We narrowed to 4,220 results for: PRS
-
Plasmid#8855DepositorAvailable SinceAug. 3, 2005AvailabilityAcademic Institutions and Nonprofits only
-
pWZ414-F14
Plasmid#23063DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 deleted amino acids 3-29Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
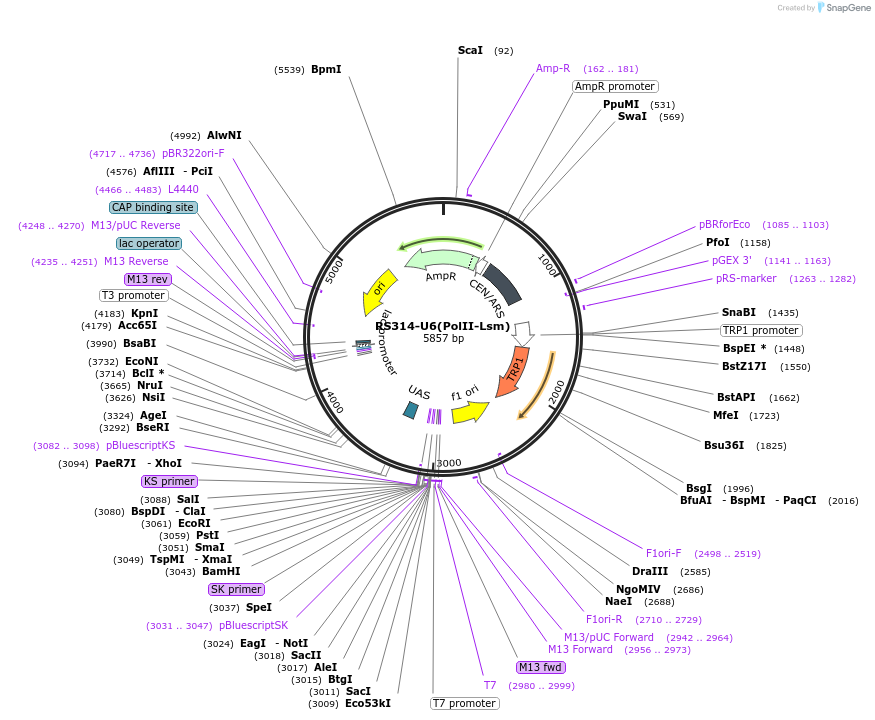
RS314-U6(PolII-Lsm)
Plasmid#200772PurposePol II expression of U6 LsmDepositorInsertGal-SNR6-II
ExpressionBacterial and YeastAvailable SinceMay 16, 2023AvailabilityAcademic Institutions and Nonprofits only -
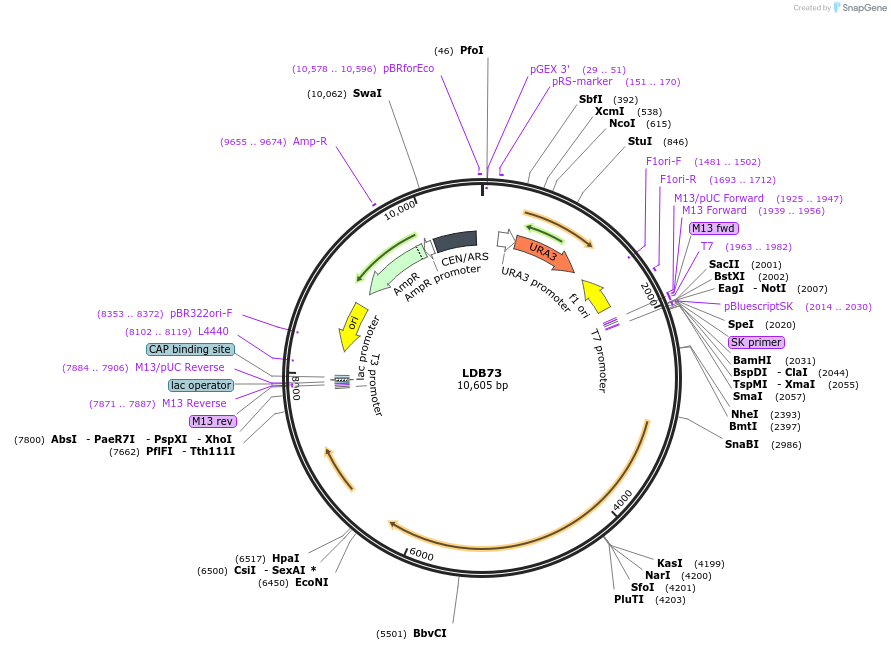
LDB73
Plasmid#185437PurposeNUP1 CEN URA3 vector for synthetic lethal sectoring assays and screeningDepositorInsertNUP1
ExpressionYeastAvailable SinceSept. 30, 2022AvailabilityAcademic Institutions and Nonprofits only -
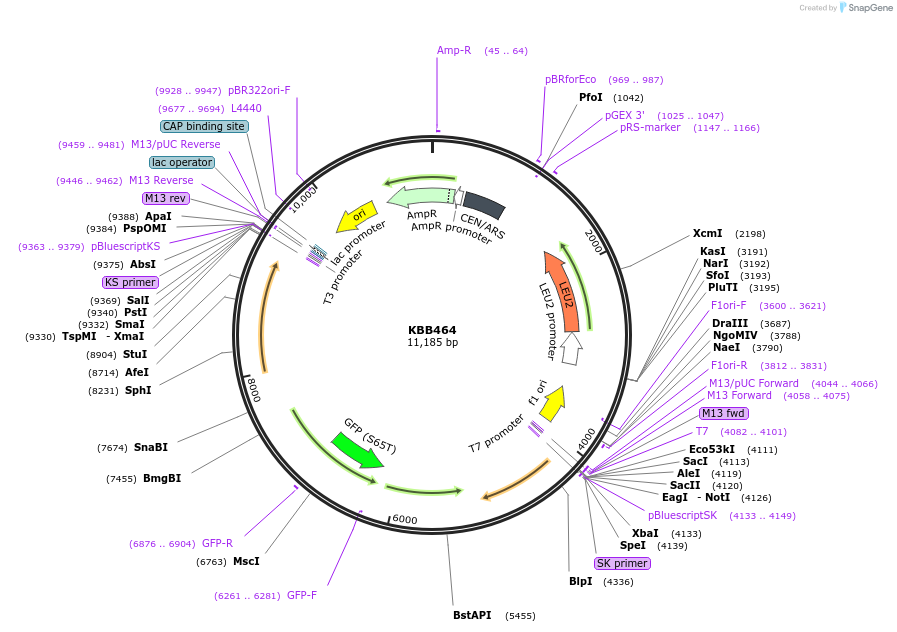
KBB464
Plasmid#185097PurposeHA-tagged POM152 with glycosylation sites mutated and with GFP at C-terminusDepositorInsertPOM152
TagsGFPExpressionYeastMutationPom152DGlyc-GFPAvailable SinceAug. 11, 2022AvailabilityAcademic Institutions and Nonprofits only -
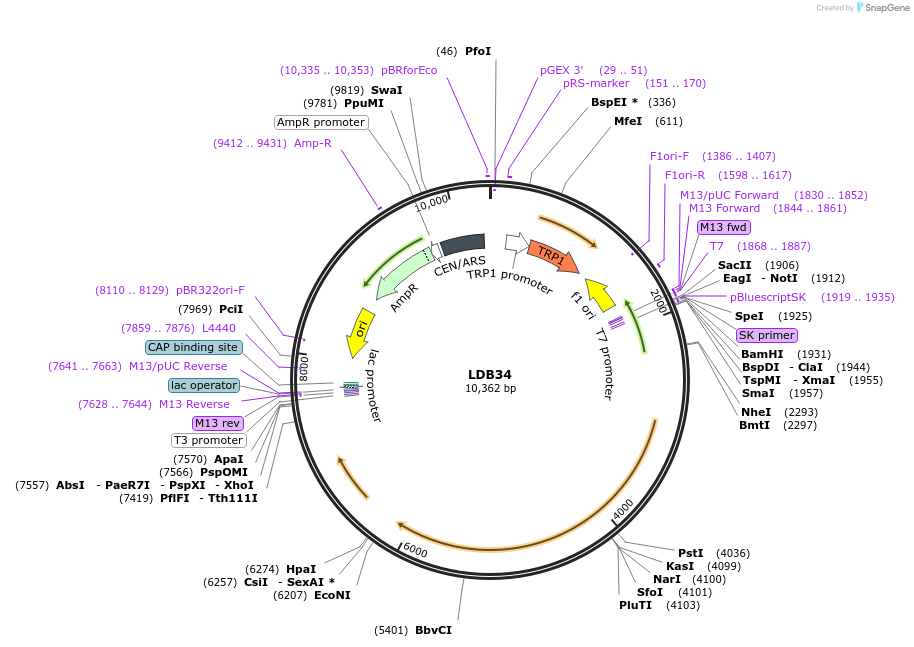
LDB34
Plasmid#185431PurposeYeast expression of Nup1 C-terminal truncation (nup1-21 allele)DepositorInsertNUP1
ExpressionYeastMutationTruncation at amino acid 1041 from the C-term of …Available SinceJuly 25, 2022AvailabilityAcademic Institutions and Nonprofits only -
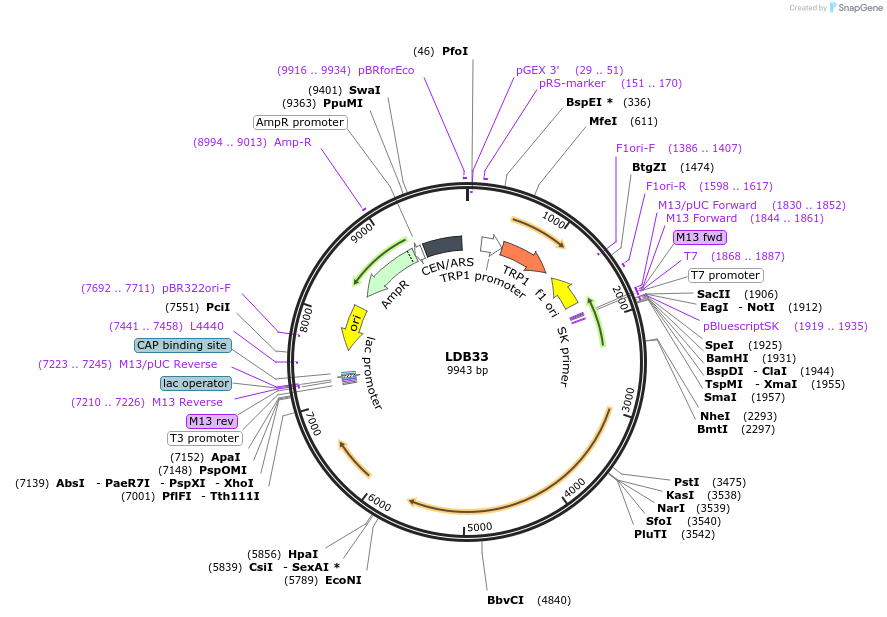
LDB33
Plasmid#185430PurposeYeast expression of Nup1 N-terminal truncation (nup1-15 allele)DepositorInsertNUP1
ExpressionYeastMutationTruncation of amino acids 3-191 from the N-term o…Available SinceJuly 21, 2022AvailabilityAcademic Institutions and Nonprofits only -
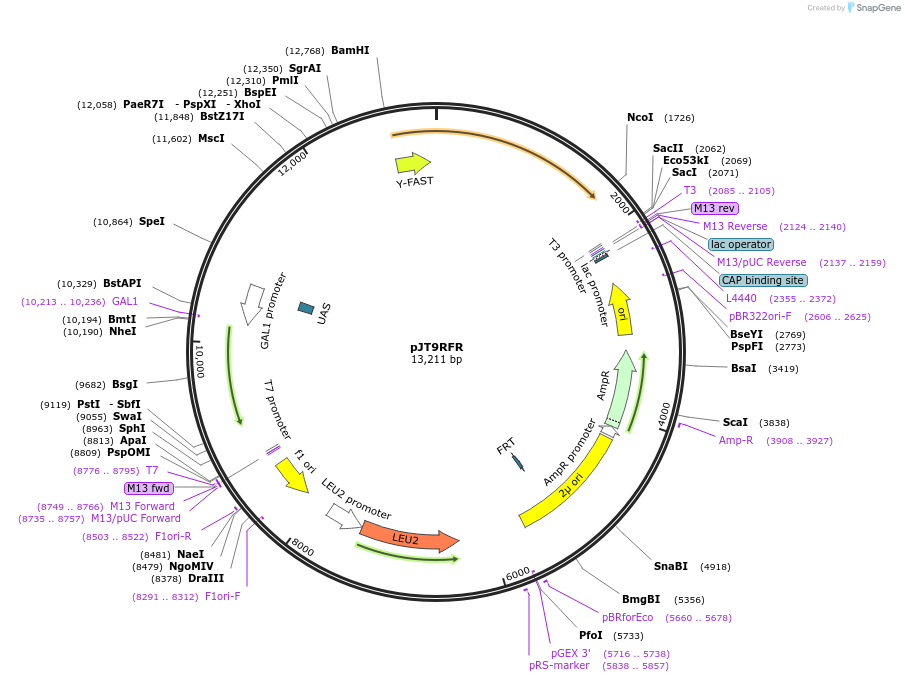
pJT9RFR
Plasmid#165067PurposeIntroducing GAL promoters-controlled sequiterpene nerolidol-producing module and Y-FAST and in S. cerevisiaeDepositorInsertT-RPL3>ERG20>P-GAL1-P-GAL2>Y-FAST>ErB1.2A-AcNES1>T-RPL41B
ExpressionYeastMutationWTAvailable SinceMarch 22, 2021AvailabilityAcademic Institutions and Nonprofits only -
pVG50
Plasmid#164708PurposeyEVenus under the control of tetOpr with ADH1t upstream of the reporterDepositorInsertADH1t - tetOpr - yEVenus - CLN2 PEST - ADH1t
ExpressionYeastAvailable SinceMarch 10, 2021AvailabilityAcademic Institutions and Nonprofits only -
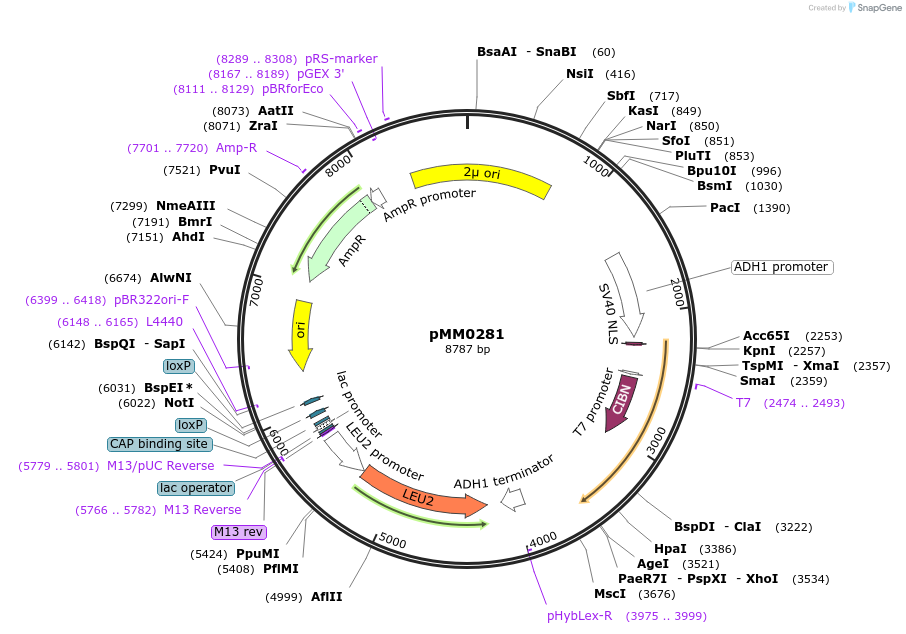
pMM0281
Plasmid#128972PurposeEncodes VP16-CIB1 under pADH1 promoterDepositorInsertpscADH1-SV40NLS-VP16-CIB1-tscADH1
ExpressionYeastAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
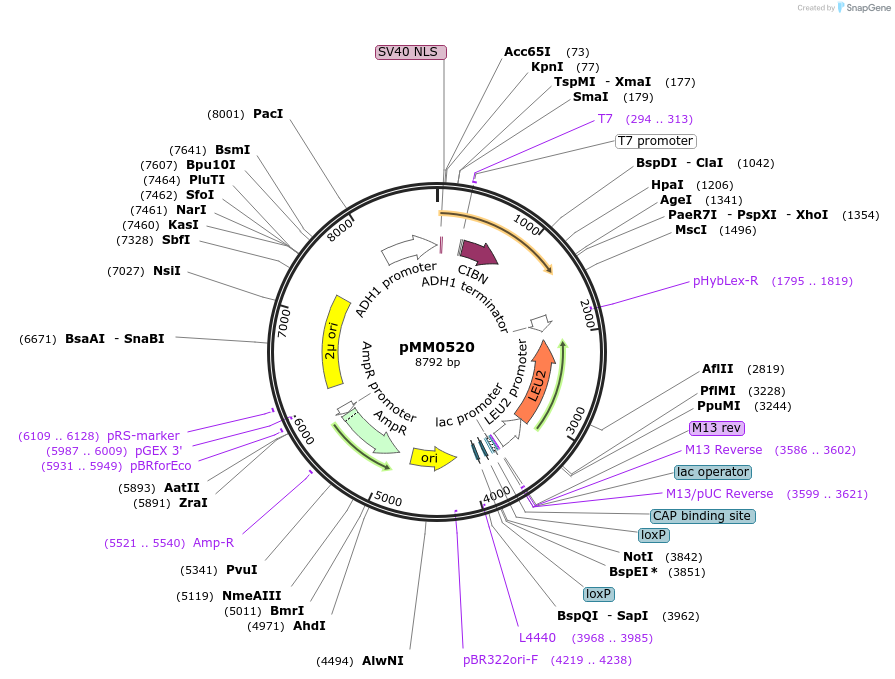
pMM0520
Plasmid#128983PurposeOverexpression plasmid for VP16-CIB1DepositorInsertSV40NLS-VP16-CIB1 LEU2 2_
ExpressionYeastAvailable SinceOct. 28, 2019AvailabilityAcademic Institutions and Nonprofits only -
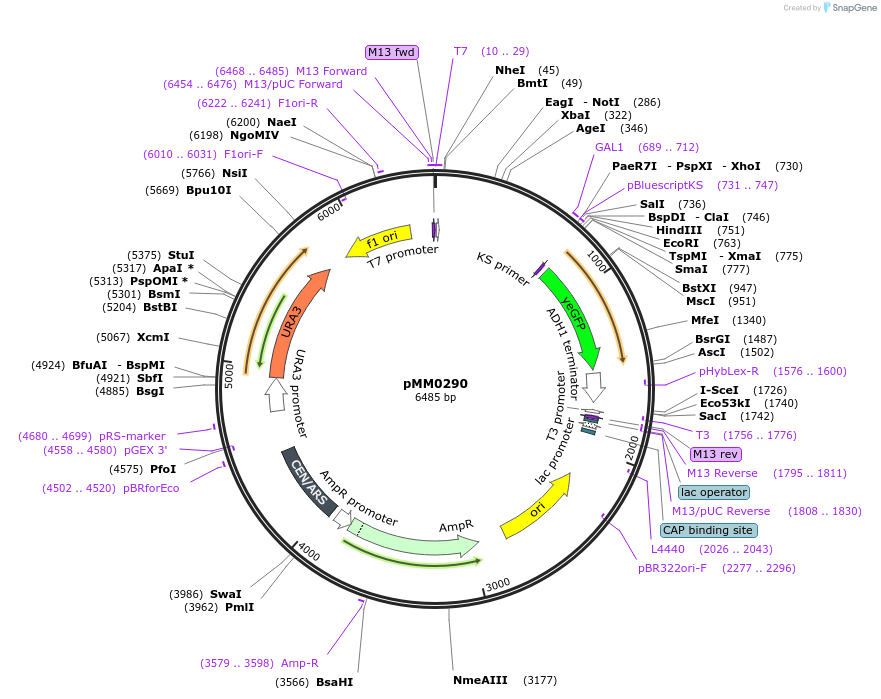
pMM0290
Plasmid#128978PurposeReporter plasmid, pZF(3BSopp)-yEVENUSDepositorInsertpZF(3BSop)-yEVENUS
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
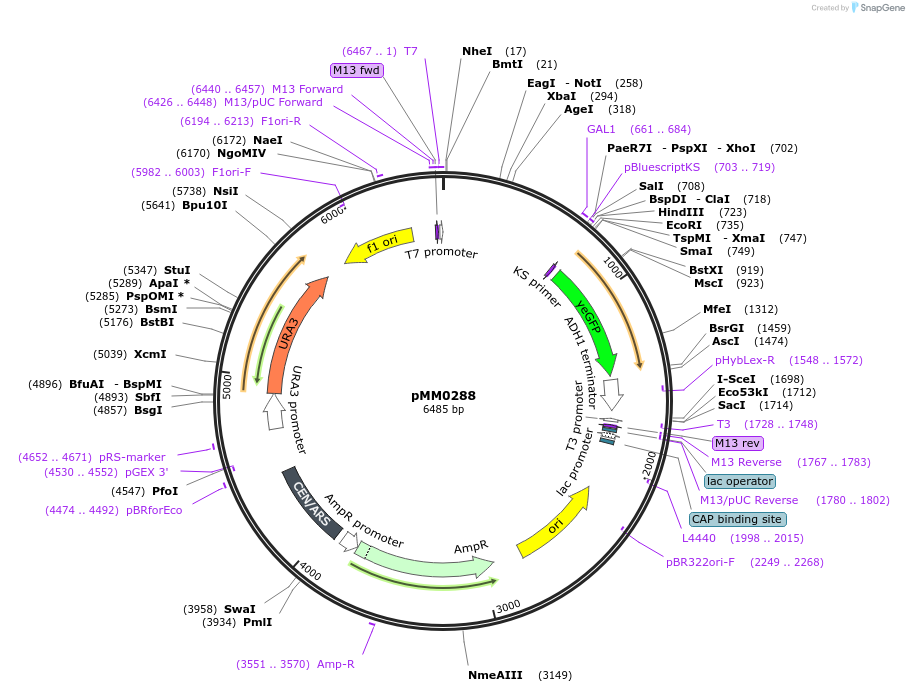
pMM0288
Plasmid#128976PurposeReporter plasmid, pZF(3BSc)-yEVENUSDepositorInsertpZF(3BSc)-yEVENUS
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
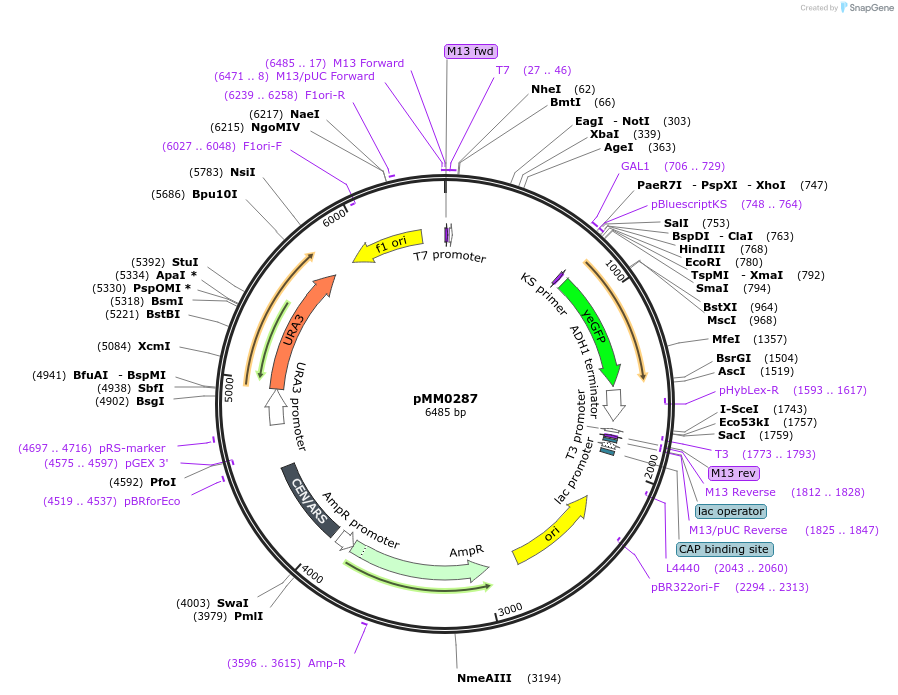
pMM0287
Plasmid#128975PurposeReporter plasmid, pZF(3BS)-yEVENUSDepositorInsertpZF(3BS)-yEVENUS
ExpressionYeastAvailable SinceOct. 16, 2019AvailabilityAcademic Institutions and Nonprofits only -
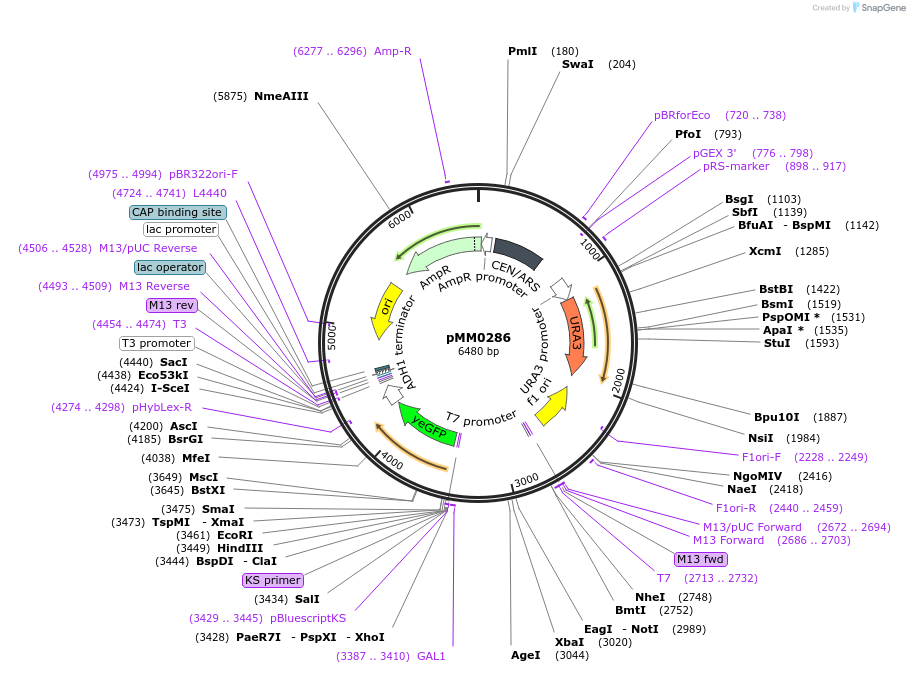
pMM0286
Plasmid#128974PurposeReporter plasmid, pZF(3oBS)-yEVENUSDepositorInsertpZF(3oBS)-yEVENUS
ExpressionYeastAvailable SinceOct. 9, 2019AvailabilityAcademic Institutions and Nonprofits only -
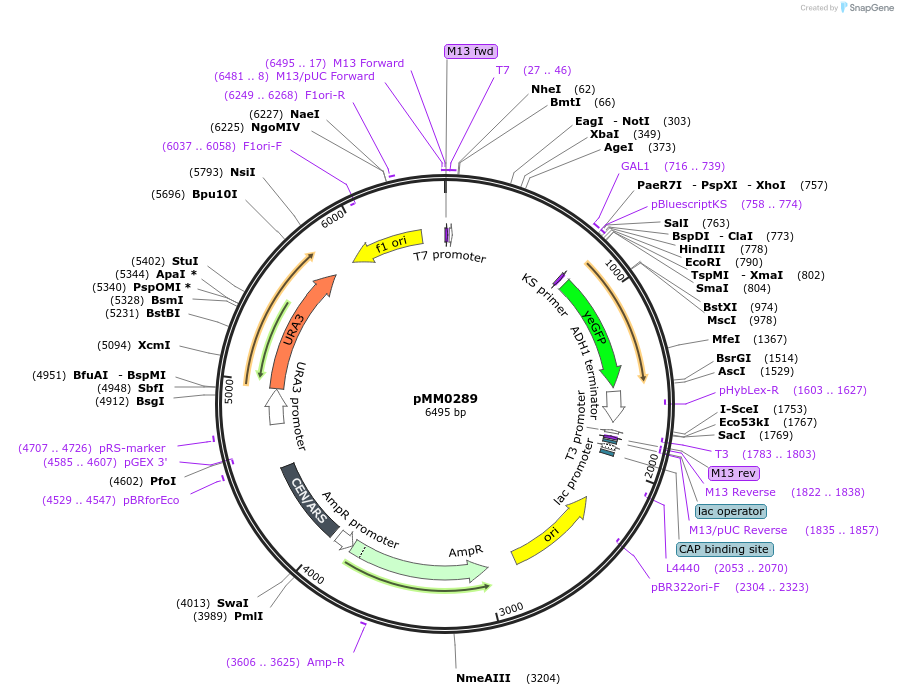
pMM0289
Plasmid#128977PurposeReporter plasmid, pZF(4BSc)-yEVENUSDepositorInsertpZF(4BSc)-yEVENUS
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
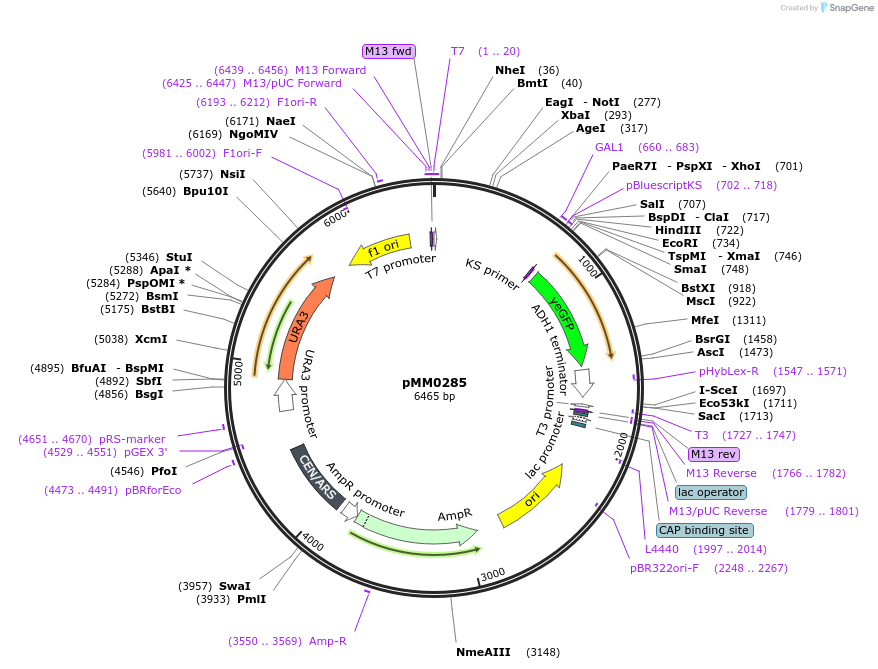
pMM0285
Plasmid#128973PurposeReporter plasmid, pZF(1BS)-yEVENUSDepositorInsertpZF(1BS)-yEVENUS
ExpressionYeastAvailable SinceOct. 7, 2019AvailabilityAcademic Institutions and Nonprofits only -
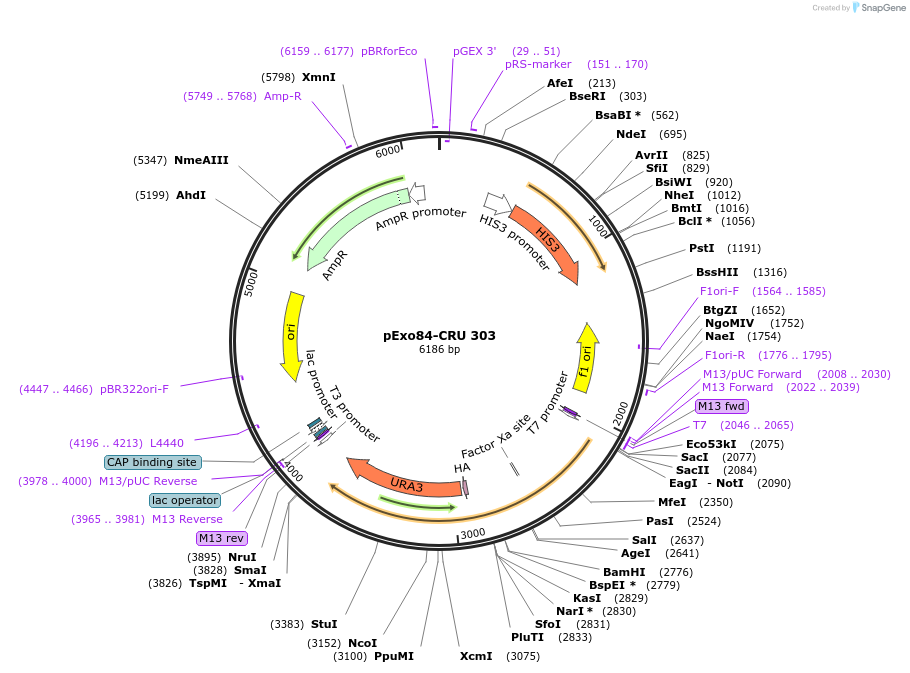
pExo84-CRU 303
Plasmid#131165PurposeFor C-terminal genomic integration of Cub-R-URA3, HIS3 complements the histidine auxotrophy of S. cerevisiae his mutantDepositorInsertScExo84
TagsCRUExpressionYeastAvailable SinceSept. 5, 2019AvailabilityAcademic Institutions and Nonprofits only -
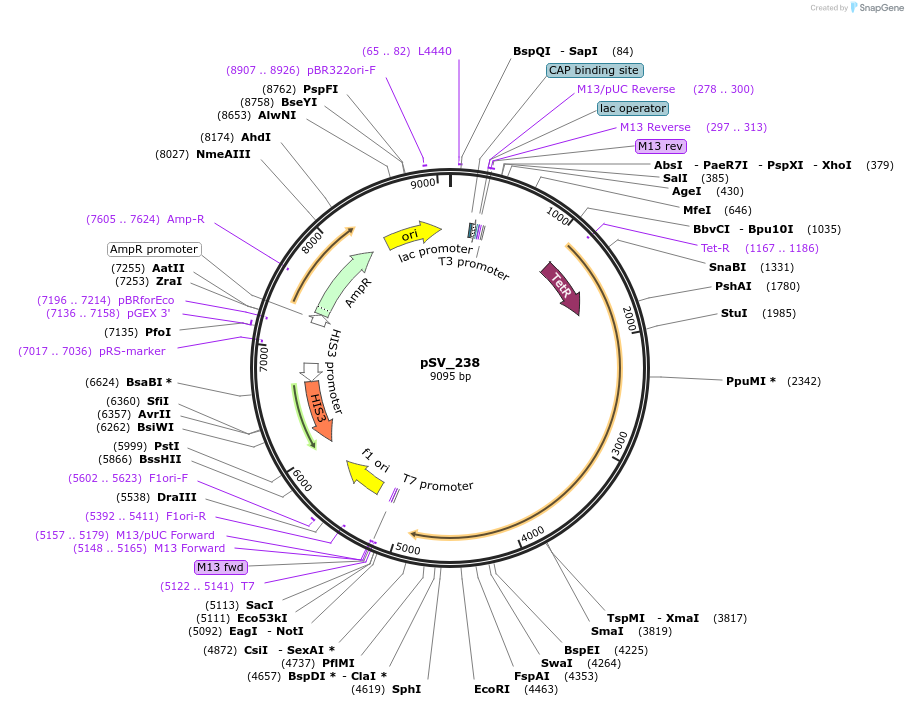
pSV_238
Plasmid#122941PurposeExpression of the repressor to control the insertional promoter.DepositorInsertrTetR.SUM1
ExpressionYeastPromoterPret2Available SinceApril 11, 2019AvailabilityAcademic Institutions and Nonprofits only -
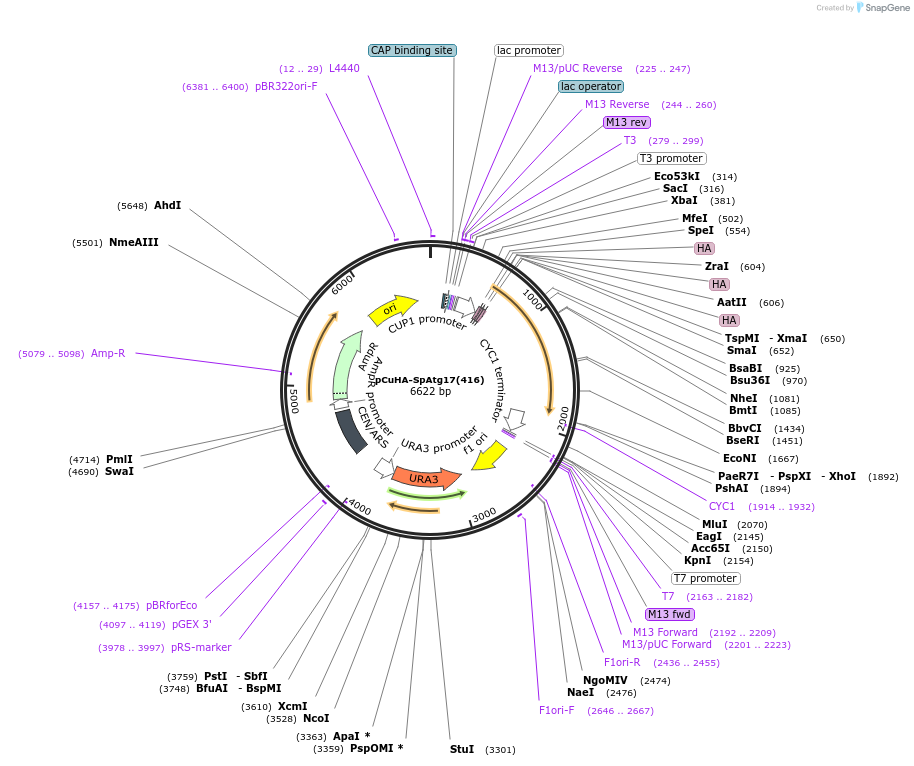
pCuHA-SpAtg17(416)
Plasmid#122079PurposeExpressing S. pombe HA-Atg17DepositorAvailable SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
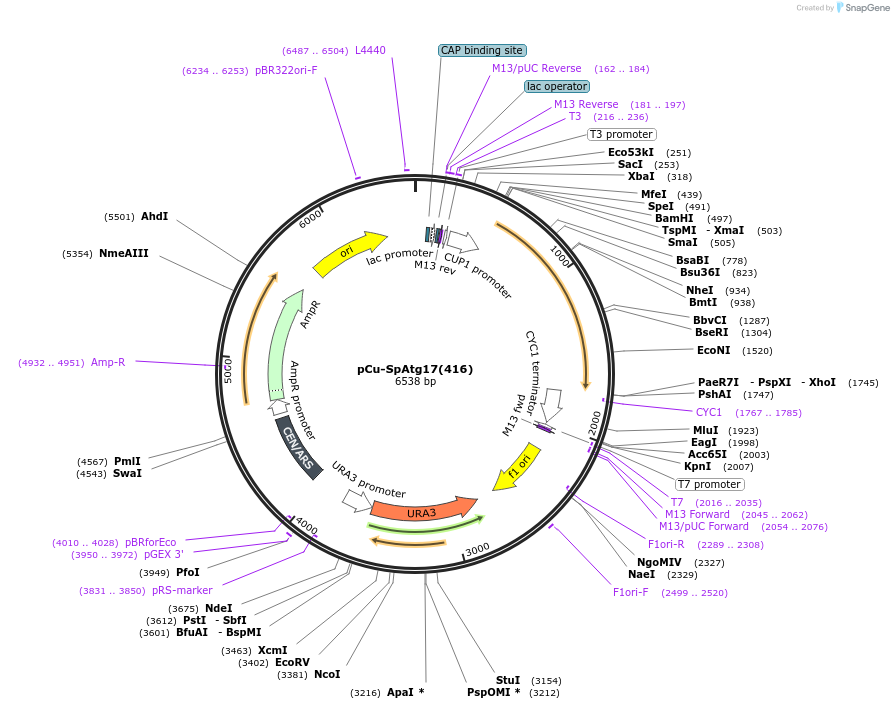
pCu-SpAtg17(416)
Plasmid#122078PurposeExpressing S. pombe Atg17DepositorInsertSpATG17 (atg17 Fission Yeast)
ExpressionYeastAvailable SinceApril 2, 2019AvailabilityAcademic Institutions and Nonprofits only -
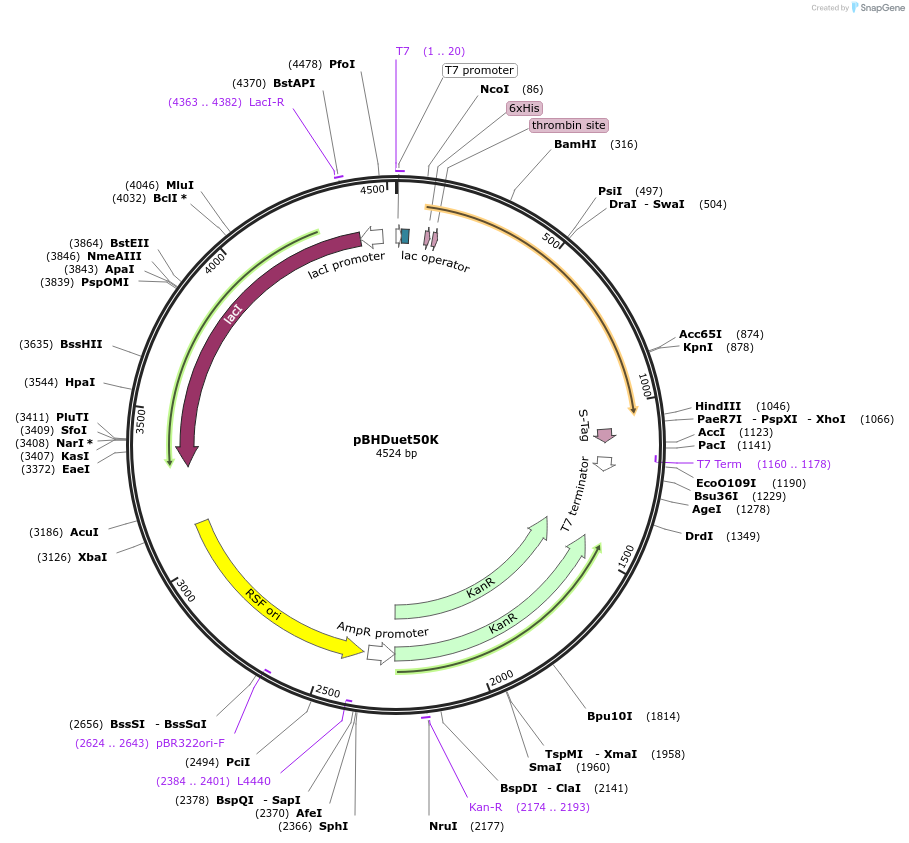
pBHDuet50K
Plasmid#118399Purposeanalysis of cis-splicing by mini inteinsDepositorInsertMvuTFIIB mini intein
ExpressionBacterialMutationincorporation of oligonucleotide during cloningAvailable SinceDec. 3, 2018AvailabilityAcademic Institutions and Nonprofits only -
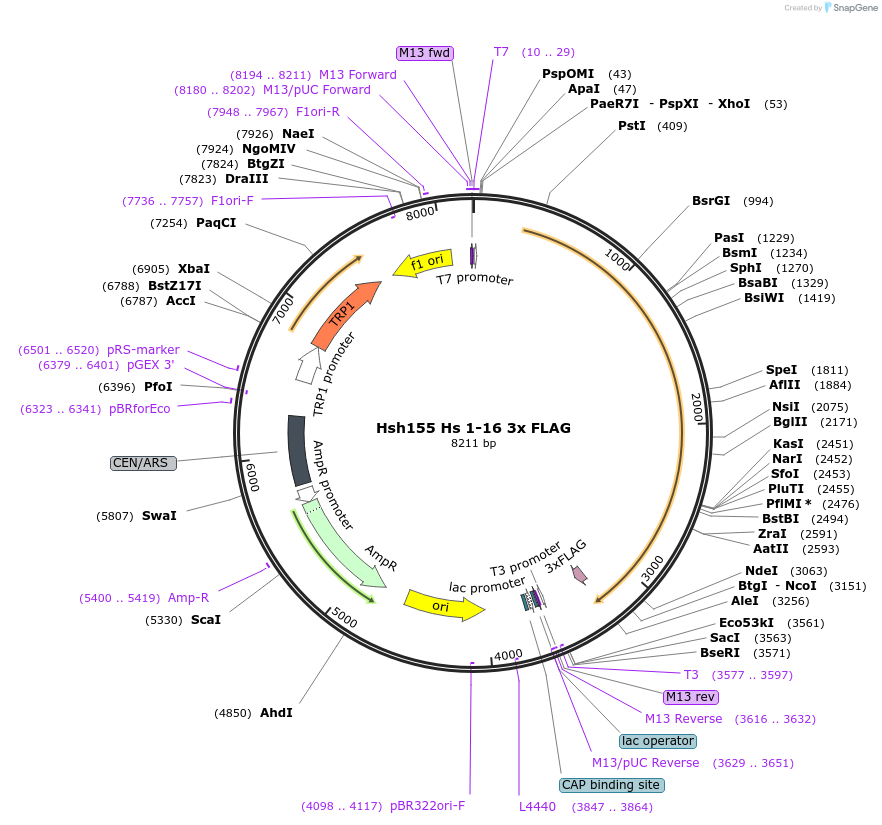
Hsh155 Hs 1-16 3x FLAG
Plasmid#111966PurposeHuman HEATs 1-16 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 1-16 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 1-16 insertedAvailable SinceSept. 11, 2018AvailabilityAcademic Institutions and Nonprofits only -
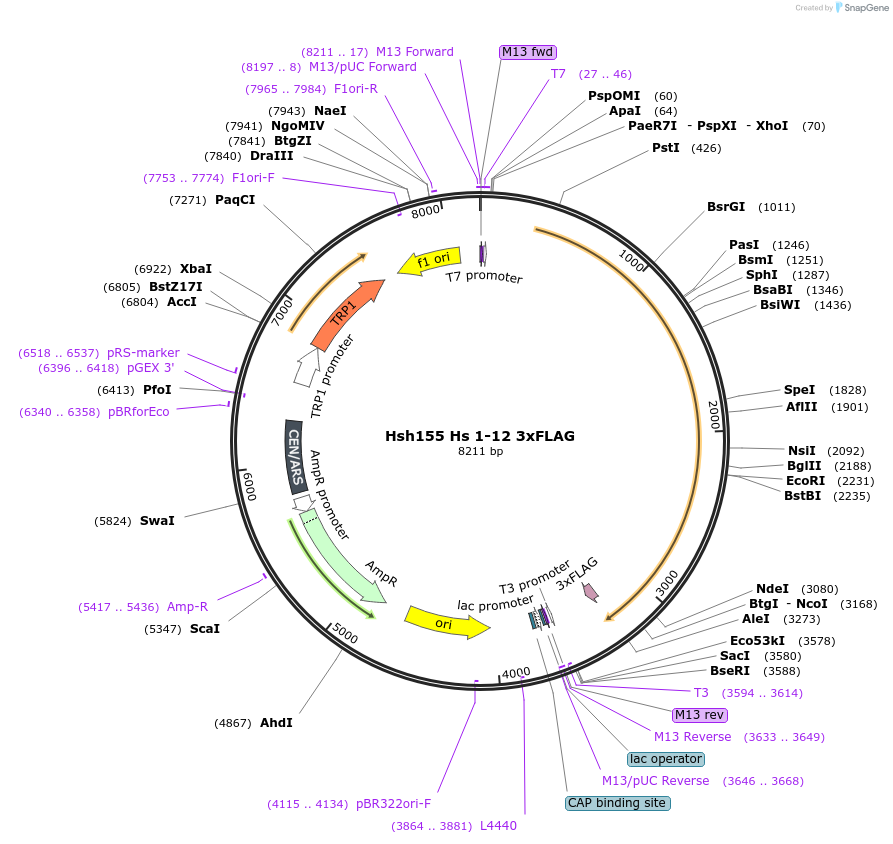
Hsh155 Hs 1-12 3xFLAG
Plasmid#111965PurposeHuman HEATs 1-12 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 1-12 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 1-12 insertedAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
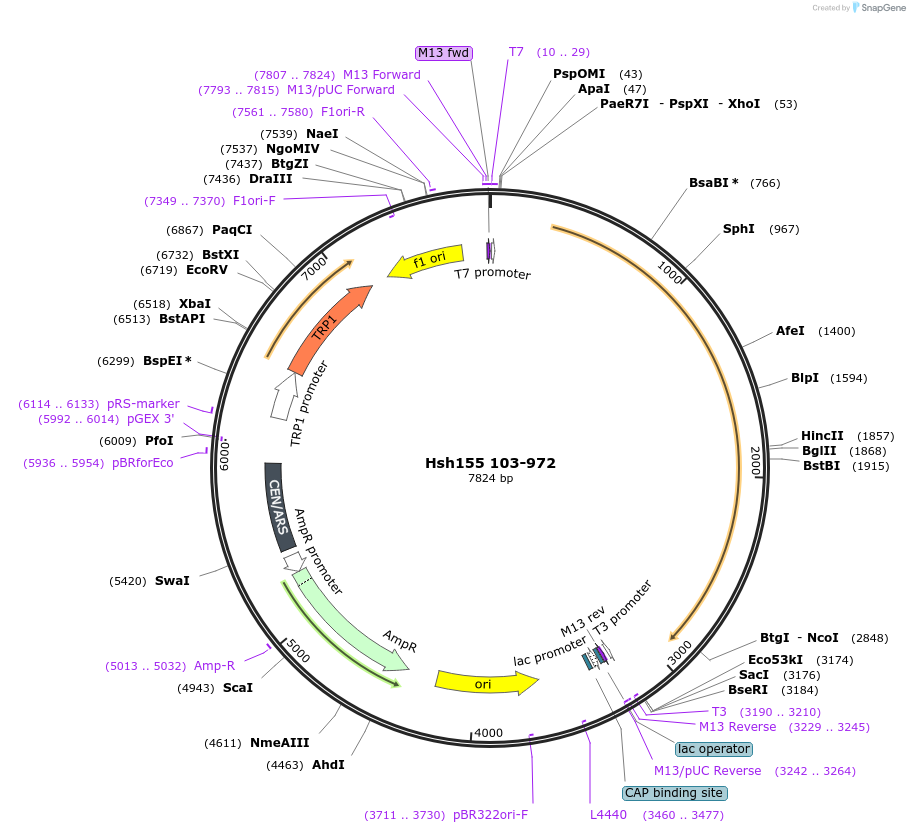
Hsh155 103-972
Plasmid#111989PurposeTruncated Hsh155 (aa 103-972)DepositorInsertHsh155 103-972
ExpressionBacterial and YeastMutation103-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
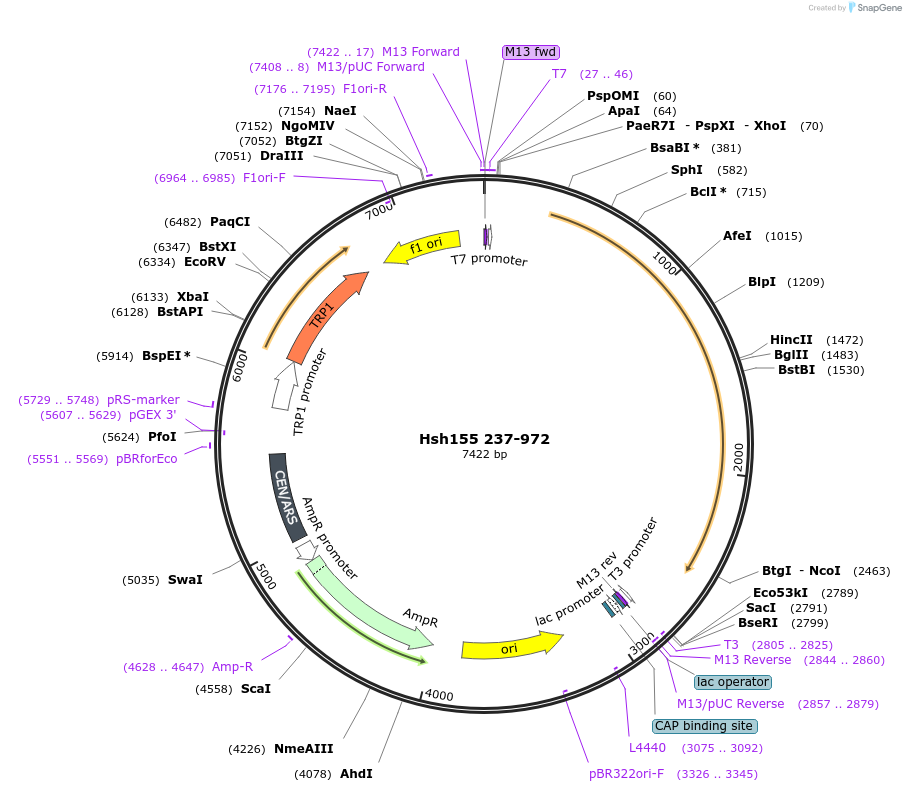
Hsh155 237-972
Plasmid#111992PurposeTruncated Hsh155 (aa 237-972)DepositorInsertHsh155 237-972
ExpressionBacterial and YeastMutation237-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
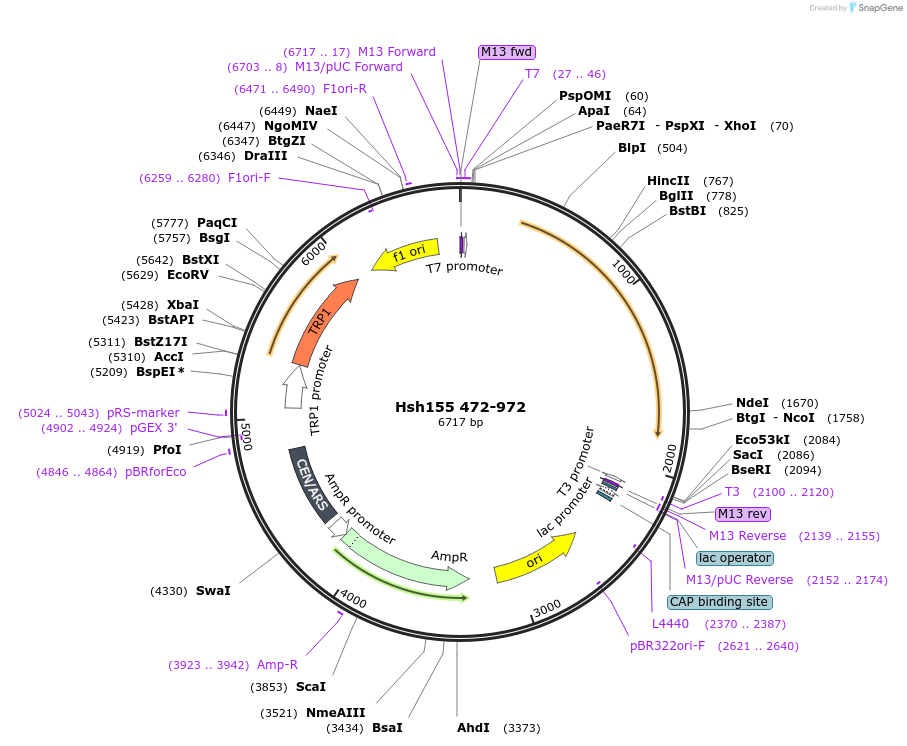
Hsh155 472-972
Plasmid#111996PurposeTruncated Hsh155 (aa 472-972)DepositorInsertHsh155 472-972
ExpressionBacterial and YeastMutation472-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
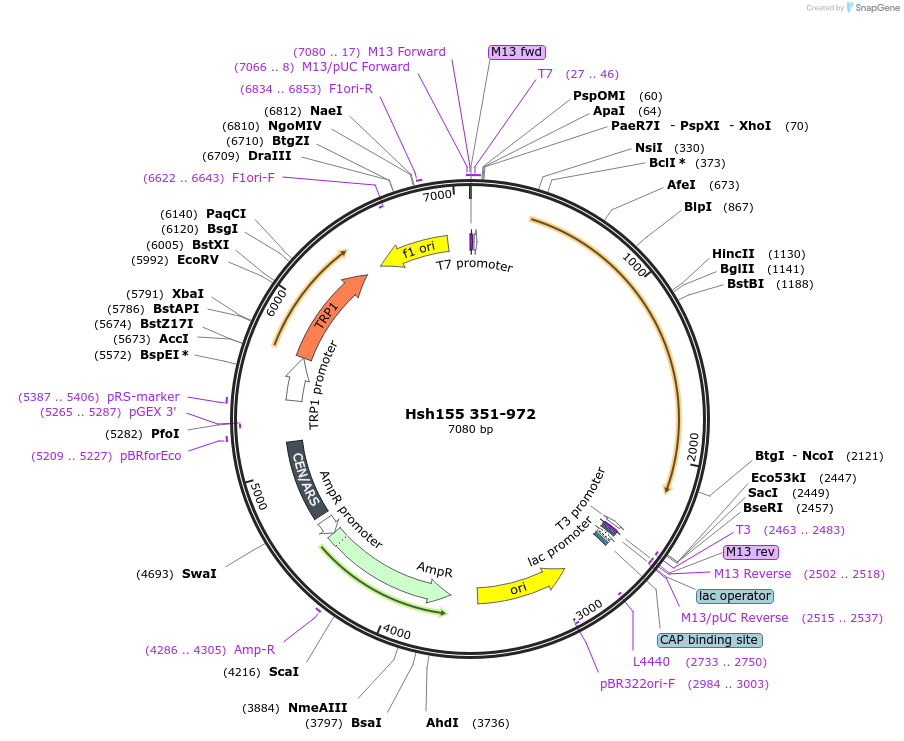
Hsh155 351-972
Plasmid#111995PurposeTruncated Hsh155 (aa 351-972)DepositorInsertHsh155 351-972
ExpressionBacterial and YeastMutation351-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
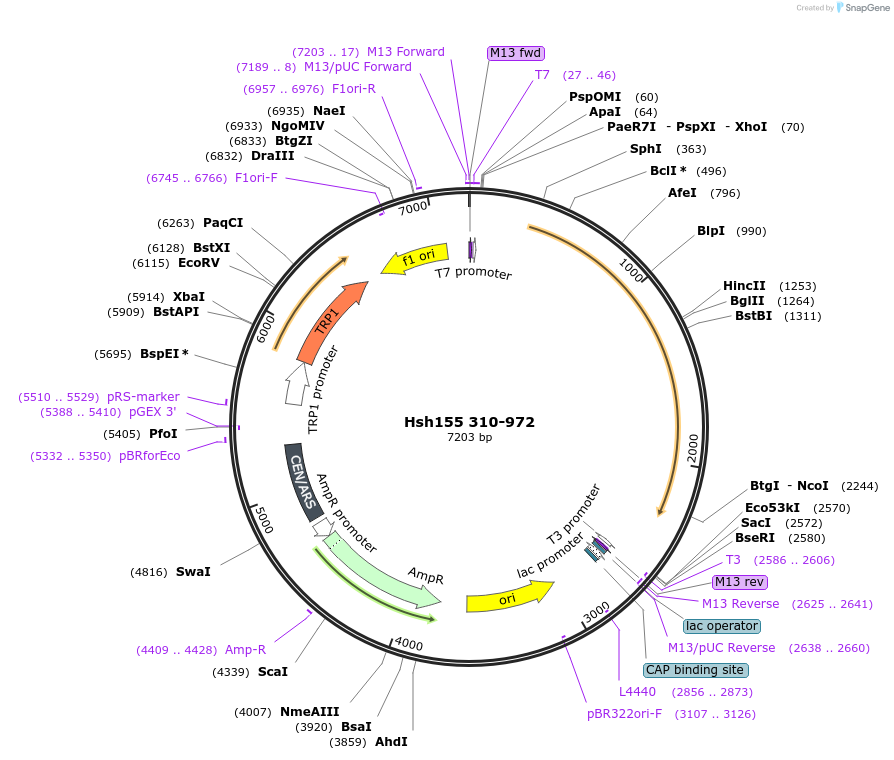
Hsh155 310-972
Plasmid#111994PurposeTruncated Hsh155 (aa 310-972)DepositorInsertHsh155 310-972
ExpressionBacterial and YeastMutation310-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
Hsh155 275-972
Plasmid#111993PurposeTruncated Hsh155 (aa 275-972)DepositorInsertHsh155 275-972
ExpressionBacterial and YeastMutation275-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
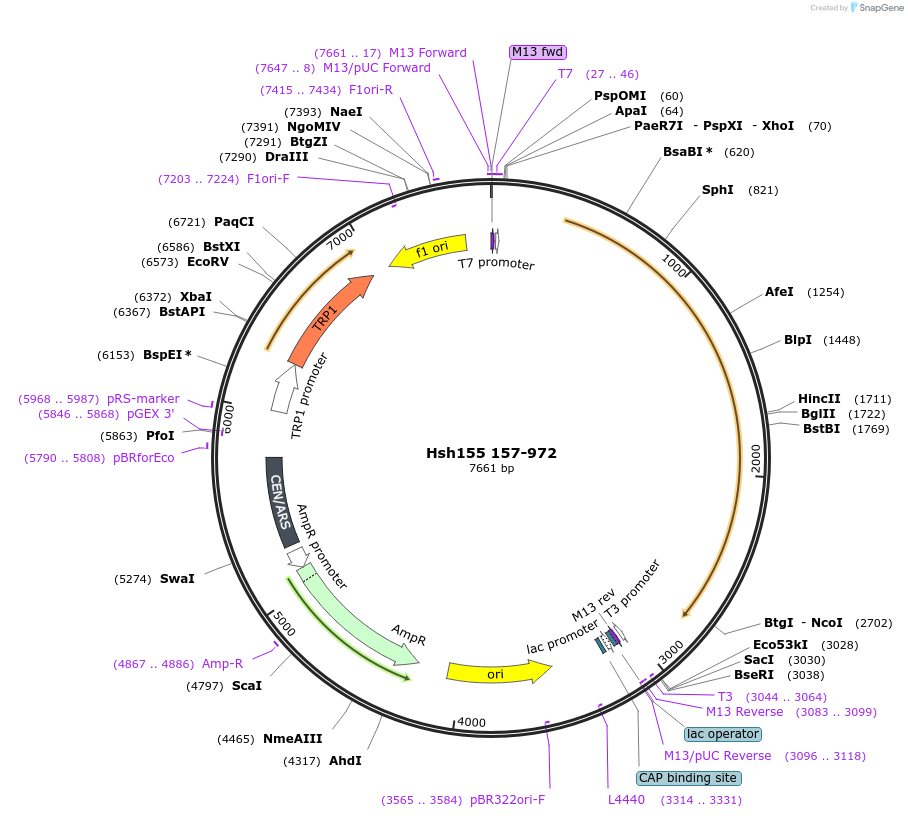
Hsh155 157-972
Plasmid#111990PurposeTruncated Hsh155 (aa 157-972)DepositorInsertHsh155 157-972
ExpressionBacterial and YeastMutation157-972 truncationAvailable SinceJune 22, 2018AvailabilityAcademic Institutions and Nonprofits only -
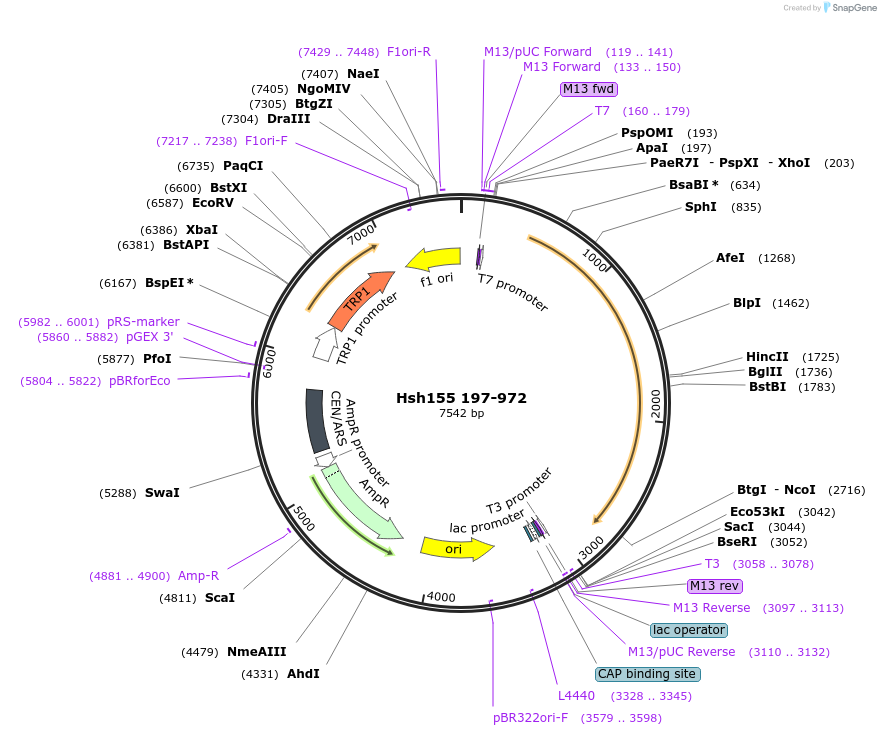
Hsh155 197-972
Plasmid#111991PurposeTruncated Hsh155 (aa 197-972)DepositorInsertHsh155 197-972
ExpressionBacterial and YeastMutation197-972 truncationAvailable SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
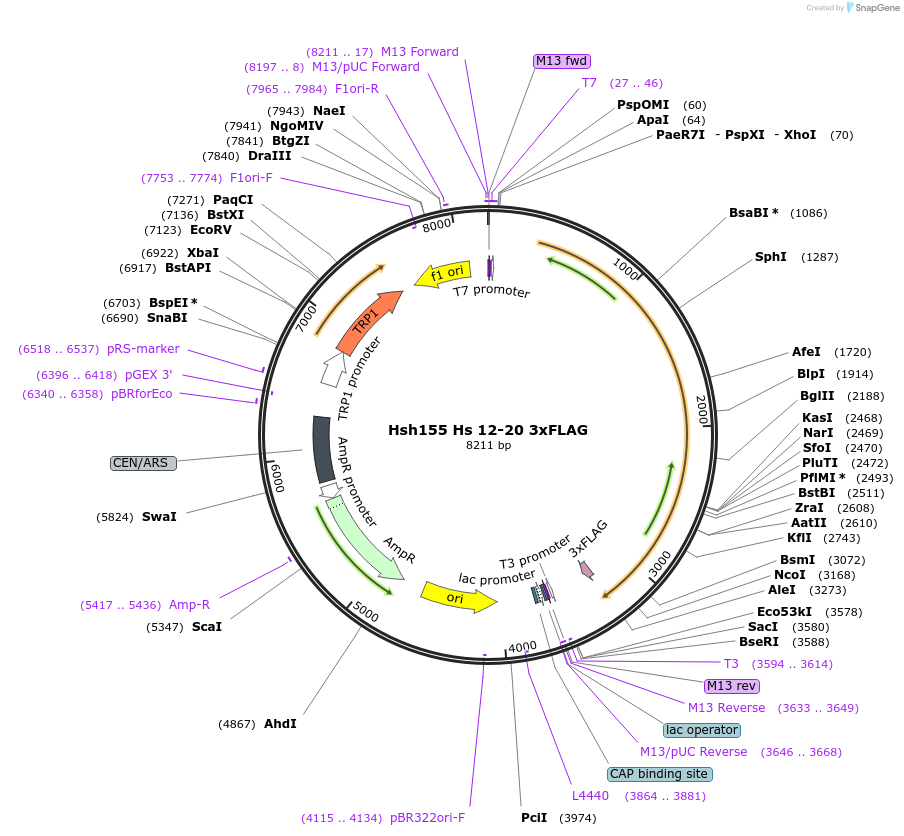
Hsh155 Hs 12-20 3xFLAG
Plasmid#111972PurposeHuman HEATs 12-20 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 12-20 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 12-20 insertedAvailable SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
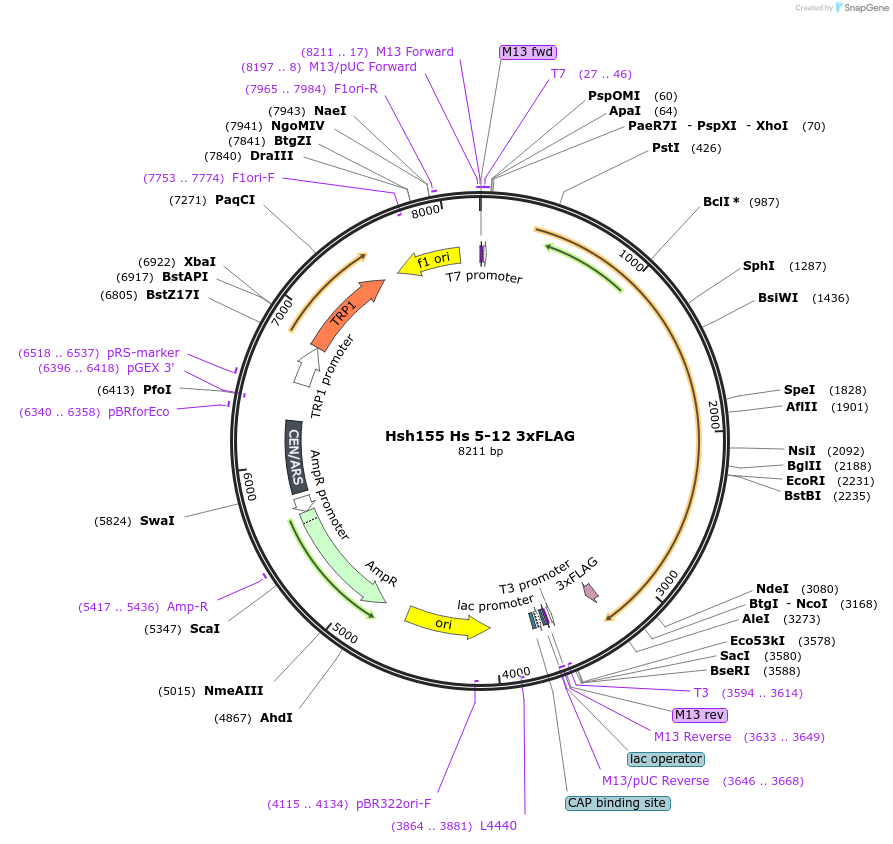
Hsh155 Hs 5-12 3xFLAG
Plasmid#111968PurposeHuman HEATs 5-12 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 5-12 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 5-12 insertedAvailable SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
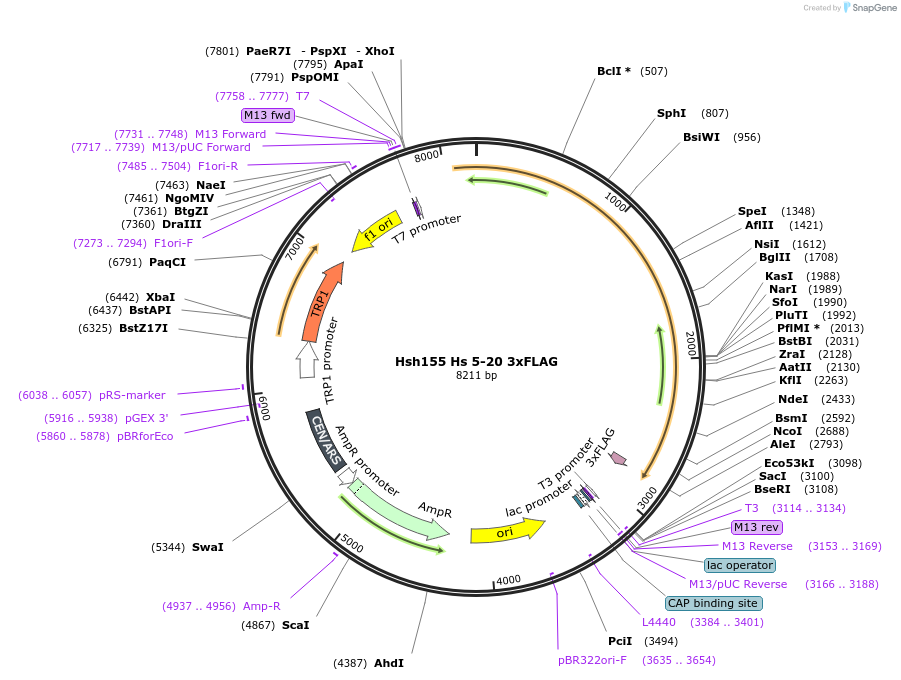
Hsh155 Hs 5-20 3xFLAG
Plasmid#111971PurposeHuman HEATs 5-20 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 5-20 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 5-20 insertedAvailable SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
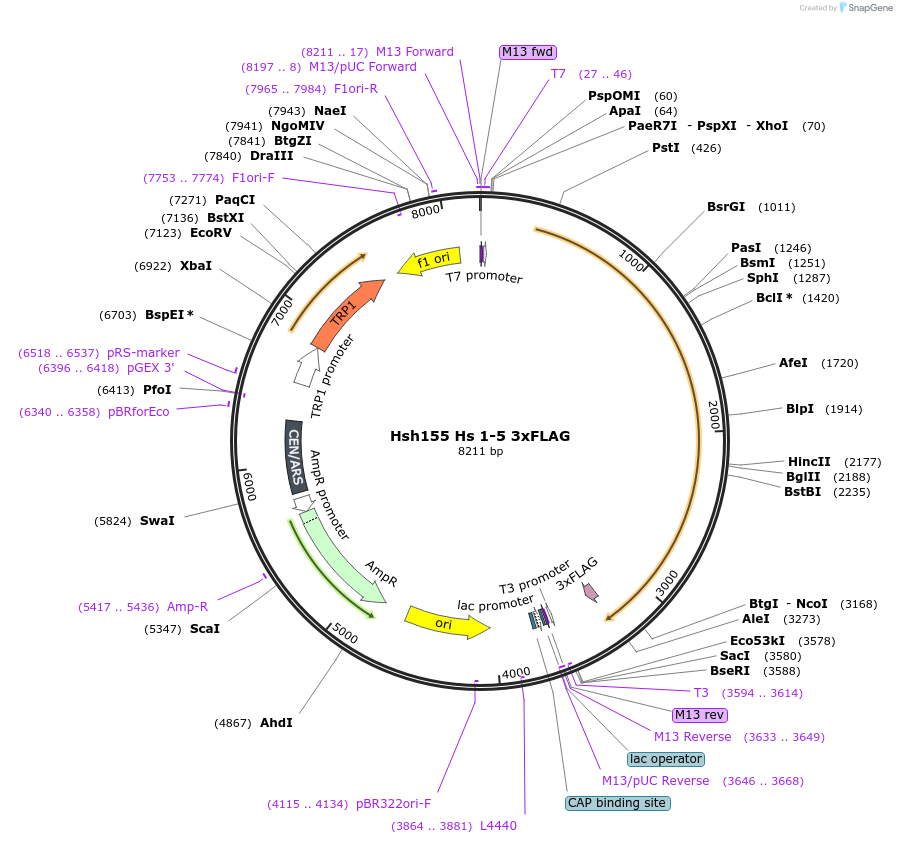
Hsh155 Hs 1-5 3xFLAG
Plasmid#111963PurposeHuman HEATs 1-5 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 1-5 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 1-5 insertedAvailable SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
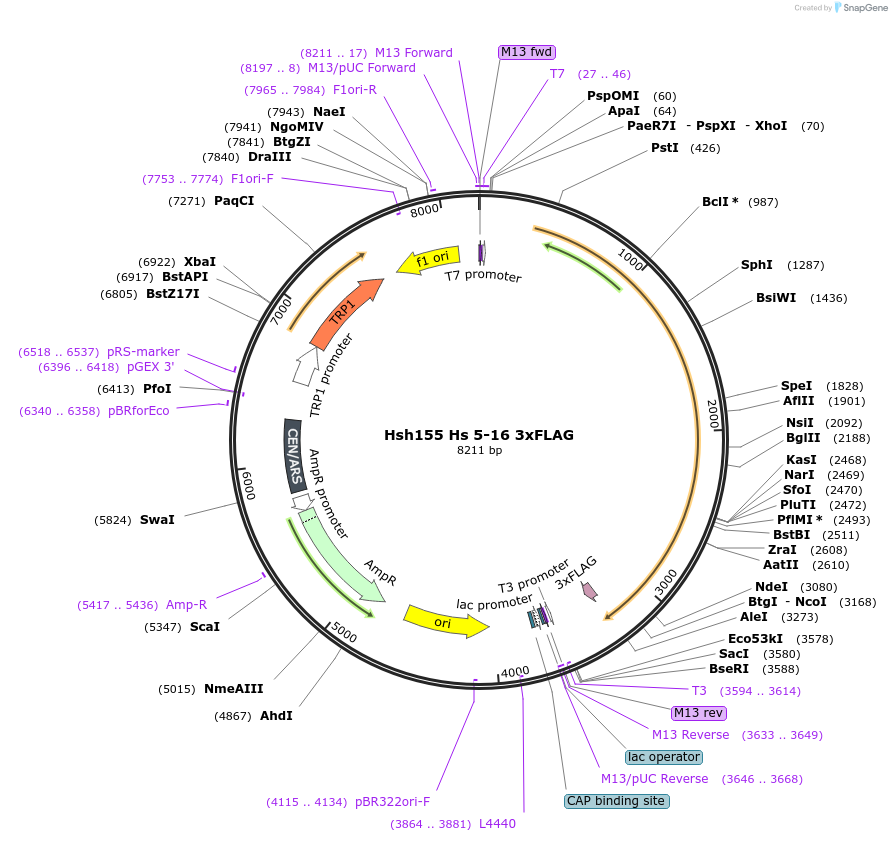
Hsh155 Hs 5-16 3xFLAG
Plasmid#111969PurposeHuman HEATs 5-16 have been inserted into Hsh155 3xFLAG.DepositorInsertHsh155 Hs 5-16 3x FLAG
Tags3xFLAGExpressionBacterial and YeastMutationHuman HEATs 5-16 insertedAvailable SinceJune 21, 2018AvailabilityAcademic Institutions and Nonprofits only -
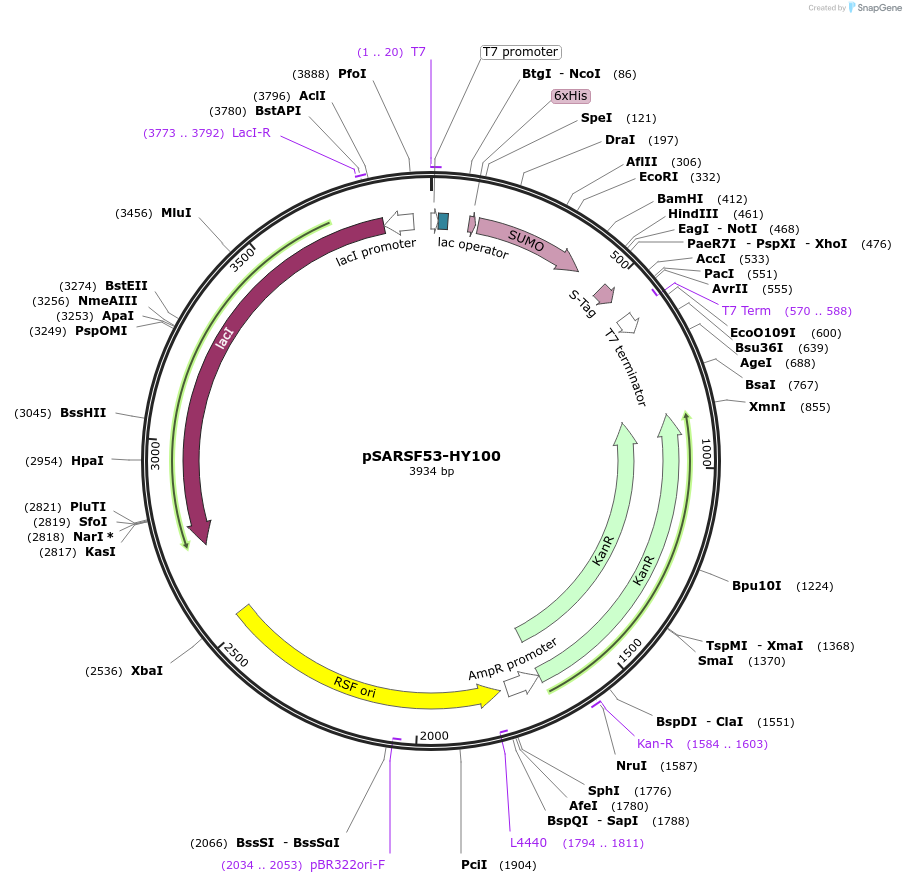
pSARSF53-HY100
Plasmid#61758PurposeNpuDnaB(delta283)N11 inteinDepositorInsertNpuDnaB(delta283)N11 intein
TagsH6-Smt3ExpressionBacterialPromoterT7Available SinceFeb. 11, 2015AvailabilityAcademic Institutions and Nonprofits only -
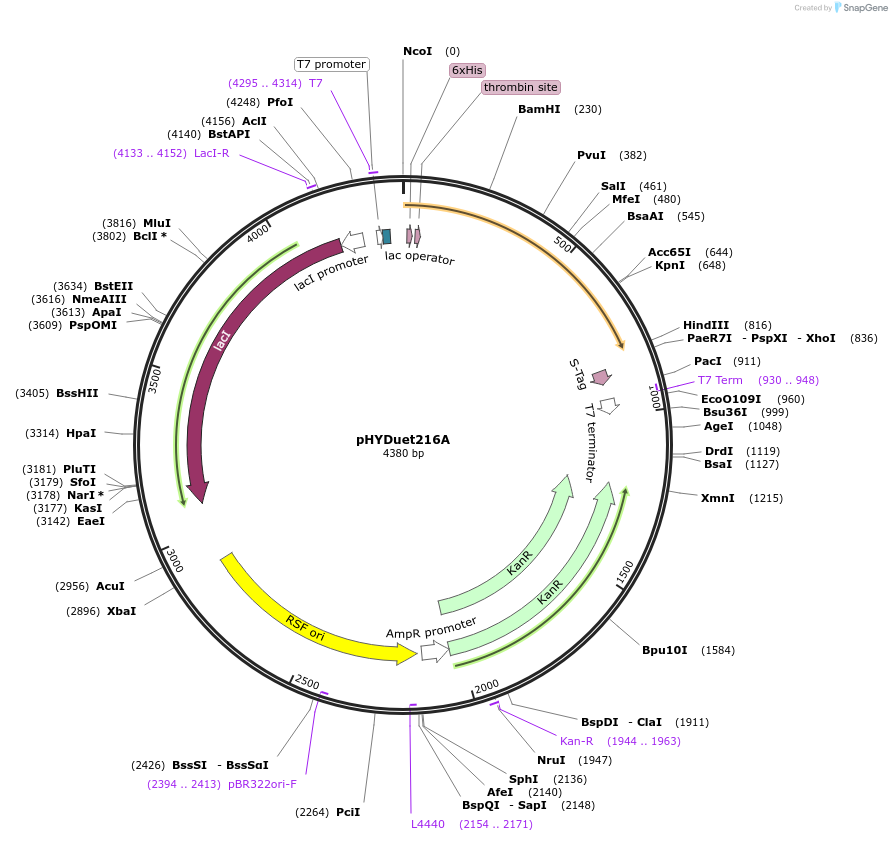
pHYDuet216A
Plasmid#45611DepositorInsertNpuDnaE delta variant
TagsGB1 and H6-GB1ExpressionBacterialMutationdeleted T76, V97, D98, L100 from NpuDnaE inteinPromoterT7Available SinceSept. 13, 2013AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F15
Plasmid#23064DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 deleted amino acids 4 - 19Available SinceMarch 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F23
Plasmid#23065DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 5 to GlutamineAvailable SinceMarch 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F33
Plasmid#23068DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysine 5 to Glutamine H4 K5QAvailable SinceMarch 3, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F47
Plasmid#23071DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysines 8 and 16 to Glutamines…Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F49
Plasmid#23073DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysines 8 and 16 to Arginine, …Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F51
Plasmid#23075DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysines 5 and 12 to Glutamine,…Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F52
Plasmid#23076DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H4 changed Lysines 5 and 12 to Arginines,…Available SinceFeb. 17, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F36
Plasmid#23069DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Lysine 14 to Glycine, H3 K14GAvailable SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
pWZ414-F43
Plasmid#23070DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Lysine 14 to Arginine, H3 K14RAvailable SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
pBM1
Plasmid#23094DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3, changed Lysine 9 to Glutamine, and Se…Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only -
pBM3
Plasmid#23096DepositorInsertyeast Histone H3-2 and Histone H4-2
ExpressionYeastMutationHistone H3 changed Serine 10 to Aspartate and Lys…Available SinceFeb. 5, 2010AvailabilityAcademic Institutions and Nonprofits only