We narrowed to 14,816 results for: NTS
-
Plasmid#139708PurposeT-DNA encoding Fungal Bioluminescent Pathway Components, complete pathway, Luz promoter is flower specific, circadian controlledDepositorInsertpSlH4:NnCPH:tSlH4, pSlRBCS2:AsNpga:tAtuG7, pAtuMas:NnH3H:tAtuMas, p35S:NnHisps:t35S, pPhODO1:NnLuz:tAtuOcs
UseLuciferaseExpressionPlantPromotervariousAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
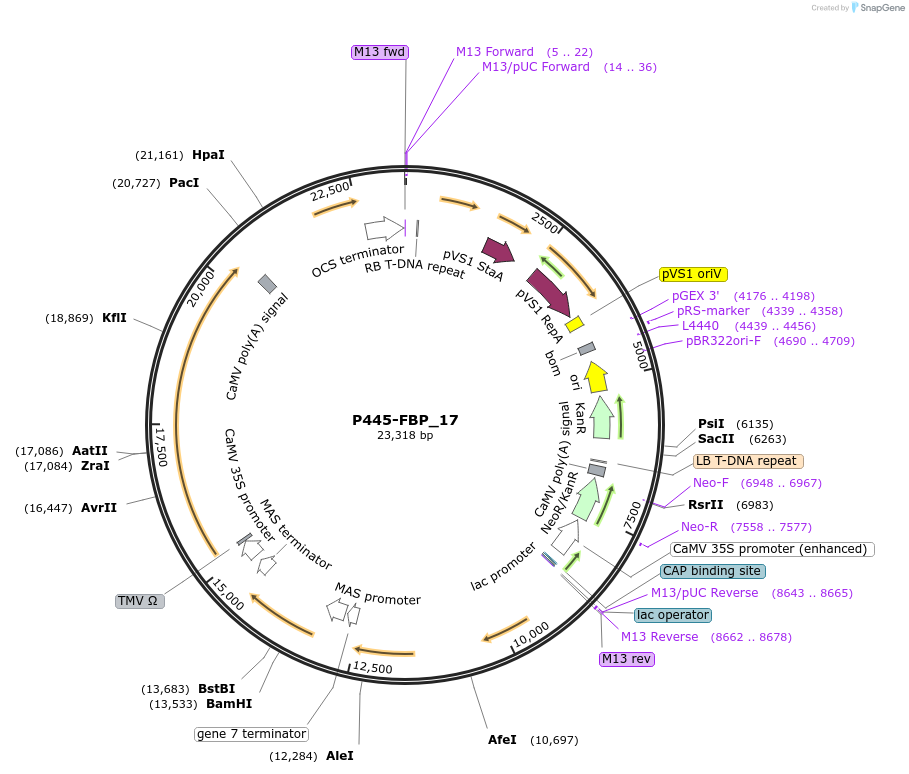
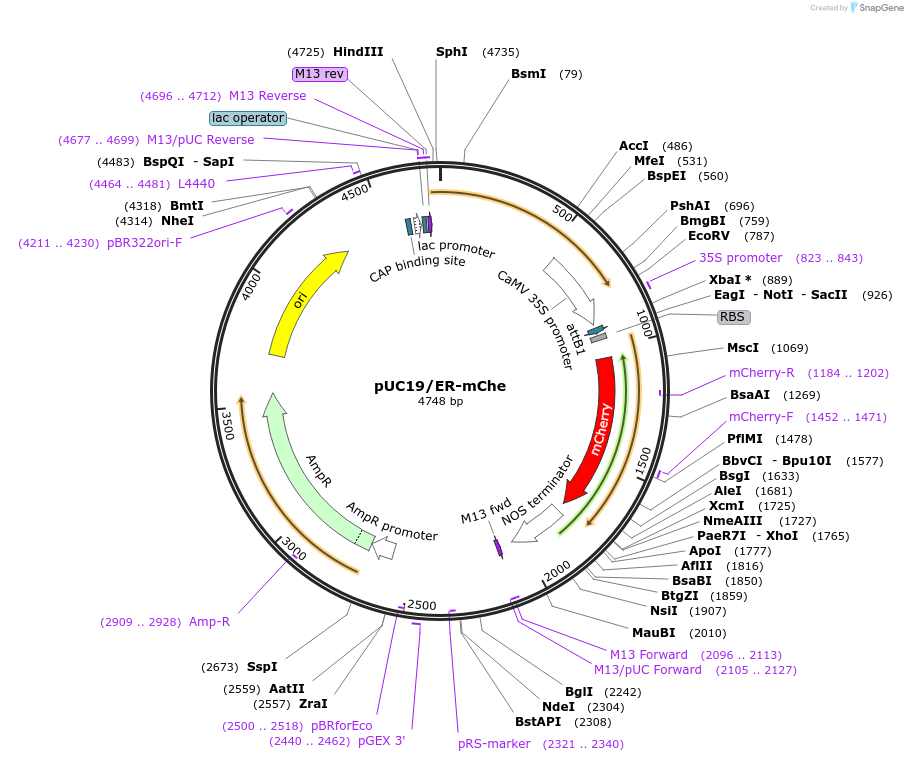
pUC19/ER-mChe
Plasmid#183163PurposeConstruct for transient expression of endoplasmic reticulum-targeted mCherry in plants.DepositorInsertER-mCherry
Tags25 aa ER signal of Glycine soja kunitz-type tryps…ExpressionPlantAvailable SinceApril 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
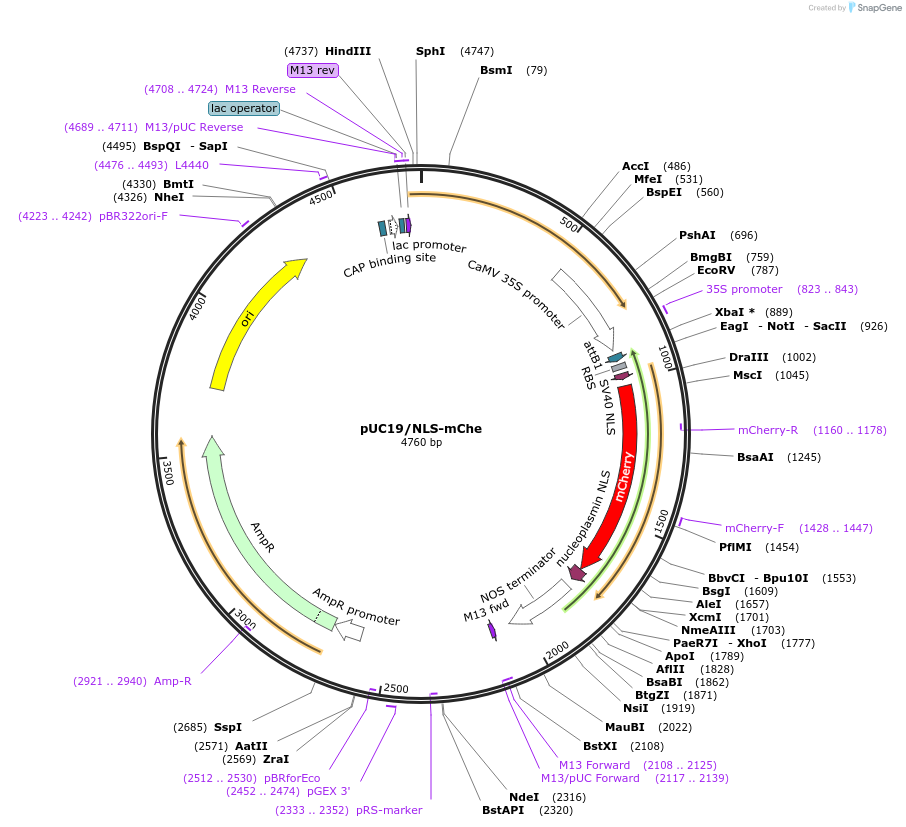
pUC19/NLS-mChe
Plasmid#183162PurposeConstruct for transient expression of cell nucleus-targeted mCherry in plants.DepositorInsertNLS-mCherry
Tags16 aa nuclear localization signal and 16 aa nucl…ExpressionPlantAvailable SinceApril 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
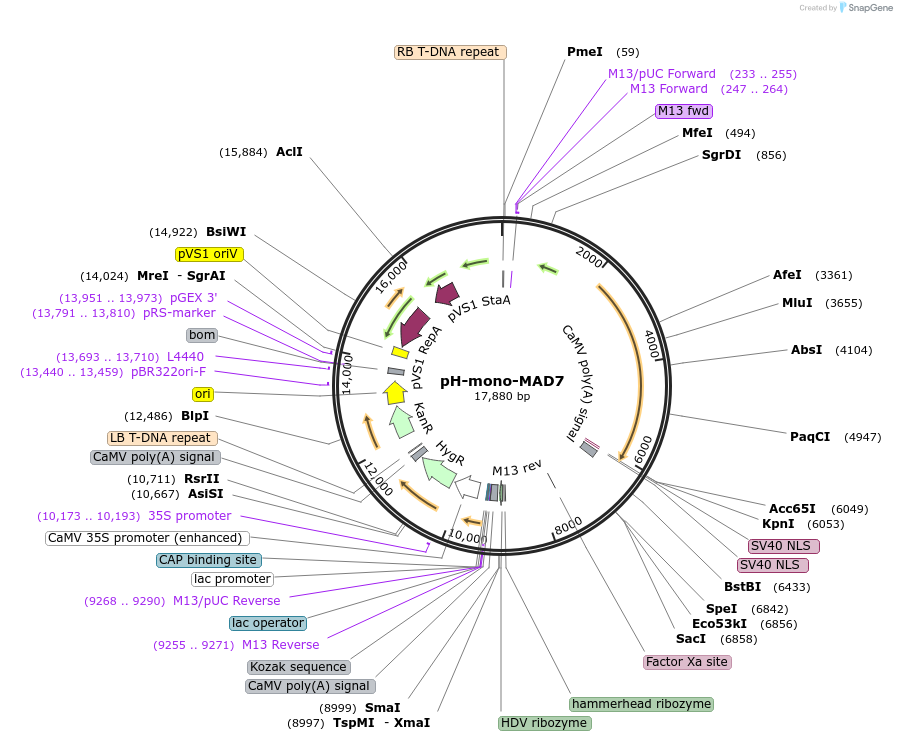
pH-mono-MAD7
Plasmid#170134PurposeFor plant prime editing in rice plants or monocotyledons protoplastsDepositorInsertMAD7(ErCas12a)
UseCRISPRExpressionPlantPromotermaize Ubiquitin-1, switchgrass Ubiquitin-1Available SinceSept. 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
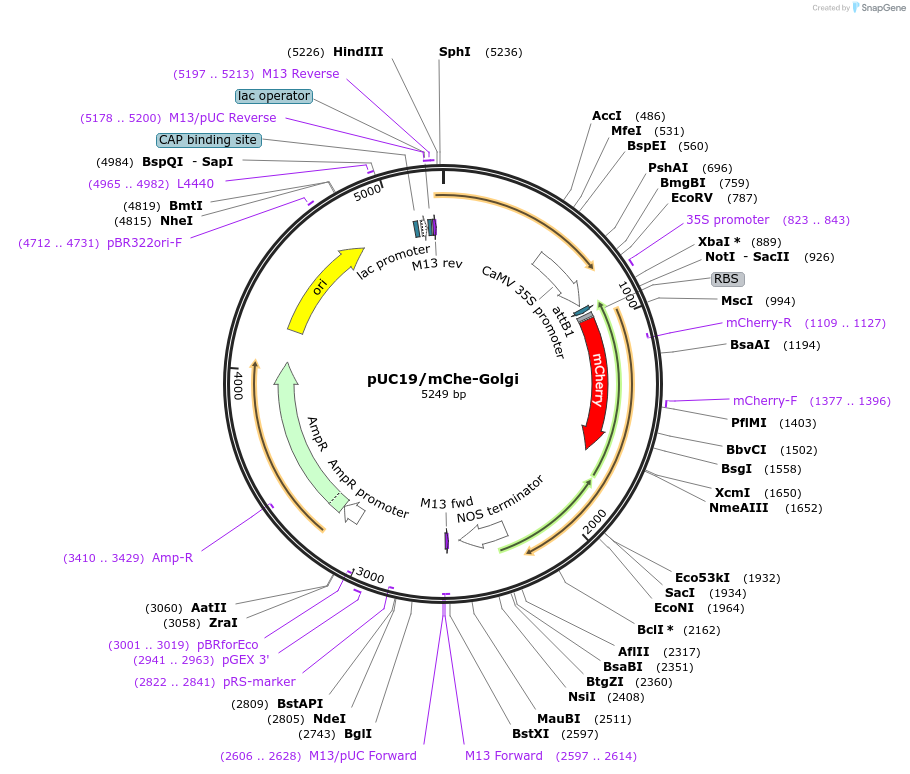
pUC19/mChe-Golgi
Plasmid#183169PurposeConstruct for transient expression of Golgi apparatus-targeted mCherry in plants.DepositorInsertmCherry-Golgi
TagsAtRER1B, A. thaliana endoplasmic reticulum retrie…ExpressionPlantAvailable SinceApril 26, 2022AvailabilityAcademic Institutions and Nonprofits only -
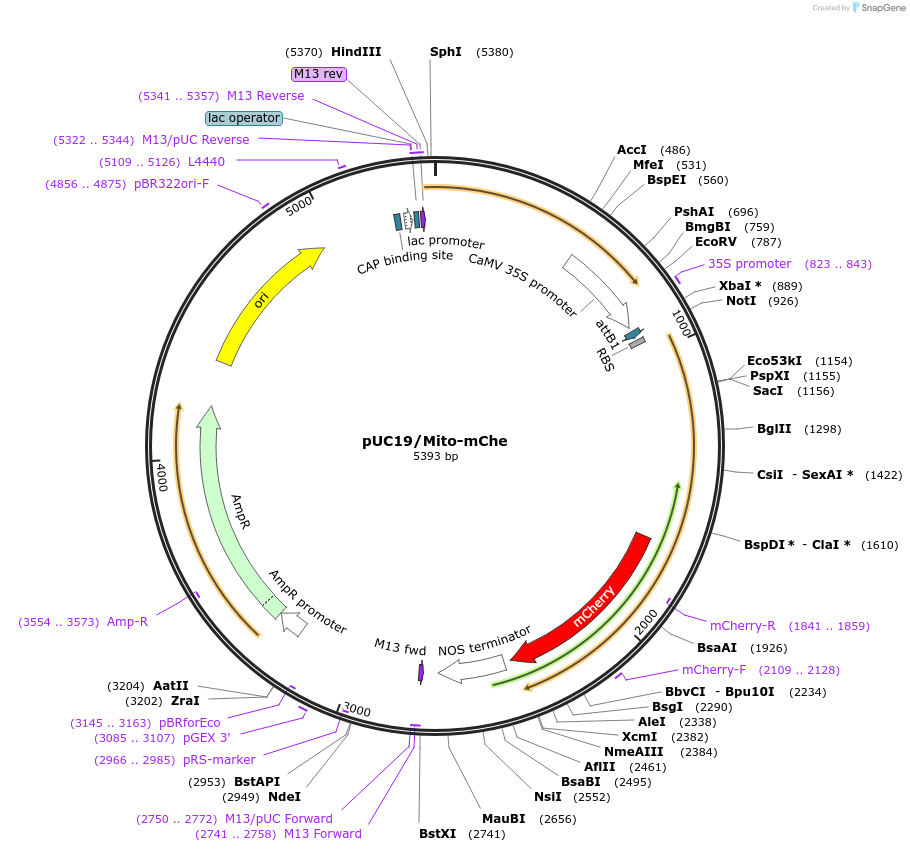
pUC19/Mito-mChe
Plasmid#183167PurposeConstruct for transient expression of mitochondrion-targeted mCherry in plants.DepositorInsertMito-mCherry
TagsOsCOX11, Oryza sativa cytochrome c oxidase assemb…ExpressionPlantAvailable SinceAug. 1, 2022AvailabilityAcademic Institutions and Nonprofits only -
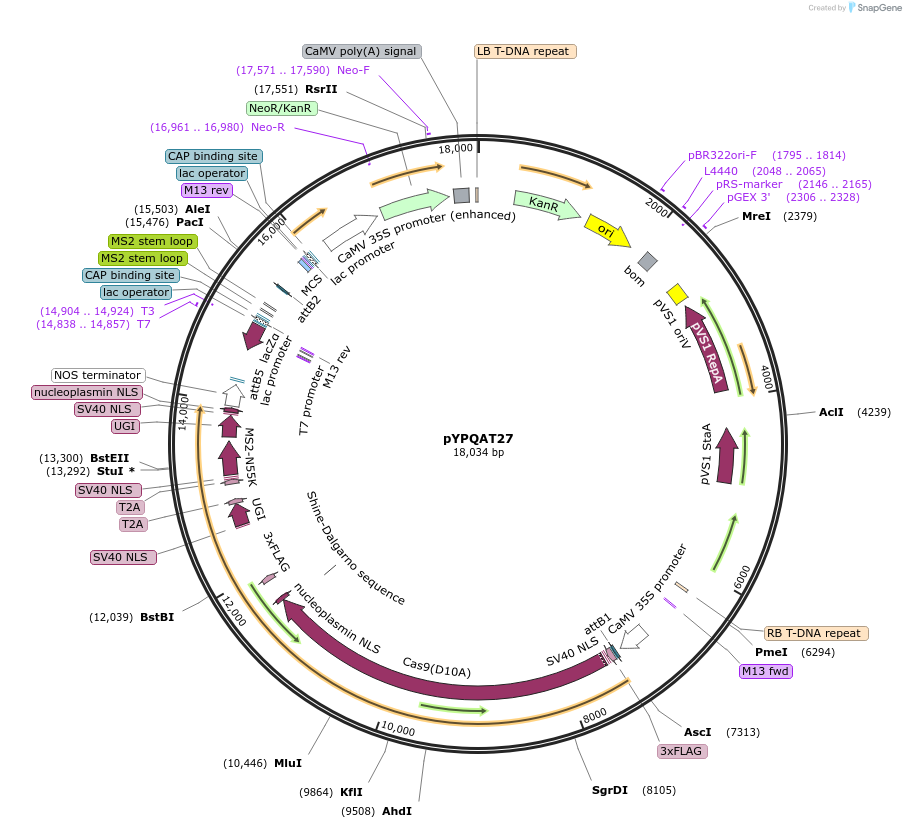
pYPQAT27
Plasmid#223399PurposeT-DNA vector for PmCDA1-CBE_V04 based C-to-T base editing for dicot plants; NGG PAM; 5' editing window; PmCDA1-CBE_V04 was driven by 2x35s and the sgRNA was driven by AtU3; Kanamycin for plant select.DepositorInsert2x35s-SpCas9D10A-PmCDA-UGI-T2A-MS2-UGI-AtU3-gRNA scaffold 2.0
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
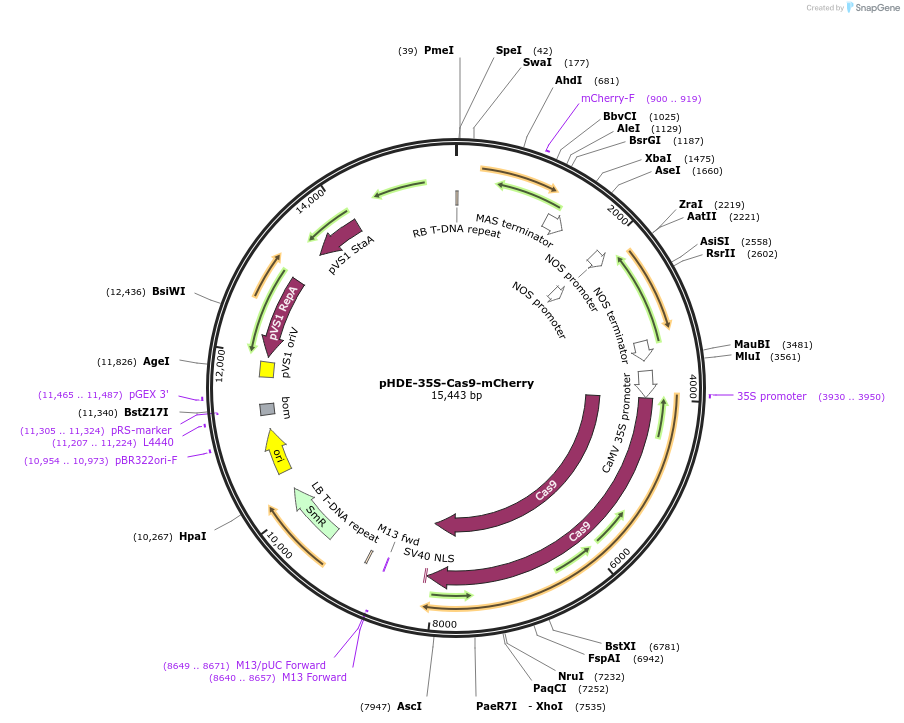
pHDE-35S-Cas9-mCherry
Plasmid#78931PurposeFor CRISPR/Cas9 mediated gene editing in Arabidopsis; provides a visual screen for Cas9-free plantsDepositorTypeEmpty backboneUseCRISPRExpressionPlantAvailable SinceJuly 12, 2016AvailabilityAcademic Institutions and Nonprofits only -
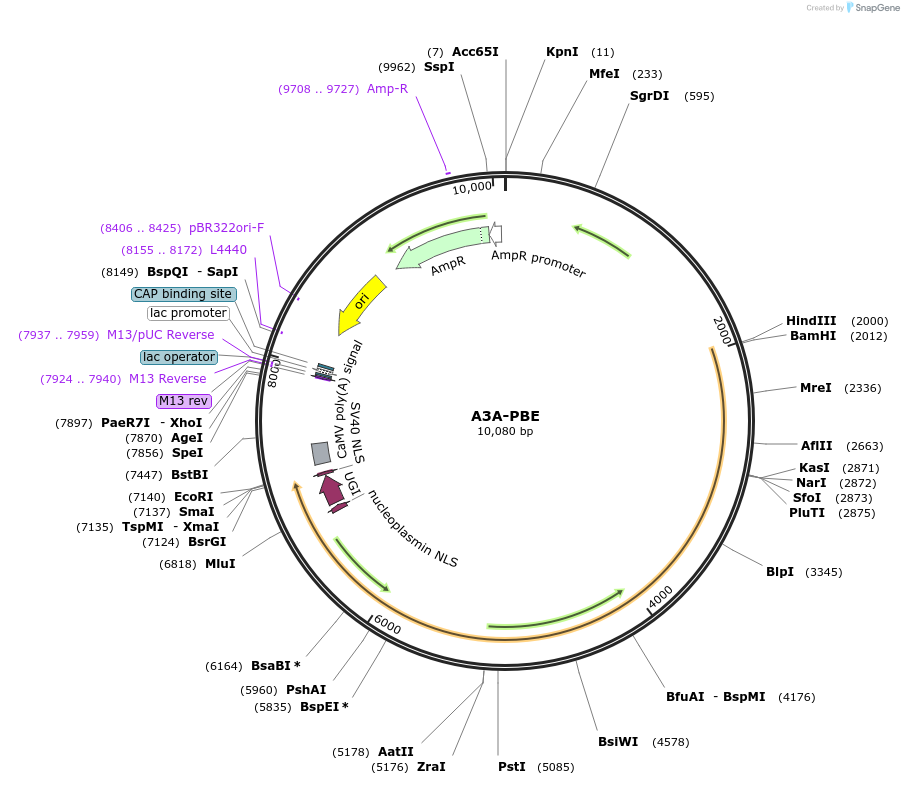
A3A-PBE
Plasmid#119768PurposeConversion of C to T in wheat plants or monocotyledons protoplastsDepositorInsertAPOBEC3A-nCas9-NLS-UGI-NLS
TagsNLS (flanking UGI)ExpressionPlantMutationnCas9 has a mutation D10A and E945K in wild type …Promotermaize Ubiquitin-1Available SinceJune 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
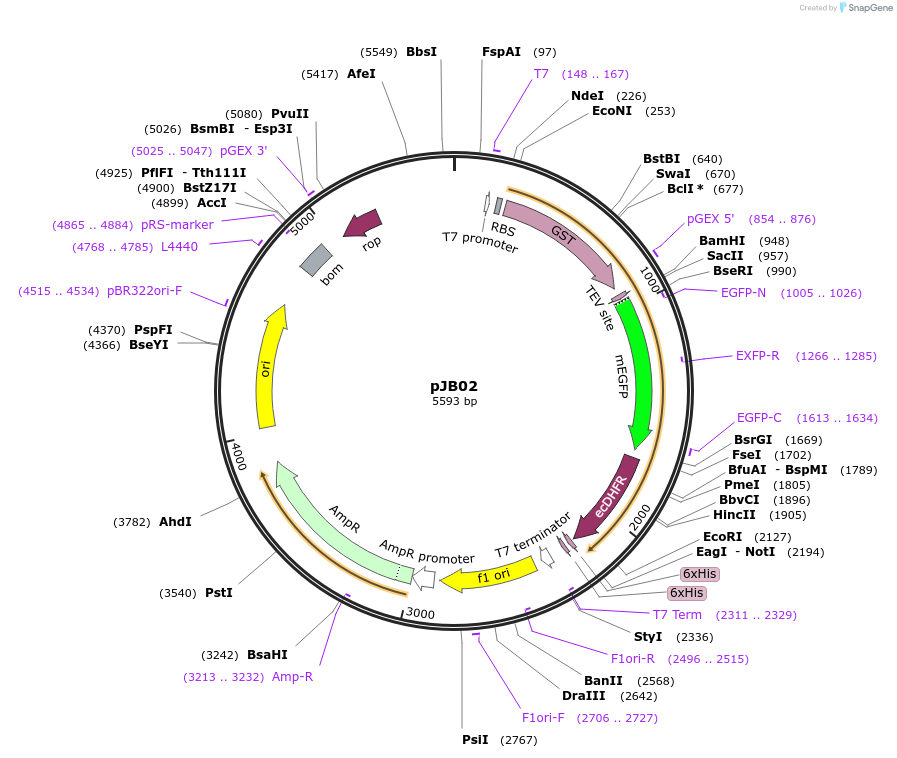
pJB02
Plasmid#111959PurposeExpresses GST-GFP-DHFR; construct generated for pull-down and localization experimentsDepositorInsertsmEGFP
eDHFR
TagsDHFRExpressionBacterialMutationeDHFR;residues 2-159 and enhanced GFP with monome…PromoterT7Available SinceAug. 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
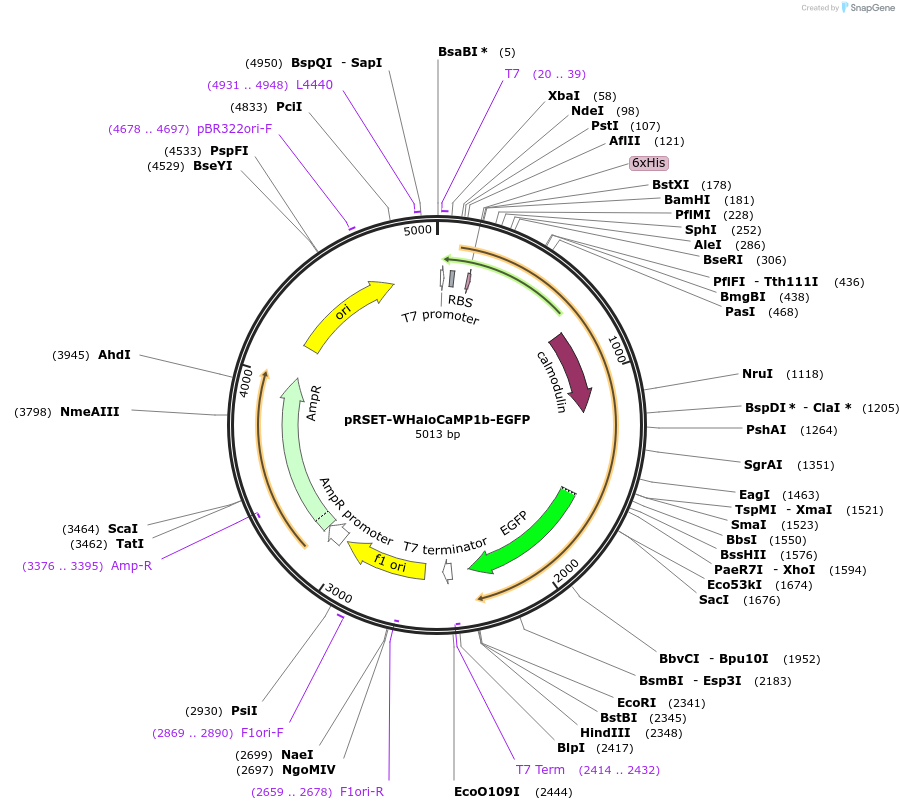
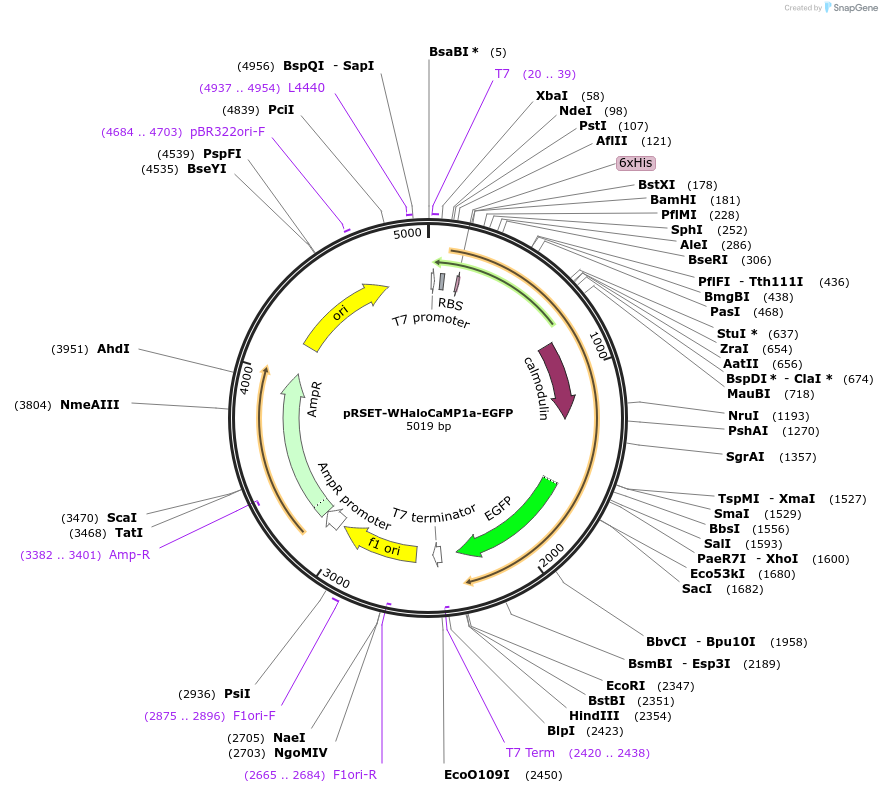
pRSET-WHaloCaMP1b-EGFP
Plasmid#205305PurposeExpression of WHaloCaMP1b-EGFP fusion in E. coliDepositorInsertWHaloCaMP1b-EGFP
ExpressionBacterialAvailable SinceAug. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
mCherry-Vimentin-C-18
Plasmid#55157PurposeLocalization: Intermediate Filaments, Excitation: 587, Emission: 610DepositorAvailable SinceSept. 17, 2014AvailabilityAcademic Institutions and Nonprofits only -
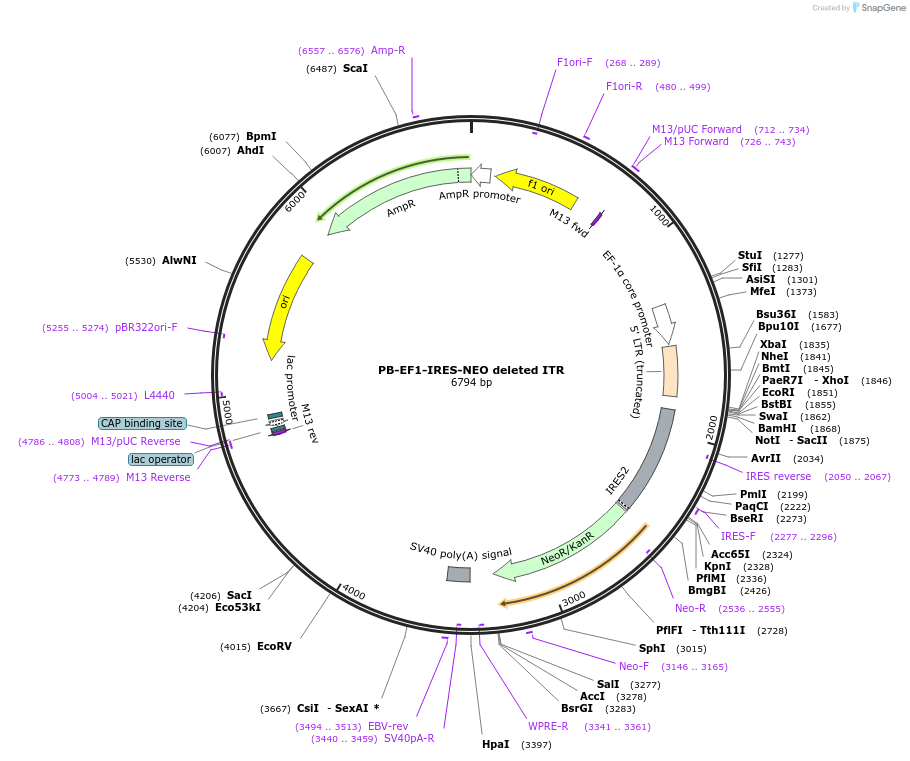
PB-EF1-IRES-NEO deleted ITR
Plasmid#67906PurposePiggyBac transposon encoding Neomycin resistance with deleted inverted terminal repeatDepositorInsertNeoR (neoR Synthetic)
ExpressionMammalianAvailable SinceJune 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
pRSET-WHaloCaMP1a-EGFP
Plasmid#205303PurposeExpression of WHaloCaMP1a-EGFP fusion in E. coliDepositorInsertWHaloCaMP1a-EGFP
ExpressionBacterialAvailable SinceAug. 18, 2023AvailabilityAcademic Institutions and Nonprofits only -
pROF417
Plasmid#155335PurposeConstitutive expression LFY-VP16-2A-RLuc in plantsDepositorInsertPCaMV35S-LFY-VP16-NLS-2A-RLuc-Tnos
UseLuciferase and Synthetic BiologyExpressionPlantAvailable SinceOct. 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
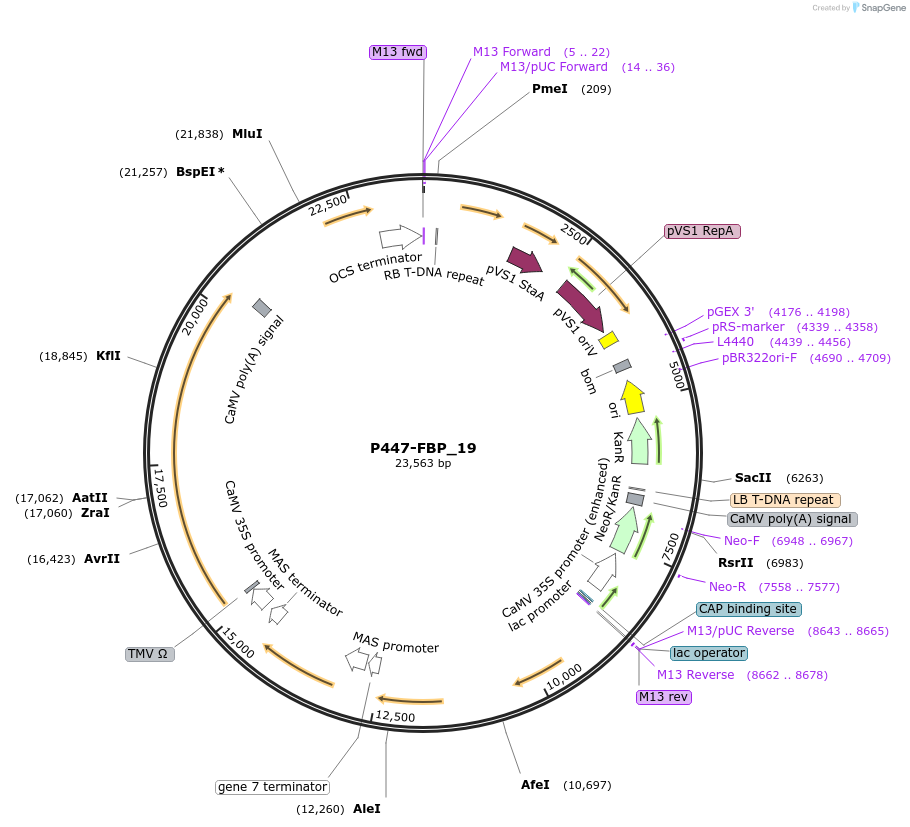
P447-FBP_19
Plasmid#139710PurposeT-DNA encoding Fungal Bioluminescent Pathway Components, complete pathway, Luz promoter is ABA responsiveDepositorInsertpSlH4:NnCPH:tSlH4, pSlRBCS2:AsNpga:tAtuG7, pAtuMas:NnH3H:tAtuMas, p35S:NnHisps:t35S, pAtRAB18:NnLuz:tAtuOcs
UseLuciferaseExpressionPlantPromotervariousAvailable SinceMay 29, 2020AvailabilityAcademic Institutions and Nonprofits only -
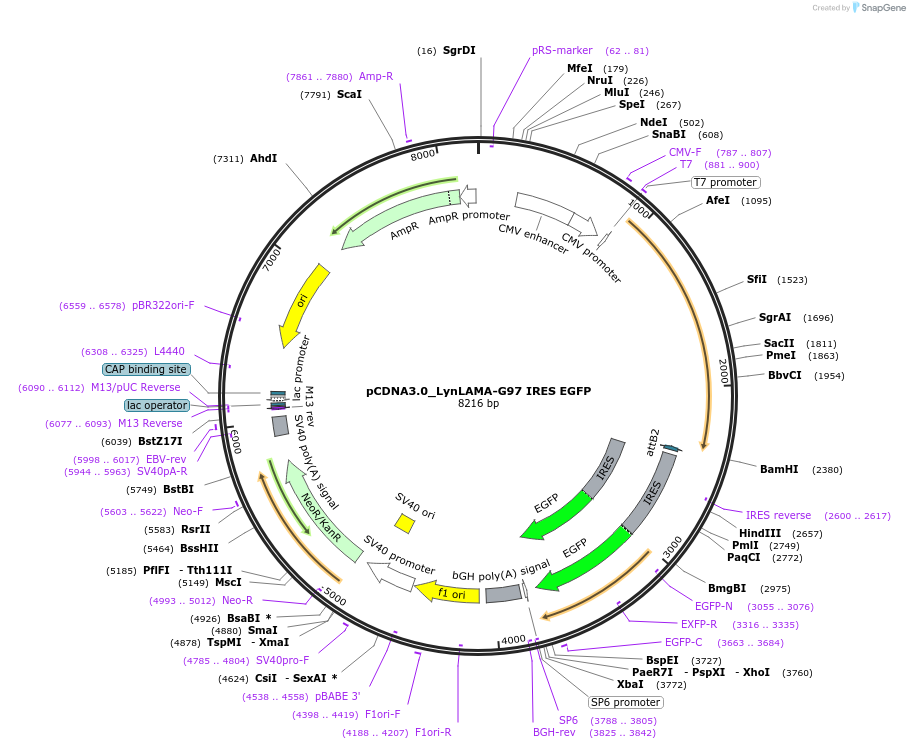
pCDNA3.0_LynLAMA-G97 IRES EGFP
Plasmid#130711PurposePlasmid encoding outer inner membrane localization of LAMA-G97 for GFP as a SNAP-Tag-fusion and EGFPDepositorInsertLyn11-SNAP-Tag-LAMA-G97 IRES EGFP
TagsLyn11 and SNAP-TagExpressionMammalianAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
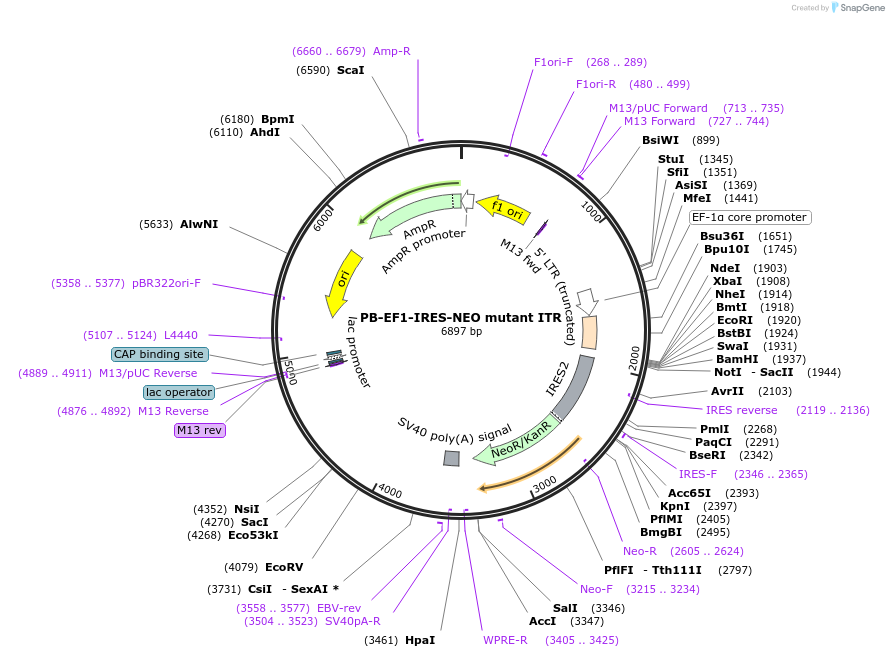
PB-EF1-IRES-NEO mutant ITR
Plasmid#67905PurposePiggyBac transposon encoding Neomycin resistance with inverted terminal repeat mutationDepositorInsertNeoR (neoR Synthetic)
ExpressionMammalianAvailable SinceJune 27, 2016AvailabilityAcademic Institutions and Nonprofits only -
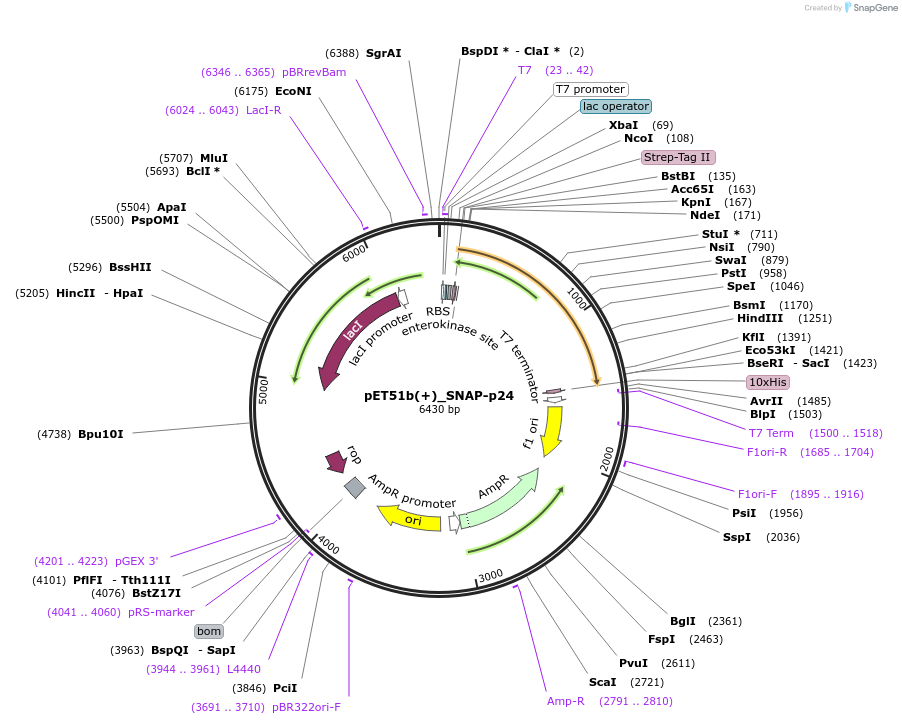
pET51b(+)_SNAP-p24
Plasmid#130718PurposePlasmid encoding the bacterial expression of a SNAP-fusion of p24 HIV Capsid proteinDepositorInsertSNAP-Tag-p24
TagsHisx10 and StrepTagExpressionBacterialMutationDeleted first Met, Ala and Last Ser from the Enha…Available SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
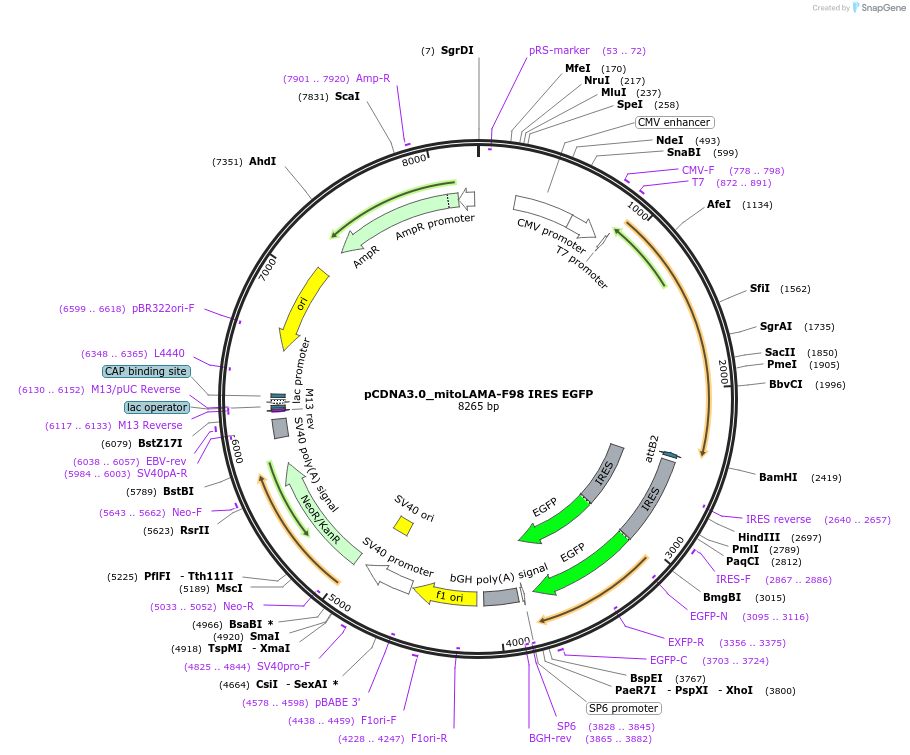
pCDNA3.0_mitoLAMA-F98 IRES EGFP
Plasmid#130706PurposePlasmid encoding outer mitochondrial localization of LAMA-F98 for GFP as a SNAP-Tag-fusion and EGFPDepositorInsertNTOM20-SNAP-Tag-LAMA-F98 IRES EGFP
TagsNTOM20 and SNAP-TagExpressionMammalianAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
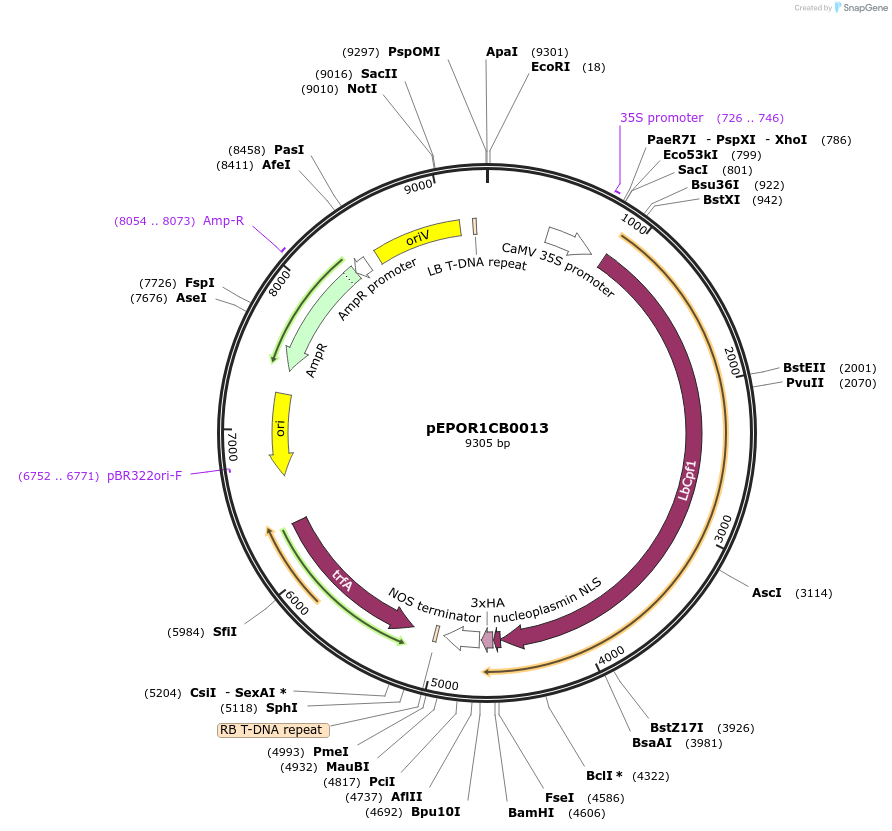
pEPOR1CB0013
Plasmid#117549PurposeLevel 1 Golden Gate Cassette: LbCas12a expression cassette for dicotyledonous plantsDepositorInsert2x35S+5'UTR OMEGA (pICH51288) + LbCas12a (with stop codon) (pEPOR0SP0007)+ Nos (pICH41421)
ExpressionPlantPromoter2x35S+5'UTR OMEGA (pICH51288)Available SinceNov. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
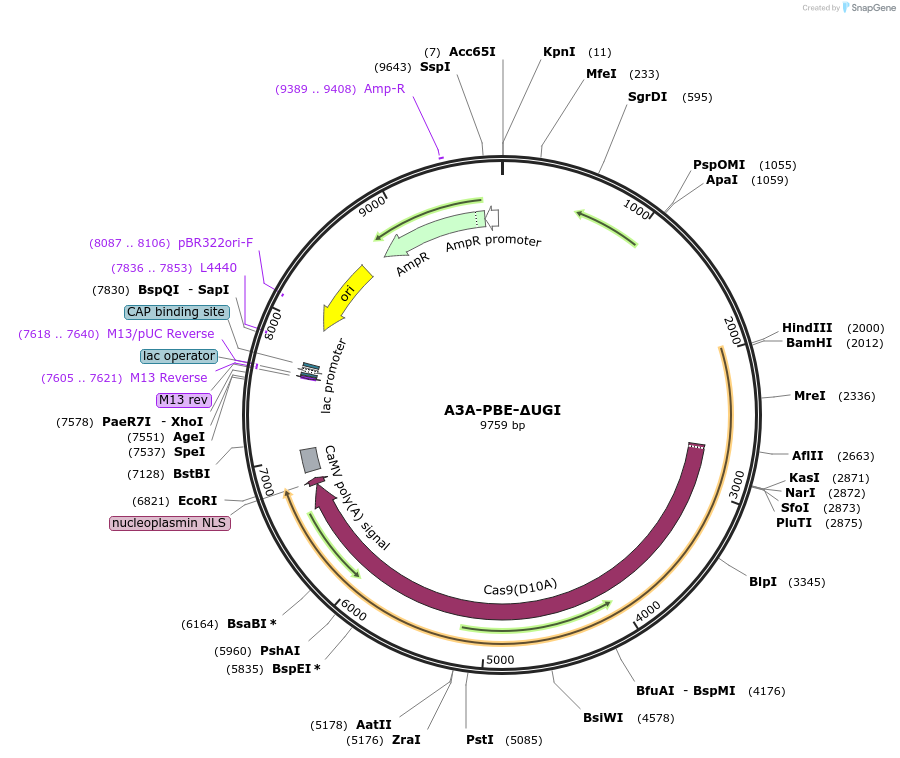
A3A-PBE-ΔUGI
Plasmid#119770PurposeConversion of C to T in wheat plants or monocotyledons protoplasts without uracil glycosylase inhibitor (UGI)DepositorInsertAPOBEC3A-nCas9-ΔUGI-NLS
TagsNLSExpressionPlantMutationnCas9 has a mutation D10A in wild type SpCas9Promotermaize Ubiquitin-1Available SinceDec. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
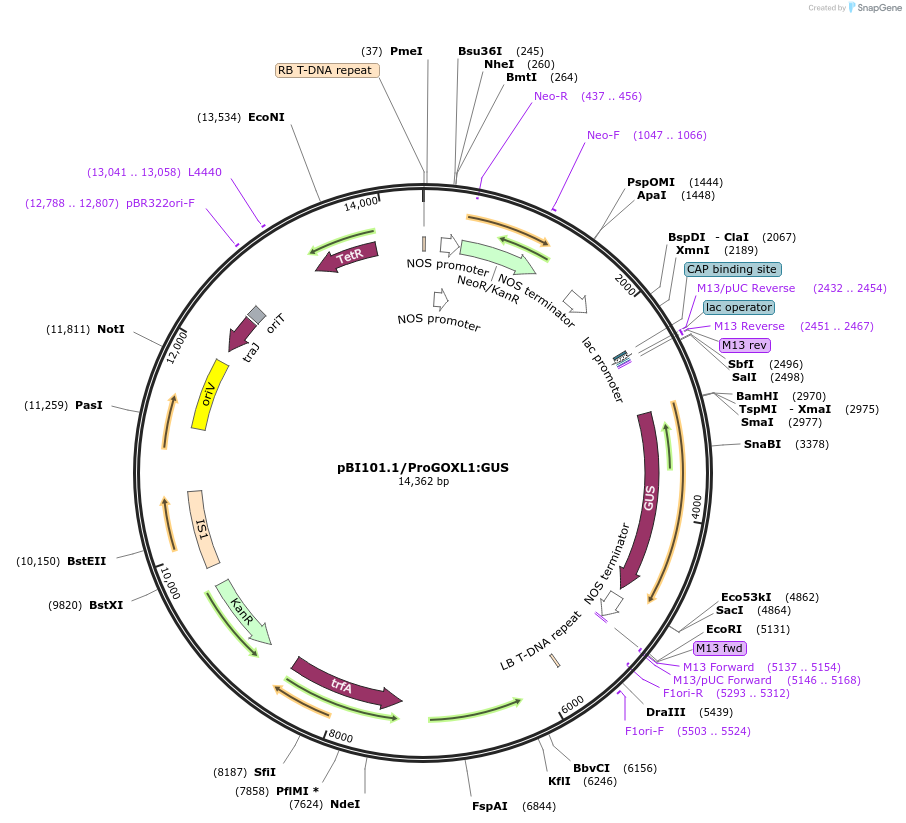
pBI101.1/ProGOXL1:GUS
Plasmid#175557PurposeBinary vector for Agrobacterium-mediated plant transformation - Promoter:GUS reporter for expression analysis of GOXL genesDepositorInsertProGOXL1
TagsGUSExpressionPlantPromoterGOXL1Available SinceJan. 13, 2022AvailabilityIndustry, Academic Institutions, and Nonprofits -
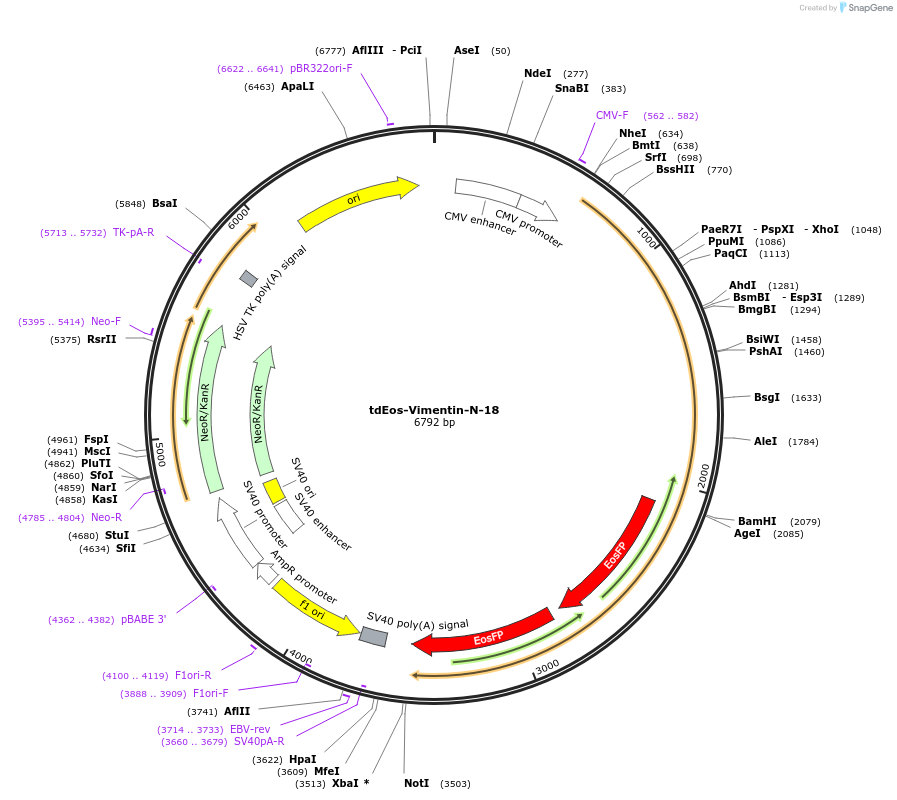
tdEos-Vimentin-N-18
Plasmid#57690PurposeLocalization: Intermediate Filaments, Excitation: 505 / 569, Emission: 516 / 581DepositorAvailable SinceJan. 13, 2015AvailabilityIndustry, Academic Institutions, and Nonprofits -
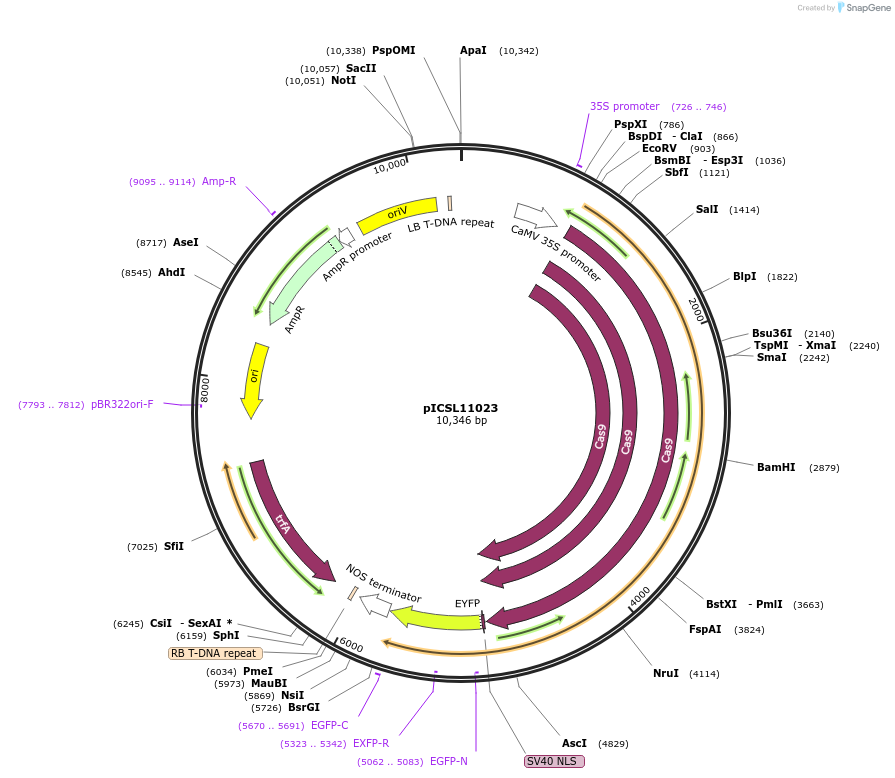
pICSL11023
Plasmid#117542PurposeLevel 1 Golden Gate Cassette: Cas9 expression cassette for dicotyledonous plantsDepositorInsert2x35S+5'UTR OMEGA (pICH51288) + SpCas9-h (no stop codon)(pICSL90005) + YFP (pICSL50005)+ Nos (pICH41421)
ExpressionPlantPromoter2x35S_OMEGA(pICH51288)Available SinceNov. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
pGEX-2TK-WW-CR-CT
Plasmid#18069DepositorAvailable SinceMay 29, 2008AvailabilityAcademic Institutions and Nonprofits only -
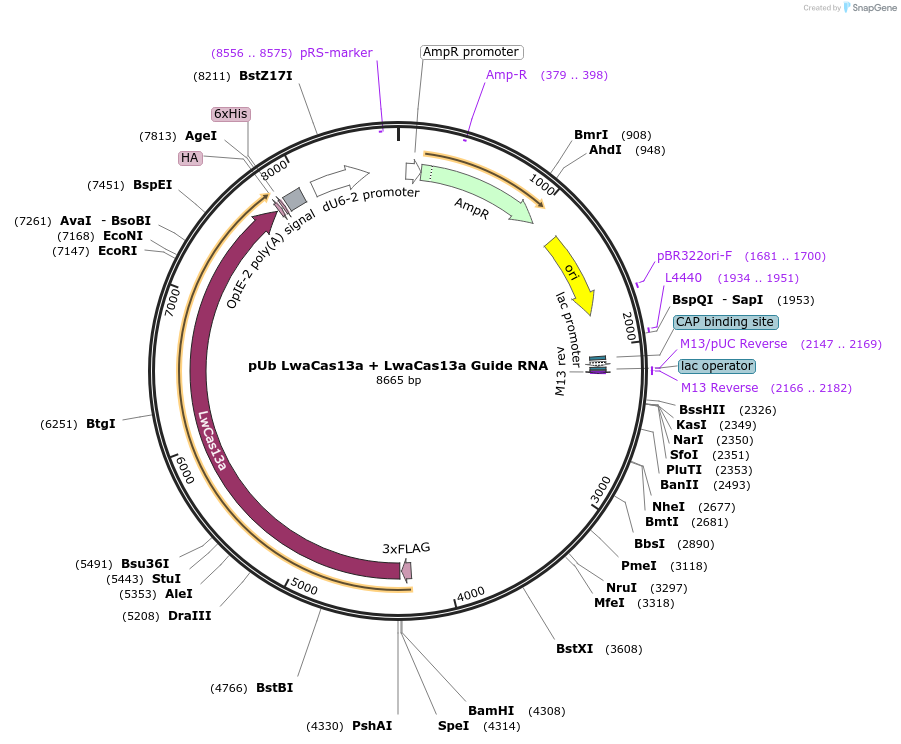
pUb LwaCas13a + LwaCas13a Guide RNA
Plasmid#176307PurposeExpresses LwaCas13a and its associated guide RNA in Drosophila cellsDepositorInsertLwaCas13a
TagsHA tagExpressionInsectMutationE656G, I1060V, D1076G- please see depositor comme…PromoterUbi-p63eAvailable SinceMarch 7, 2022AvailabilityAcademic Institutions and Nonprofits only -
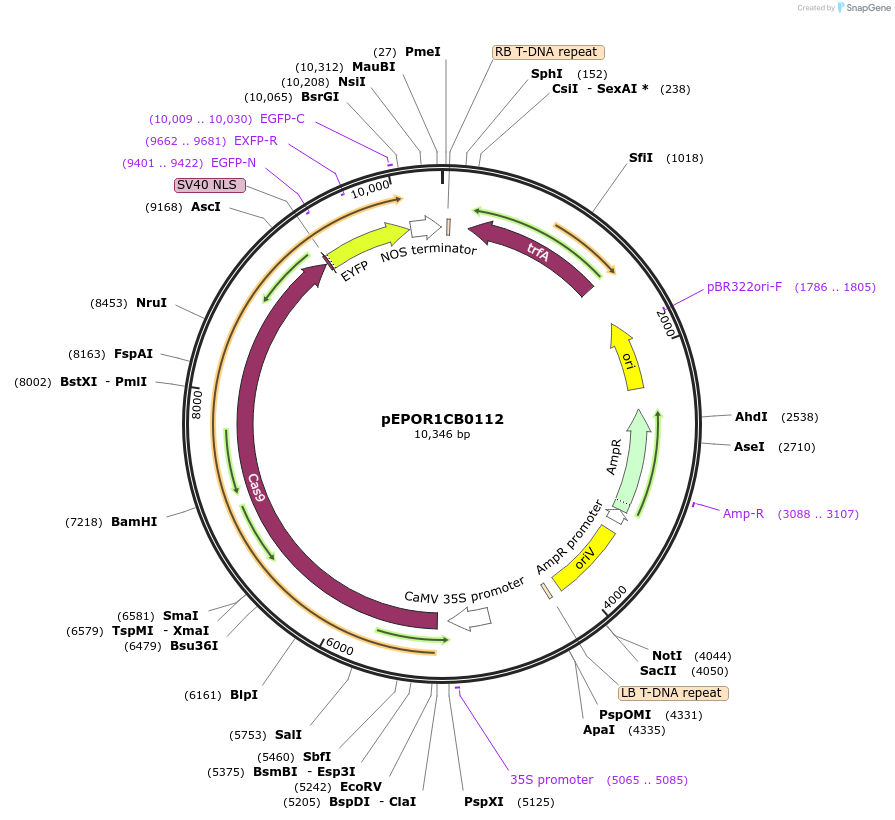
pEPOR1CB0112
Plasmid#117548PurposeLevel 1 Golden Gate Cassette: xCas9 3.7 expression cassette for dicotyledonous plantsDepositorInsert2x35S+5'UTR OMEGA (pICH51288) + Sp-xCas9 3.7 (no stop codon) (pEPOR0SP0011) + YFP (pICSL50005) + Nos (pICH41421)
ExpressionPlantPromoter2x35S+5'UTR OMEGA (pICH51288)Available SinceNov. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
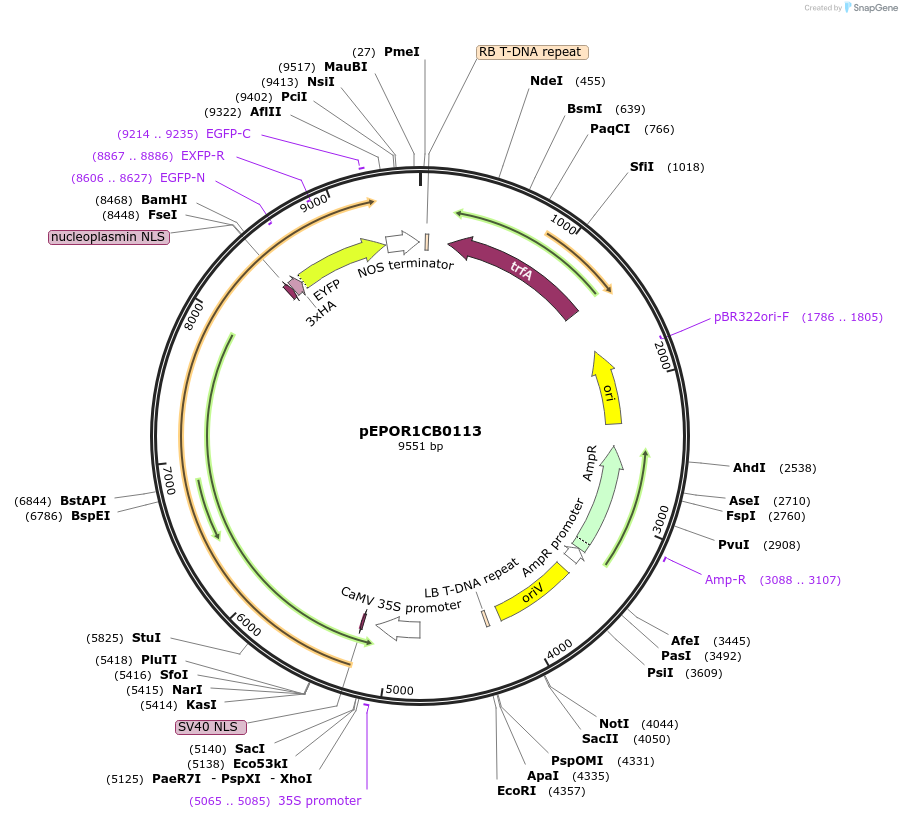
pEPOR1CB0113
Plasmid#117552PurposeLevel 1 Golden Gate Cassette: eSaCas9 expression cassette for dicotyledonous plantsDepositorInsert2x35S+5'UTR OMEGA (pICH51288) + eSaCas9 (no stop codon) (pEPOR0SP0012) + YFP (pICSL50005) + Nos (pICH41421)
ExpressionPlantPromoter2x35S+5'UTR OMEGA (pICH51288)Available SinceNov. 13, 2018AvailabilityAcademic Institutions and Nonprofits only -
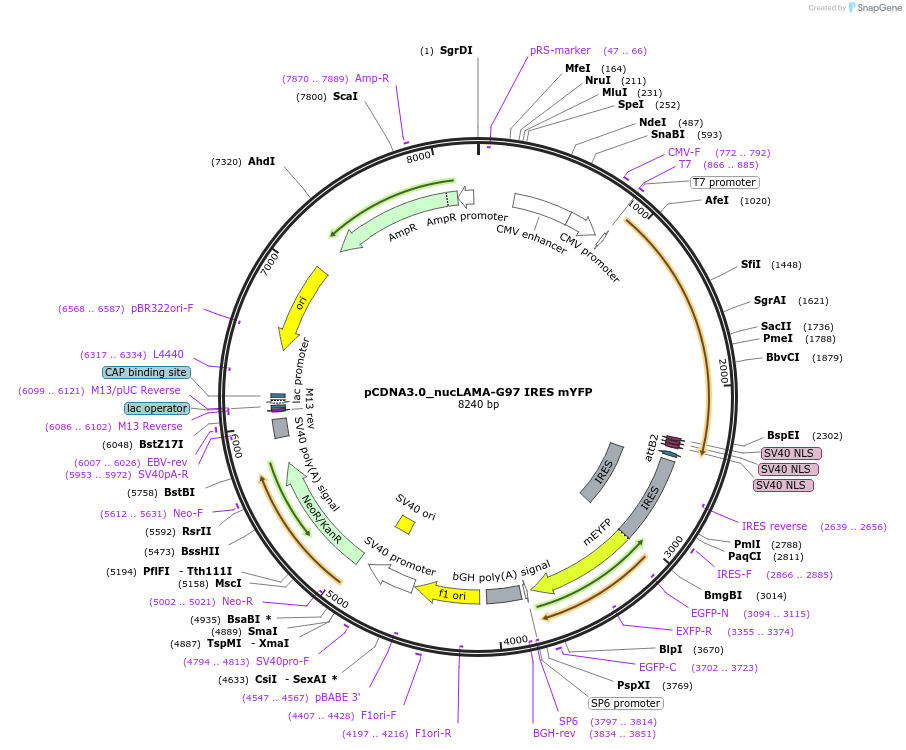
pCDNA3.0_nucLAMA-G97 IRES mYFP
Plasmid#130713PurposePlasmid encoding nuclear localization of LAMA-G97 for GFP as a SNAP-Tag-fusion and mYFPDepositorInsertSNAP-Tag-LAMA-G97-NLS IRES mYFP
TagsNLS and SNAP-TagExpressionMammalianAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
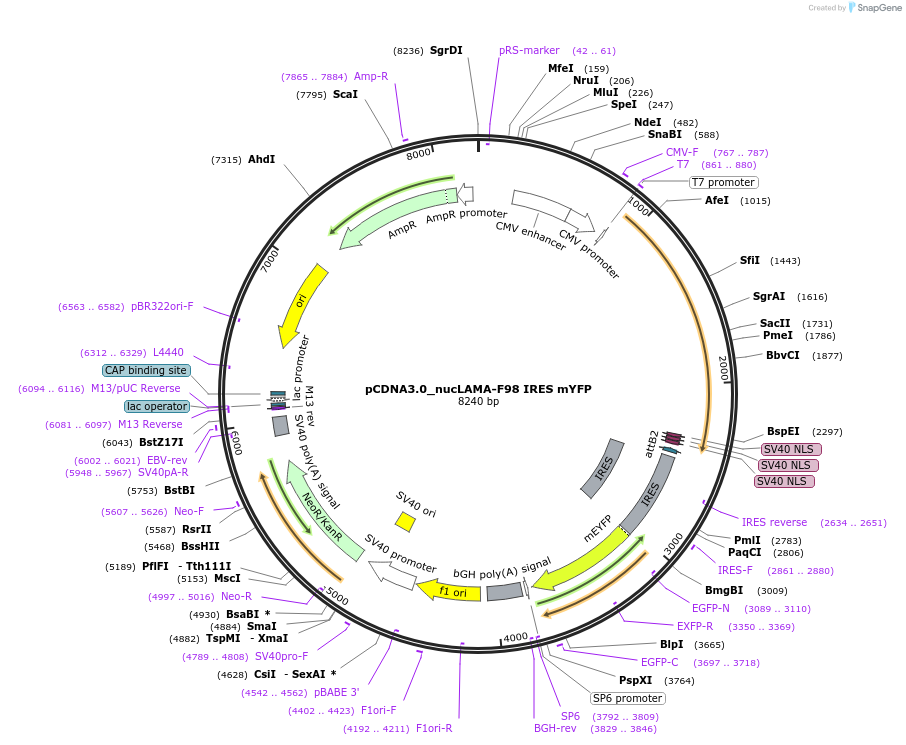
pCDNA3.0_nucLAMA-F98 IRES mYFP
Plasmid#130712PurposePlasmid encoding nuclear localization of LAMA-F98 for GFP as a SNAP-Tag-fusion and mYFPDepositorInsertSNAP-Tag-LAMA-F98-NLS IRES mYFP
TagsNLS and SNAP-TagExpressionMammalianAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
pCDNA3.0_mitoLAMA-G97 IRES EGFP
Plasmid#130707PurposePlasmid encoding outer mitochondrial localization of LAMA-G97 for GFP as a SNAP-Tag-fusion and EGFPDepositorInsertNTOM20-SNAP-Tag-LAMA-G97 IRES EGFP
TagsNTOM20 and SNAP-TagExpressionMammalianAvailable SinceAug. 26, 2019AvailabilityAcademic Institutions and Nonprofits only -
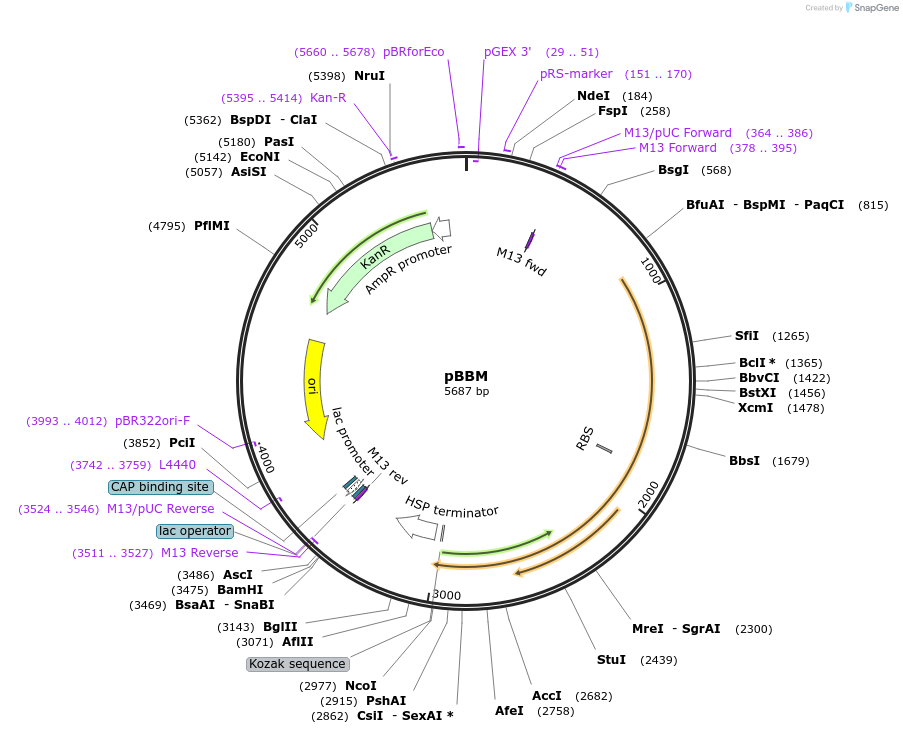
pBBM
Plasmid#243675PurposeExpresses a maize optimized CDS of BABY BOOM in plants using the Cestrum Yellow Leaf Curling Virus (CmYLCV) 9.11 promoter and an Arabidopsis HSP terminatorDepositorInsertMaize optimized CDS of BABY BOOM
ExpressionPlantAvailable SinceNov. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
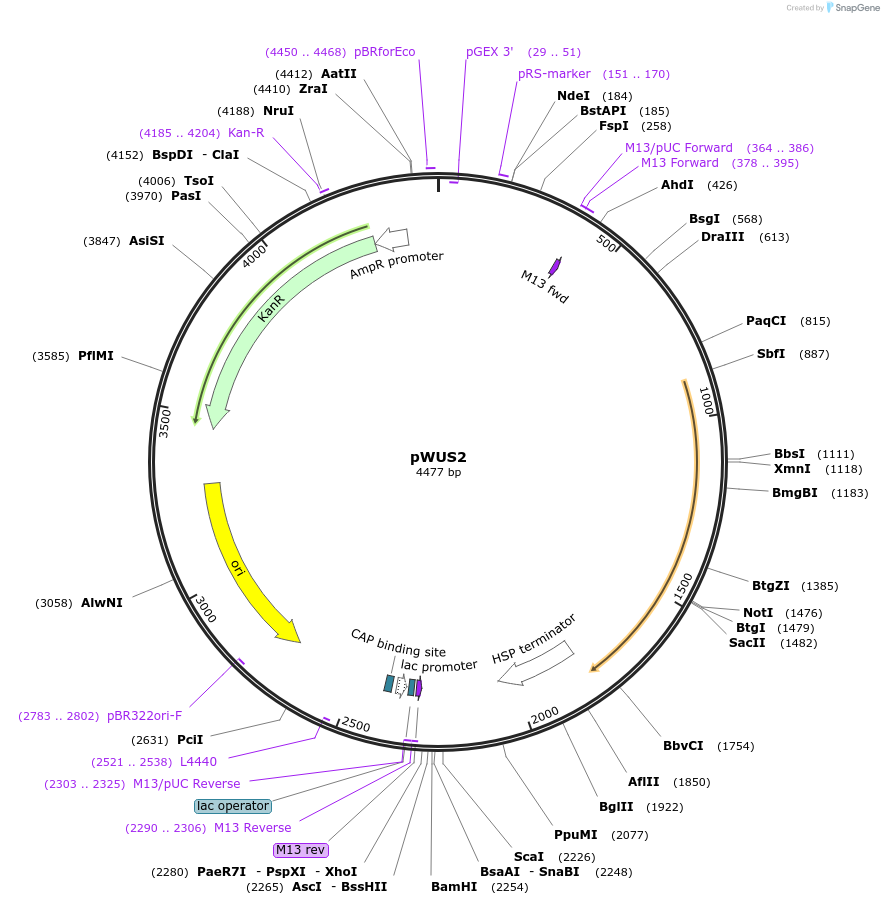
pWUS2
Plasmid#243676PurposeExpresses a maize optimized CDS of WUSCHEL2 in plants using the Cestrum Yellow Leaf Curling Virus (CmYLCV) 9.11 promoter and an Arabidopsis HSP terminatorDepositorInsertMaize optimized CDS of WUSHCEL2
ExpressionPlantAvailable SinceNov. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
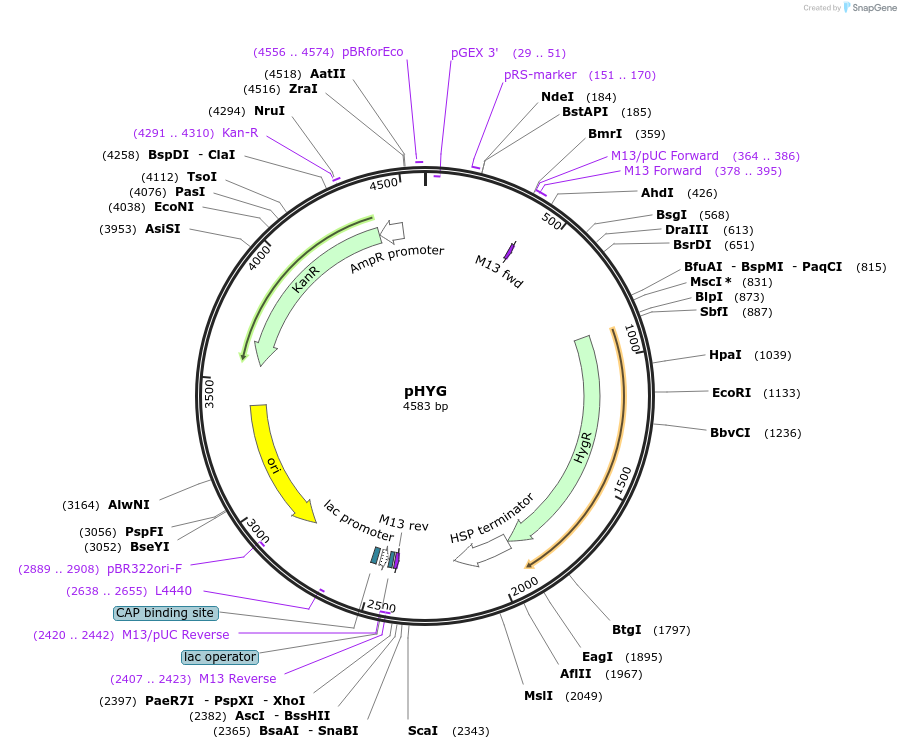
pHYG
Plasmid#243678PurposeExpresses the HYGROMYCIN PHOSPHOTRANSFERASE (HPT) CDS in plants using the Cestrum Yellow Leaf Curling Virus (CmYLCV) 9.11 promoter and an Arabidopsis HSP terminatorDepositorInsertHYGROMYCIN PHOSPHOTRANSFERASE (HPT) CDS
ExpressionPlantAvailable SinceNov. 4, 2025AvailabilityAcademic Institutions and Nonprofits only -
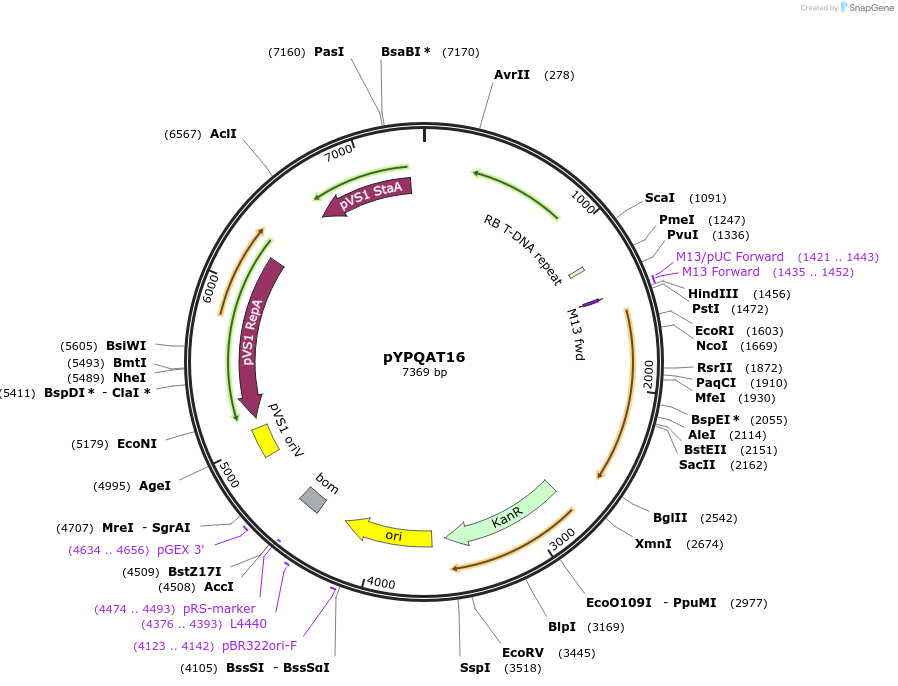
pYPQAT16
Plasmid#223388PurposeT-DNA vector for A3A/Y130F-CBE_V01 based C-to-T base editing for monocot plants; NGG PAM; wide working window; A3A/Y130F-CBE_V01 was driven by ZmUbi1 and the sgRNA was driven by OsU3 promoter.DepositorInsertZmUbi-hA3A-Y130F-SpCas9-D10A-UGI-OsU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceMay 28, 2025AvailabilityAcademic Institutions and Nonprofits only -
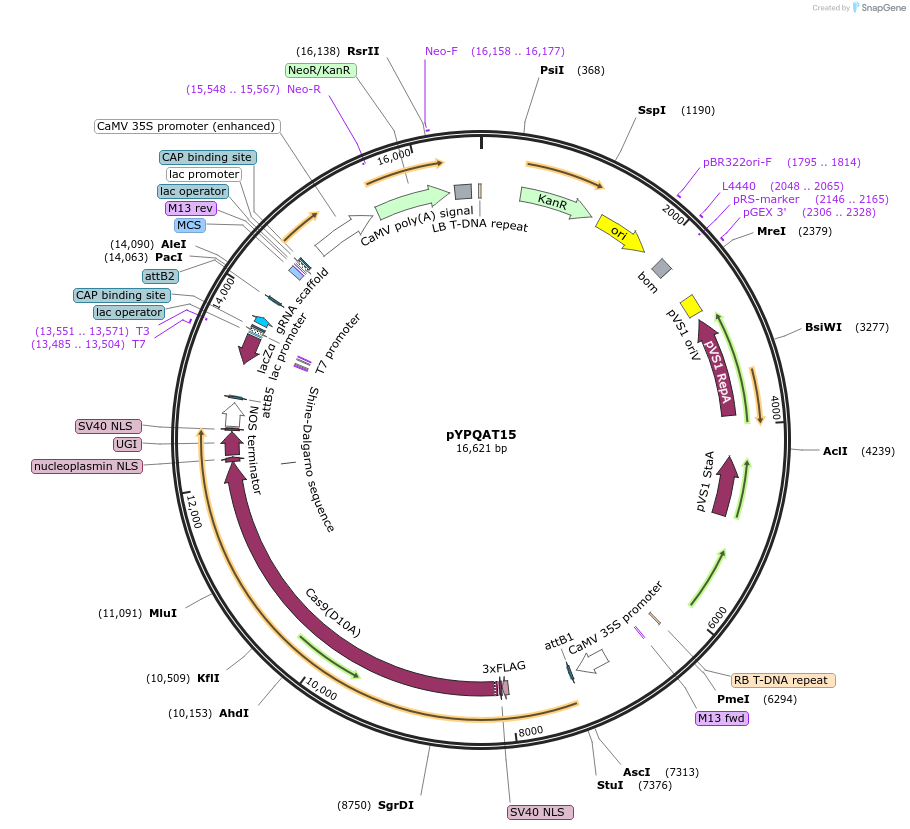
pYPQAT15
Plasmid#223387PurposeT-DNA vector for A3A/Y130F-CBE_V01 based C-to-T base editing for dicot plants; NGG PAM; wide working window; A3A/Y130F-CBE_V01 was driven by 2x35s and the sgRNA was driven by AtU3 promoter.DepositorInsert2x35s-hA3A-Y130F-SpCas9-D10A-UGI-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
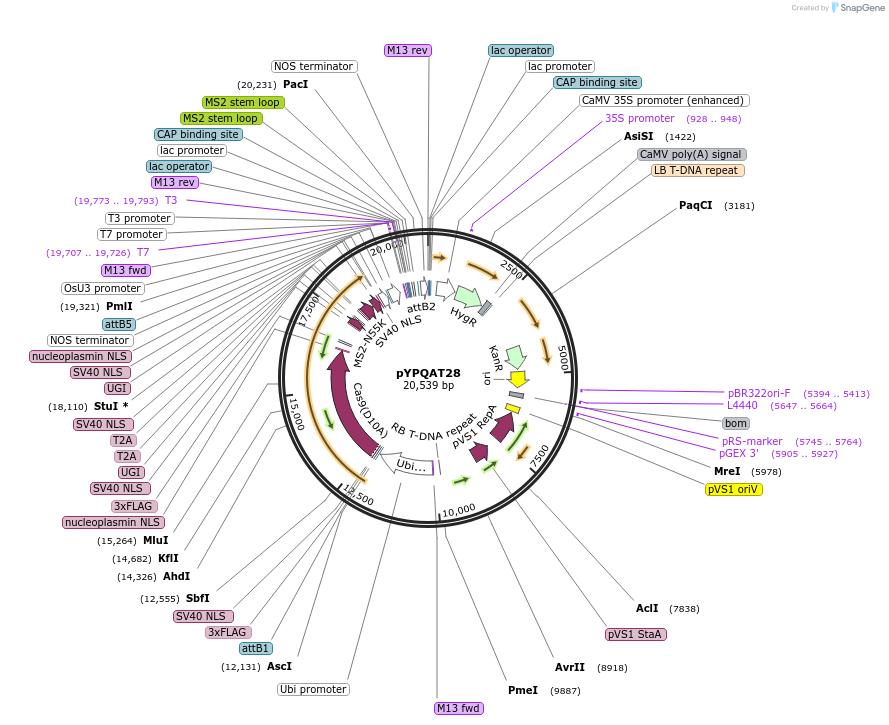
pYPQAT28
Plasmid#223400PurposeT-DNA vector for PmCDA1-CBE_V04 based C-to-T base editing for monocot plants; NGG PAM; 5' editing window; PmCDA1-CBE_V04 was driven by ZmUbi1 and sgRNA was driven by OsU3; Hygromycin for plant select.DepositorInsertZmUbi-SpCas9D10A-PmCDA-UGI-T2A-MS2-UGI-OsU3-gRNA scaffold 2.0
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
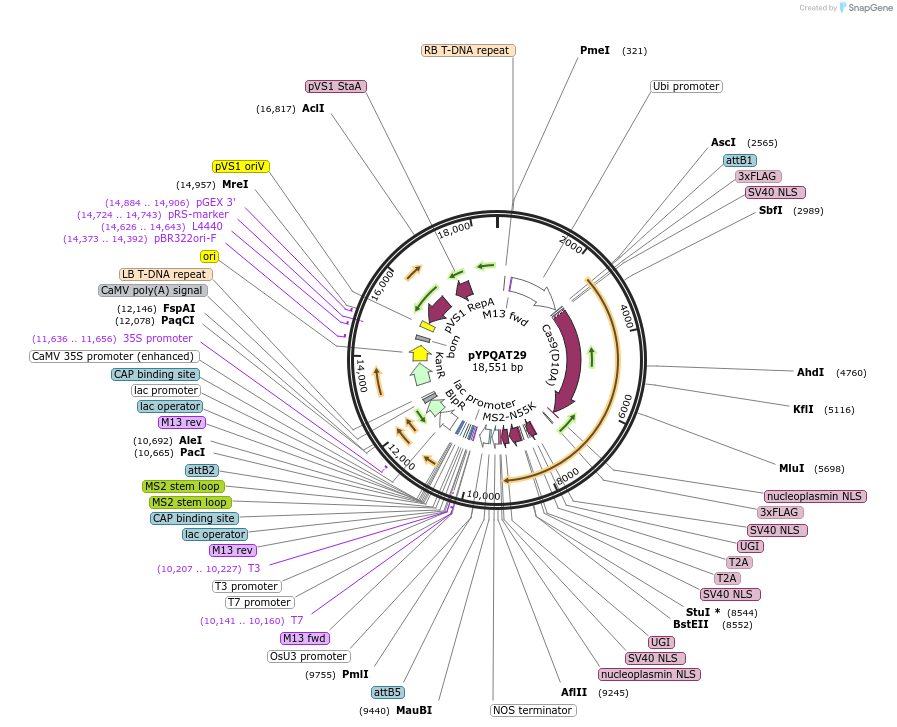
pYPQAT29
Plasmid#223401PurposeT-DNA vector for PmCDA1-CBE_V04 based C-to-T base editing for monocot plants; NGG PAM; 5' editing window; PmCDA1-CBE_V04 was driven by ZmUbi1 and the sgRNA was driven by OsU3; BASTA for plant select.DepositorInsertZmUbi-SpCas9D10A-PmCDA-UGI-T2A-MS2-UGI-OsU3-gRNA scaffold 2.0
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only -
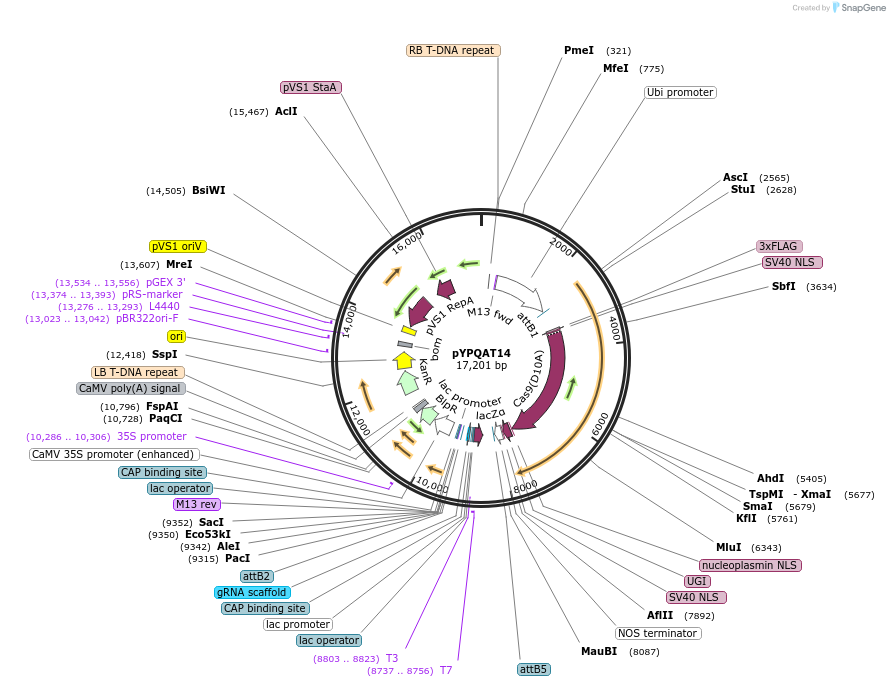
pYPQAT14
Plasmid#223386PurposeT-DNA vector for A3A/Y130F-CBE_V01 based C-to-T base editing for dicot plants; NGG PAM; wide working window; A3A/Y130F-CBE_V01 was driven by ZmUbi1 and sgRNA was by AtU3; BASTA for plant selection.DepositorInsertZmUbi-hA3A-Y130F-SpCas9-D10A-UGI-AtU3-gRNA scaffold
UseCRISPRTags3x FLAG and NLSExpressionPlantAvailable SinceApril 10, 2025AvailabilityAcademic Institutions and Nonprofits only