We narrowed to 2,760 results for: FAS
-
Plasmid#198645PurposeEasy-MISE toolkit pEM-plasmid containing PGK1 promoter with EF protruding endsDepositorInsertpPGK1
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
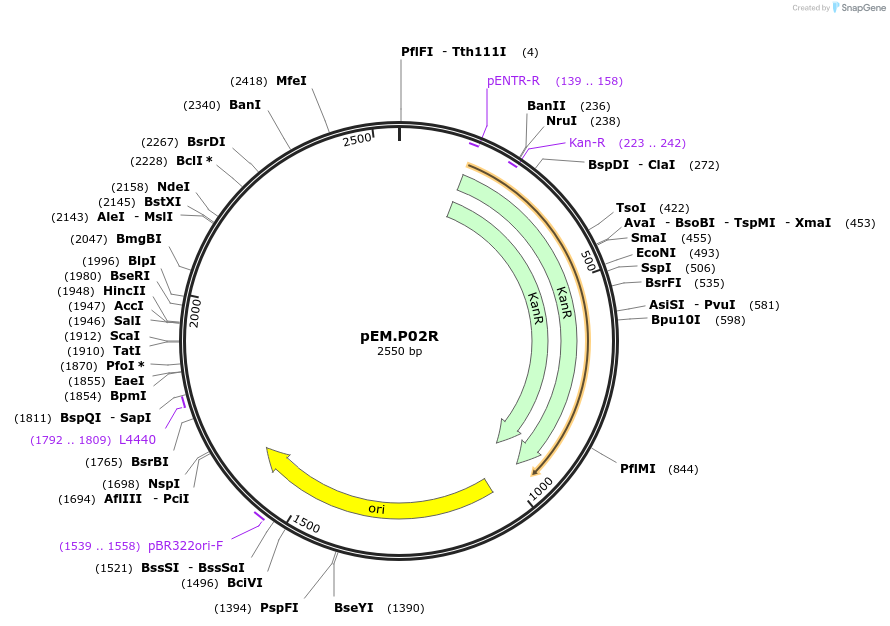
pEM.P02R
Plasmid#198644PurposeEasy-MISE toolkit pEM-plasmid containing ENO2 promoter with FG protruding endsDepositorInsertpENO2
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
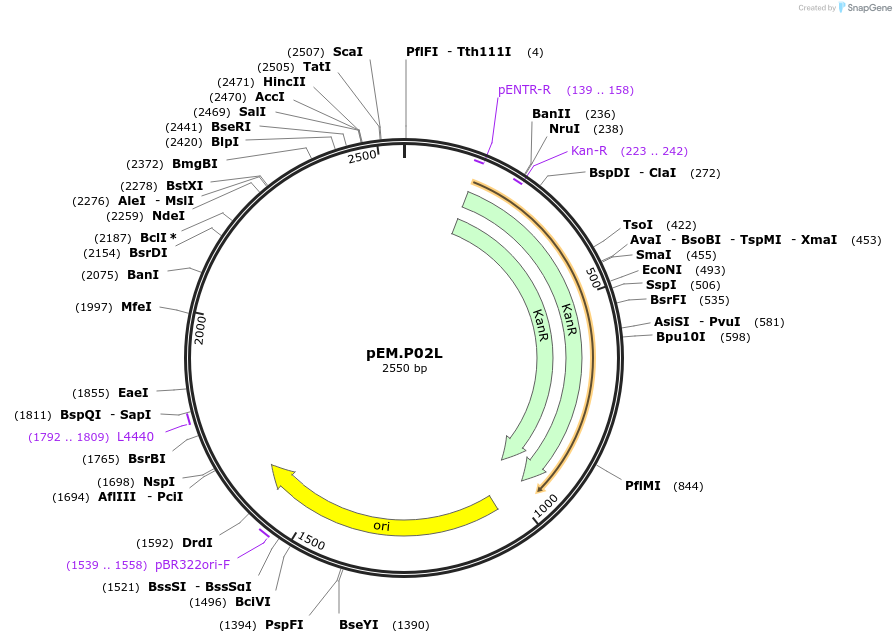
pEM.P02L
Plasmid#198643PurposeEasy-MISE toolkit pEM-plasmid containing ENO2 promoter with EF protruding endsDepositorInsertpENO2
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
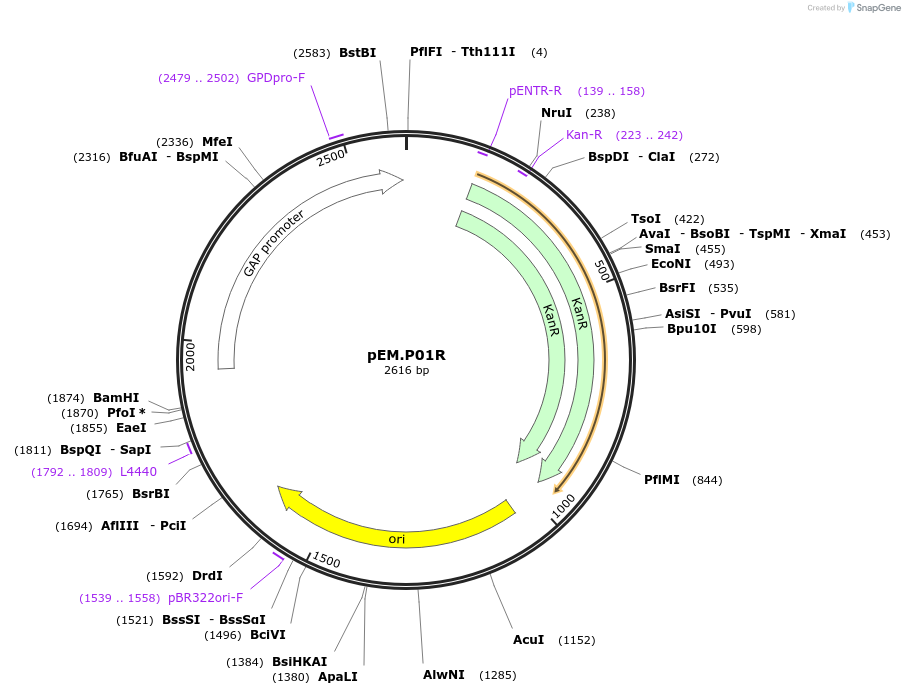
pEM.P01R
Plasmid#198642PurposeEasy-MISE toolkit pEM-plasmid containing TDH3 promoter with FG protruding endsDepositorInsertpTDH3
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
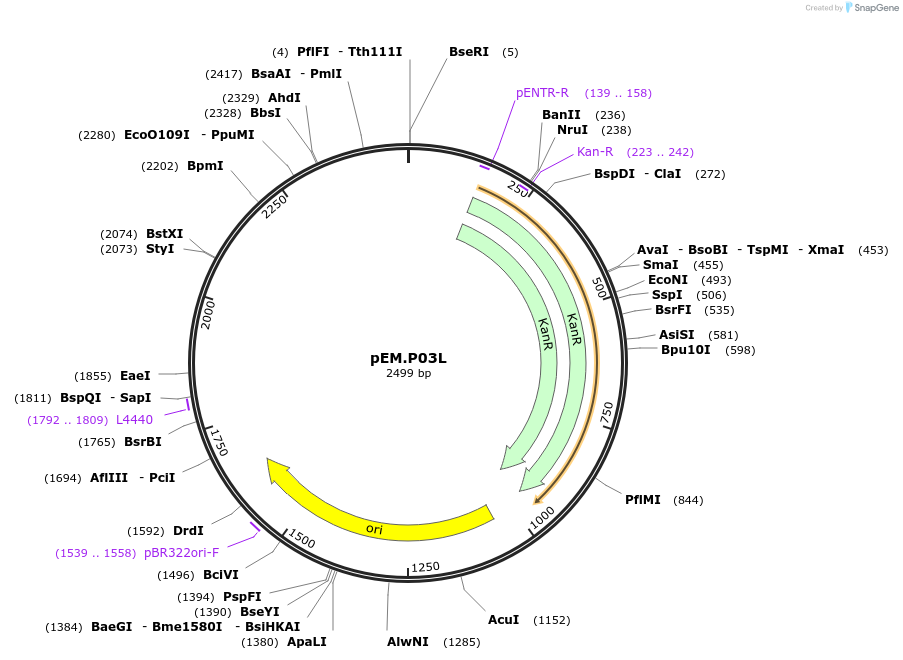
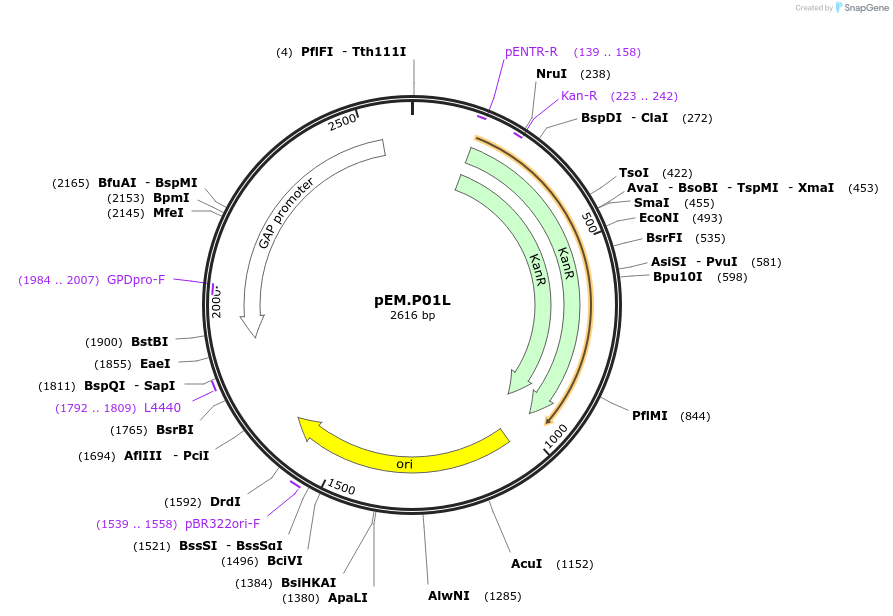
pEM.P01L
Plasmid#198641PurposeEasy-MISE toolkit pEM-plasmid containing TDH3 promoter with EF protruding endsDepositorInsertpTDH3
UseSynthetic BiologyExpressionYeastAvailable SinceMay 15, 2023AvailabilityAcademic Institutions and Nonprofits only -
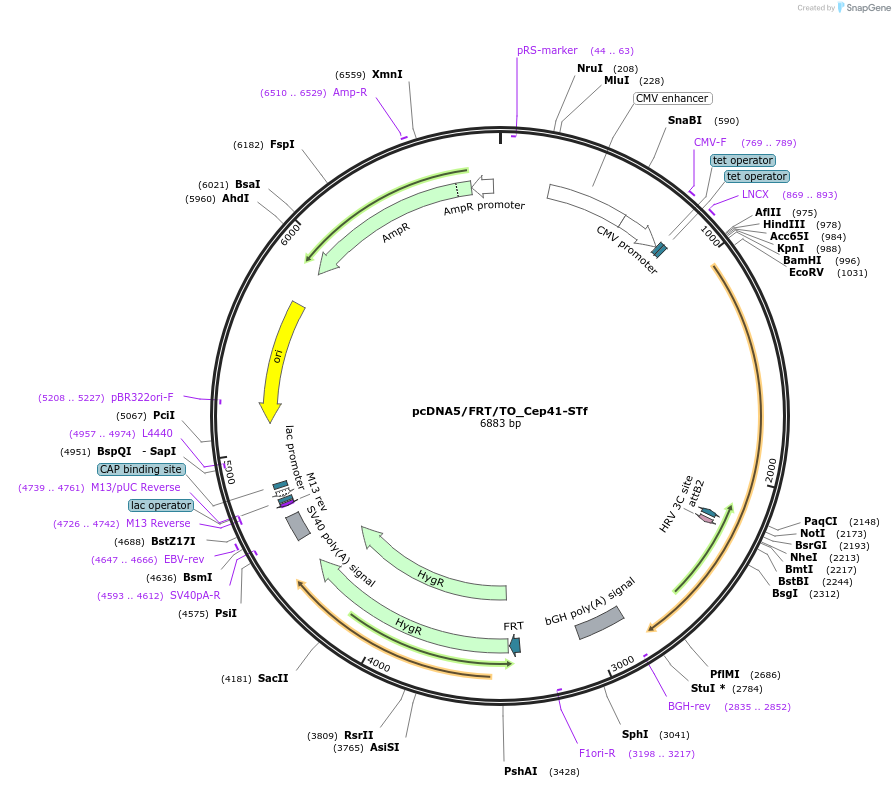
pcDNA5/FRT/TO_Cep41-STf
Plasmid#182010PurposeCMV overexpression of SNAP-tag fast in mammalian cell lines associated to microtubulesDepositorInsertCep41-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
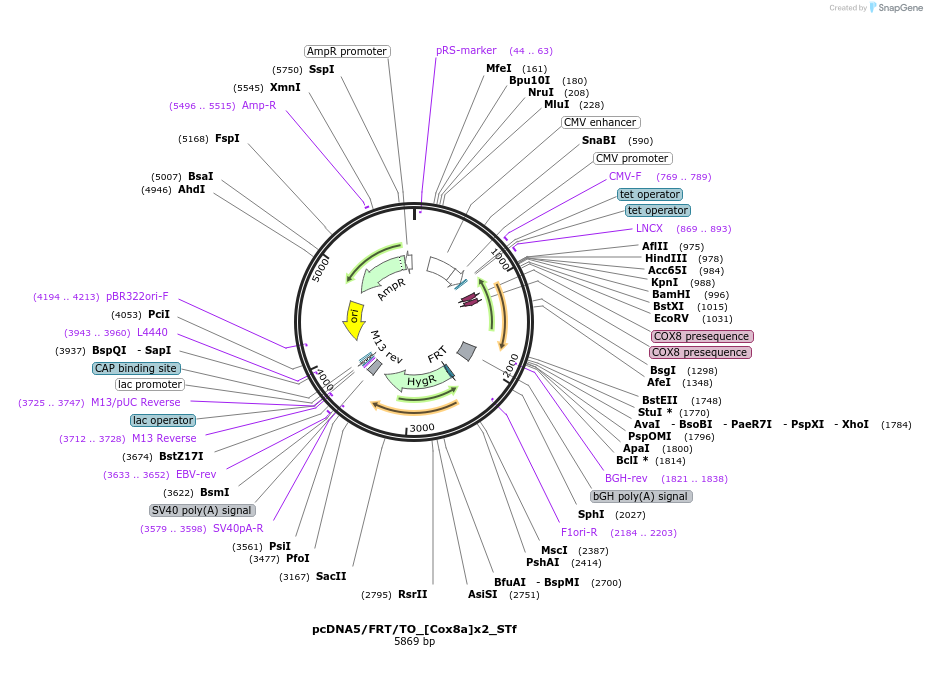
pcDNA5/FRT/TO_[Cox8a]x2_STf
Plasmid#182003PurposeCMV overexpression of SNAP-tag fast in the mitochondrial matrix of mammalian cell linesDepositorInsert[Cox8a]x2_STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
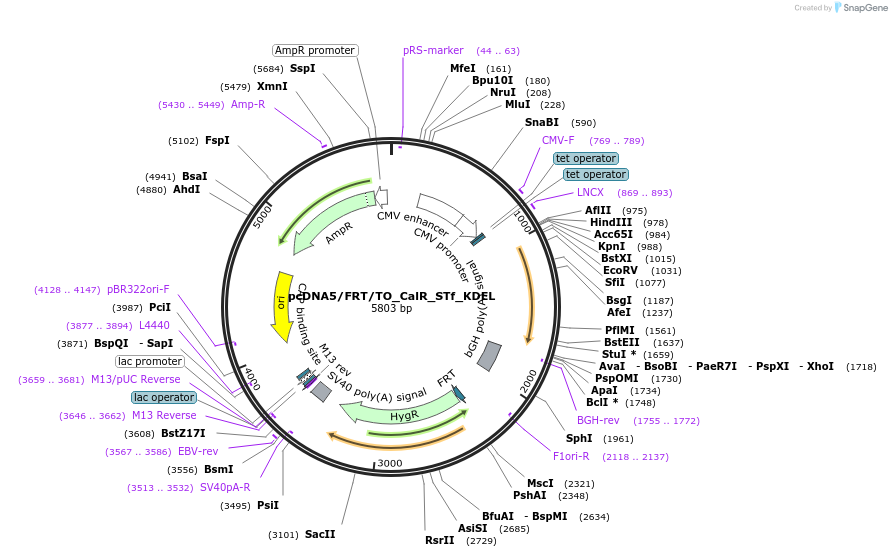
pcDNA5/FRT/TO_CalR_STf_KDEL
Plasmid#182004PurposeCMV overexpression of SNAP-tag fast in the endoplasmic reticulum of mammalian cell linesDepositorInsertCalR_STf_KDEL
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
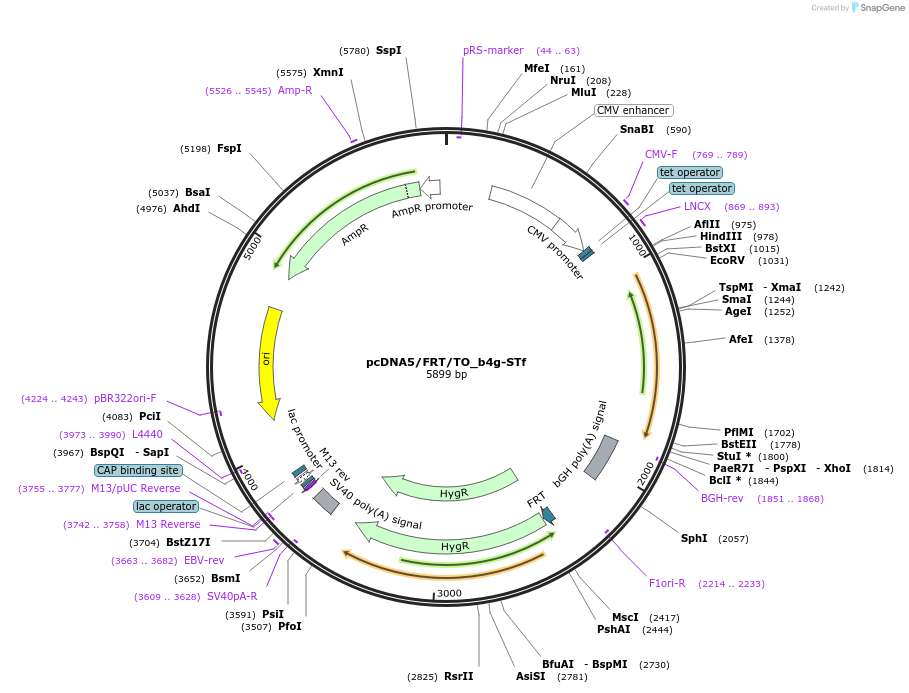
pcDNA5/FRT/TO_b4g-STf
Plasmid#182005PurposeCMV overexpression of SNAP-tag fast in the golgi apparatus of mammalian cell linesDepositorInsertb4g-STf
ExpressionMammalianMutationFast mutation on SNAP-tag E30RPromoterCMVAvailable SinceNov. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
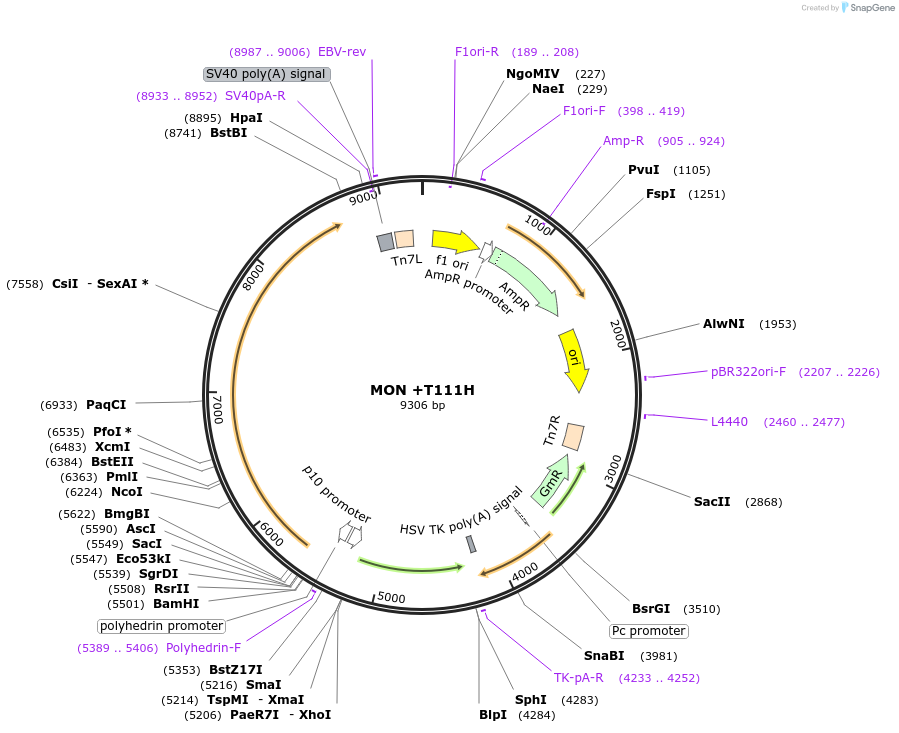
MON +T111H
Plasmid#187432PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged threonine at 117 to histidinePromoterPH and p10Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
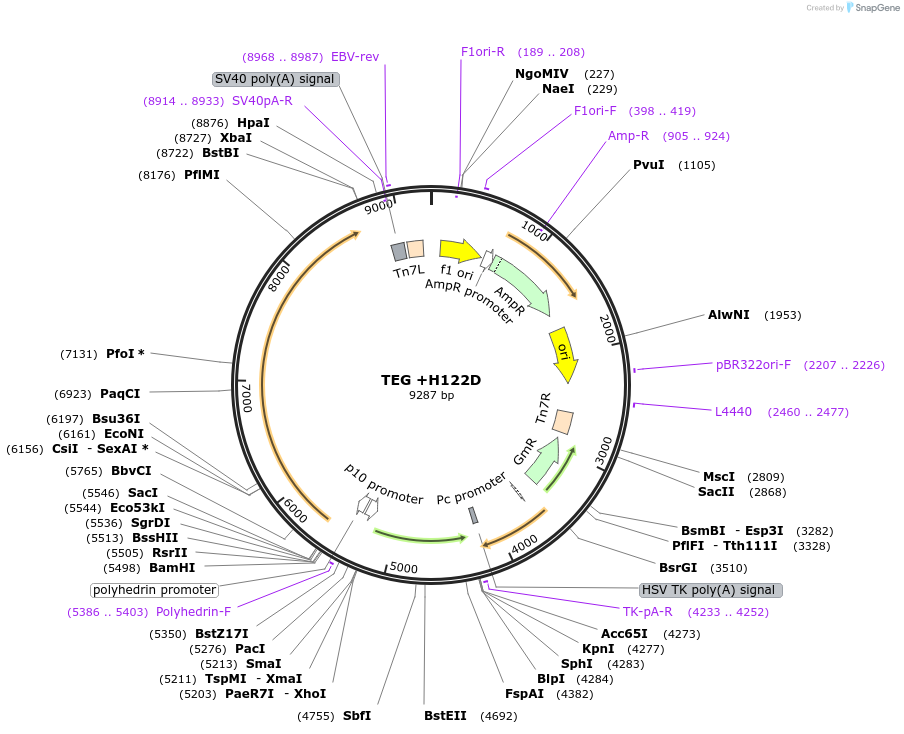
TEG +H122D
Plasmid#187434PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged histidine at 128 to aspartic acidPromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
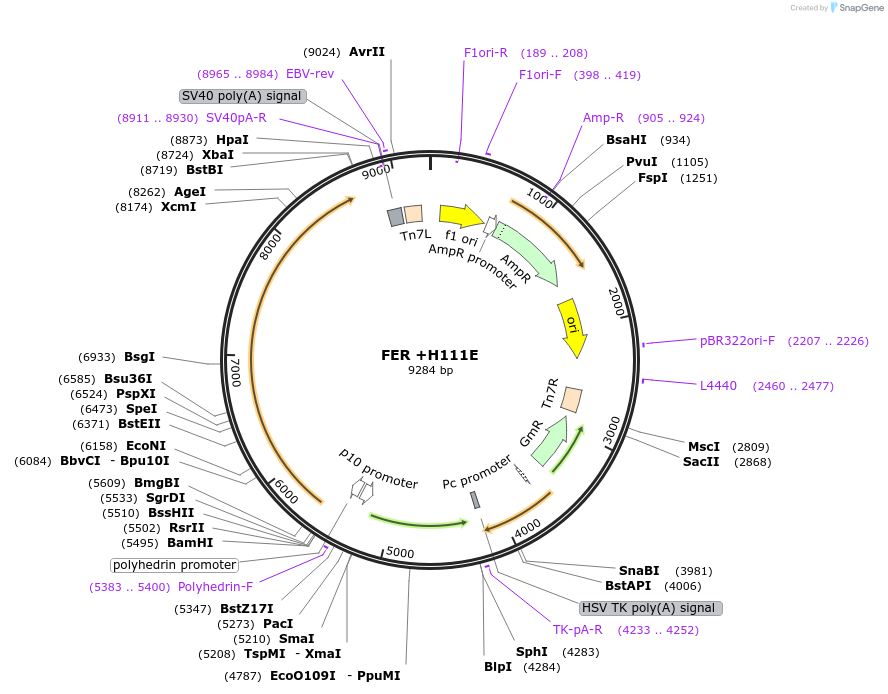
FER +H111E
Plasmid#187429PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged histidine at 117 to glutamic acidPromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
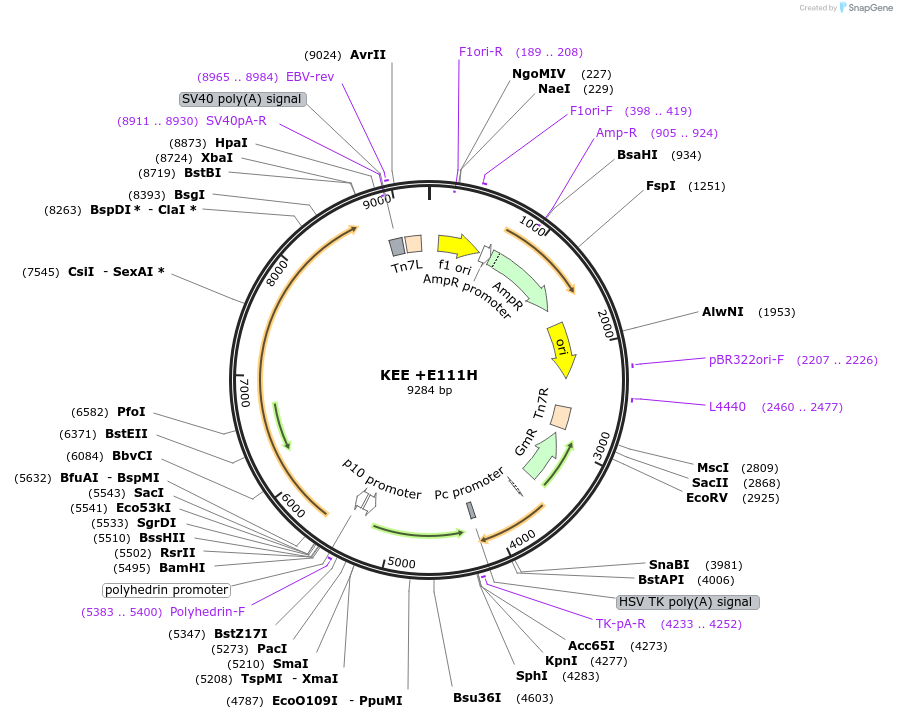
KEE +E111H
Plasmid#187431PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged glutamic acid at 117 to histidinePromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
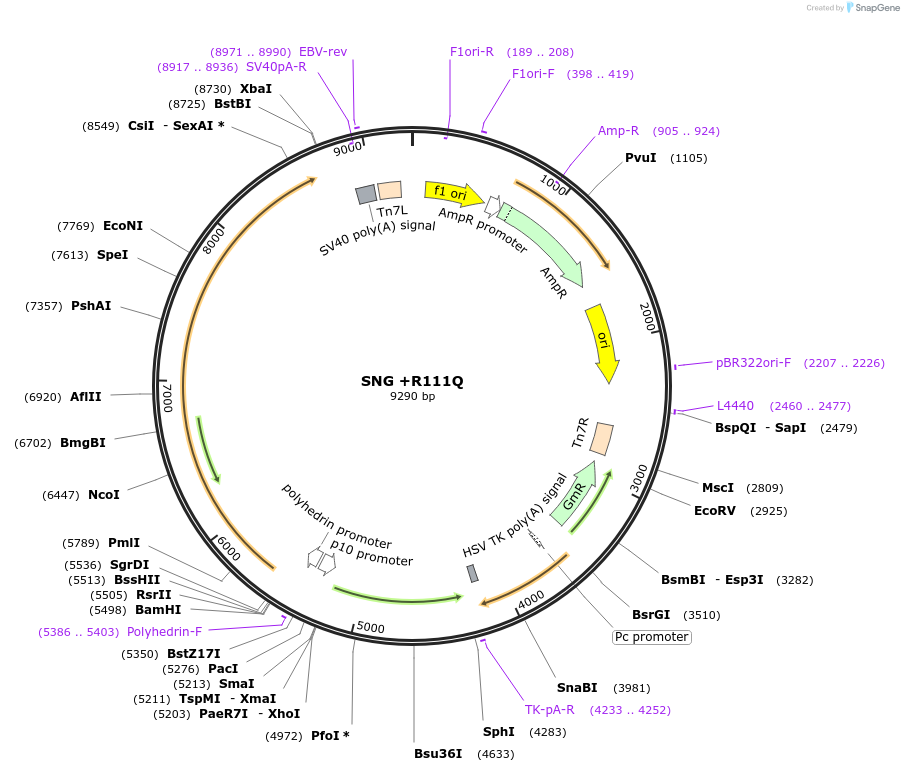
SNG +R111Q
Plasmid#187437PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged arginine at 117 to glutaminePromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
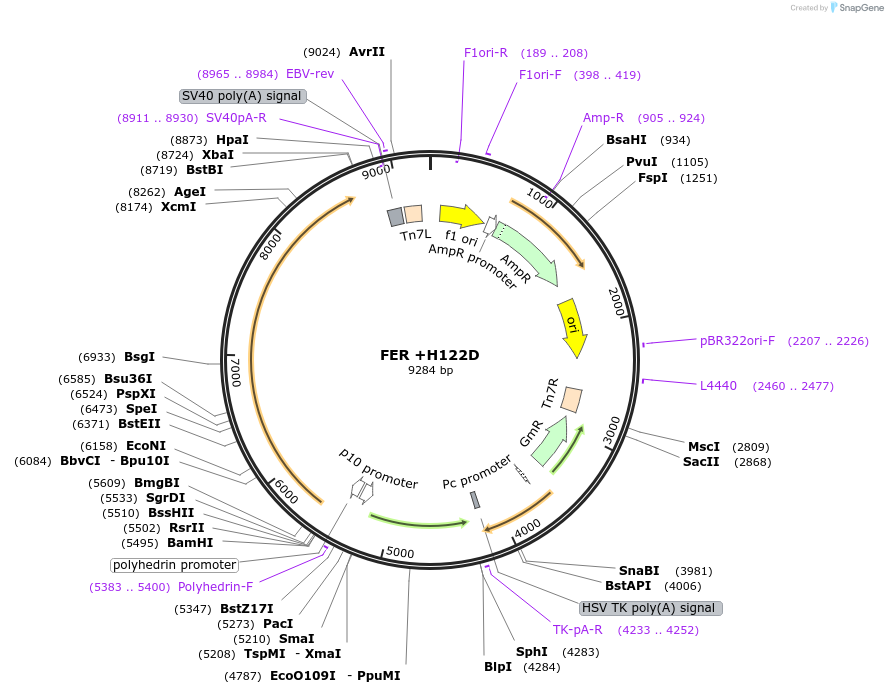
FER +H122D
Plasmid#187428PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged histidine at 128 to aspartic acidPromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
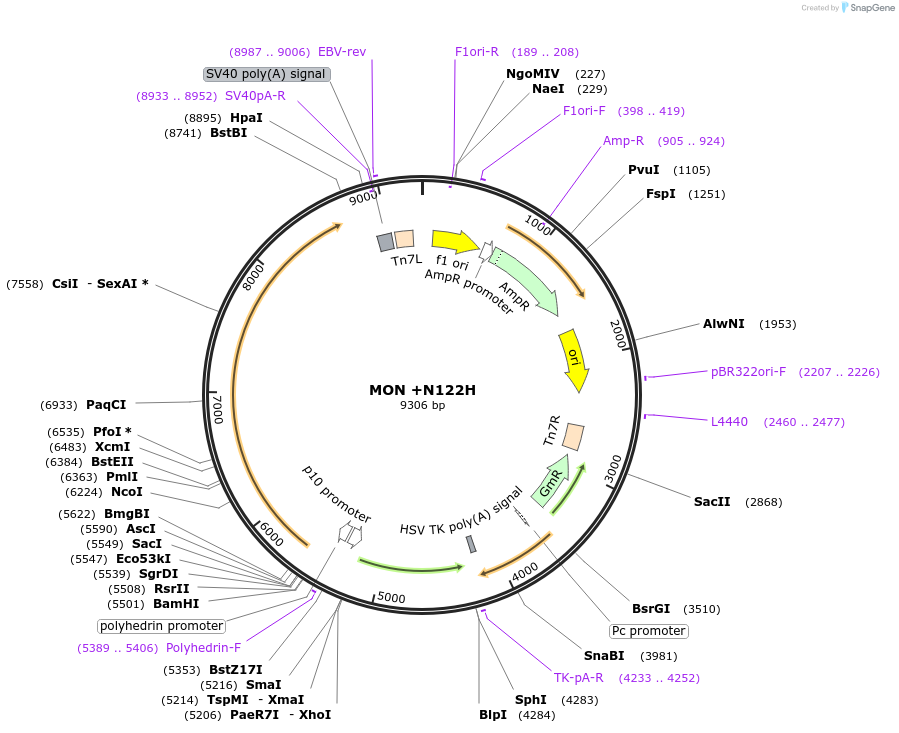
MON +N122H
Plasmid#187433PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged asparagine at 128 to histidinePromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
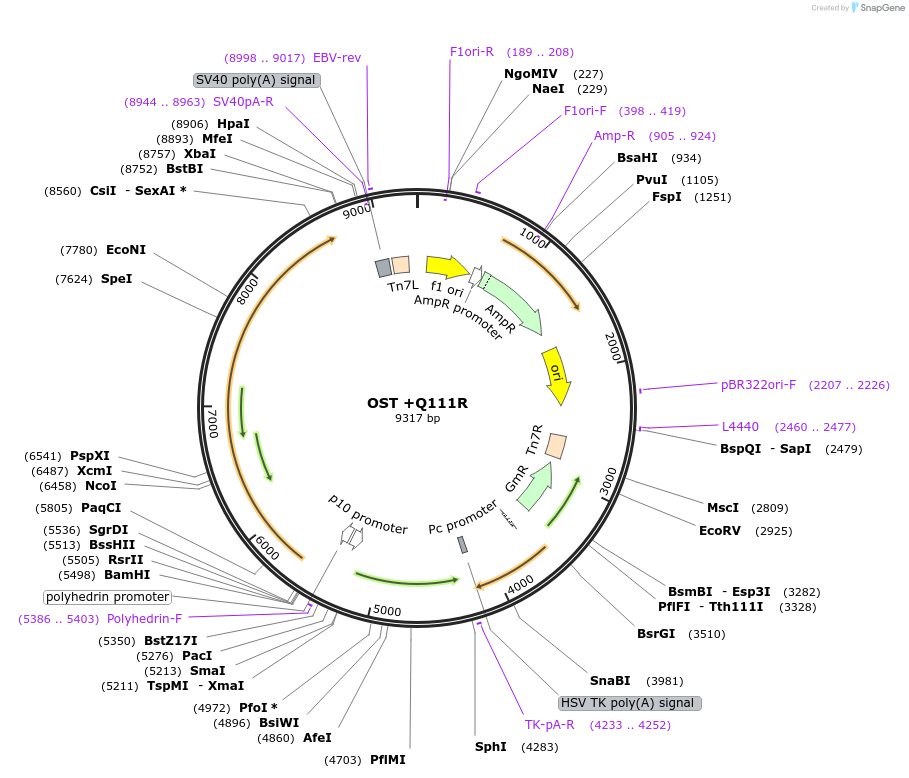
OST +Q111R
Plasmid#187436PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged glutamine at 116 to argininePromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
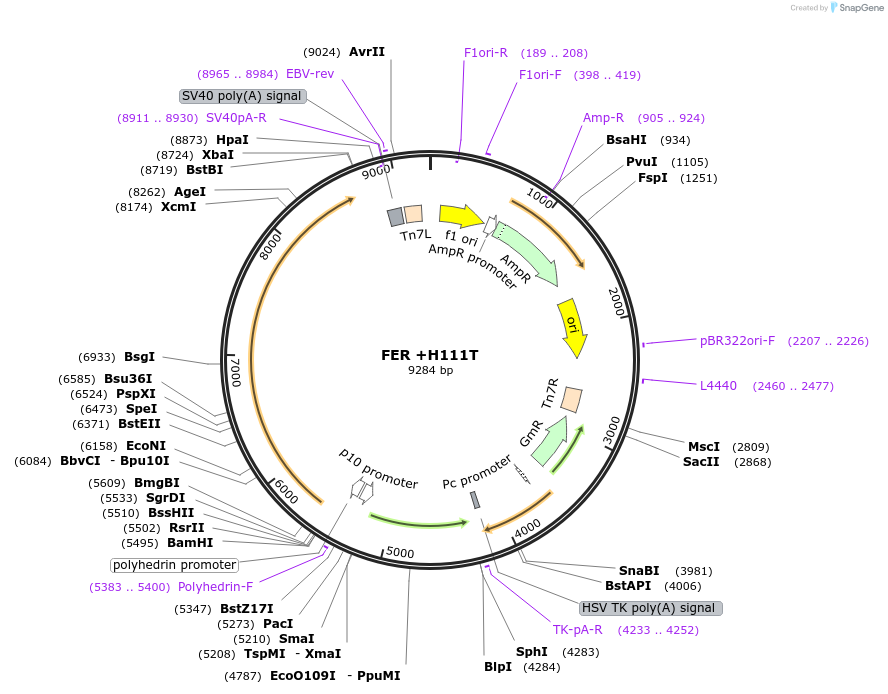
FER +H111T
Plasmid#187430PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged histidine at 117 to threoninePromoterPH and p10Available SinceOct. 12, 2022AvailabilityAcademic Institutions and Nonprofits only -
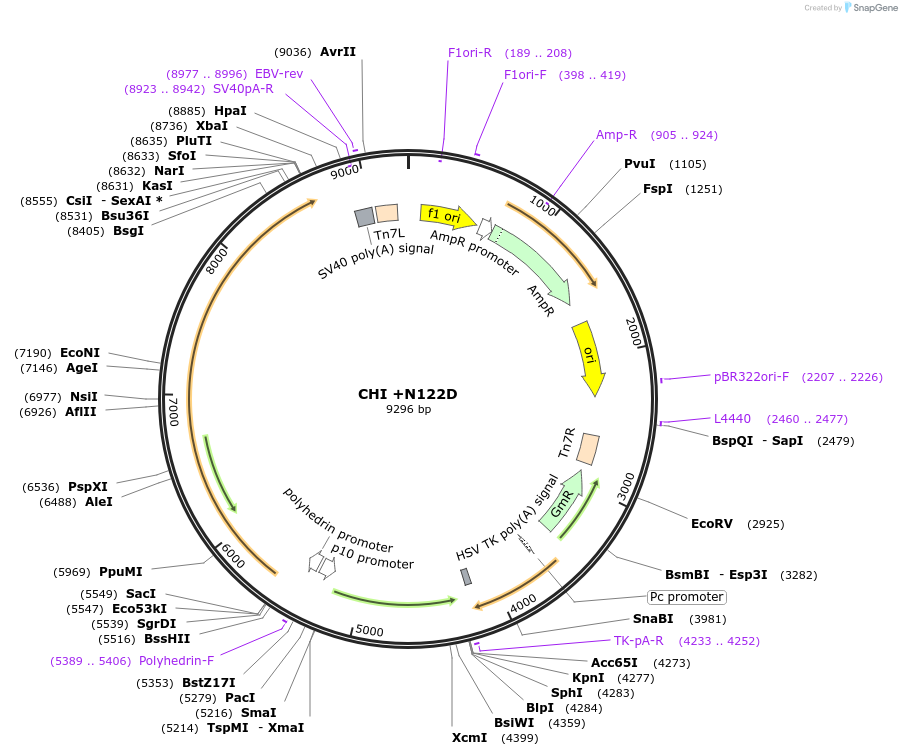
CHI +N122D
Plasmid#187427PurposeExpresses Na,K-ATPase (ATP1A1 and ATP1B1 in pFastBac Dual vector) in insect cellsDepositorInsertsATP1A1
ATP1B1
ExpressionInsectMutationchanged asparagine at 130 to aspartic acidPromoterPH and p10Available SinceOct. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
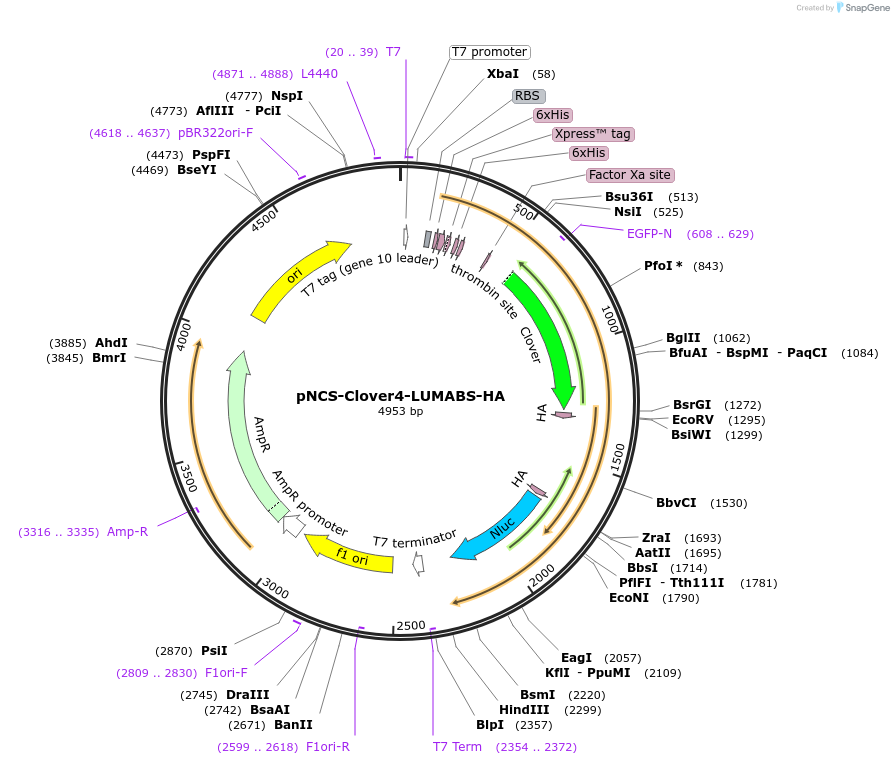
pNCS-Clover4-LUMABS-HA
Plasmid#183486PurposeTo express the Clover4-LUMABS-HA sensor for Avian Influenza Virus antibody detection.DepositorInsertClover4-LUMABS-HA
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
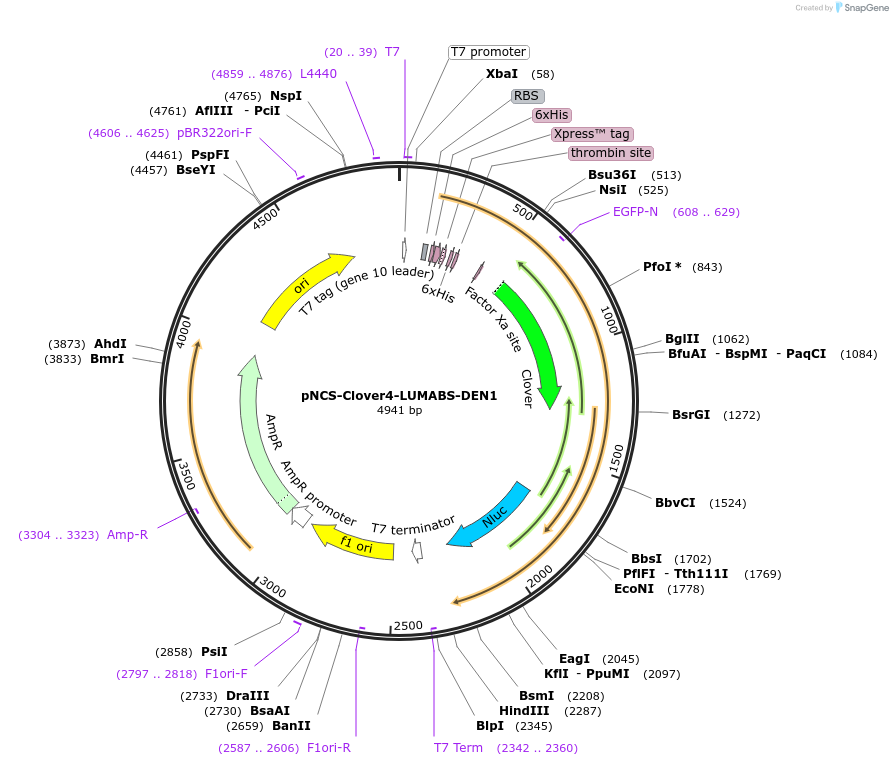
pNCS-Clover4-LUMABS-DEN1
Plasmid#183487PurposeTo express the Clover4-LUMABS-DEN1 sensor for Dengue Virus antibody dection.DepositorInsertClover4-LUMABS-DEN1
ExpressionBacterialAvailable SinceAug. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
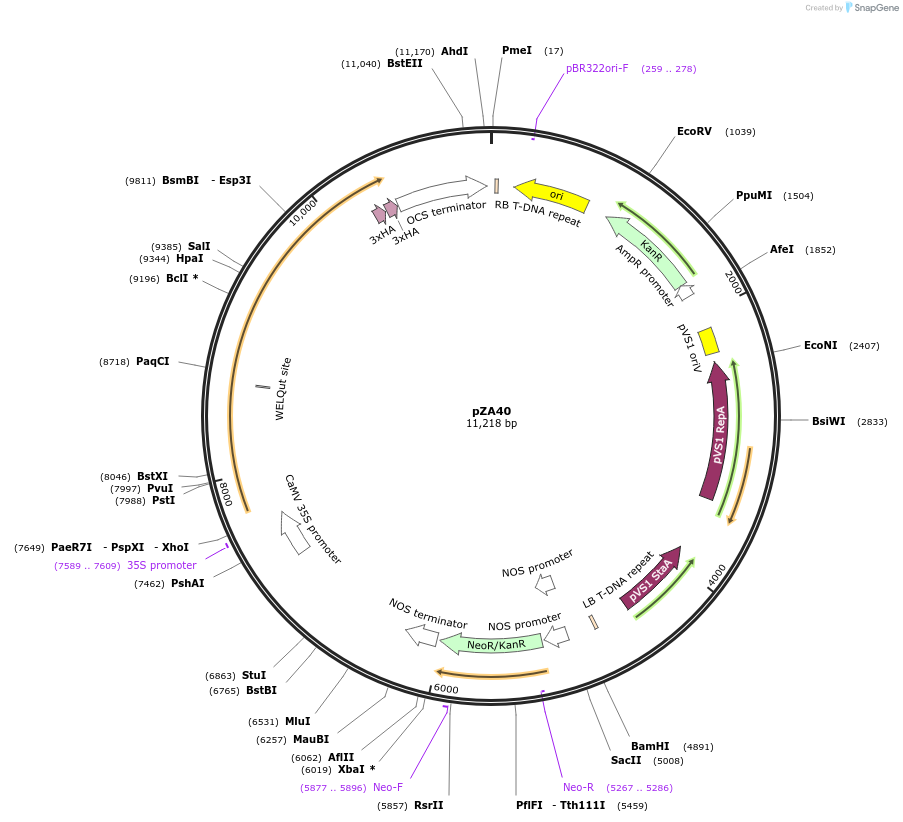
pZA40
Plasmid#158518PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationL17E, D481VAvailable SinceNov. 16, 2020AvailabilityAcademic Institutions and Nonprofits only -
-
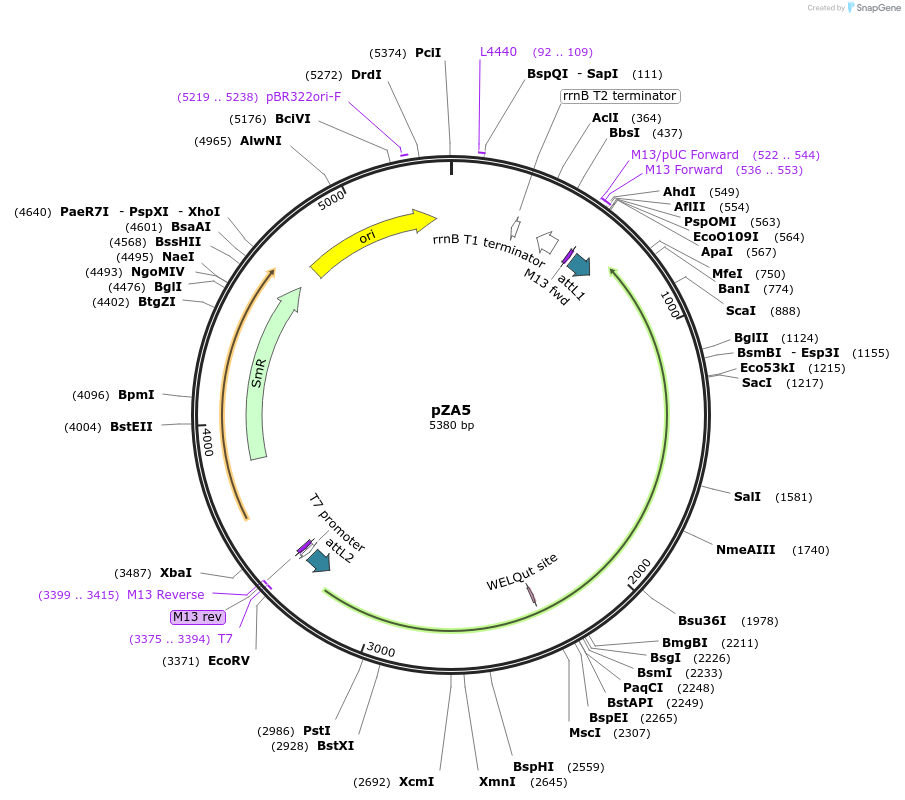
pZA5
Plasmid#158483PurposeEntry clone for Golden Gate assembly. Golden gate compatible N. benthamiana ZAR1 with STOP codonDepositorInsertZAR1
UseGolden gate entry vectorMutationSTOP codonAvailable SinceAug. 27, 2020AvailabilityAcademic Institutions and Nonprofits only -
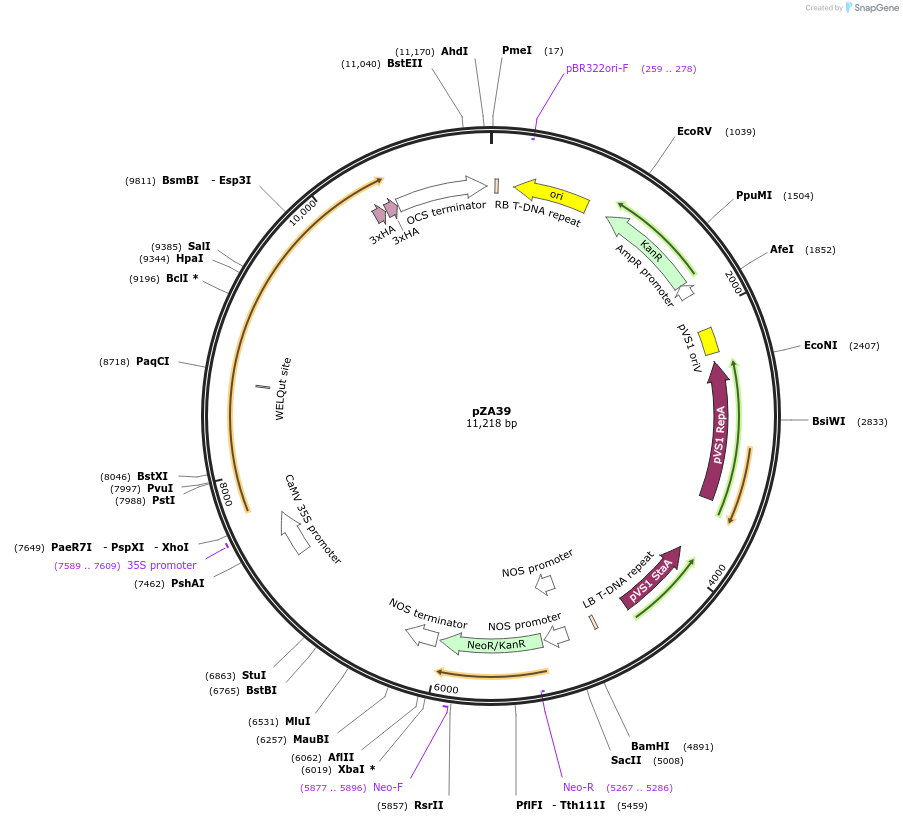
pZA39
Plasmid#158517PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 L17E-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationL17EAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
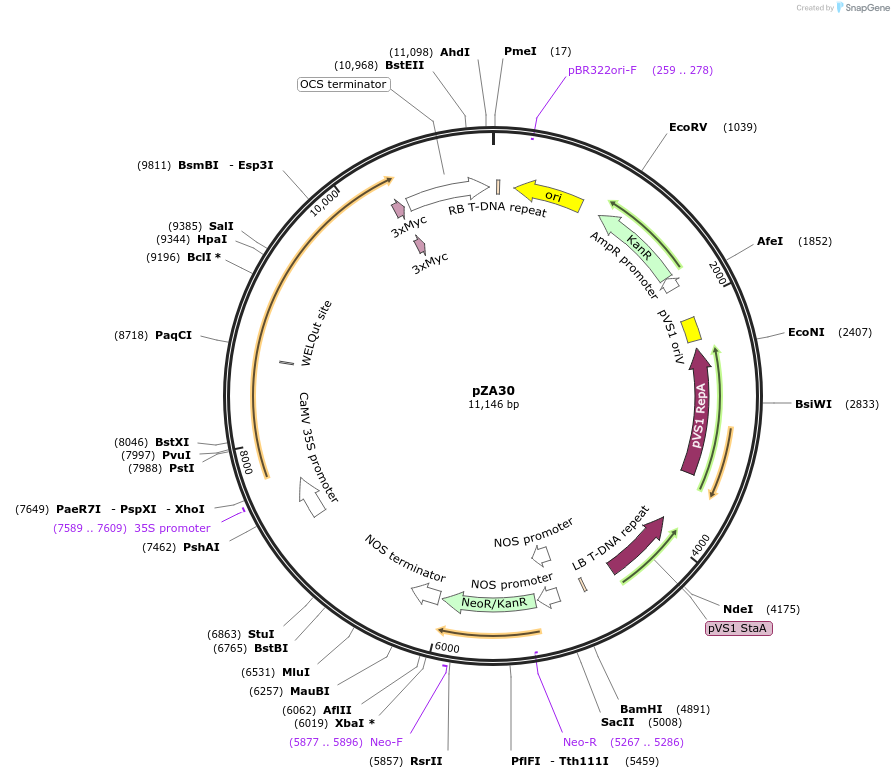
pZA30
Plasmid#158508PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
Tags4xMycExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
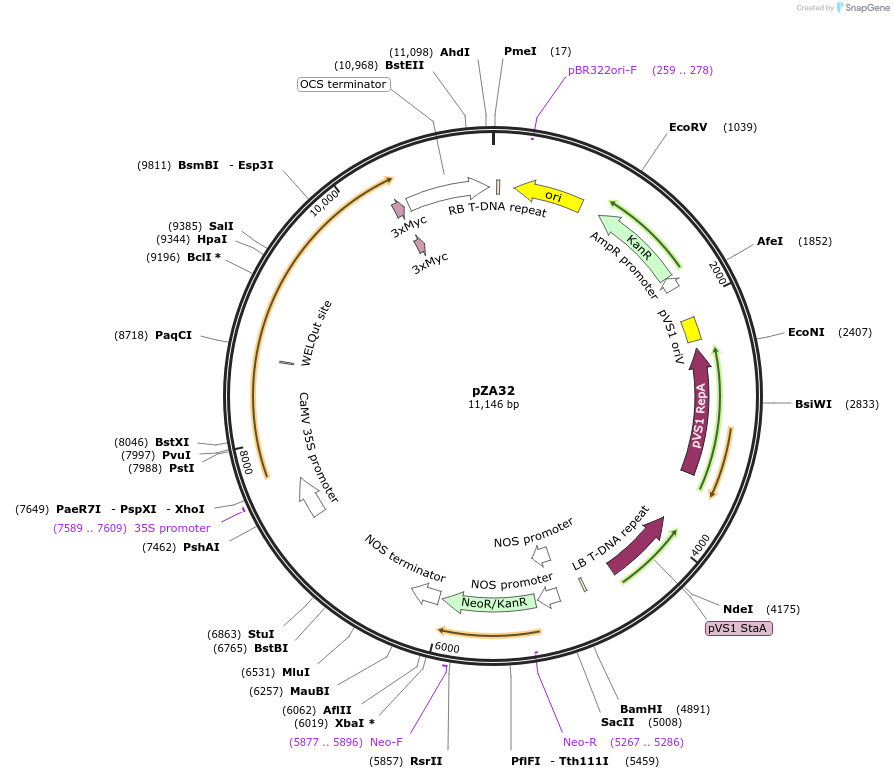
pZA32
Plasmid#158510PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-4xMyc. Km plant selection marker.DepositorInsertZAR1-4xMyc
Tags4xMycExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
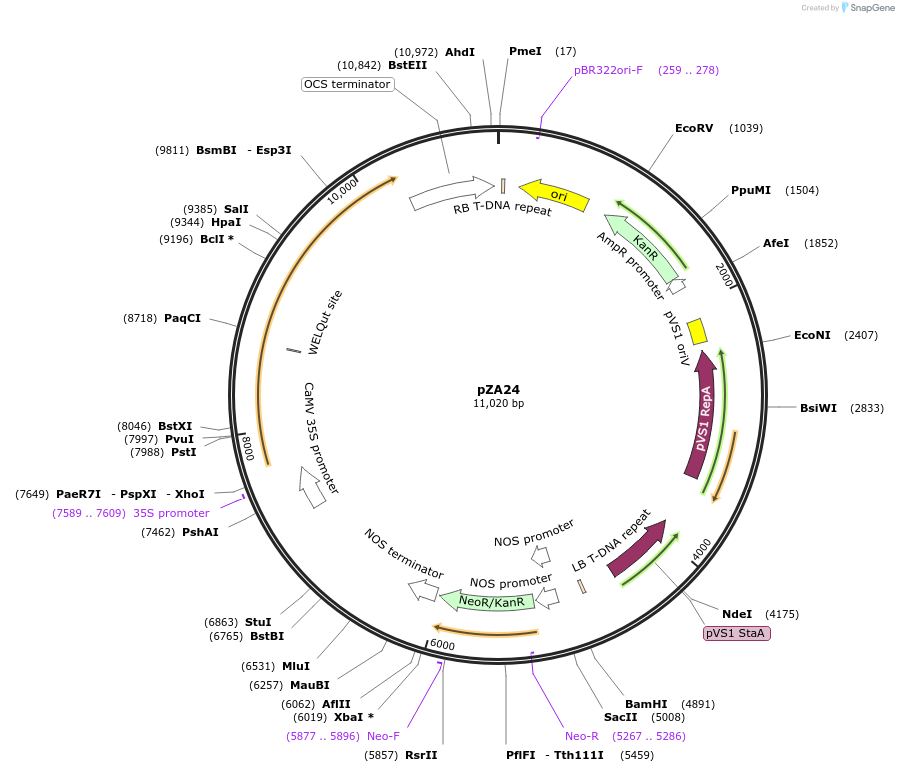
pZA24
Plasmid#158502PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E. Km plant selection marker.DepositorInsertZAR1
ExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
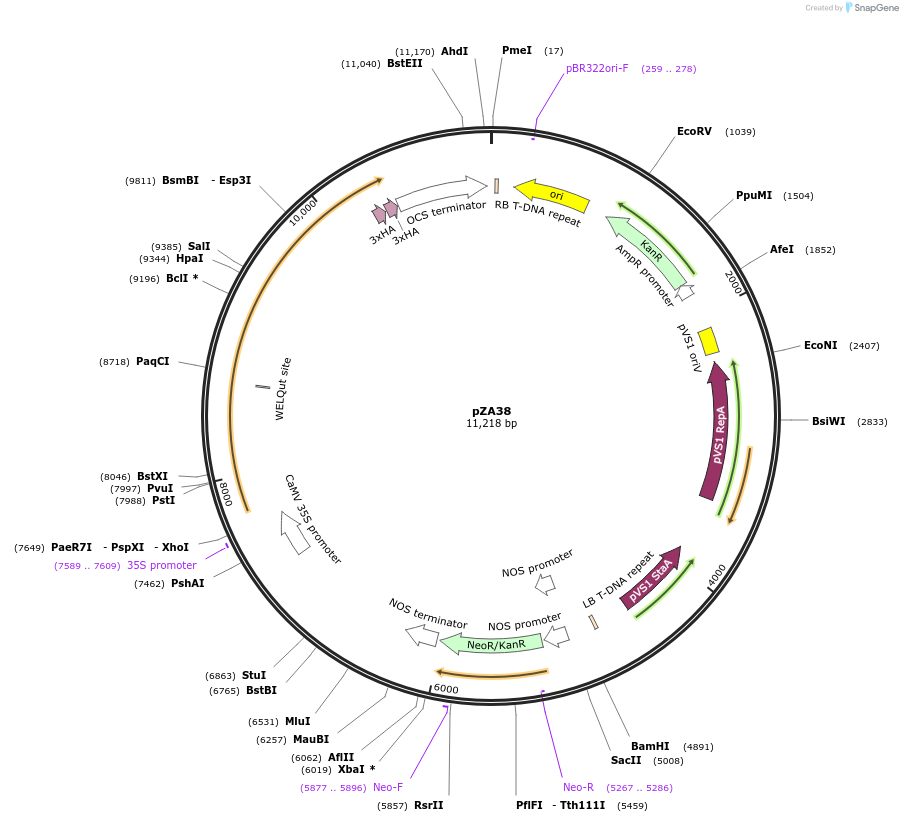
pZA38
Plasmid#158516PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-6xHA. Km plant selection marker.DepositorInsertZAR1-6xHA
Tags6xHAExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
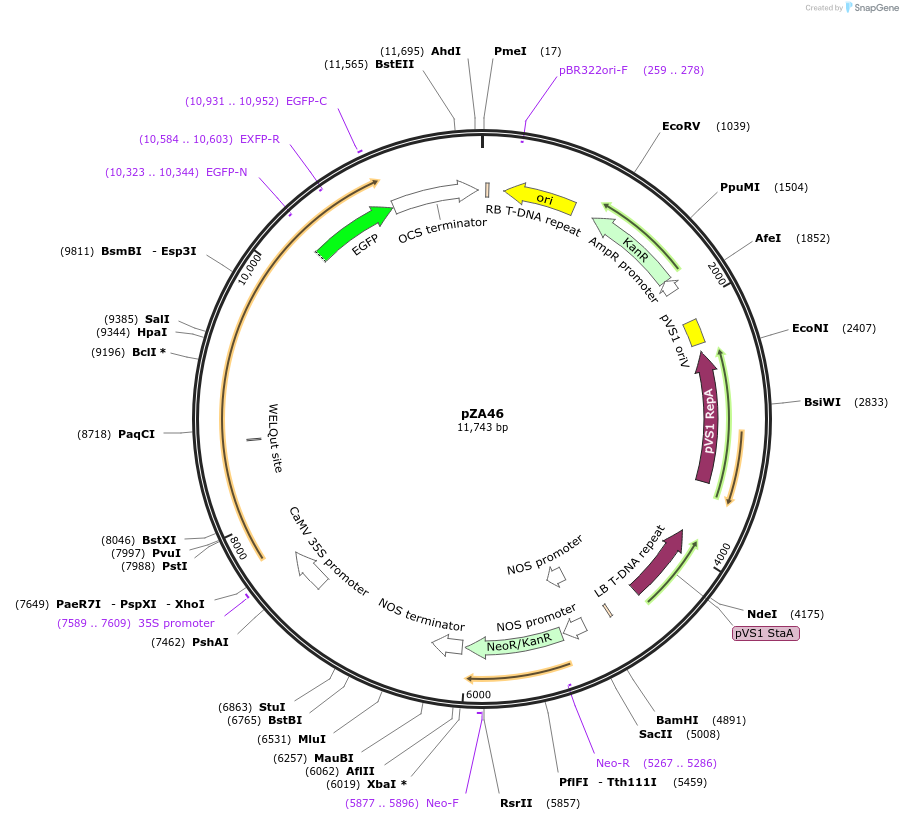
pZA46
Plasmid#158524PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationD481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
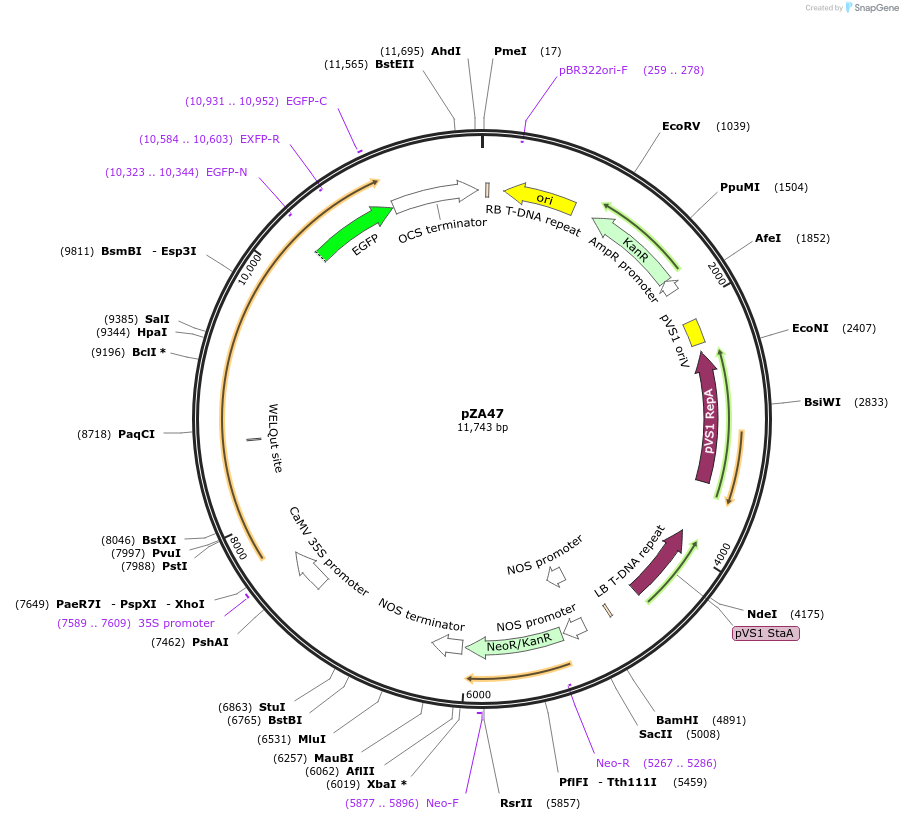
pZA47
Plasmid#158525PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 L17E-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationL17EAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
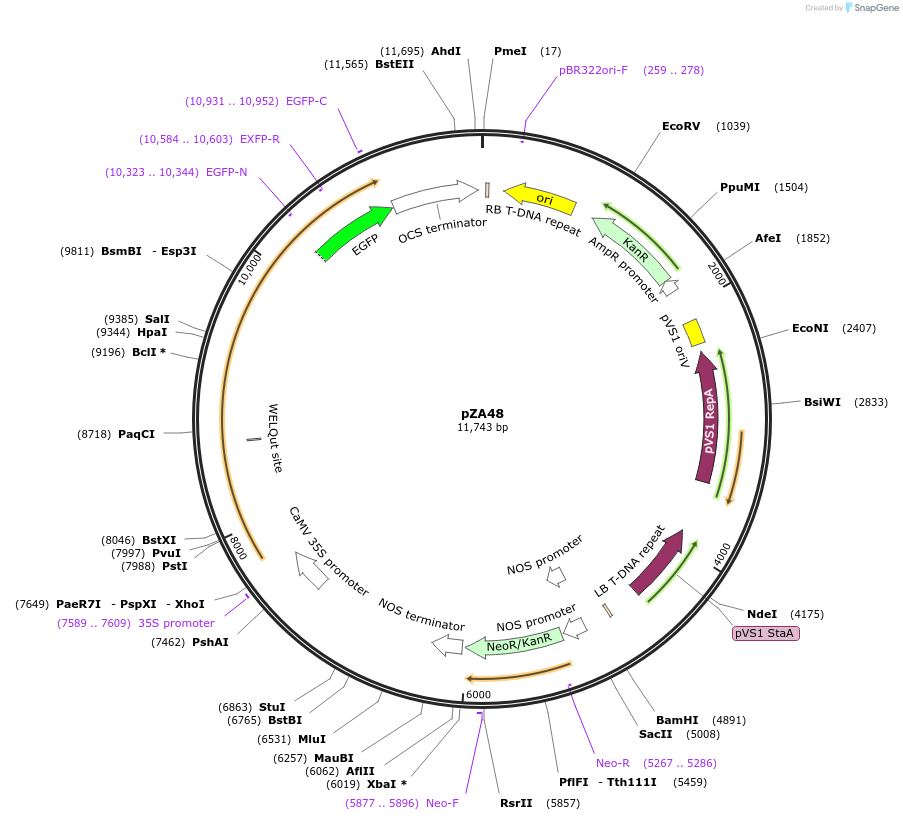
pZA48
Plasmid#158526PurposeIn planta gene expression of Golden gate compatible N. benthamiana ZAR1 D481V/L17E-eGFP. Km plant selection marker.DepositorInsertZAR1-eGFP
TagseGFPExpressionPlantMutationL17E, D481VAvailable SinceAug. 26, 2020AvailabilityAcademic Institutions and Nonprofits only -
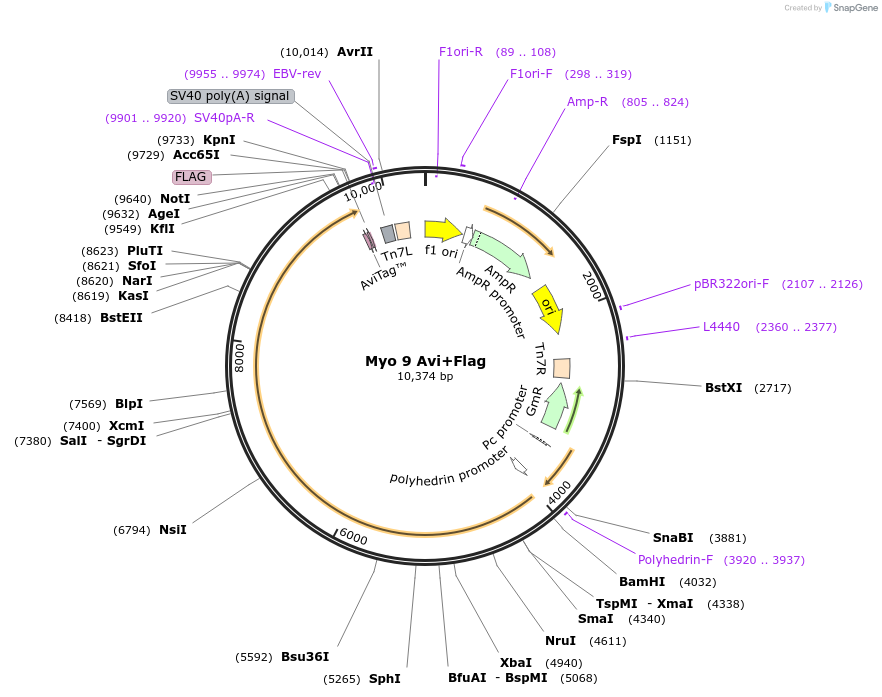
Myo 9 Avi+Flag
Plasmid#134969PurposeExpression and purification of c.elegans Myo 9 (Hum 7) using baculovirus from pFastBac1DepositorAvailable SinceJan. 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
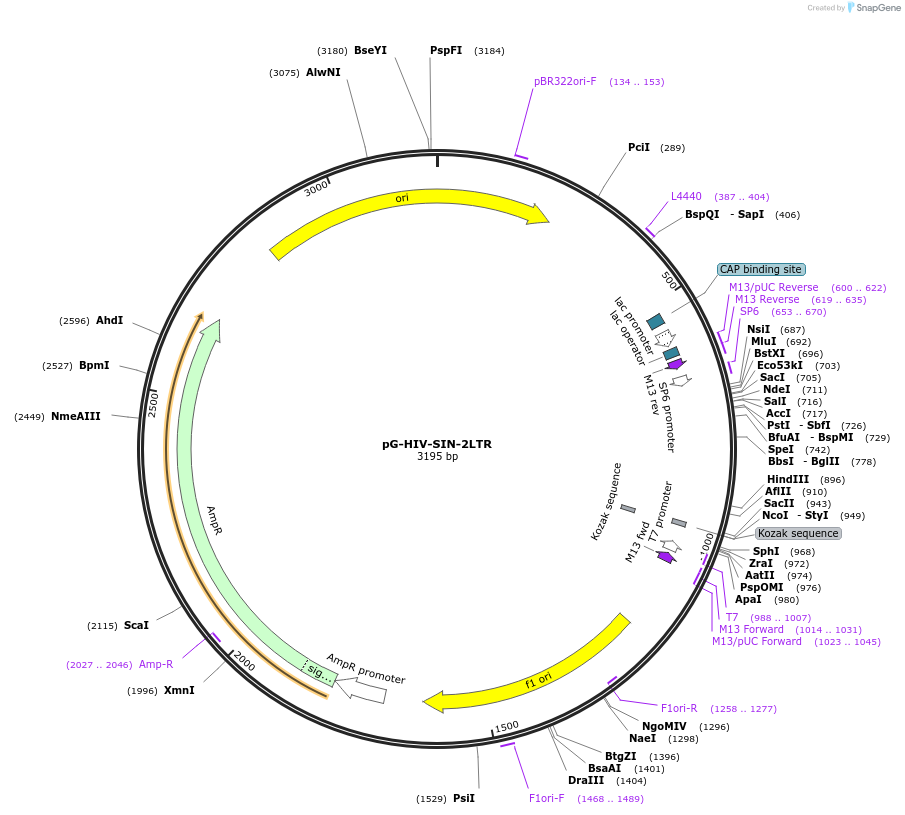
pG-HIV-SIN-2LTR
Plasmid#104591PurposeStandard for qPCRDepositorInsert2LTR circle junction of self-inactivating HIV-based vectors
UseCloning vectorAvailable SinceJan. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
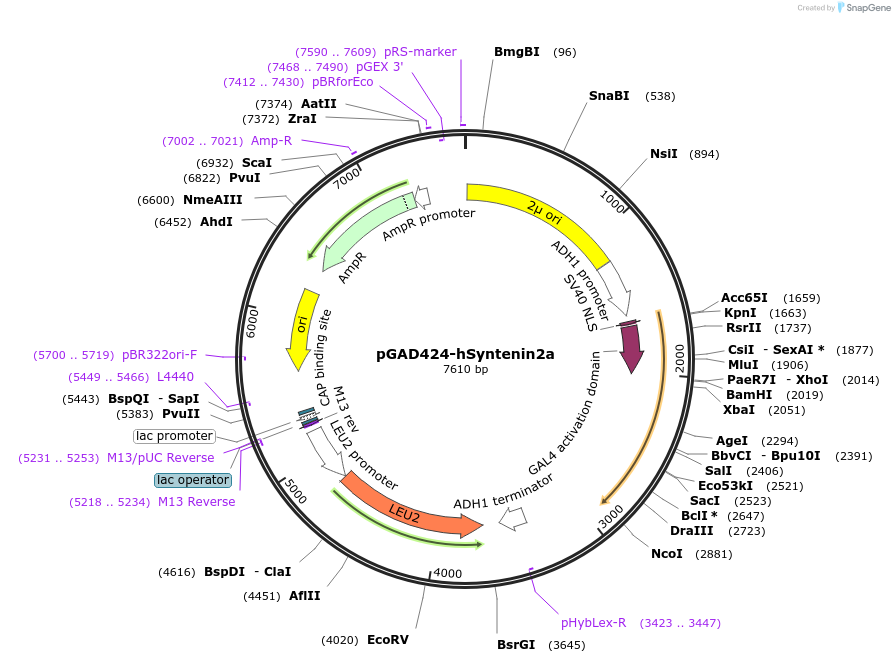
pGAD424-hSyntenin2a
Plasmid#89426PurposeTwo-hybrid systemDepositorAvailable SinceMay 3, 2017AvailabilityAcademic Institutions and Nonprofits only -
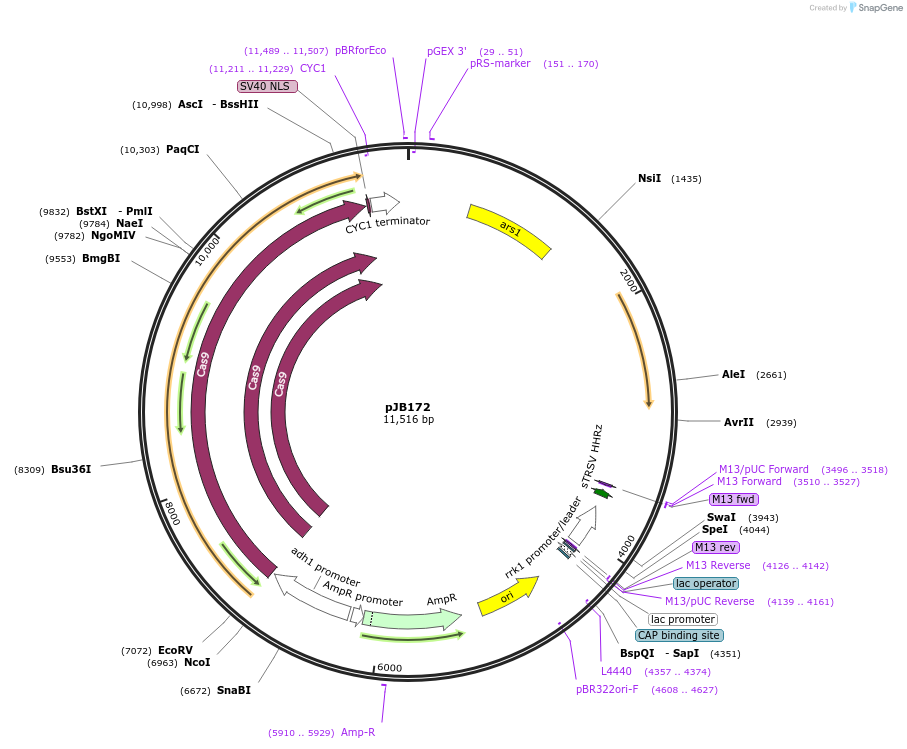
pJB172
Plasmid#86992PurposeCRISPR/Cas9 in fission yeast using fluoride selection and targetting pil1DepositorInsertgRNA targeting pil1
ExpressionYeastAvailable SinceFeb. 22, 2017AvailabilityAcademic Institutions and Nonprofits only -
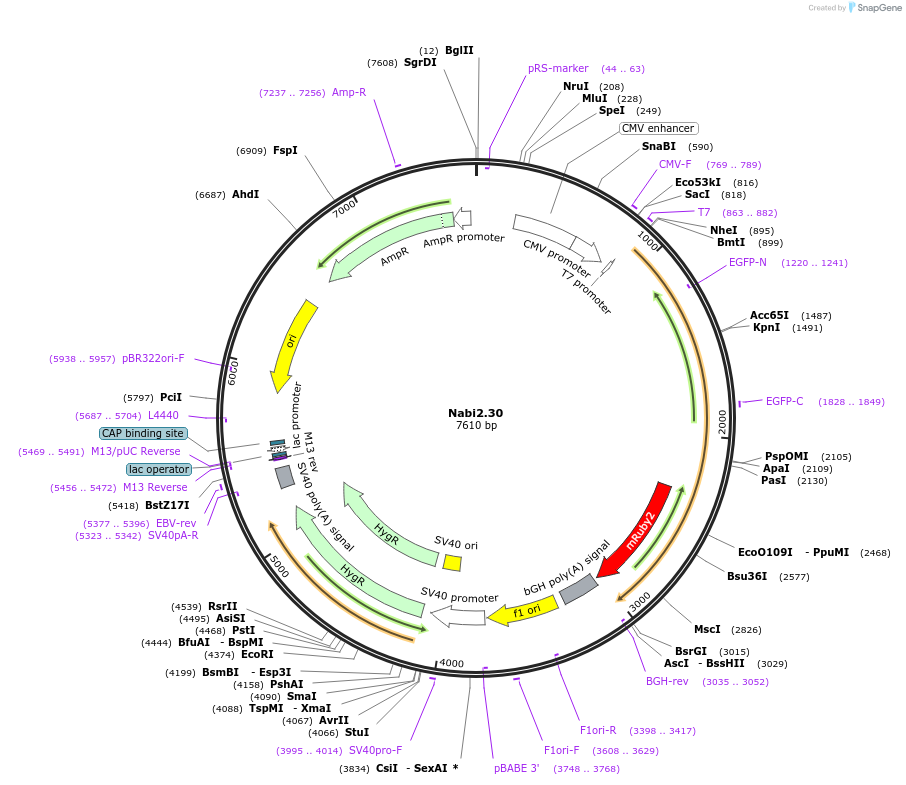
Nabi2.30
Plasmid#73798PurposeNabi2.30 is a FRET (Förster Resonance Energy Transfer)-based protein voltage sensor. This probe contains Clover and mRuby2 inserted at different locations in the Ciona voltage sensitive domain.DepositorInsertNabi2.30
ExpressionMammalianAvailable SinceApril 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
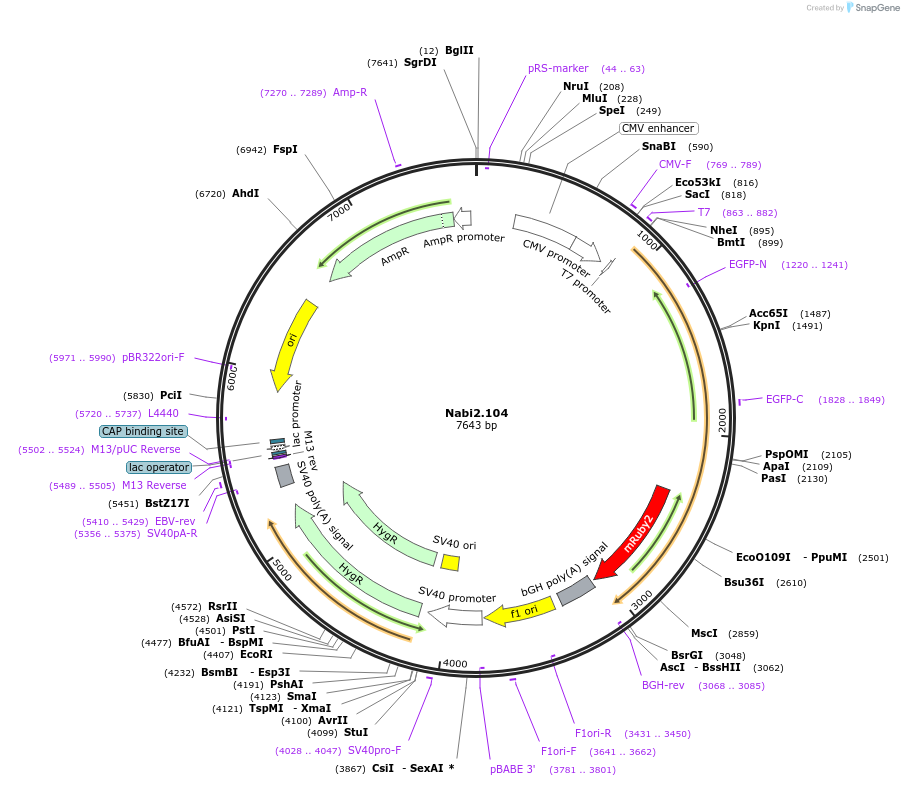
Nabi2.104
Plasmid#73799PurposeNabi2.104 is a FRET (Förster Resonance Energy Transfer)-based protein voltage sensor. This probe contains Clover and mRuby2 inserted at different locations in the Ciona voltage sensitive domain.DepositorInsertNabi2.104
ExpressionMammalianAvailable SinceApril 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
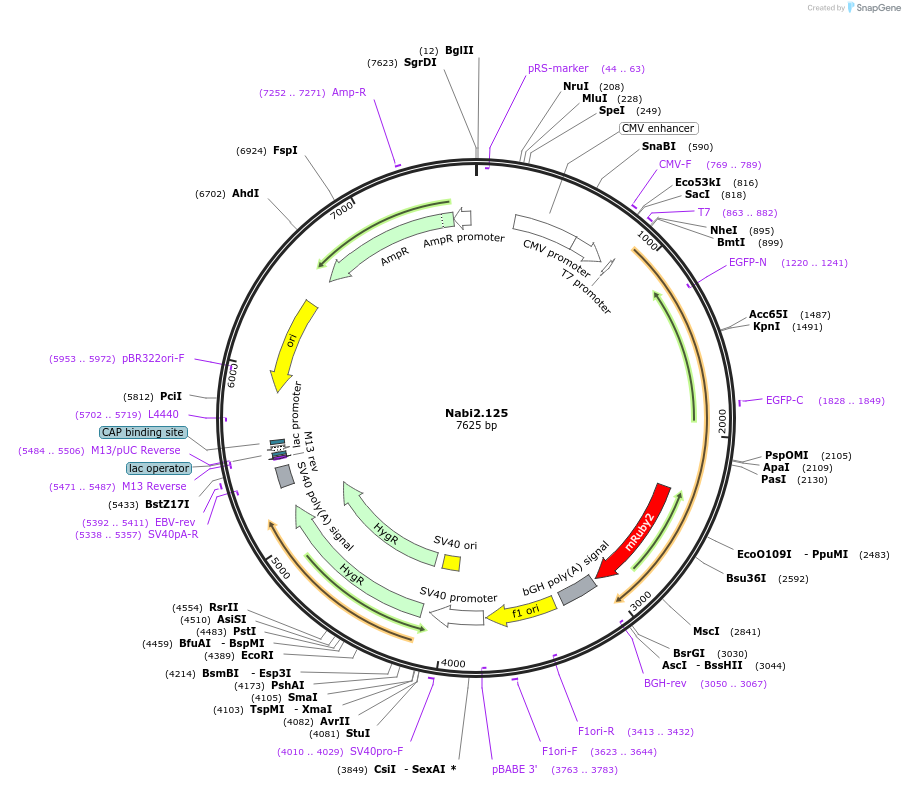
Nabi2.125
Plasmid#73800PurposeNabi2.125 is a FRET (Förster Resonance Energy Transfer)-based protein voltage sensor. This probe contains Clover and mRuby2 inserted at different locations in the Ciona voltage sensitive domain.DepositorInsertNabi2.125
ExpressionMammalianAvailable SinceApril 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
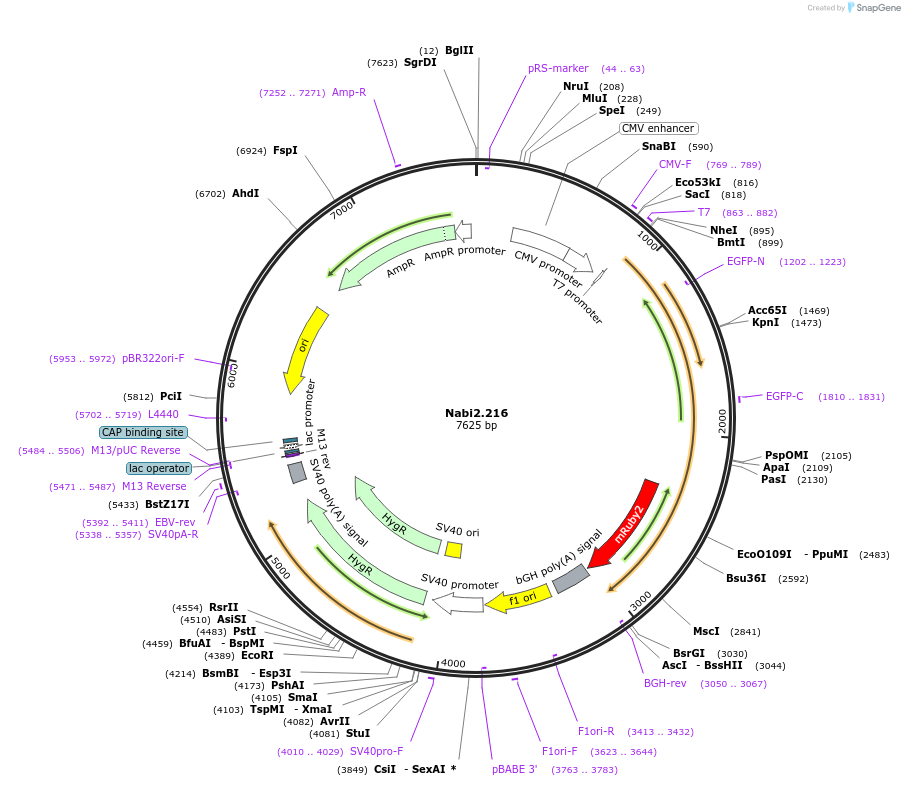
Nabi2.216
Plasmid#73801PurposeNabi2.216 is a FRET (Förster Resonance Energy Transfer)-based protein voltage sensor. This probe contains Clover and mRuby2 inserted at different locations in the Ciona voltage sensitive domain.DepositorInsertNabi2.216
ExpressionMammalianAvailable SinceApril 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
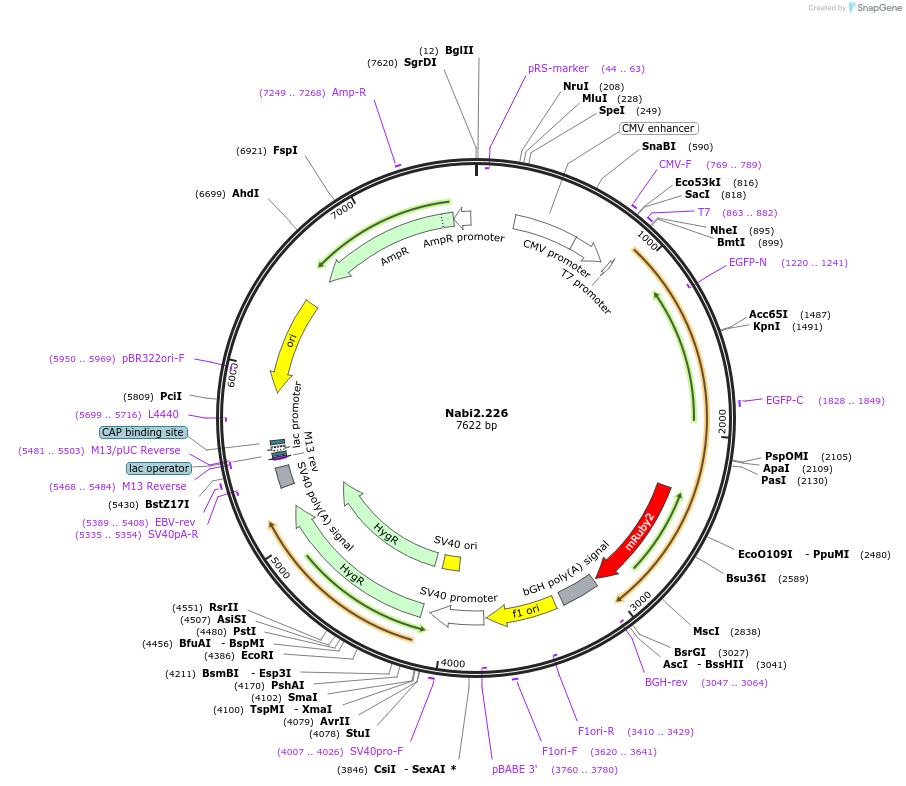
Nabi2.226
Plasmid#73802PurposeNabi2.226 is a FRET (Förster Resonance Energy Transfer)-based protein voltage sensor. This probe contains Clover and mRuby2 inserted at different locations in the Ciona voltage sensitive domain.DepositorInsertNabi2.226
ExpressionMammalianAvailable SinceApril 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
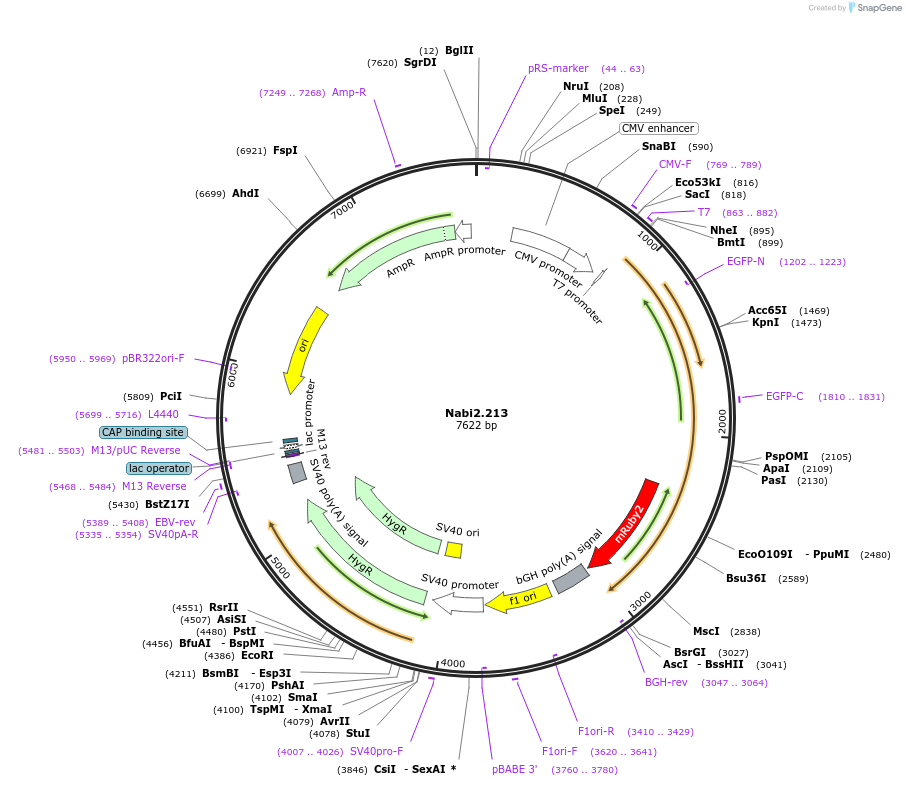
Nabi2.213
Plasmid#73757PurposeNabi2.213 is a FRET (Förster Resonance Energy Transfer)-based protein voltage sensor. This probe contains Clover and mRuby2 inserted at different locations in the Ciona voltage sensitive domain.DepositorInsertNabi2.213
ExpressionMammalianAvailable SinceApril 7, 2016AvailabilityAcademic Institutions and Nonprofits only -
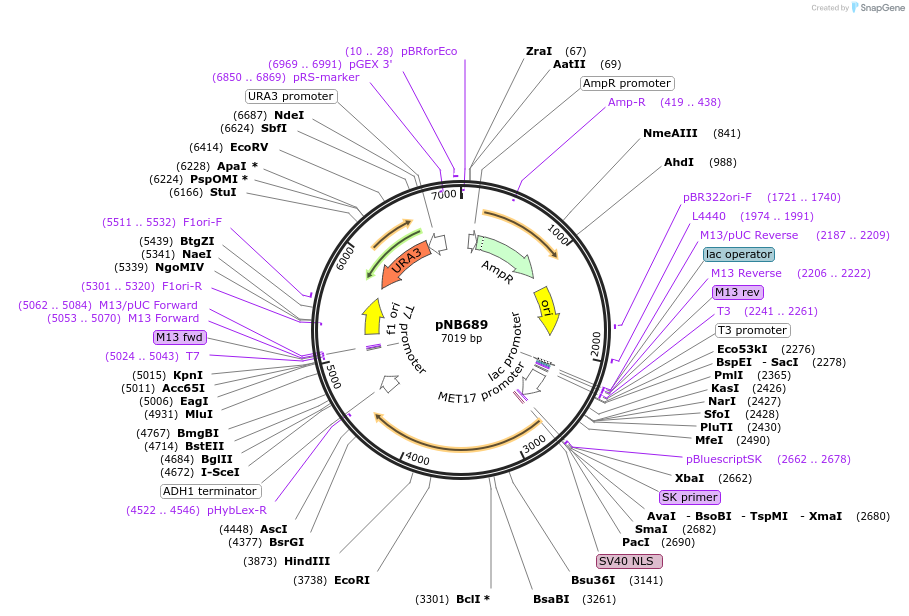
pNB689
Plasmid#60133PurposeGreen beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA), driven by MET17 promoter.DepositorInsertGrLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterMET17prAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
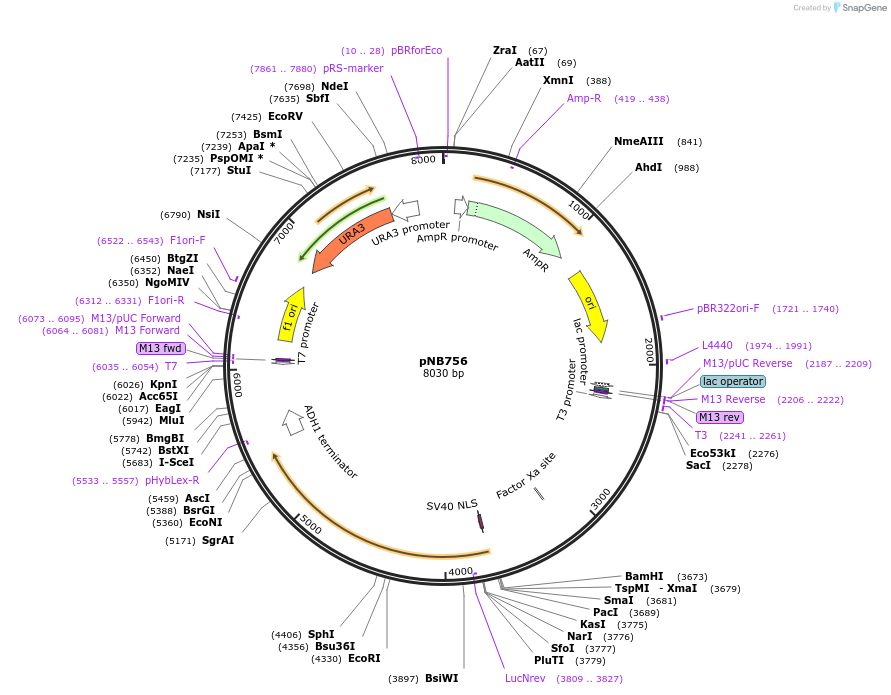
pNB756
Plasmid#60149PurposeYellow beetle luciferase with nuclear localization signal (NLS) and ClpXP degron tag (ssrA), driven by LEU1 promoter.DepositorInsertYeLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterLEU1prAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
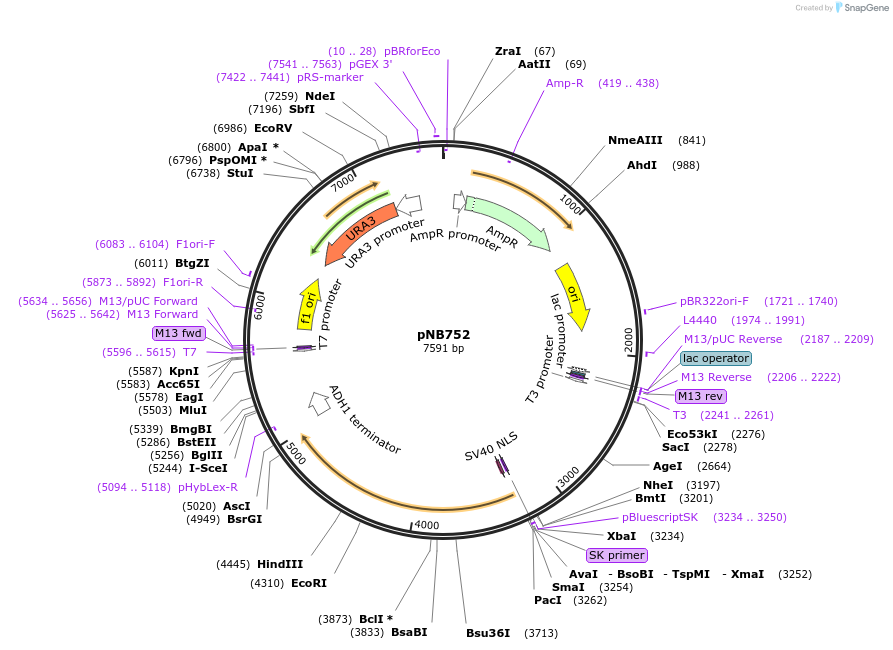
pNB752
Plasmid#60155PurposeGreen beetle luciferase with nuclear localization signal (NLS) and adjustable degron tag (ssrA), driven by ADE17 promoter.DepositorInsertGrLuc
TagsClpXP degron and Nuclear localization signal (NLS…ExpressionYeastPromoterADE17prAvailable SinceOct. 31, 2014AvailabilityAcademic Institutions and Nonprofits only -
pUAST myc Rab35 CA
Plasmid#58382Purposeexpression of drosophila myc-Rab35 constitutively active mutantDepositorInsertRab35 (Rab35 Fly)
TagsmycExpressionInsectMutationQ67L (constitutive active)PromoterGAL4Available SinceJuly 21, 2014AvailabilityAcademic Institutions and Nonprofits only -
pUAST YFP Rab35 CA
Plasmid#58381Purposeexpression of drosophila YFP-Rab35 constitutively active mutantDepositorInsertRab35 (Rab35 Fly)
TagsYFPExpressionInsectMutationQ67L (constitutive active)PromoterGAL4Available SinceJuly 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
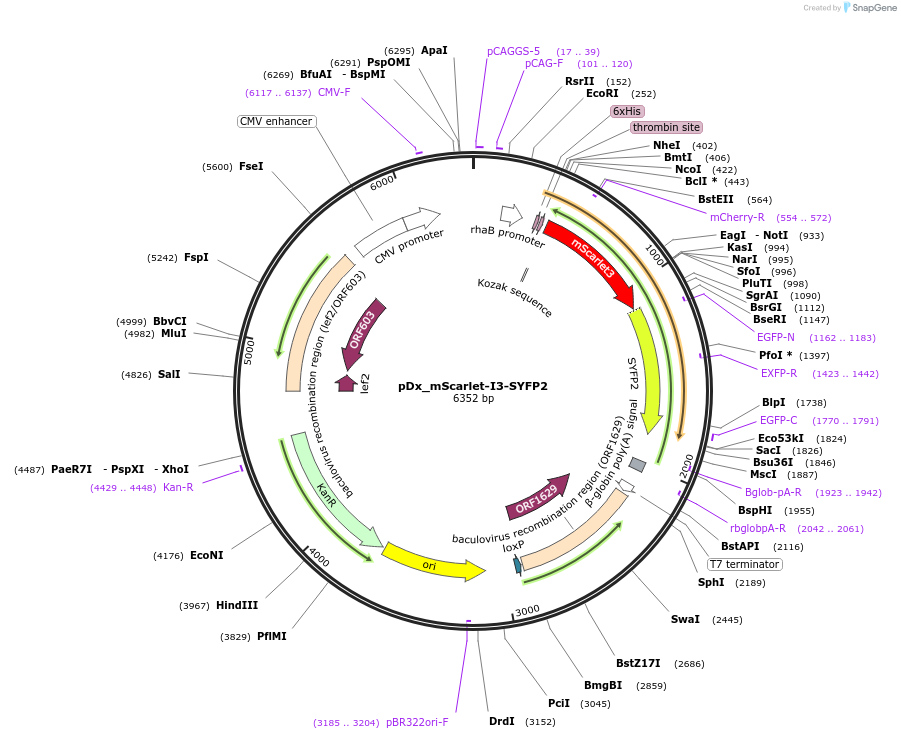
pDx_mScarlet-I3-SYFP2
Plasmid#189763PurposeDual expression vector for bacteria and mammalian cells producing mScarlet-I3-SYFP2 fusion protein for FRET analysisDepositorInsertmScarlet-I3-SYFP2
Tags6xHisExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only -
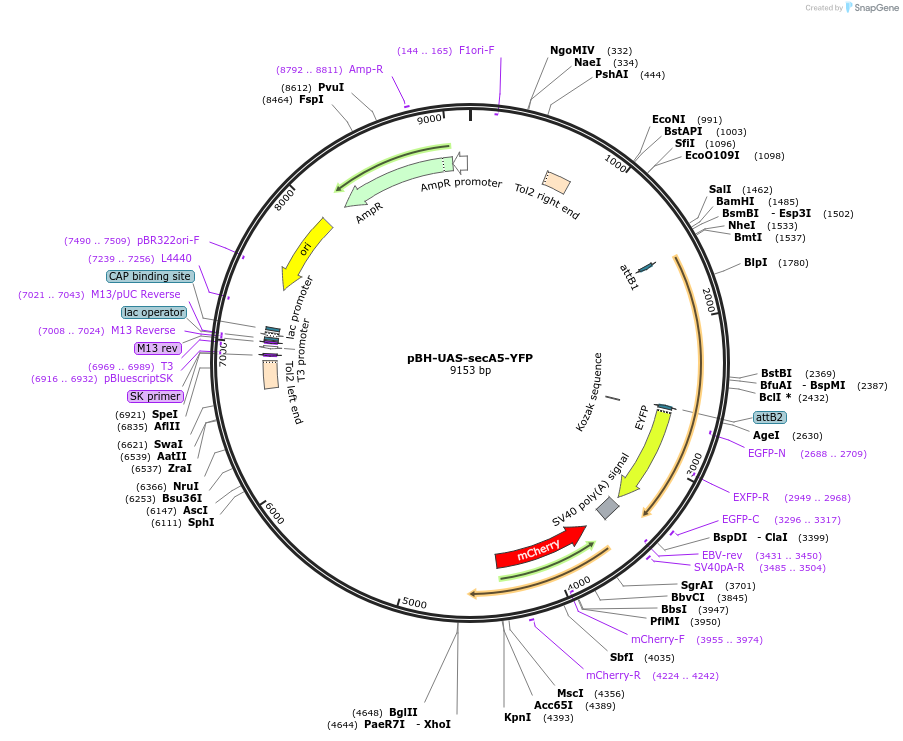
pBH-UAS-secA5-YFP
Plasmid#32359PurposeLive imaging of apoptotic cells in zebrafishDepositorAvailable SinceSept. 19, 2011AvailabilityAcademic Institutions and Nonprofits only -
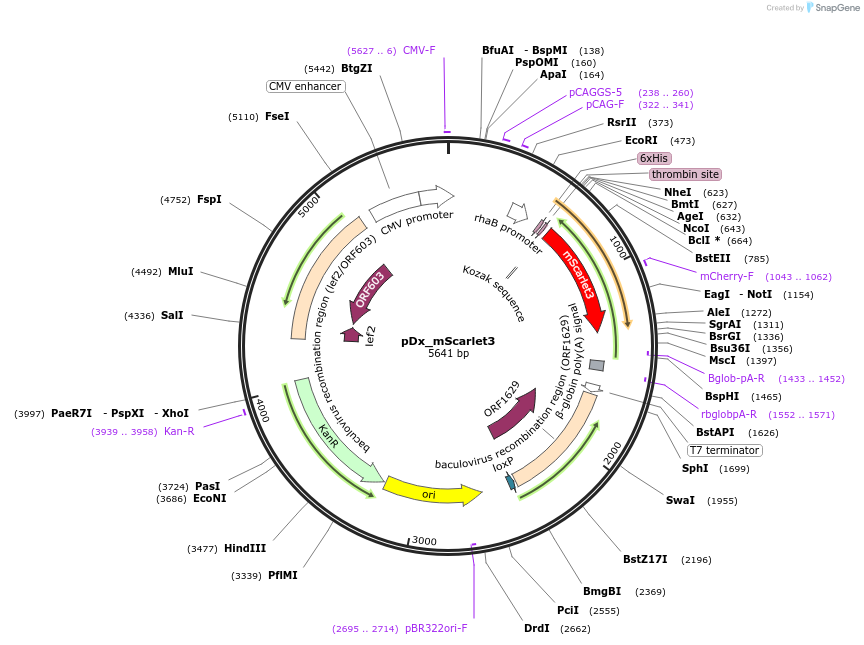
pDx_mScarlet3
Plasmid#189754PurposeDual expression vector for bacteria and mammalian cells producing mScarlet3 red fluorescent proteinDepositorInsertmScarlet3
Tags6xHisExpressionBacterial and MammalianPromoterCMV & RhaAvailable SinceMarch 21, 2023AvailabilityAcademic Institutions and Nonprofits only