We narrowed to 17,478 results for: igh
-
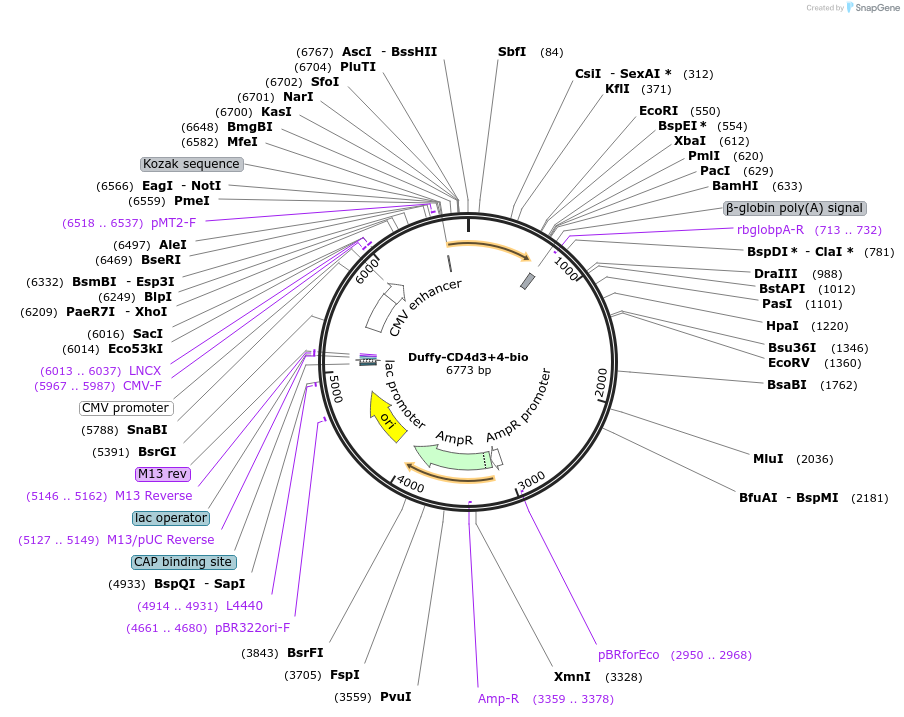
Plasmid#73122PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding Duffy with rat CD4d3+4-bioDepositorInsertDuffy (ACKR1 Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
bio-CD4d3+4-TMEM24
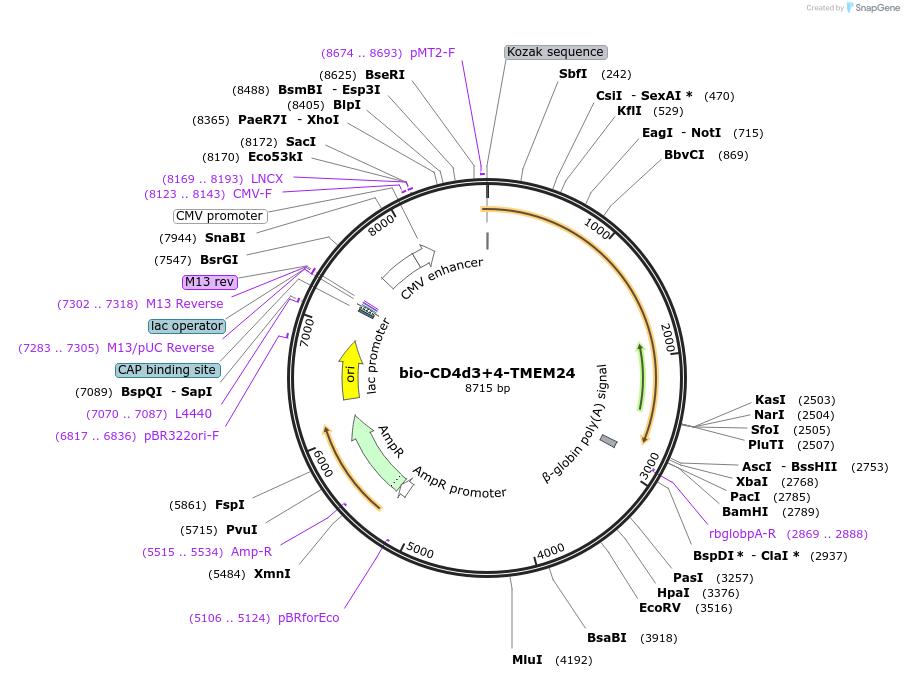
Plasmid#73092PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding TMEM24 with bio-rat CD4d3+4DepositorInsertTMEM24 (C2CD2L Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
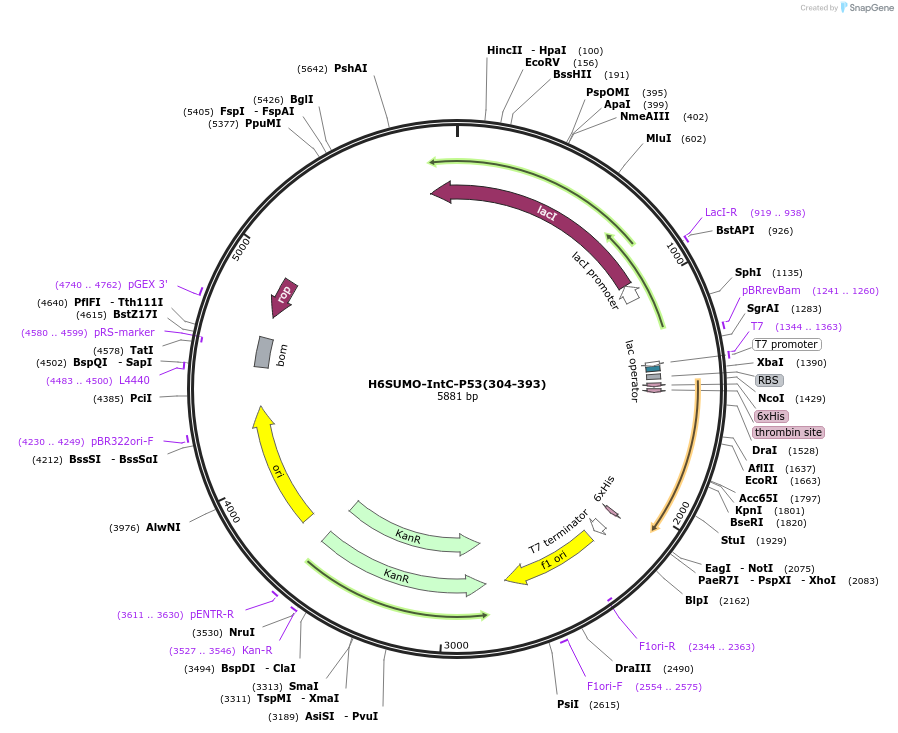
H6SUMO-IntC-P53(304-393)
Plasmid#200316Purposebacterial expression of P53 TET-CTD for segmental labelingDepositorInsertp53 (304-393) (TP53 Human)
TagsHis6-SUMO and designed intein CExpressionBacterialPromoterT7Available SinceMay 31, 2023AvailabilityAcademic Institutions and Nonprofits only -
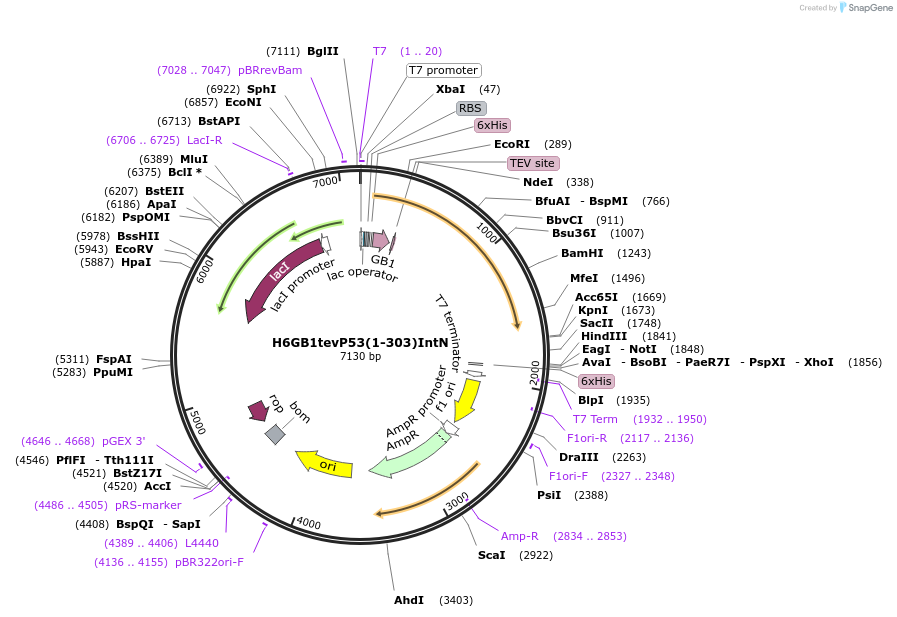
H6GB1tevP53(1-303)IntN
Plasmid#200312PurposeBacterial expression of P53 NTAD-DBD for segmental labelingDepositorInsertp53 (TP53 Human)
TagsHis6GB1tev and designed intein NExpressionBacterialMutationM133L, V203A, N239Y, N268DPromoterT7Available SinceApril 26, 2023AvailabilityAcademic Institutions and Nonprofits only -
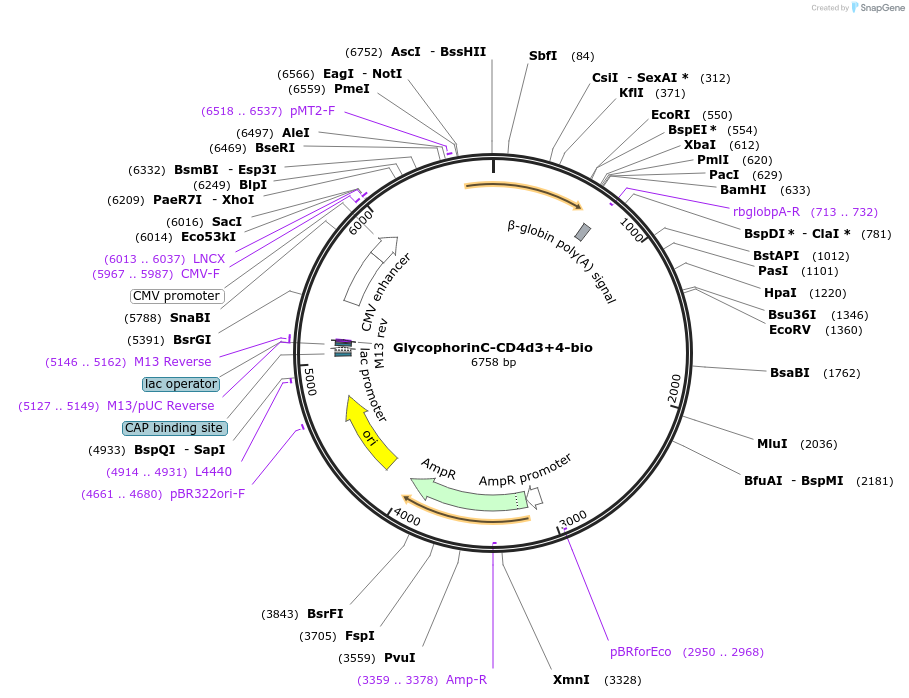
GlycophorinC-CD4d3+4-bio
Plasmid#73096PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding GlycophorinC with rat CD4d3+4-bioDepositorInsertGlycophorin C (GYPC Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 18, 2016AvailabilityAcademic Institutions and Nonprofits only -
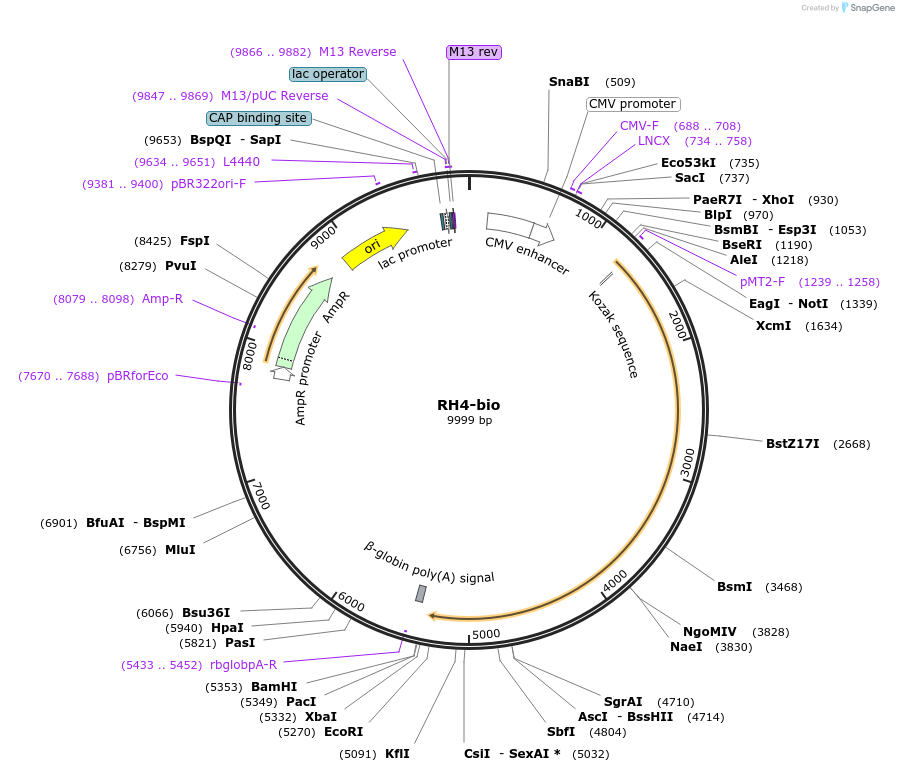
RH4-bio
Plasmid#47779PurposeExpresses enzymatically monobiotinylated partial RH4 ectodomain with no N-linked glycans upon cotransfection with BirA in mammalian cells. Contains a C-terminal rat Cd4 d3+4 tag.DepositorInsertCodon-optimised RH4 (partial)
Tagsenzymatic biotinylation sequence and rat Cd4 d3+4ExpressionMammalianMutationExogenous signal peptide of mouse origin, changed…PromoterCMVAvailable SinceFeb. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
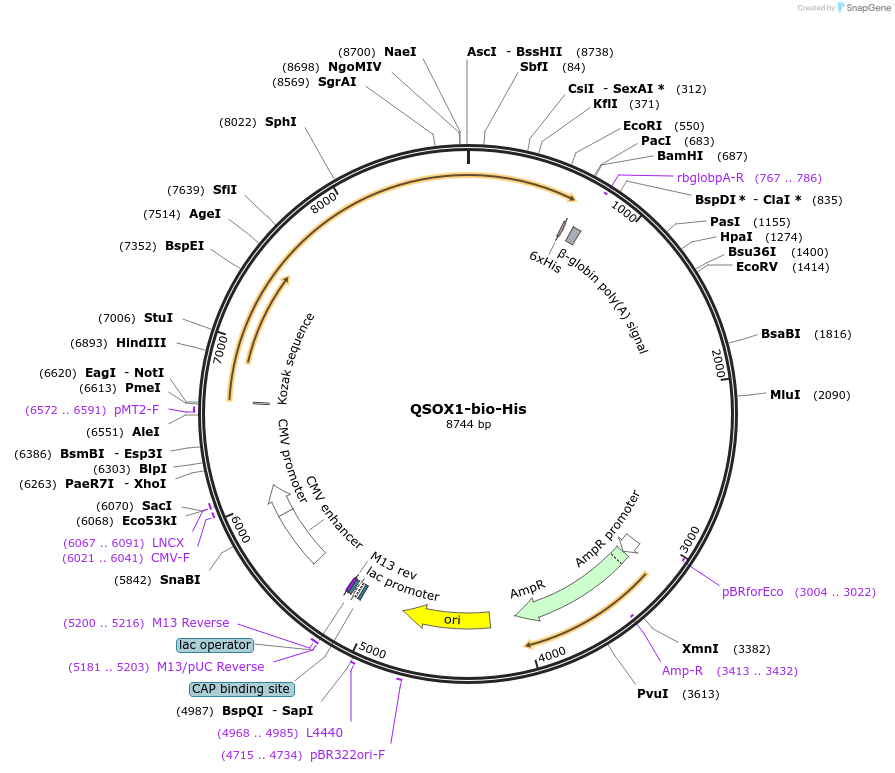
QSOX1-bio-His
Plasmid#51825PurposeExpresses full-length Sulfhydryl oxidase 1 precursor ectodomain in mammalian cells. C-terminal rat Cd4d3+4 tag, biotinylation sequence and His tag.DepositorInsertQSOX1 (QSOX1 Human)
TagsHis tag, enzymatic biotinylation sequence, and ra…ExpressionMammalianPromoterCMVAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
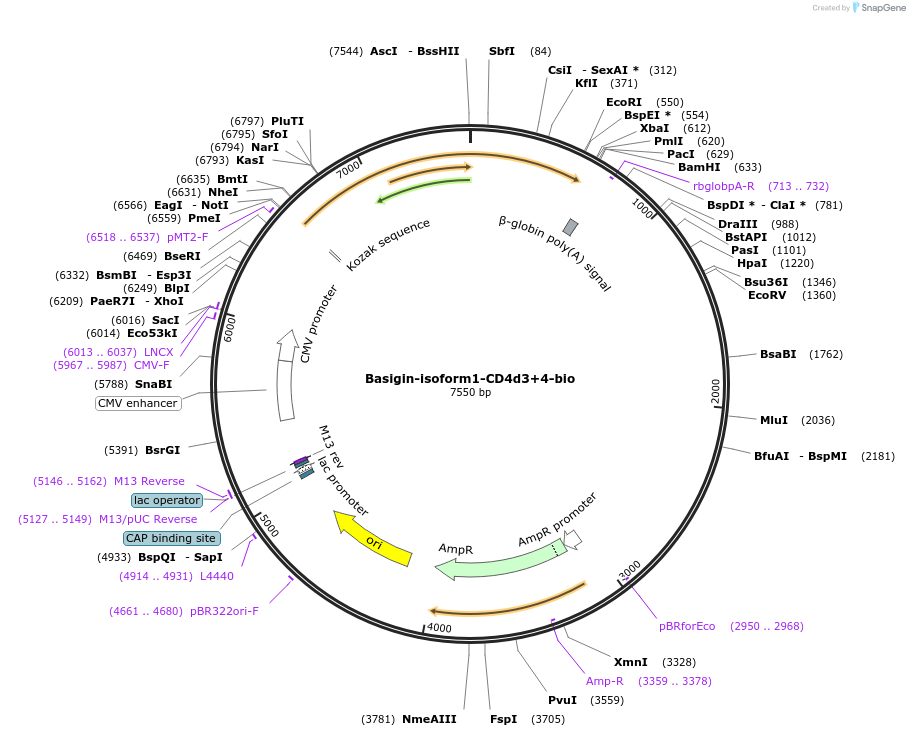
Basigin-isoform1-CD4d3+4-bio
Plasmid#73100PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding Basigin-isoform1 with rat CD4d3+4-bioDepositorInsertBasigin isoform 1 (BSG Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
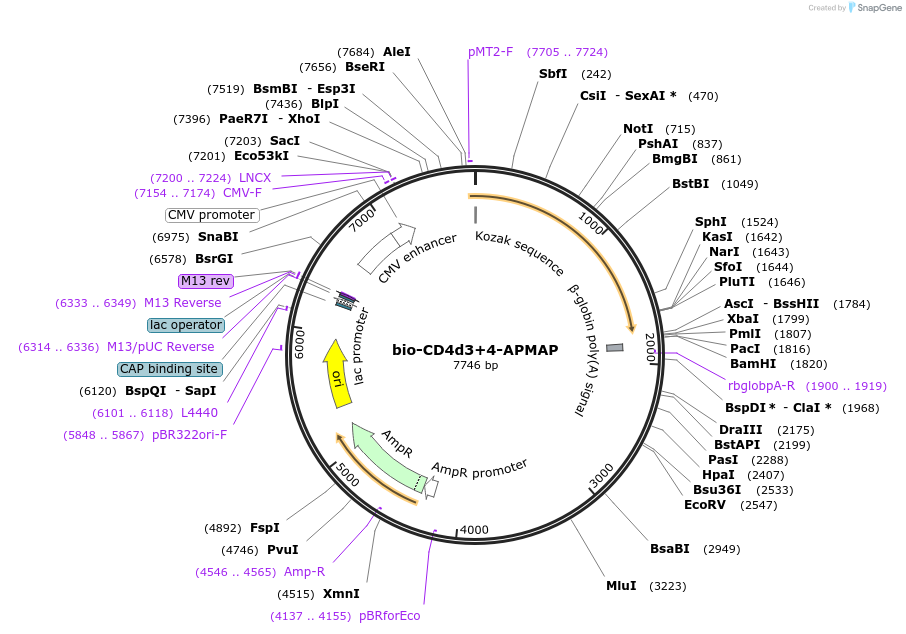
bio-CD4d3+4-APMAP
Plasmid#73091PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding APMAP with bio-rat CD4d3+4DepositorInsertAPMAP (APMAP Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
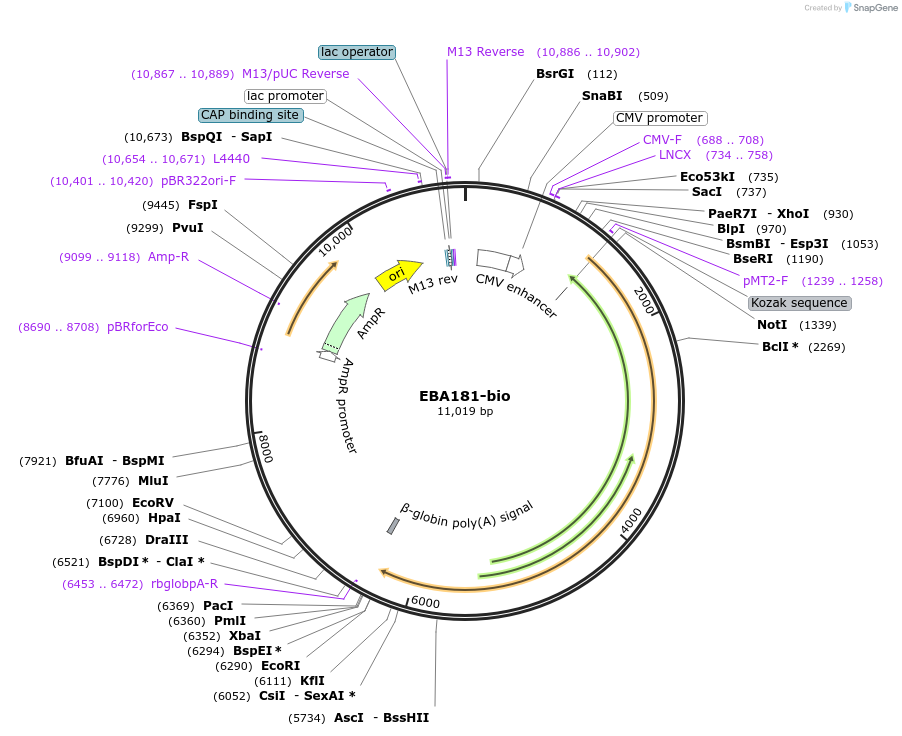
EBA181-bio
Plasmid#47744PurposeExpresses enzymatically monobiotinylated full-length EBA181 ectodomain with no N-linked glycans upon cotransfection with BirA in mammalian cells. Contains a C-terminal rat Cd4 d3+4 tag.DepositorInsertCodon-optimised EBA181
Tagsenzymatic biotinylation sequence and rat Cd4 d3+4ExpressionMammalianMutationExogenous signal peptide of mouse origin, changed…PromoterCMVAvailable SinceFeb. 4, 2014AvailabilityAcademic Institutions and Nonprofits only -
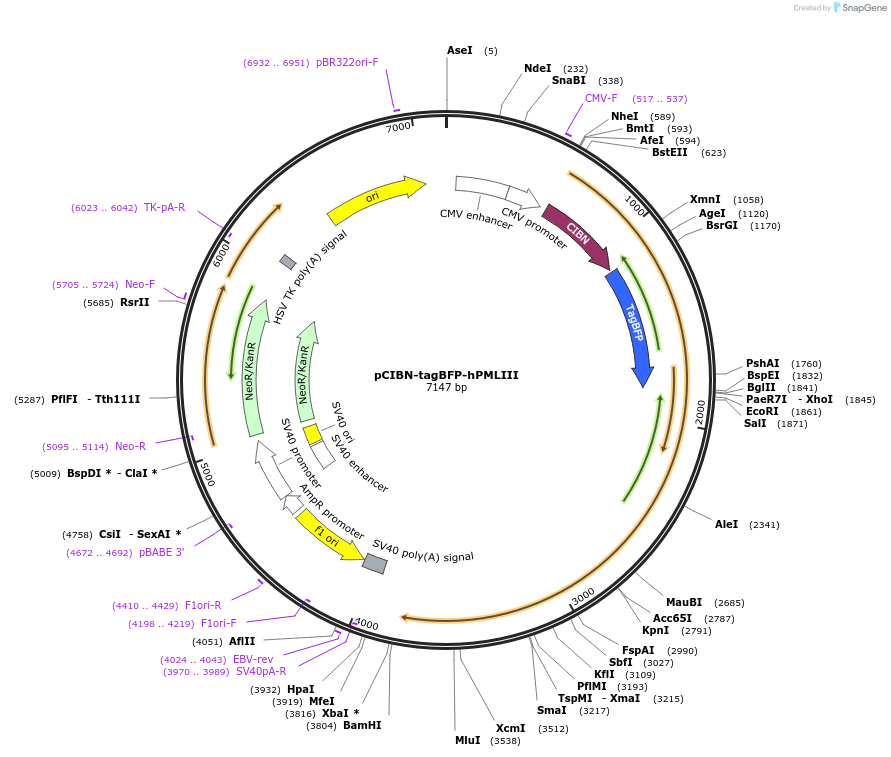
pCIBN-tagBFP-hPMLIII
Plasmid#103807PurposeBLInCR 'Localizer' construct that marks the PML nuclear bodies and is targeted by a PHR-tagged effector upon illumination with blue lightDepositorAvailable SinceJan. 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
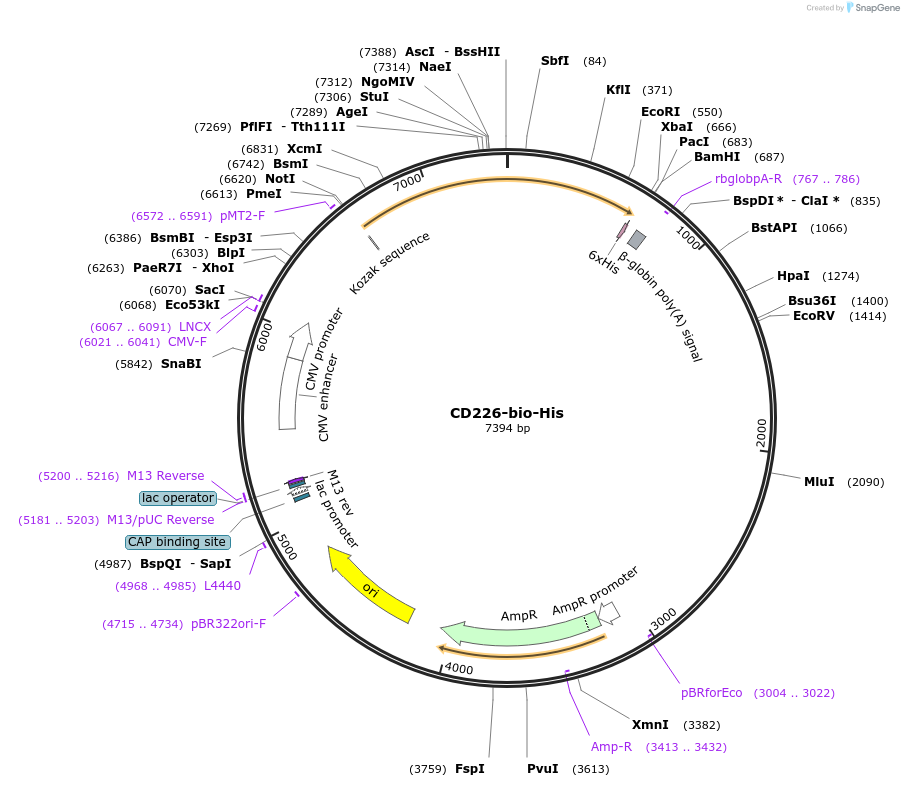
CD226-bio-His
Plasmid#51646PurposeExpresses full-length CD226 antigen precursor ectodomain in mammalian cells. C-terminal rat Cd4d3+4 tag, biotinylation sequence and His tag.DepositorInsertCD226 (CD226 Human)
TagsHis tag, enzymatic biotinylation sequence, and ra…ExpressionMammalianPromoterCMVAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
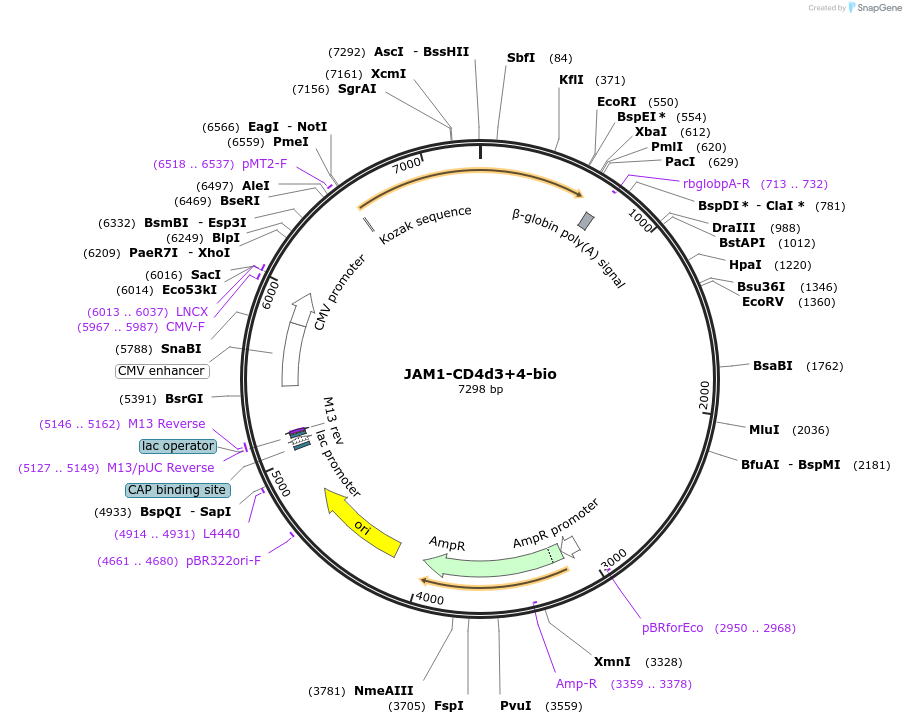
JAM1-CD4d3+4-bio
Plasmid#73110PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding JAM1 with rat CD4d3+4-bioDepositorInsertJAM1 (F11R Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 28, 2016AvailabilityAcademic Institutions and Nonprofits only -
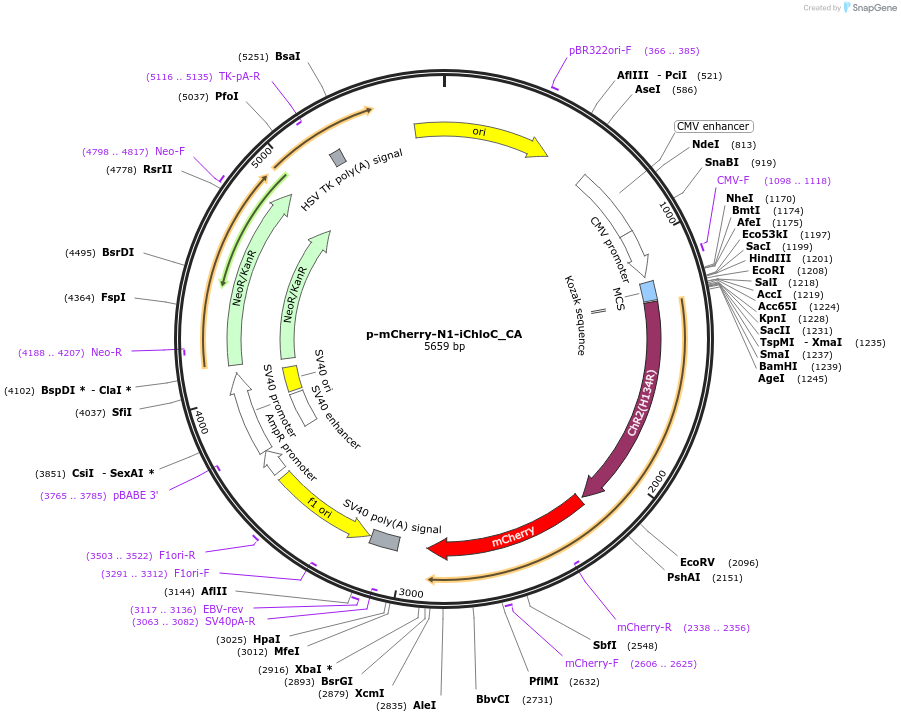
p-mCherry-N1-iChloC_CA
Plasmid#98171Purposeslow cycling (open for minutes) step-function artificial anion conducting channelrhodopsin (aACR). High light sensitivity. Activation with blue to green light, inactivation max with 605 nm (accelarates closure to ms). Codon optimized for mammalian expression.DepositorInsertSynthetic construct iChloC_C128A gene
TagsmCherryExpressionMammalianMutationE83Q, E90R, E101S, C128A, T159C, D156NPromoterCMV (+enhancer)Available SinceJuly 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
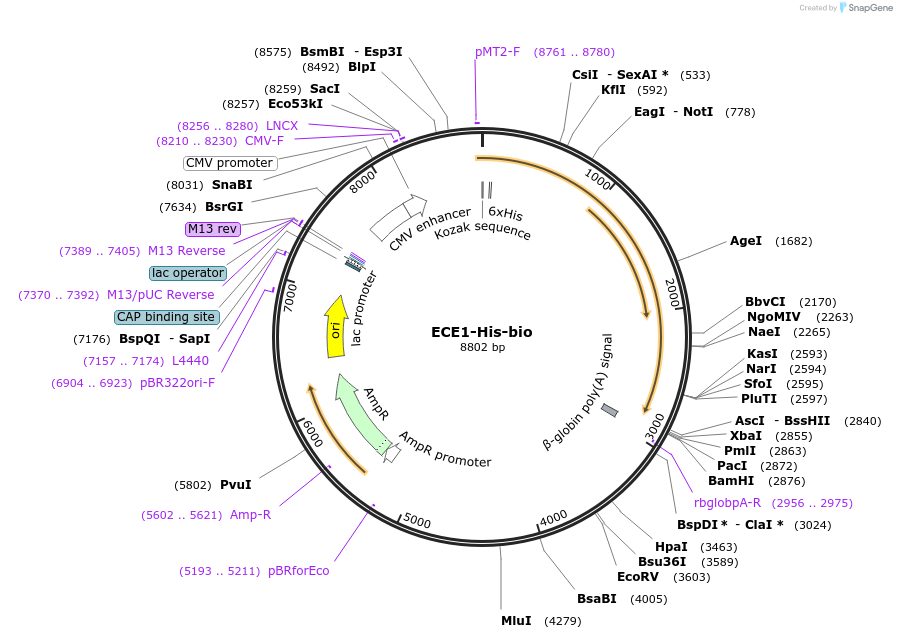
ECE1-His-bio
Plasmid#53361PurposeExpresses full-length Endothelin converting enzyme 1 ectodomain in mammalian cells. N-terminal His tag, biotinylation sequence and rat Cd4d3+4 tag.DepositorInsertECE1 (ECE1 Human)
TagsHis tag, enzymatic biotinylation sequence, and ra…ExpressionMammalianPromoterCMVAvailable SinceFeb. 27, 2015AvailabilityAcademic Institutions and Nonprofits only -
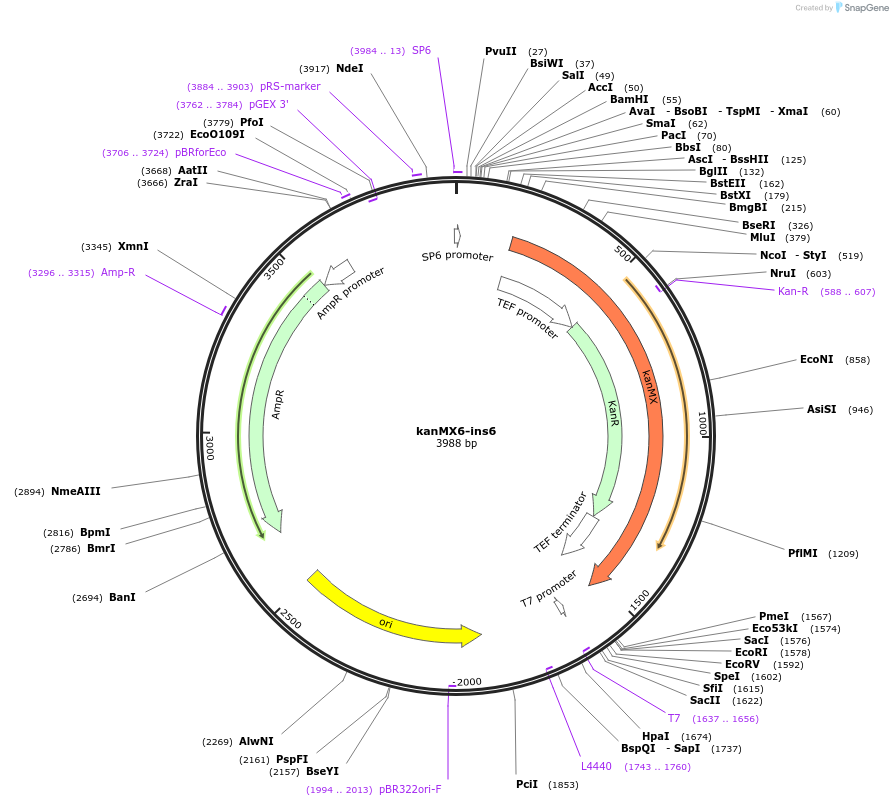
kanMX6-ins6
Plasmid#195043PurposepFA6a derived selection cassette 5' flanked with tDEG1 transcription terminator, allows genome modification without disruption of insertion neighboring genes by transcription interferenceDepositorInsertKanR
UseYeast genomic targetingTagsS. cerevisiae DEG1 terminator, A. gossypii TEF pr…Available SinceFeb. 9, 2023AvailabilityAcademic Institutions and Nonprofits only -
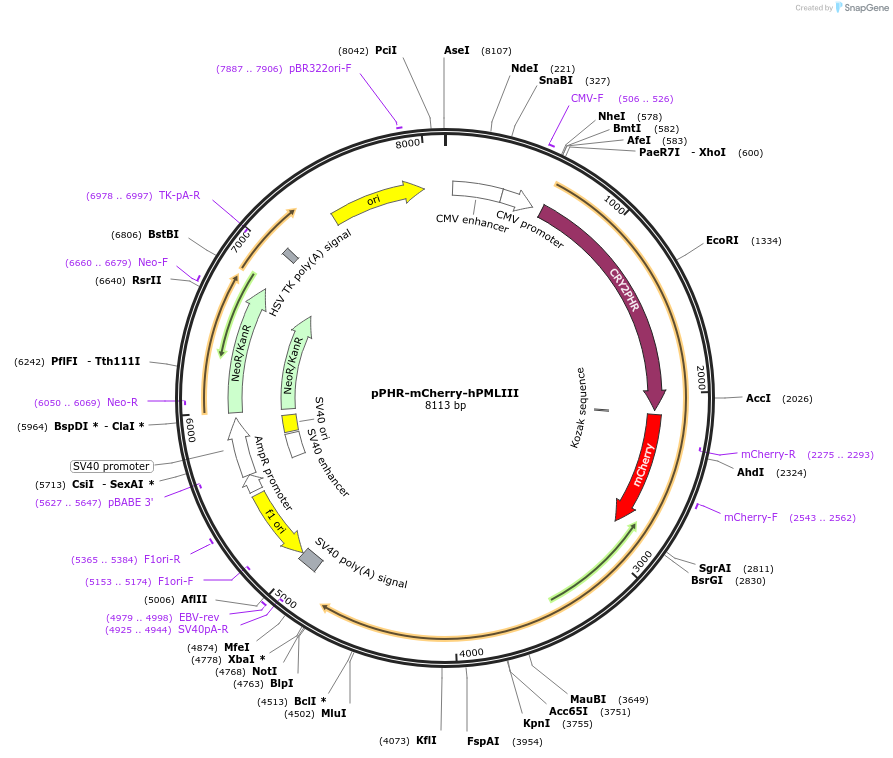
pPHR-mCherry-hPMLIII
Plasmid#103824PurposeSoluble BLInCR effector that is recruited to 'localizer' sites upon blue light illuminationDepositorAvailable SinceDec. 1, 2017AvailabilityAcademic Institutions and Nonprofits only -
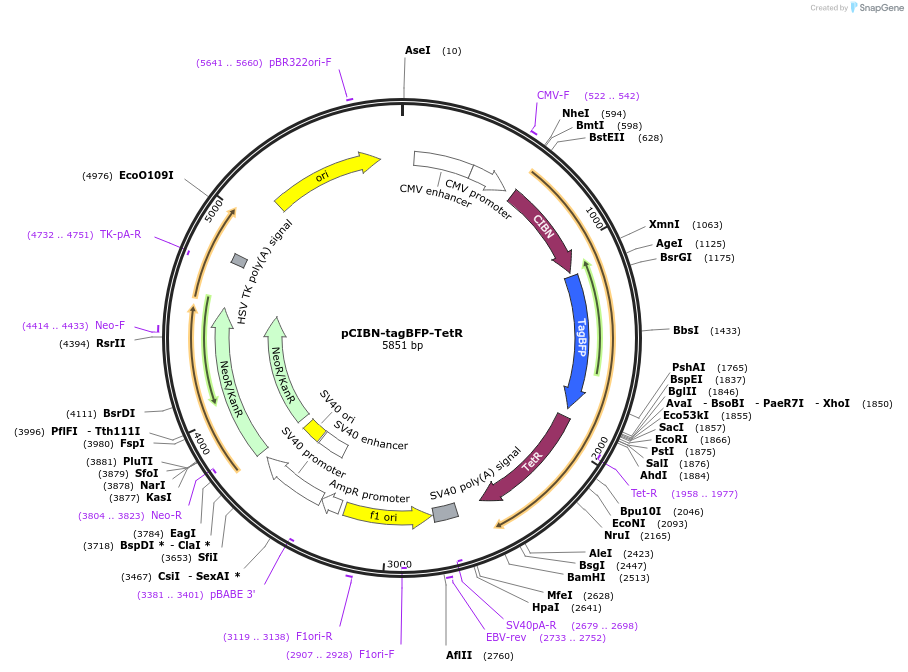
pCIBN-tagBFP-TetR
Plasmid#103797PurposeBLInCR 'Localizer' construct that marks tetO arrays and is targeted by a PHR-tagged effector upon illumination with blue lightDepositorExpressionMammalianMutationTetR: A4G (M2V)PromoterCMVAvailable SinceJan. 9, 2018AvailabilityAcademic Institutions and Nonprofits only -
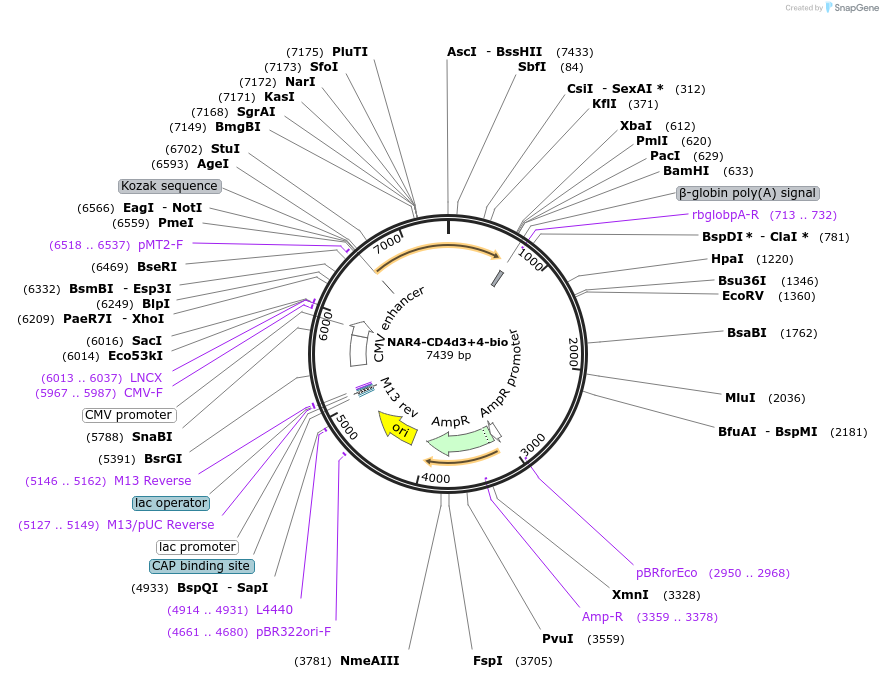
NAR4-CD4d3+4-bio
Plasmid#73118PurposeEXPRESs plasmid for human erythrocyte surface proteins encoding NAR4 with rat CD4d3+4-bioDepositorInsertNAR4 (ART4 Human)
Tagsbiotinylation peptide and ratCD4d3+4ExpressionMammalianPromoterCMVAvailable SinceMarch 29, 2016AvailabilityAcademic Institutions and Nonprofits only -
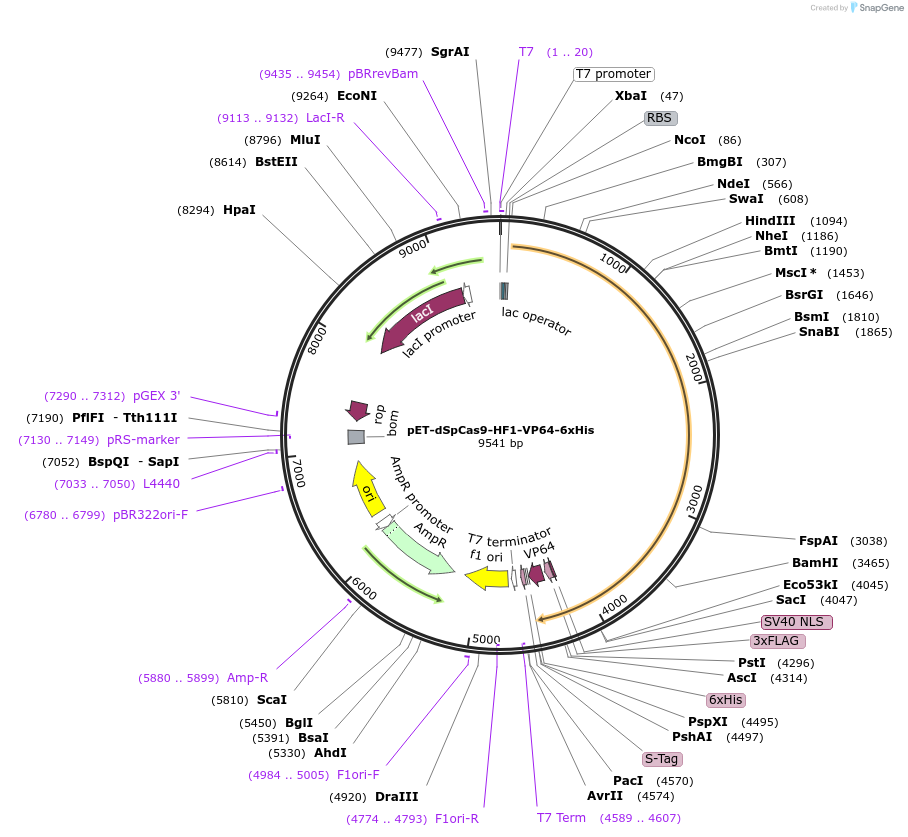
pET-dSpCas9-HF1-VP64-6xHis
Plasmid#92118PurposeExpression of dead/inactive increased fidelity SpCas9-HF1-VP64-6xHis in bacterial cellsDepositorInsertdead/inactive SpCas9-HF1-NLS-3xFLAG-VP64
UseCRISPRTags3xFLAG, 6xHis, NLS, and VP64ExpressionBacterialMutationD10A, N497A, R661A, Q695A, H840A, Q926APromoterT7Available SinceOct. 11, 2017AvailabilityAcademic Institutions and Nonprofits only