We narrowed to 8,645 results for: Set;
-
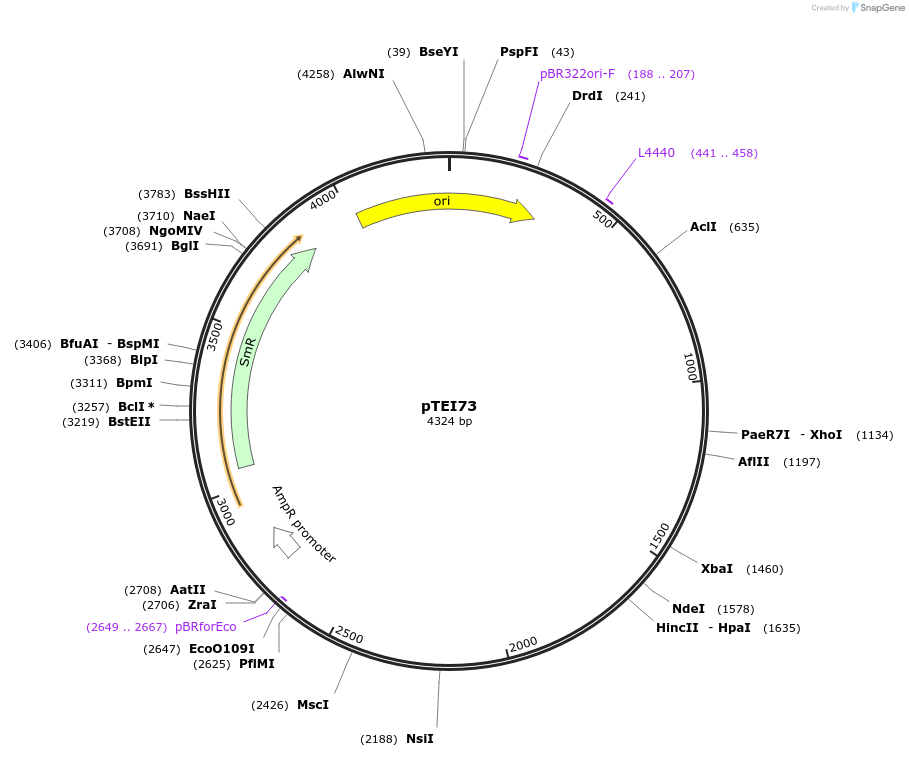
Plasmid#105373PurposeSynthetic biologyDepositorInsertpromotor (CUE1 (At5g33320; 2000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
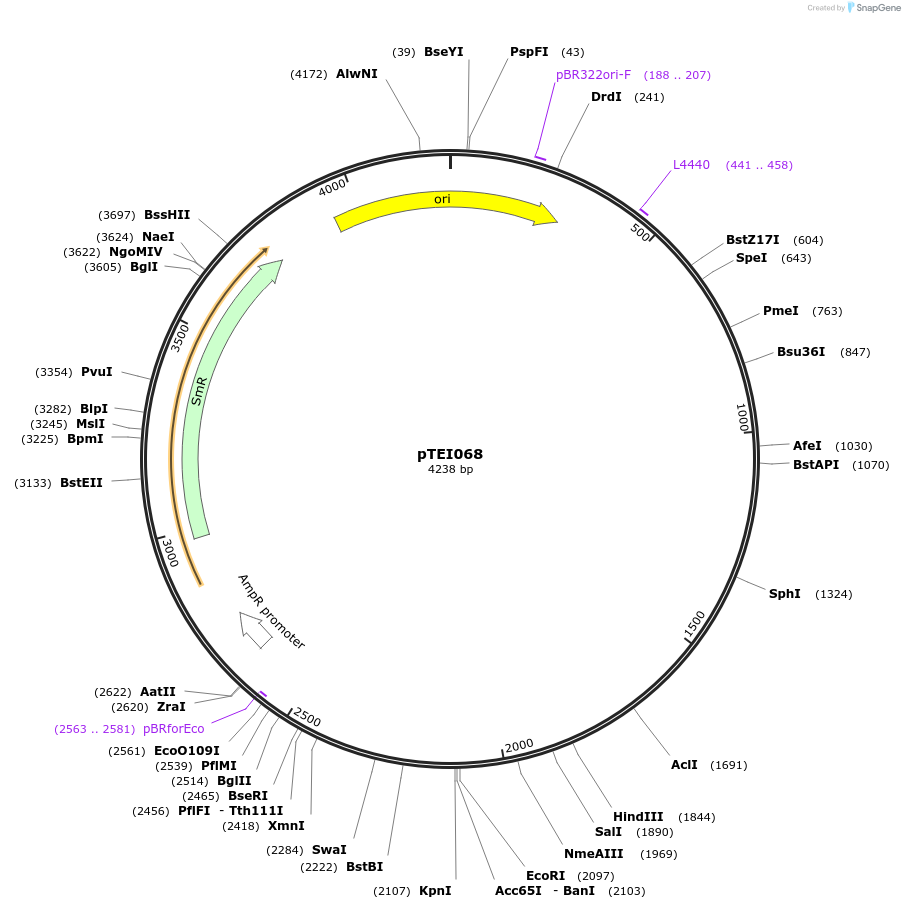
pTEI069
Plasmid#105374PurposeSynthetic biologyDepositorInsertpromotor BODYGUARD1 (At1g64670; 1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
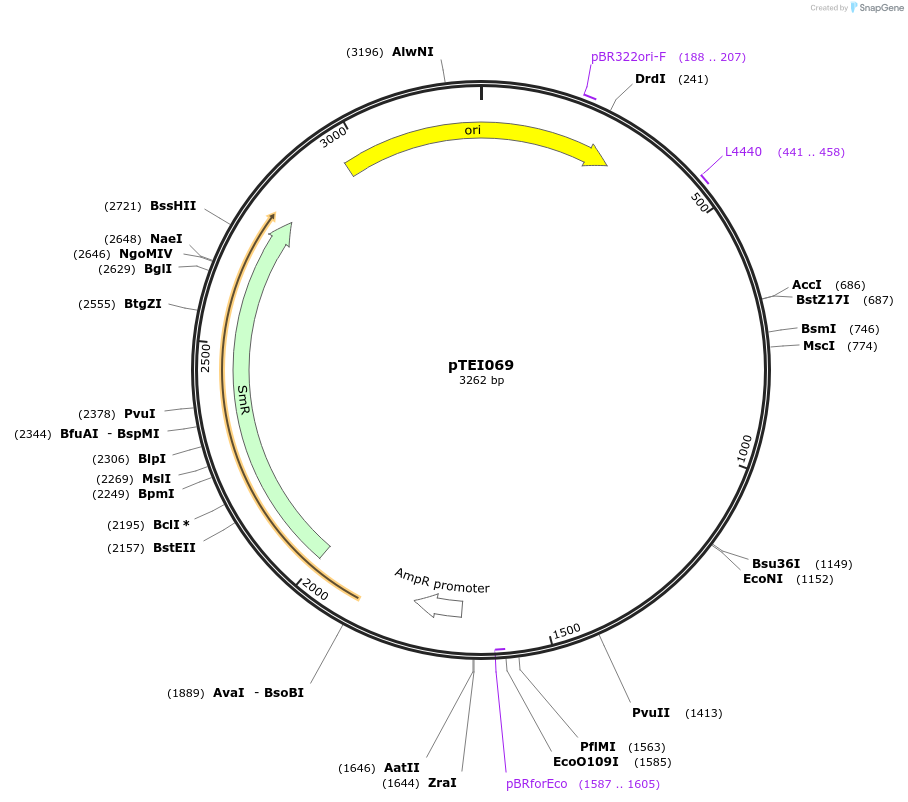
pTEI070
Plasmid#105375PurposeSynthetic biologyDepositorInsertpromotor "GC1" (At1g22690; 1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
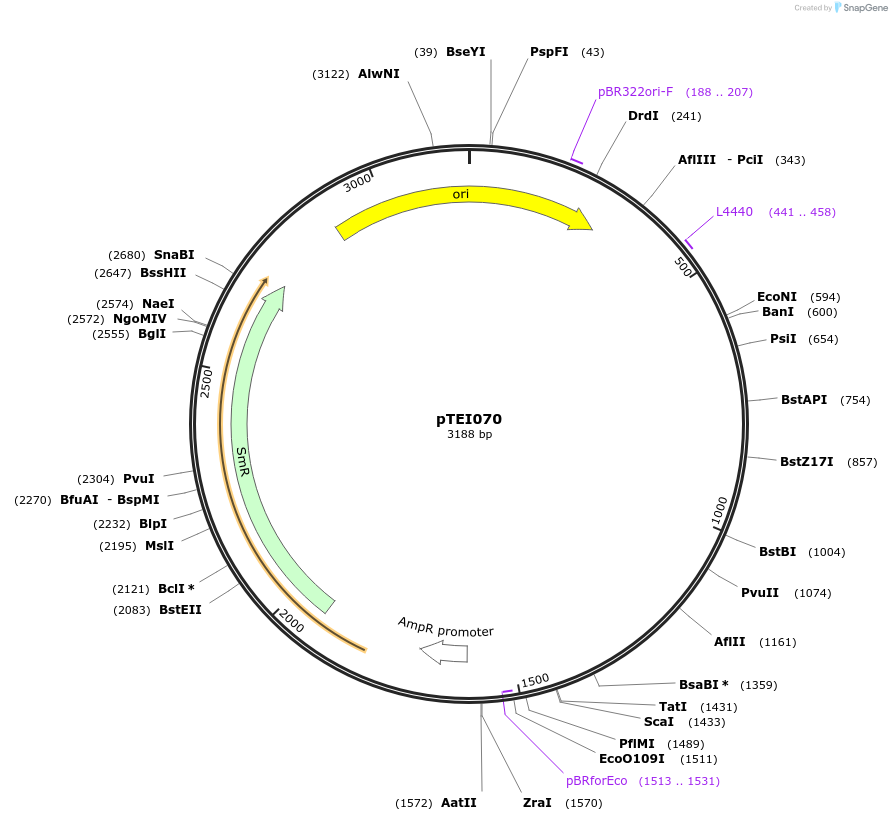
pTEI73
Plasmid#105377PurposeSynthetic biologyDepositorInsertpromotor ALMT12 (At4g17585; 2000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
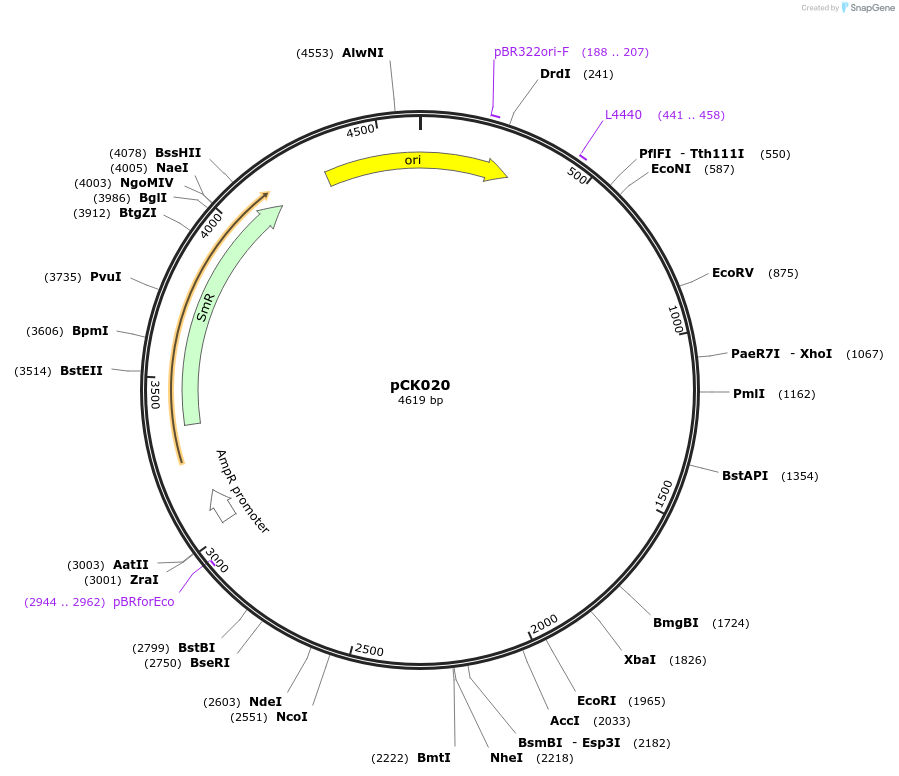
pCK020
Plasmid#105379PurposeSynthetic biologyDepositorInsertpromotor WUSCHEL (At3g11260; 2300bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
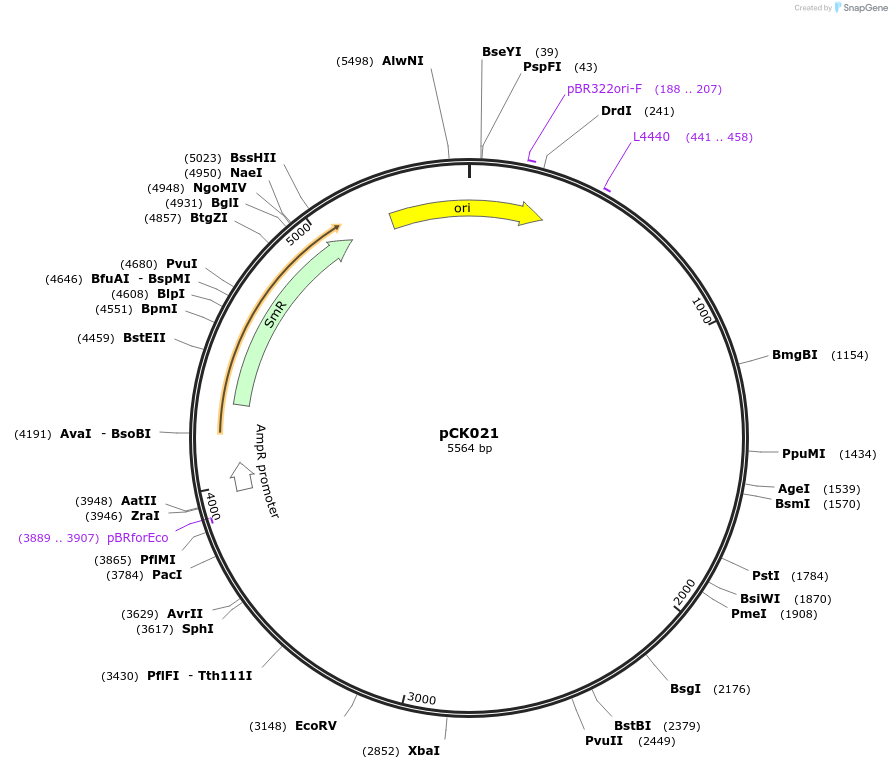
pCK021
Plasmid#105380PurposeSynthetic biologyDepositorInsertpromotor SOMBRERO (At1g79580; 3300bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
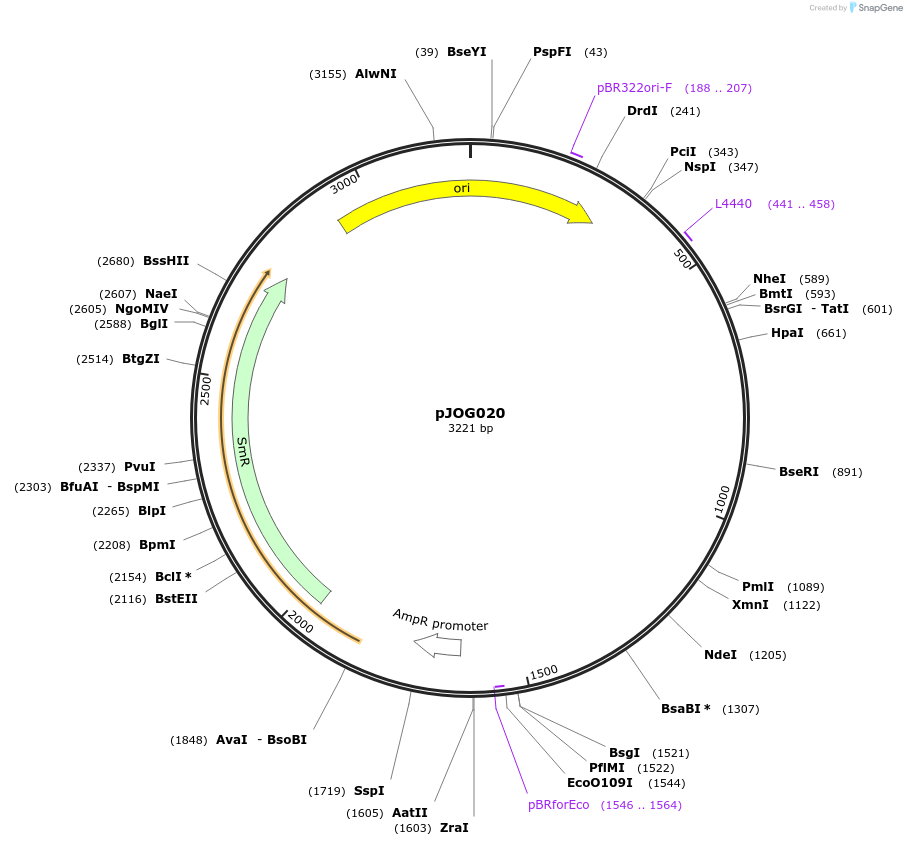
pJOG020
Plasmid#105382PurposeSynthetic biologyDepositorInsertpromotor Ubi4-2 (1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
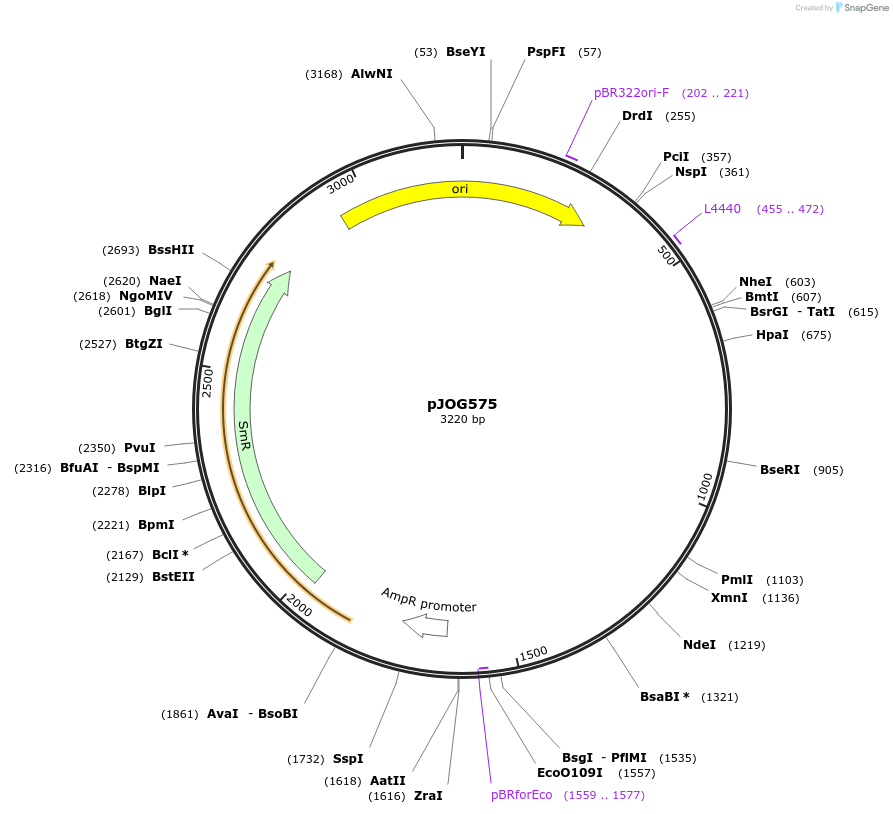
pJOG575
Plasmid#105383PurposeSynthetic biologyDepositorInsertpromotor Ubi4-2 (1000bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
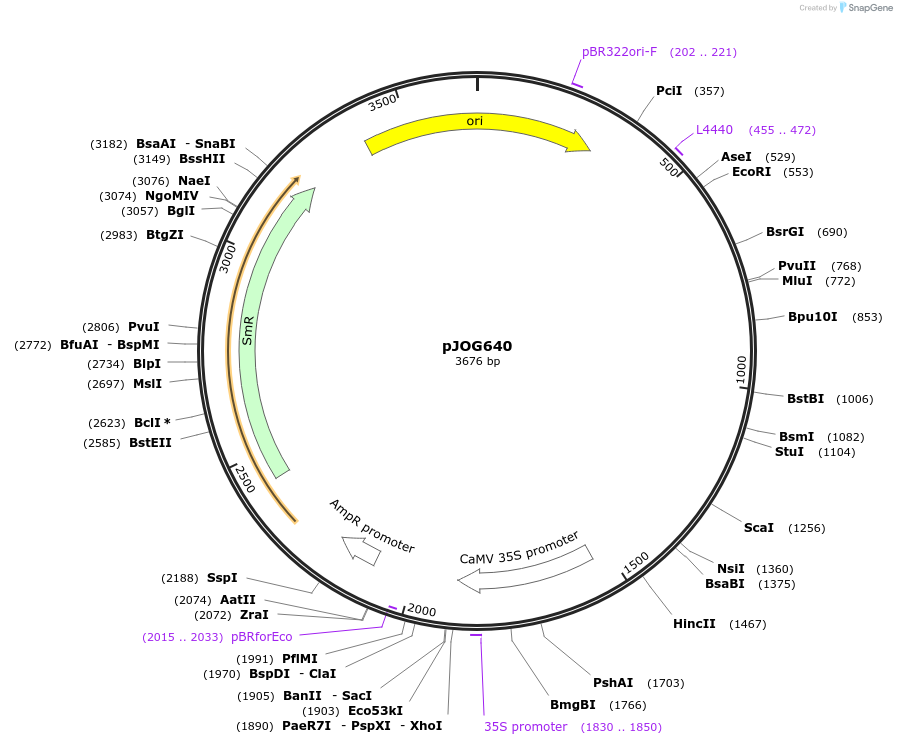
pJOG640
Plasmid#105354PurposeSynthetic biologyDepositorInsertpromotor 35S (1300bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
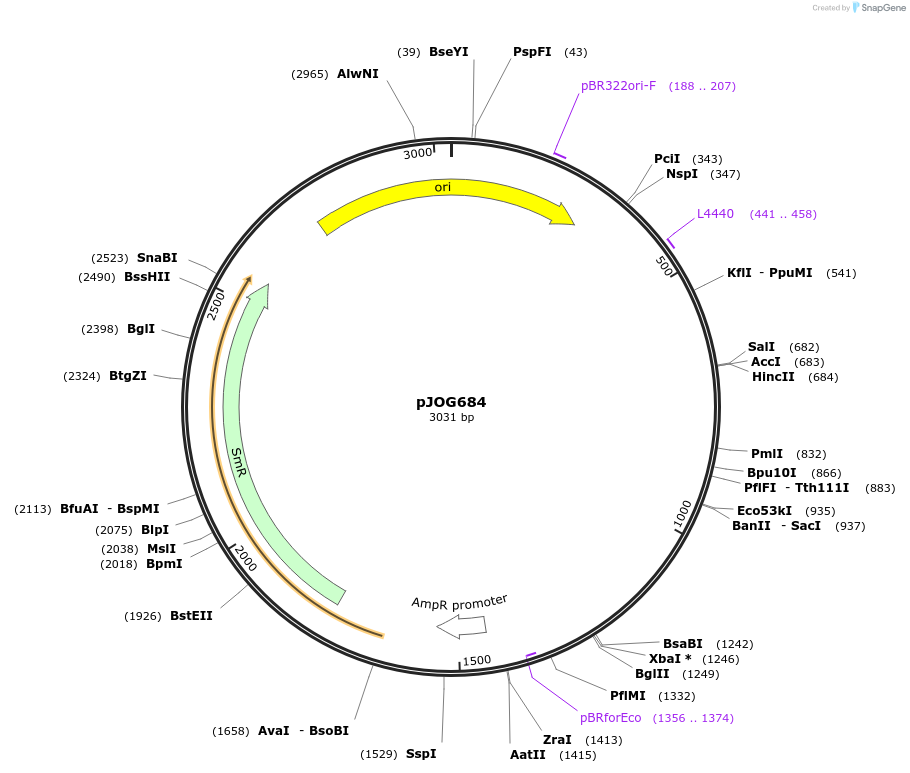
pJOG684
Plasmid#105355PurposeSynthetic biologyDepositorInsertpromotor UBQ10 (At4g05320; 800bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
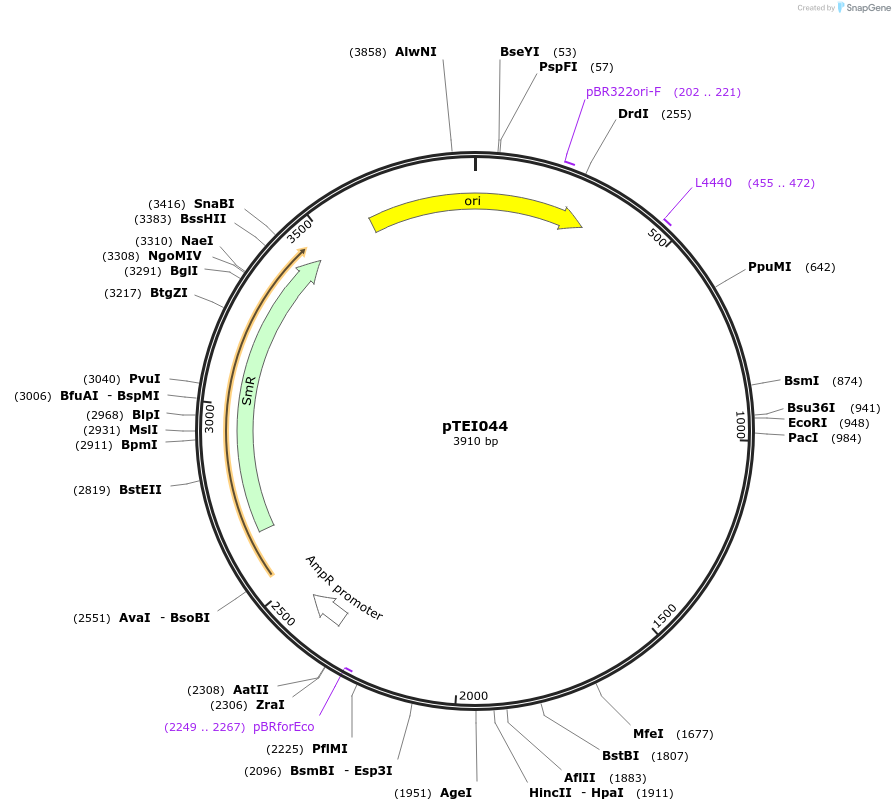
pTEI044
Plasmid#105357PurposeSynthetic biologyDepositorInsertpromotor RPS5a (At3g11940; 1700bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
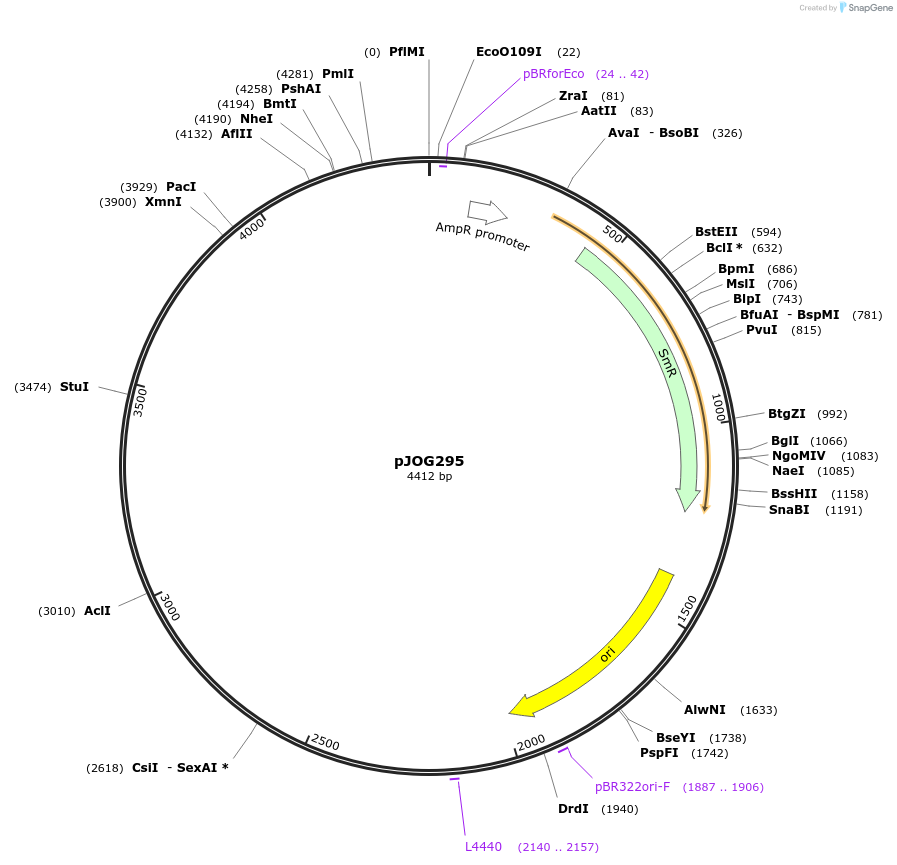
pJOG295
Plasmid#105362PurposeSynthetic biologyDepositorInsertpromotor GILT (At4g12960;2100bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
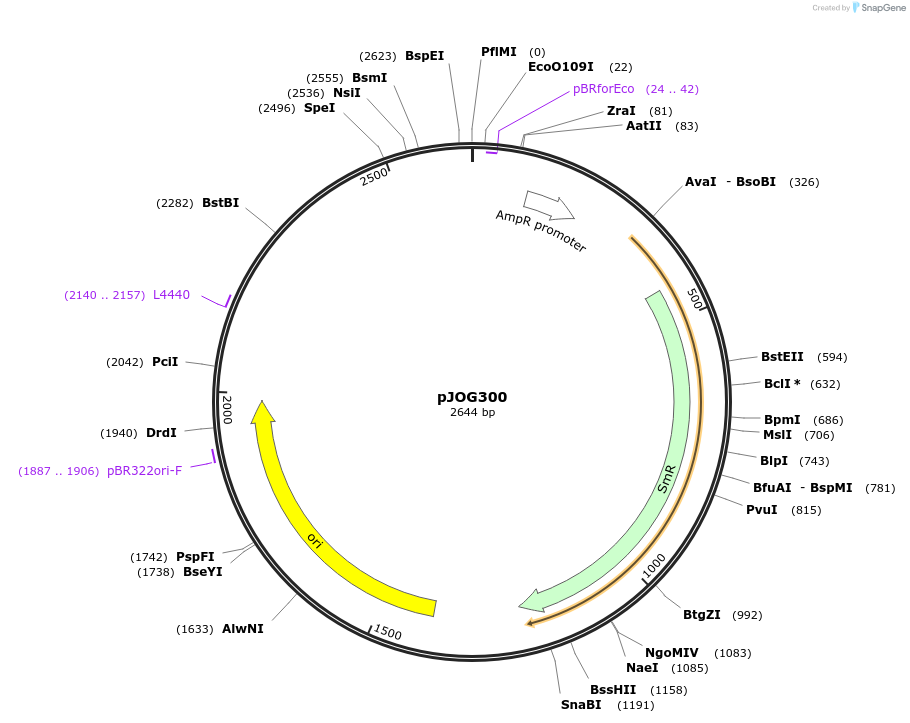
pJOG300
Plasmid#105363PurposeSynthetic biologyDepositorInsertpromotor (At1g66850;350bp)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
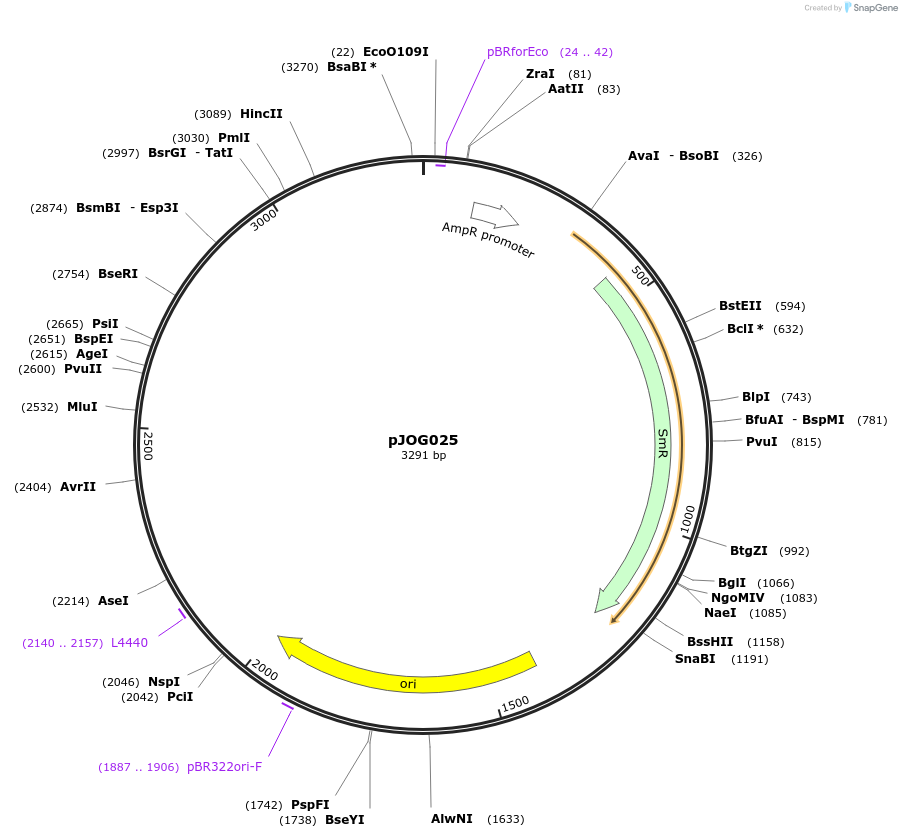
pJOG025
Plasmid#105364PurposeSynthetic biologyDepositorInsertpromotor EDS1 (At3g48090)
UseSynthetic BiologyMutationBsaI/ BpiI restriction sites removedAvailable SinceAug. 1, 2018AvailabilityAcademic Institutions and Nonprofits only -
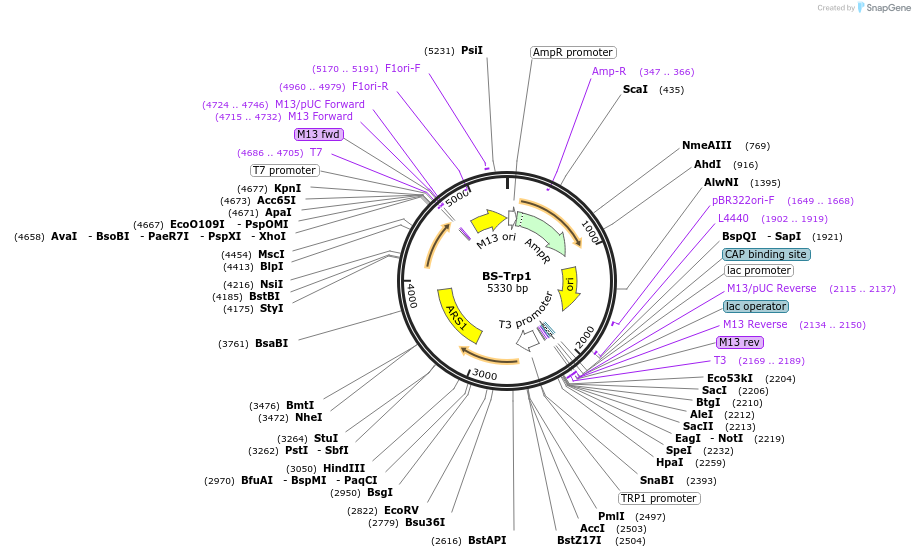
BS-Trp1
Plasmid#69196Purposecomplementation of yeast auxotrophic marker TRP1. Although this is the plasmid described in the paper, the depositor notes that it has a low integration efficiency and Plasmid 69503 should be used.DepositorInsertTRP1
ExpressionYeastAvailable SinceOct. 28, 2015AvailabilityAcademic Institutions and Nonprofits only -
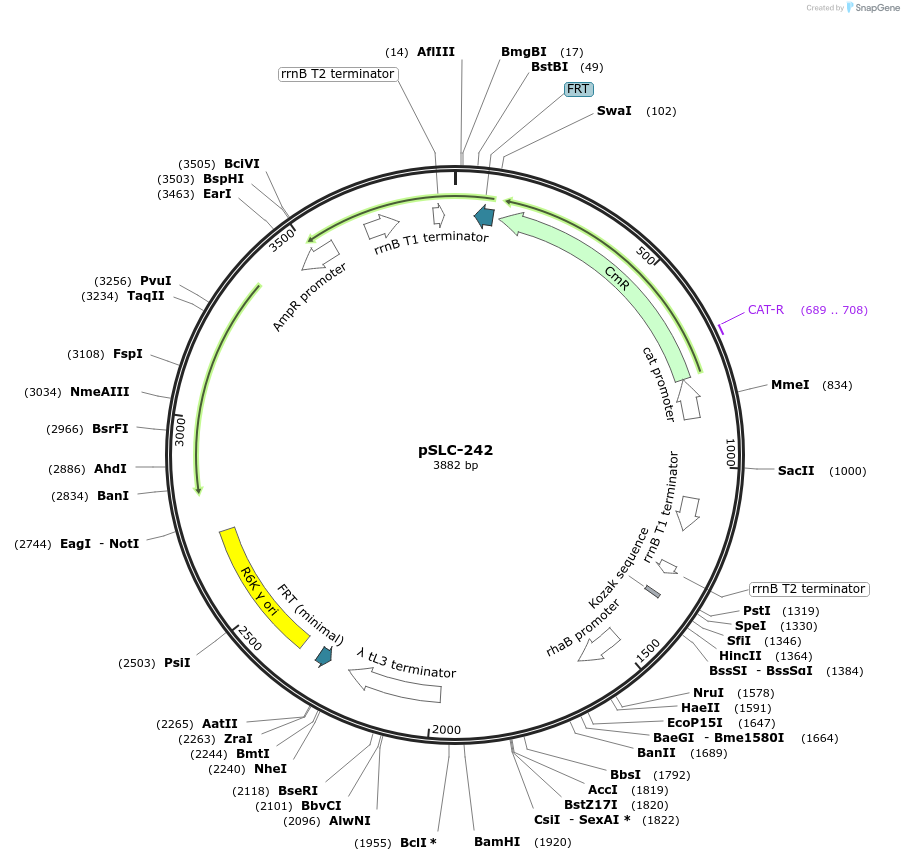
pSLC-242
Plasmid#73189PurposeTemplate plasmid which encodes chloramphenicol resistance gene for positive selection and a toxin gene (relE) under the control of rhamnose induceable promoter (PrhaB) for negative selection.DepositorInsertchloramphenicol (cat Although Ampicillin gene is present, it was inactivated while cloning.)
ExpressionBacterialPromoterchloramphenicol promoterAvailable SinceJuly 19, 2016AvailabilityAcademic Institutions and Nonprofits only -
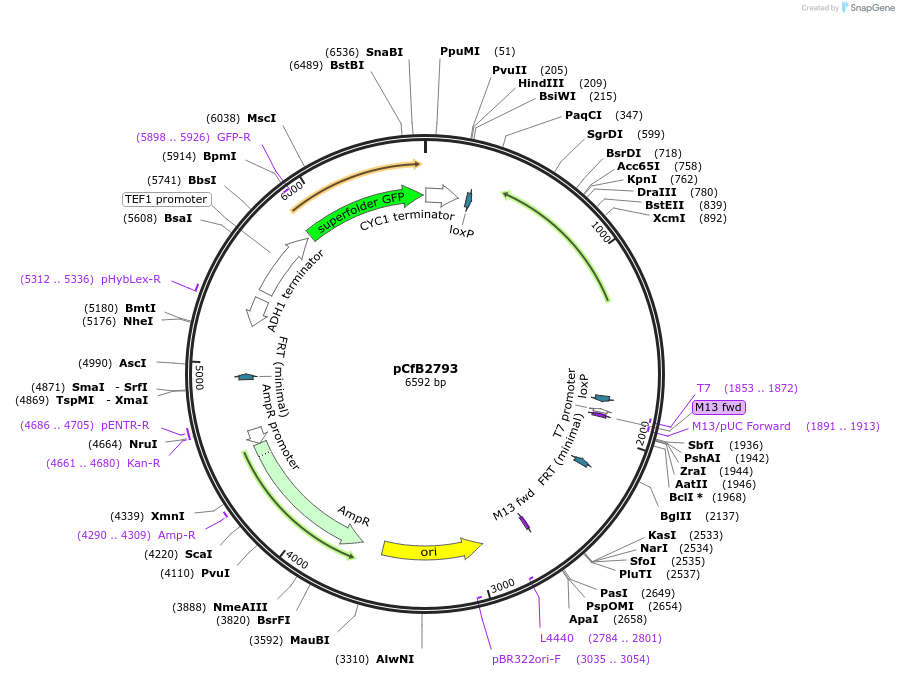
pCfB2793
Plasmid#64043PurposepCfB2793 is a vector for multiple integrations at sites sharing homology with Ty2Cons. The selective marker is Kl.URA3. Additionally it contains a PTEF1-GFP as reporter.DepositorInsertGFP
UseCre/LoxExpressionYeastPromoterTEF1 promoter and noneAvailable SinceAug. 15, 2016AvailabilityAcademic Institutions and Nonprofits only -
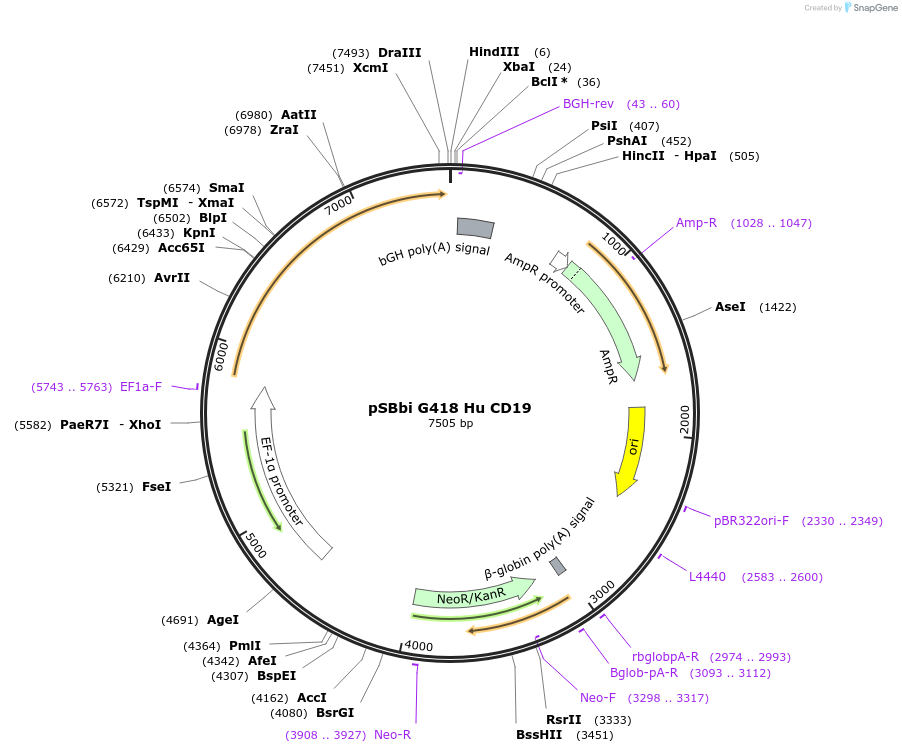
pSBbi G418 Hu CD19
Plasmid#183247PurposeExpression of human CD19 antigen in a Sleeping Beauty transposon vectorDepositorInsertCD19 (CD19 Human)
ExpressionMammalianAvailable SinceOct. 13, 2022AvailabilityAcademic Institutions and Nonprofits only -
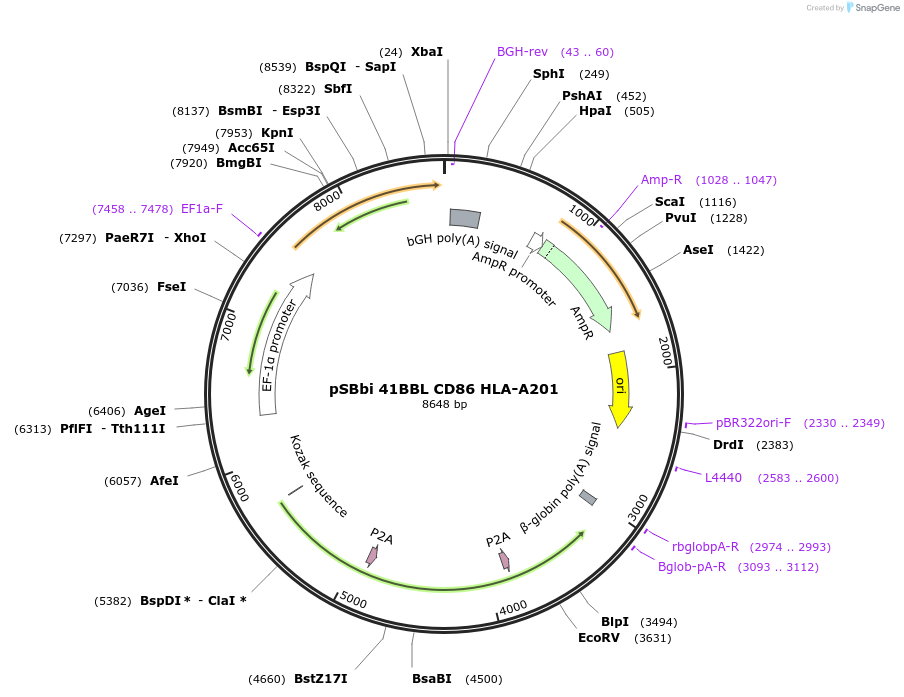
pSBbi 41BBL CD86 HLA-A201
Plasmid#183248PurposeExpression of human 41BBL, CD86 and HLA-A201 in a Sleeping Beauty transposon vector for use in artificial antigen presenting cellsDepositorInsertsExpressionMammalianAvailable SinceAug. 18, 2022AvailabilityAcademic Institutions and Nonprofits only -
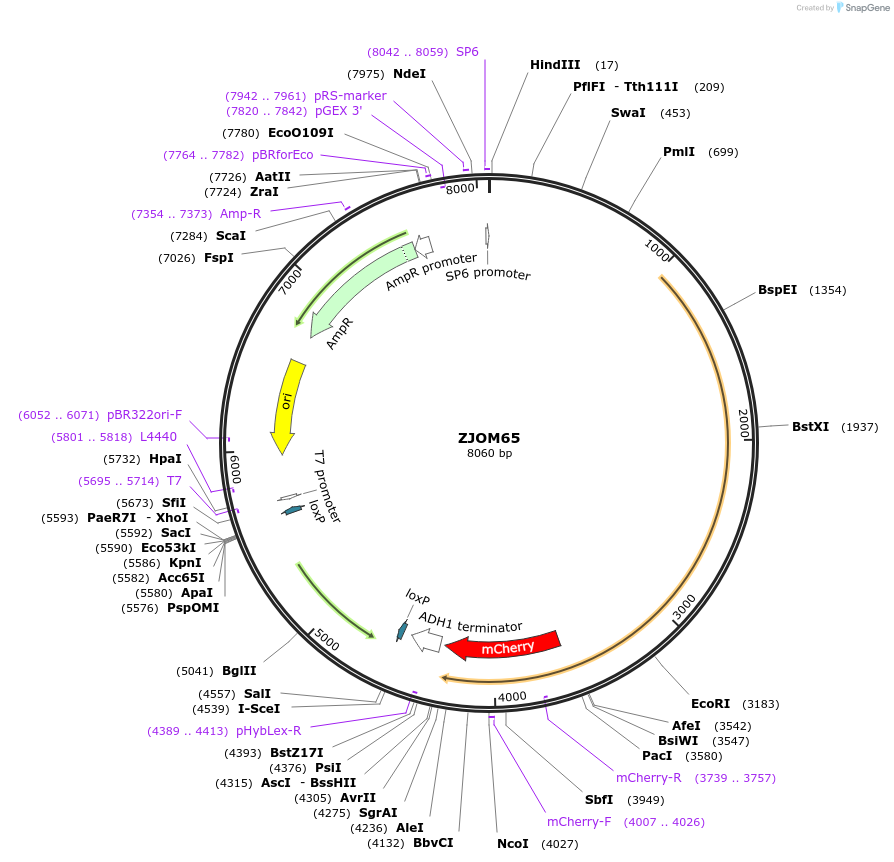
ZJOM65
Plasmid#133654PurposeIntegration of VPH1-mCherry as an additional copy in genome, use auxotrophic marker TRP1(Kluyveromyces lactis). Marker for yeast vacuole.DepositorAvailable SinceJan. 3, 2020AvailabilityAcademic Institutions and Nonprofits only