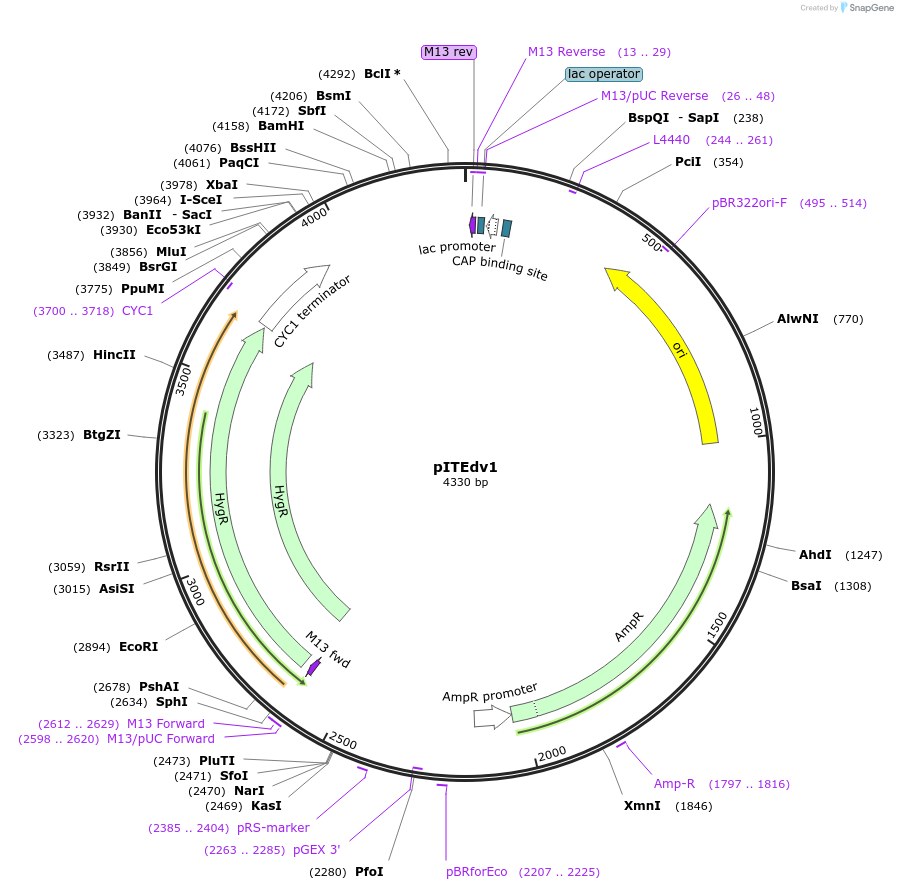
pITEdv1
Plasmid#218283PurposeUse together with with pJE13HD7 and pJE13HD7 derivatives to integrate heterologous genes at ho locusDepositorInsertHph>tAgTEF1-ISceI-Repeat1-HO(1828, 1979)
ExpressionYeastAvailable SinceMay 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
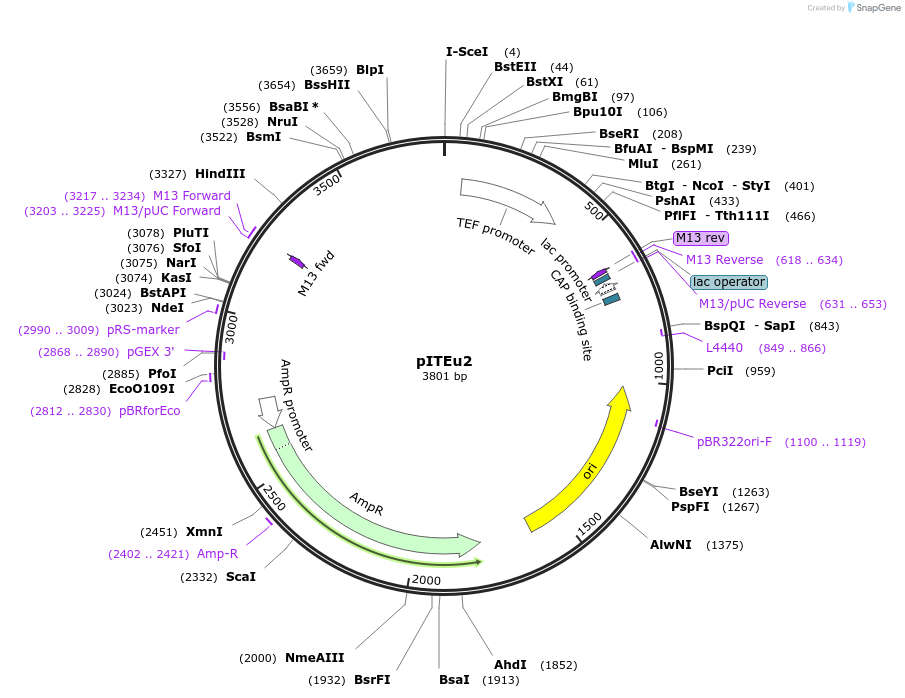
pITEu2
Plasmid#218282PurposeUse together with pToxEXP/pToxAmp plasmids to insert heterologous genes at ho locusDepositorInsertHO(-253, -1)-Repeat2-ISceI-pAgTEF1>Hph(partial)
ExpressionYeastAvailable SinceMay 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
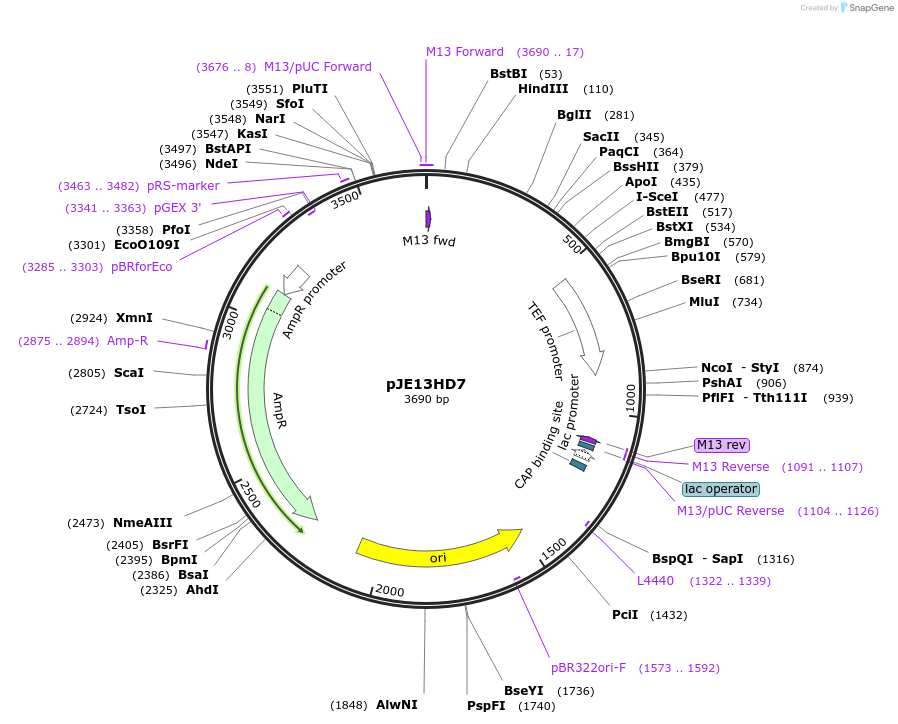
pJE13HD7
Plasmid#218281PurposeUse together with pITEdv1, or pToxEXP1/pToxAmp plasmids to insert heterologous gene(s) at ho locus.DepositorInsertHO(-253, -1)-Repeat1-ISceI-pAgTEF1>Hph(partial)
ExpressionYeastAvailable SinceMay 15, 2024AvailabilityAcademic Institutions and Nonprofits only -
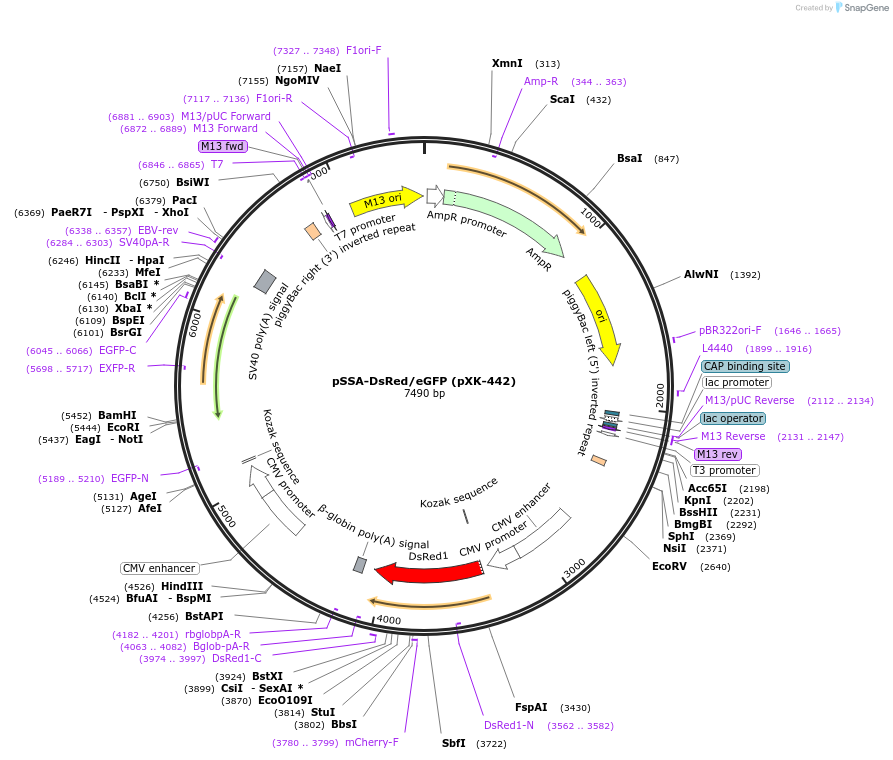
pSSA-DsRed/eGFP (pXK-442)
Plasmid#208827PurposeThe SSA-based surrogate reporter for validating the activity of different gene editing tools as well as for helping to screen gene-editing positive cells. Key Construct: CMV-DsRed-CMV-eGFP*(SSA).DepositorTypeEmpty backboneExpressionMammalianPromoterCMVAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
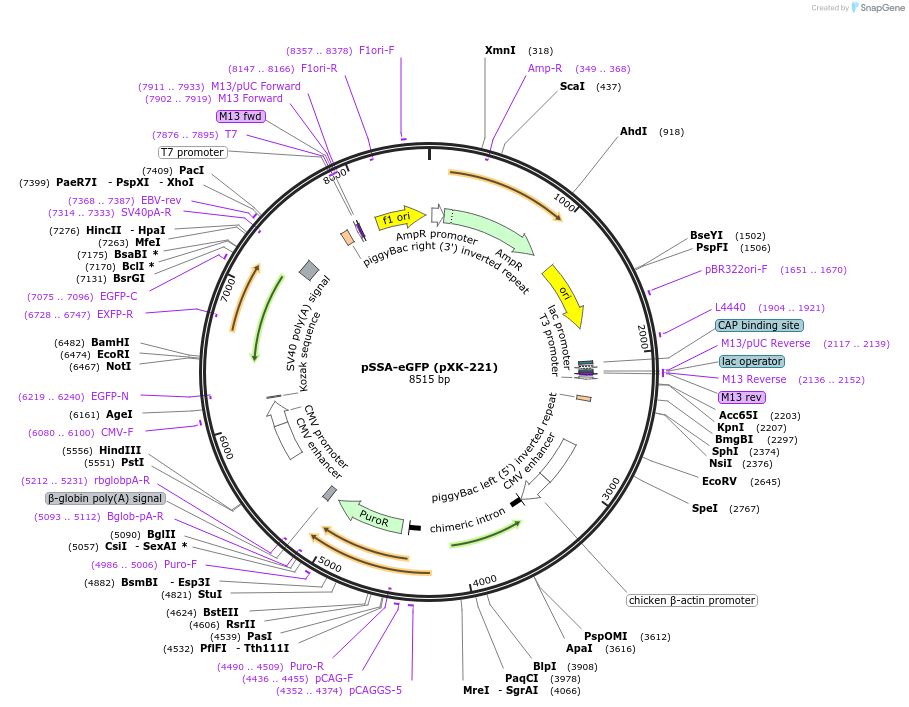
pSSA-eGFP (pXK-221)
Plasmid#208826PurposeThe SSA-based surrogate reporter for validating the activity of gene editing tools as well as for helping to screen gene-editing positive cells. Key Construct: CAG-PuroR-CMV-eGFP*(SSA).DepositorTypeEmpty backboneExpressionMammalianPromoterCMVAvailable SinceDec. 14, 2023AvailabilityAcademic Institutions and Nonprofits only -
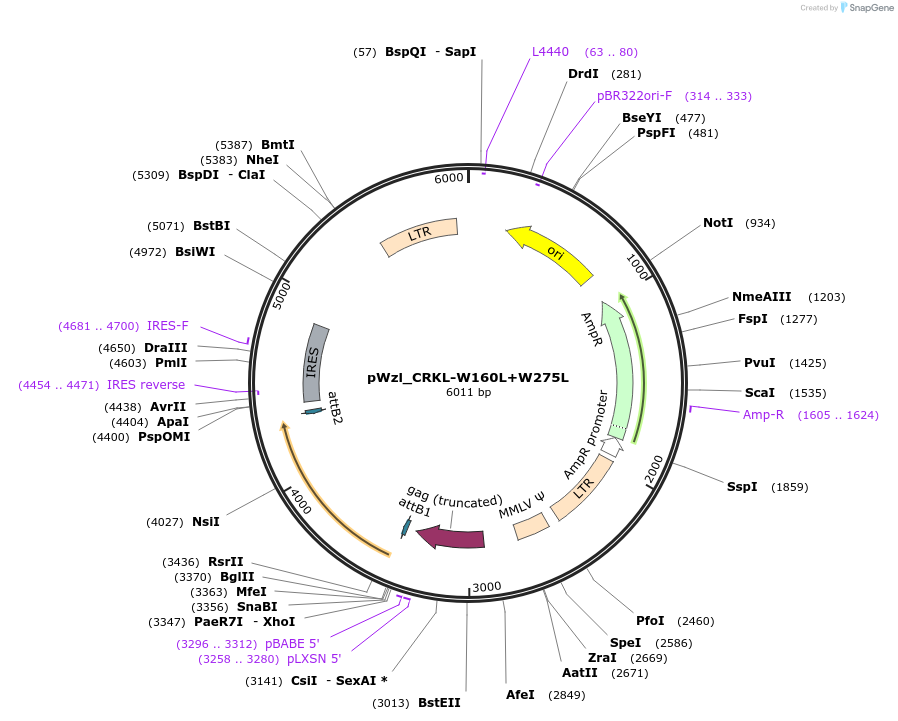
pWzl_CRKL-W160L+W275L
Plasmid#196021PurposeExpresses CRKL W160L + W275LDepositorInsertCRKL
UseRetroviralAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
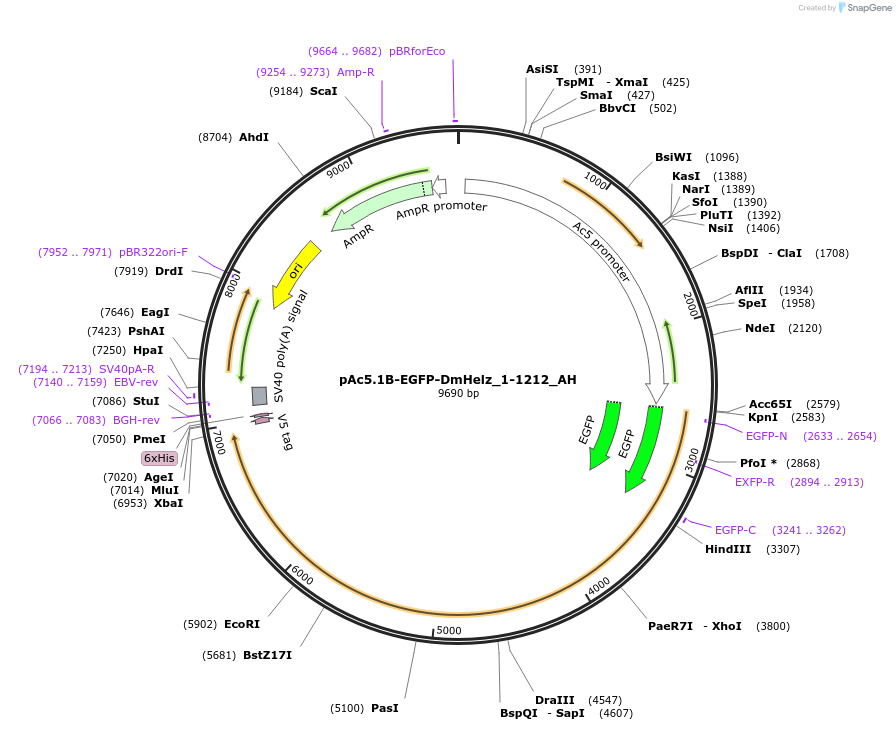
pAc5.1B-EGFP-DmHelz_1-1212_AH
Plasmid#148882PurposeInsect Expression of DmHelz_1-1212DepositorInsertDmHelz_1-1212 (CG9425 Fly)
ExpressionInsectAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
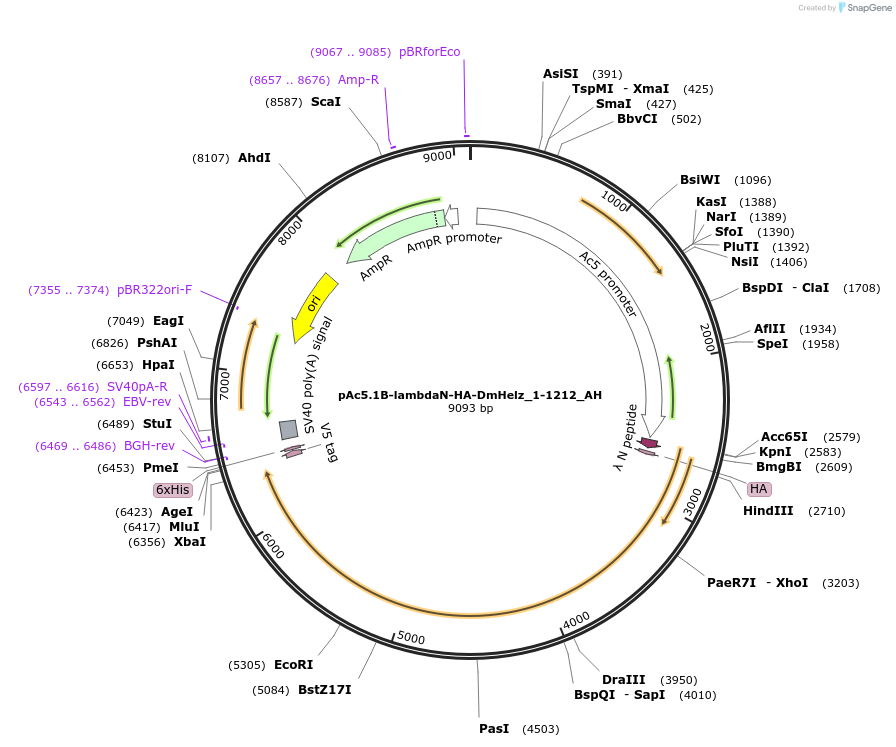
pAc5.1B-lambdaN-HA-DmHelz_1-1212_AH
Plasmid#148886PurposeInsect Expression of DmHelz_1-1212DepositorInsertDmHelz_1-1212 (CG9425 Fly)
ExpressionInsectAvailable SinceMarch 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
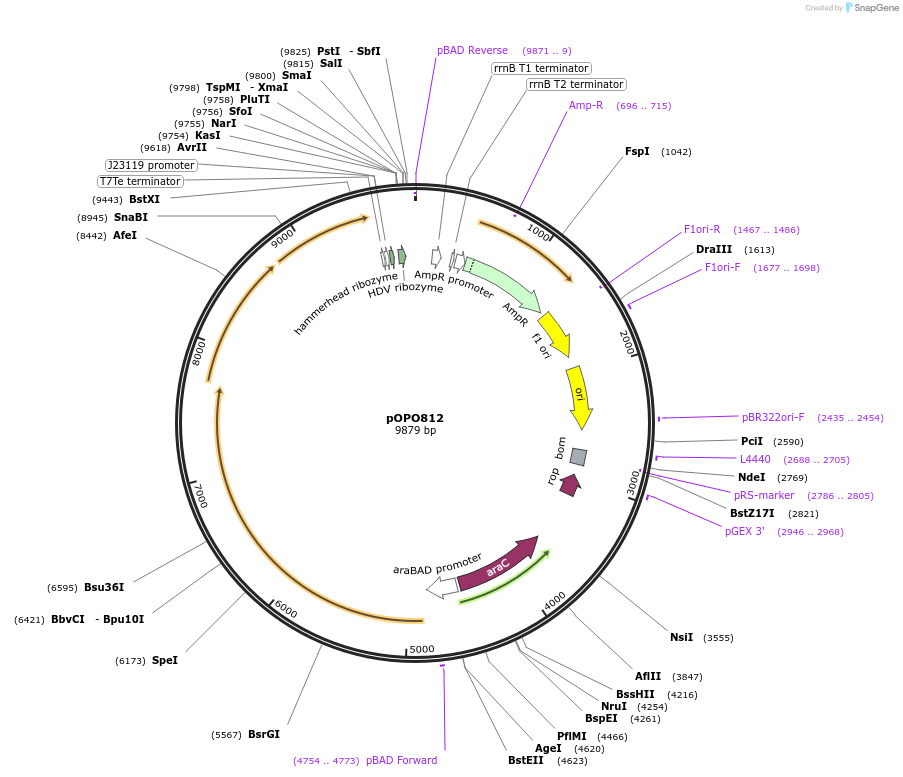
pOPO812
Plasmid#195306PurposepBAD322-MycaIDCas-ΔCas6-RGR-lacZs, arapBAD promoter expressing type I-D Cascade of McCAST (Tn7575) without cas6 gene and ribozyme flanked lacZ spacer 5.DepositorInsertMcCAST Cascade Δcas6 and ribozyme flanked lacZ spacer 5
ExpressionBacterialPromoterArabinose inducible arapBADAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
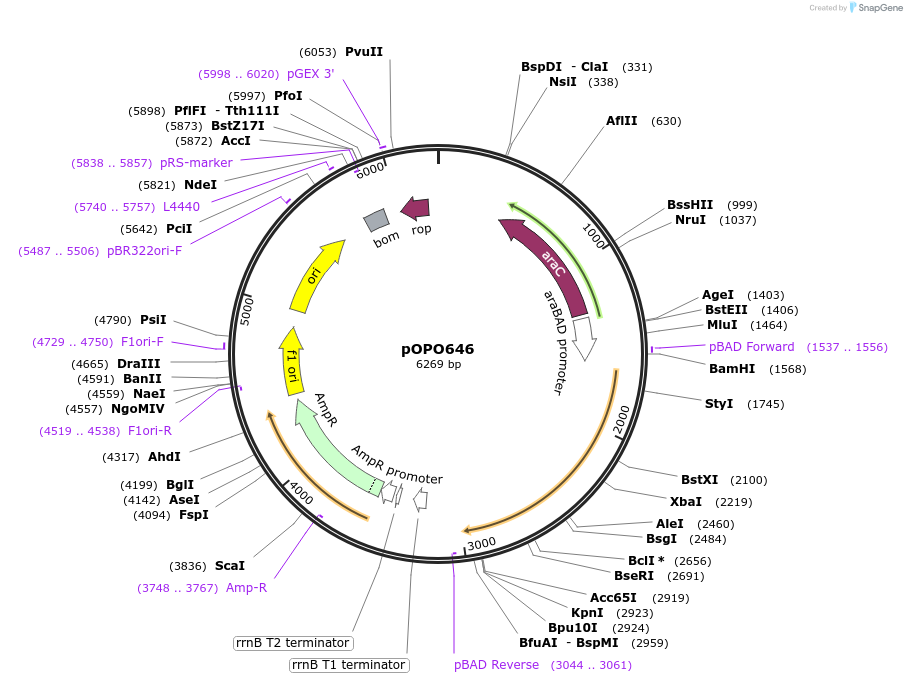
pOPO646
Plasmid#195307PurposepBAD322-MycaTnsD, arapBAD promoter expressing cyanobacterial tRNA-Leu gene targeting TnsD protein of McCAST (Tn7575).DepositorInsertTnsD of McCAST
ExpressionBacterialPromoterArabinose inducible arapBADAvailable SinceMarch 13, 2023AvailabilityAcademic Institutions and Nonprofits only -
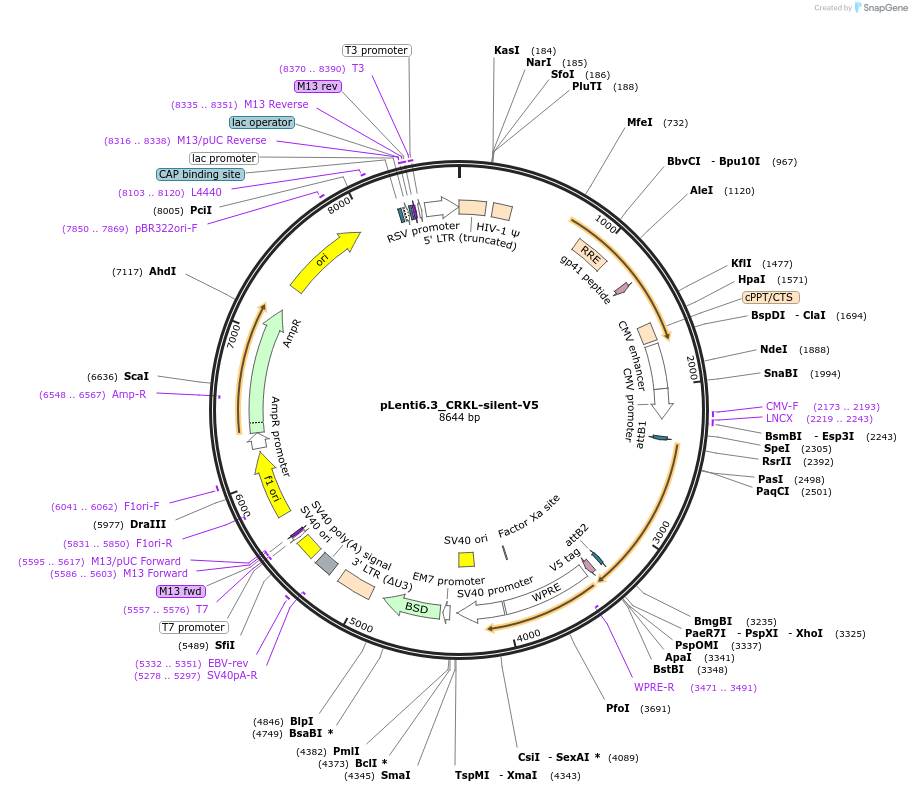
pLenti6.3_CRKL-silent-V5
Plasmid#196015PurposeExpresses CRKL-silent, V5 taggedDepositorInsertCRKL
UseLentiviralExpressionMammalianAvailable SinceMarch 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
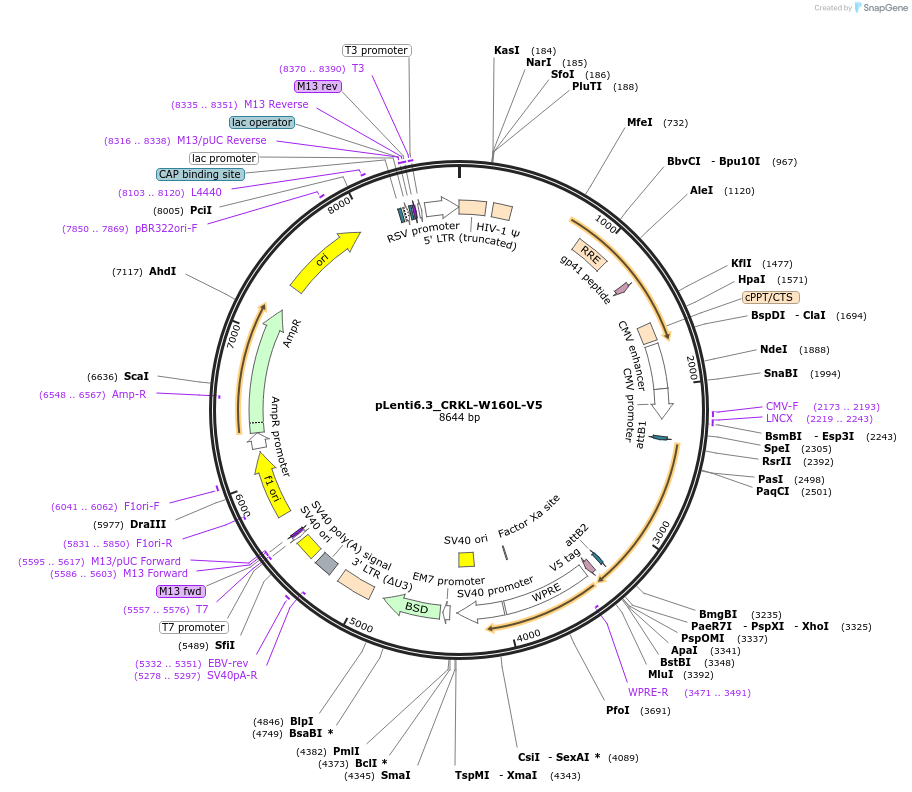
pLenti6.3_CRKL-W160L-V5
Plasmid#196017PurposeExpresses CRKL W160L, V5 taggedDepositorInsertCRKL
UseLentiviralExpressionMammalianAvailable SinceMarch 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
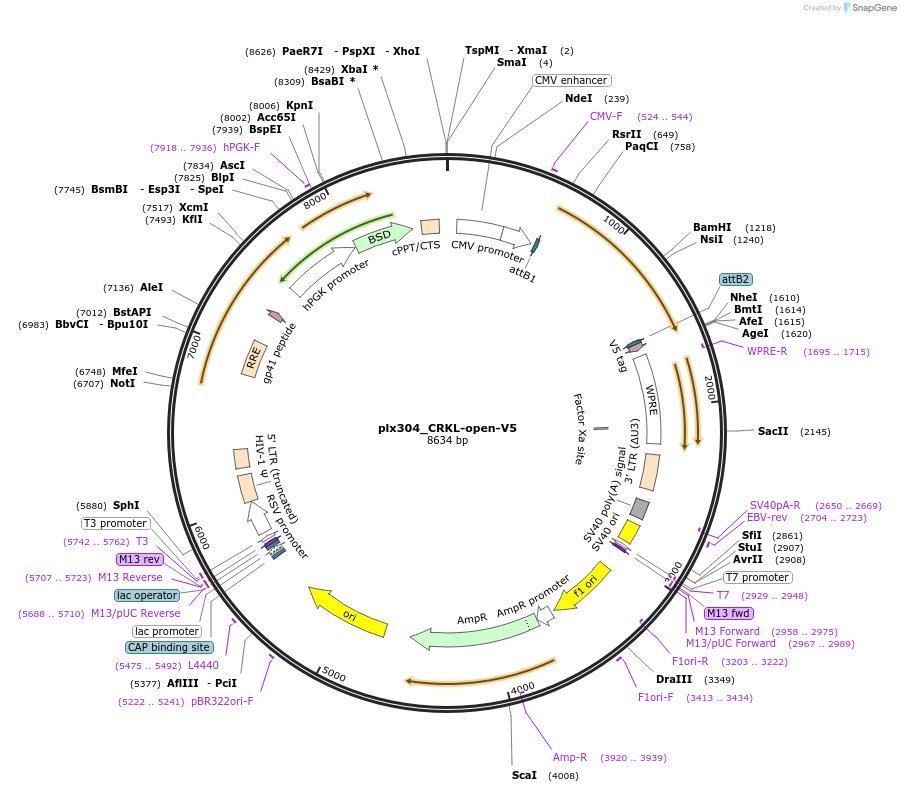
plx304_CRKL-open-V5
Plasmid#196023PurposeExpresses CRKL open, V5 taggedDepositorInsertCRKL
UseLentiviralAvailable SinceMarch 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
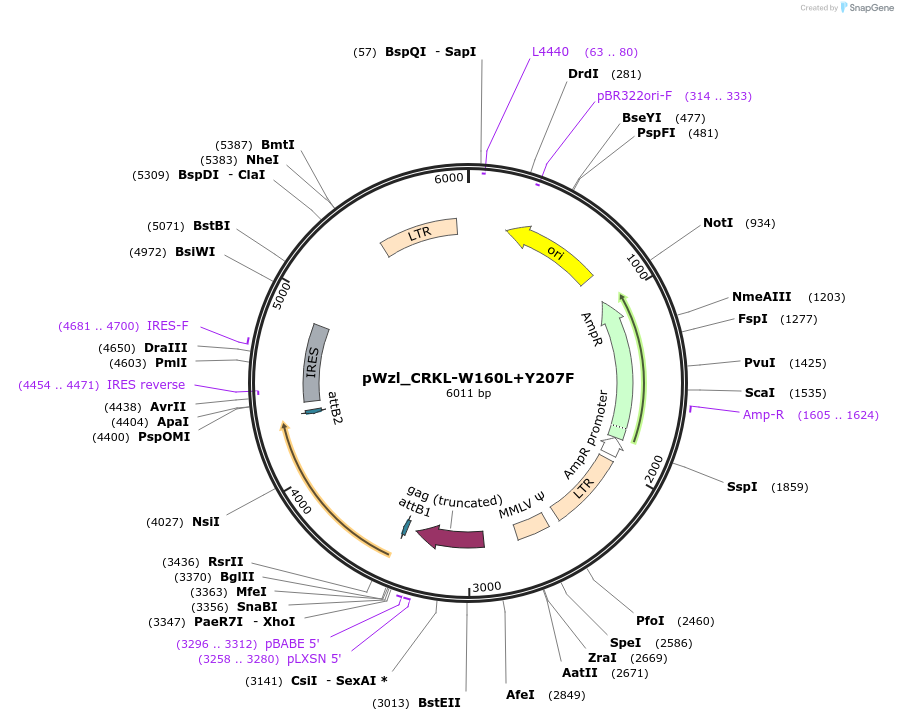
pWzl_CRKL-W160L+Y207F
Plasmid#196020PurposeExpresses Expresses CRKL W160L + Y207FDepositorInsertCRKL
UseRetroviralAvailable SinceMarch 3, 2023AvailabilityAcademic Institutions and Nonprofits only -
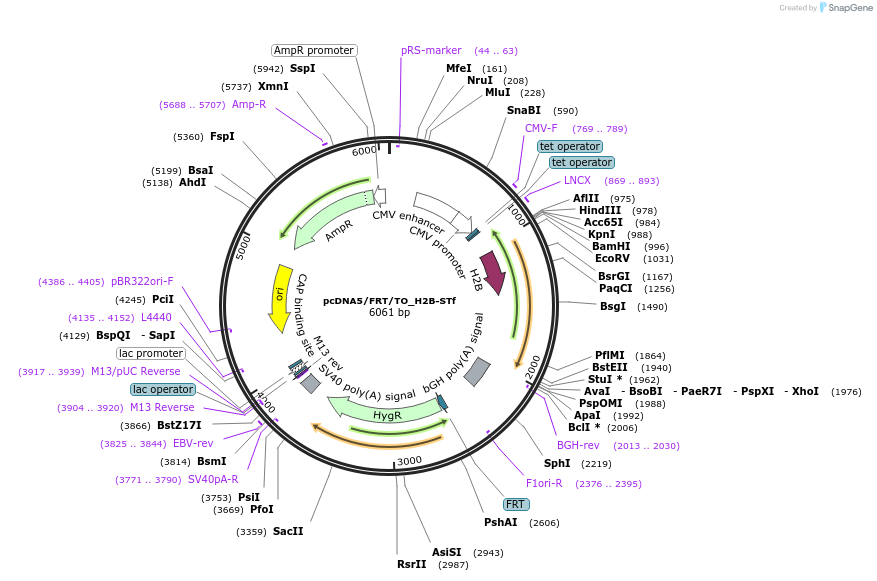
pcDNA5/FRT/TO_H2B-STf
Plasmid#181998PurposeCMV overexpression of SNAP-tag fast fusion to H2B for nuclear localization in mammalian cell linesDepositorInsertH2B-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
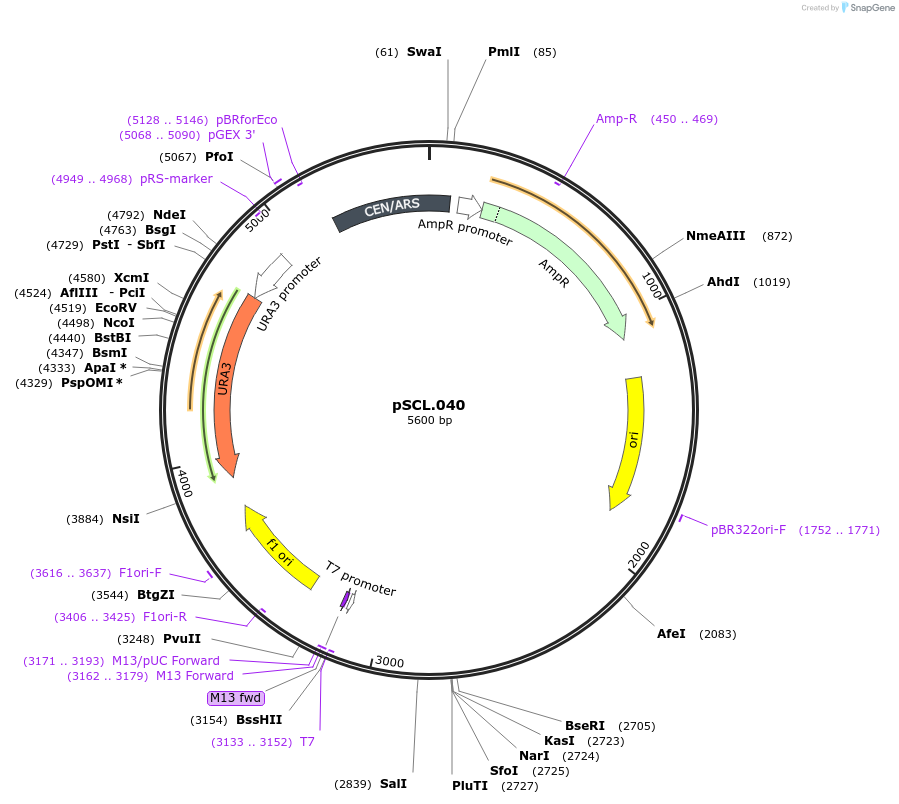
pSCL.040
Plasmid#184974PurposeTest effect of extending a1/a2 on ADE2 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, ADE2 P272X, a1/a2 length: 27 v2
ExpressionYeastMutationADE2 donor P272stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
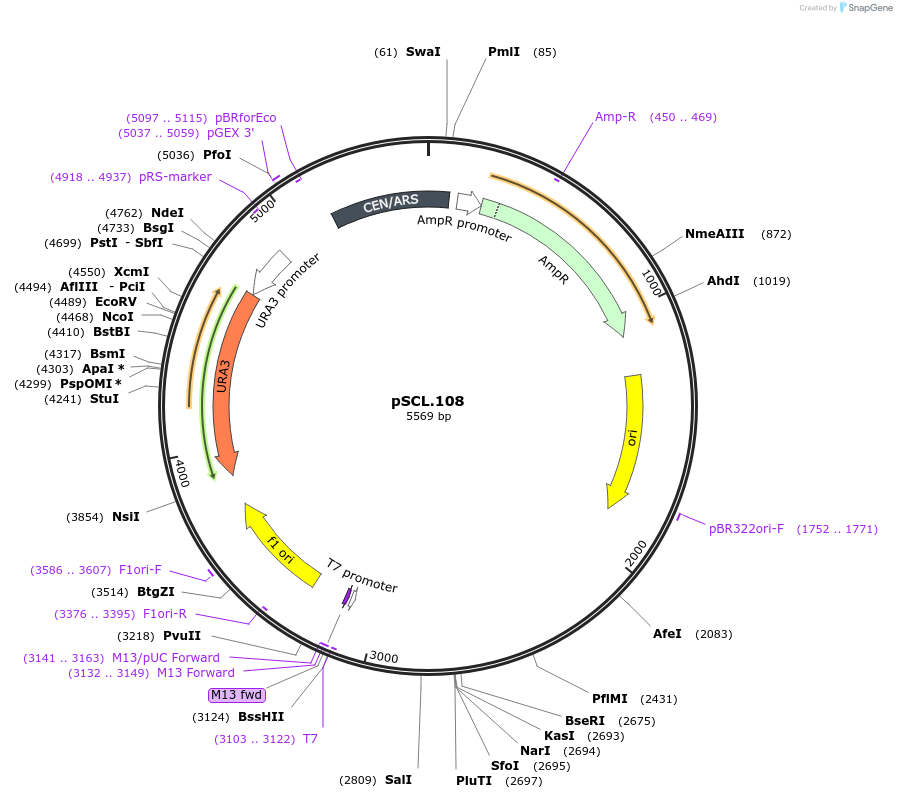
pSCL.108
Plasmid#184977PurposeExpress -Eco1 LYP1 editing ncRNA and gRNADepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 12
ExpressionYeastMutationLYP1 donor E27stopPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
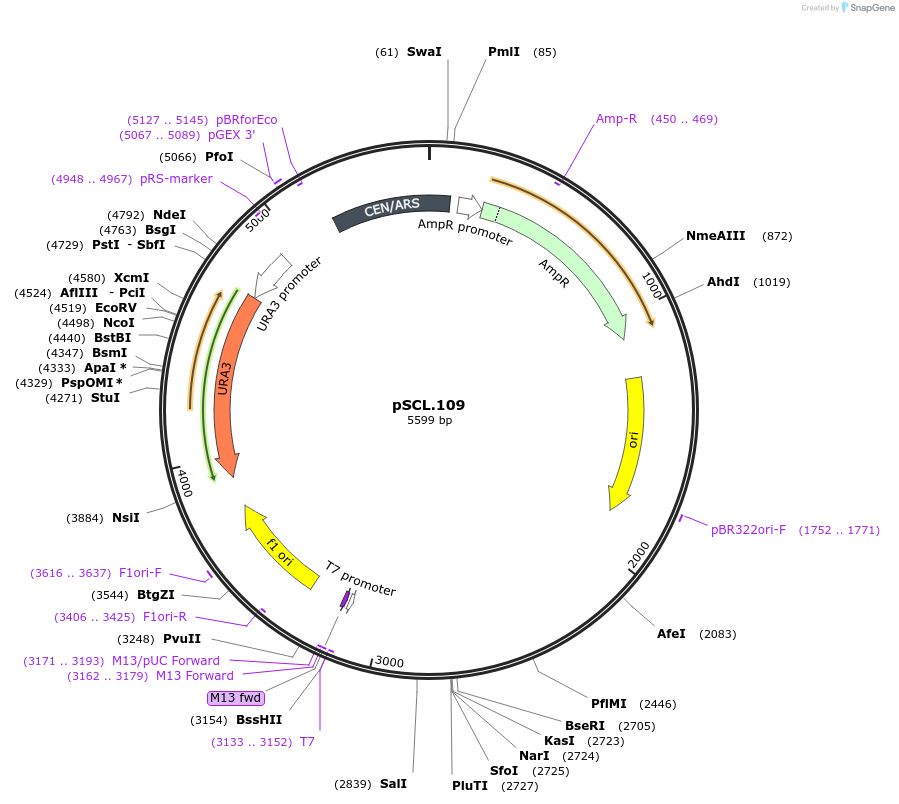
pSCL.109
Plasmid#184978PurposeTest effect of extending a1/a2 on LYP1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, LYP1 E27X, a1/a2 length: 27 v1
ExpressionYeastMutationLYP1 donor E27stop, a1/a2 length extended to 27 bpPromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
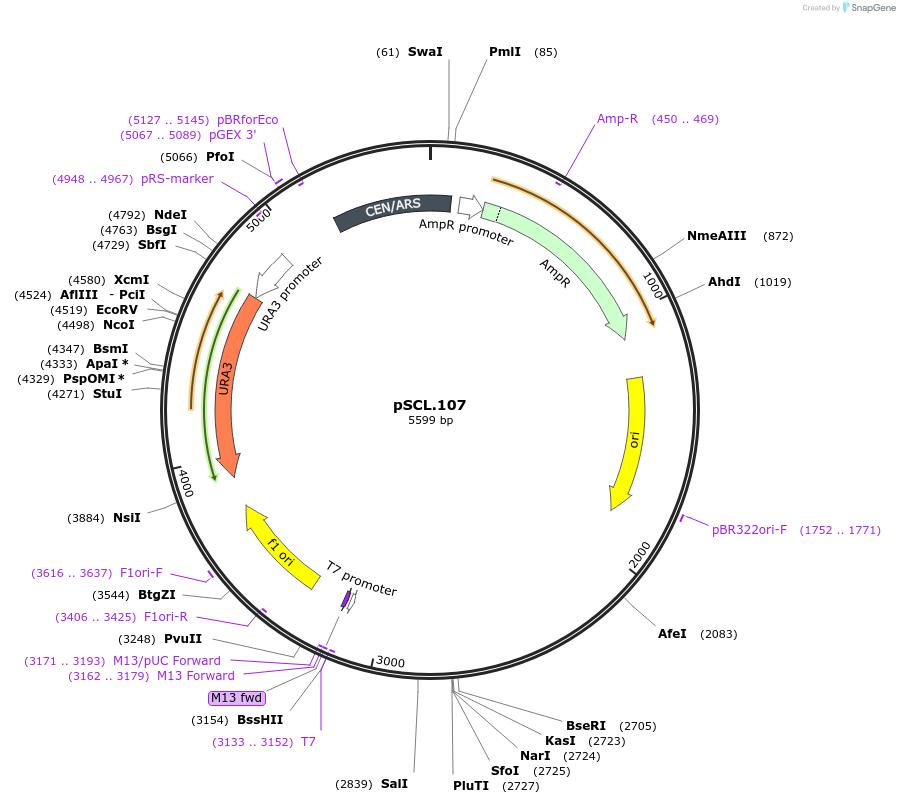
pSCL.107
Plasmid#184976PurposeTest effect of extending a1/a2 on CAN1 editing rates in yeastDepositorInsertEco1 editing ncRNA and gRNA, CAN1 G444X, a1/a2 length: 27 v1
ExpressionYeastMutationCAN1 donor G444stop, a1/a2 length extended to 27 …PromoterGal7Available SinceNov. 4, 2022AvailabilityAcademic Institutions and Nonprofits only