We narrowed to 10,309 results for: yeast
-
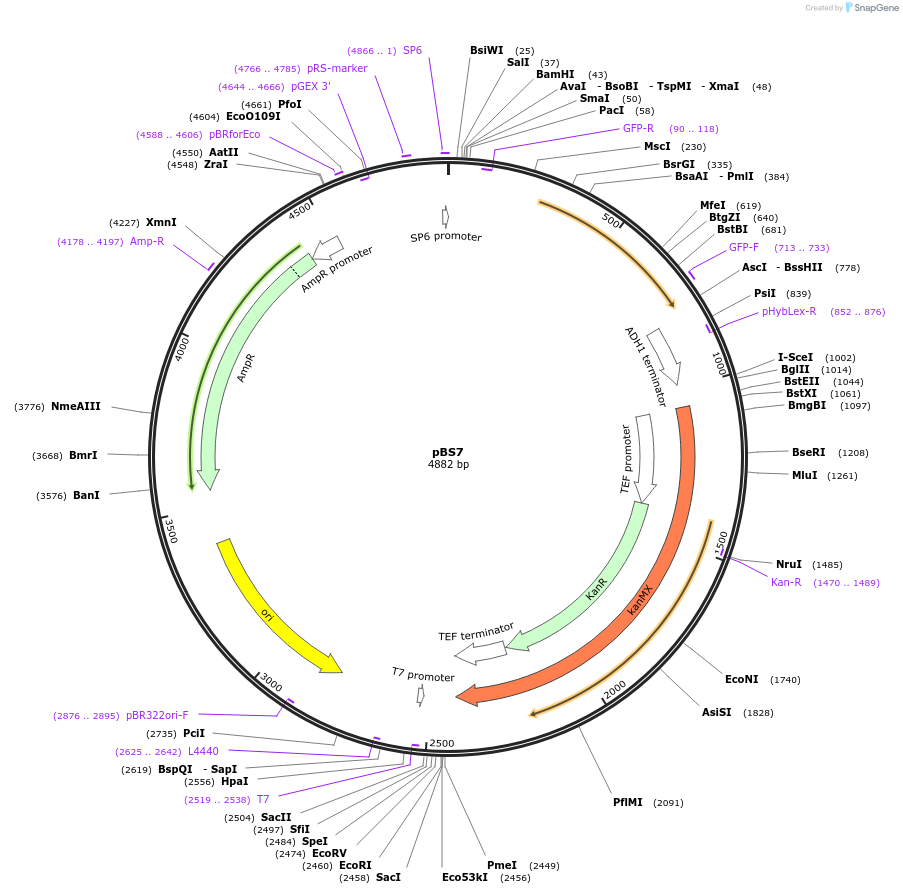
Plasmid#83774PurposeUseful for tagging yeast proteins with C-terminal Venus tag; G418 selectionDepositorInsertVenus
TagsVenusExpressionYeastMutationF46L, F64L, S65G, V68L, Q69M, S72A, Q80R, M153T, …Available SinceNov. 4, 2016AvailabilityAcademic Institutions and Nonprofits only -
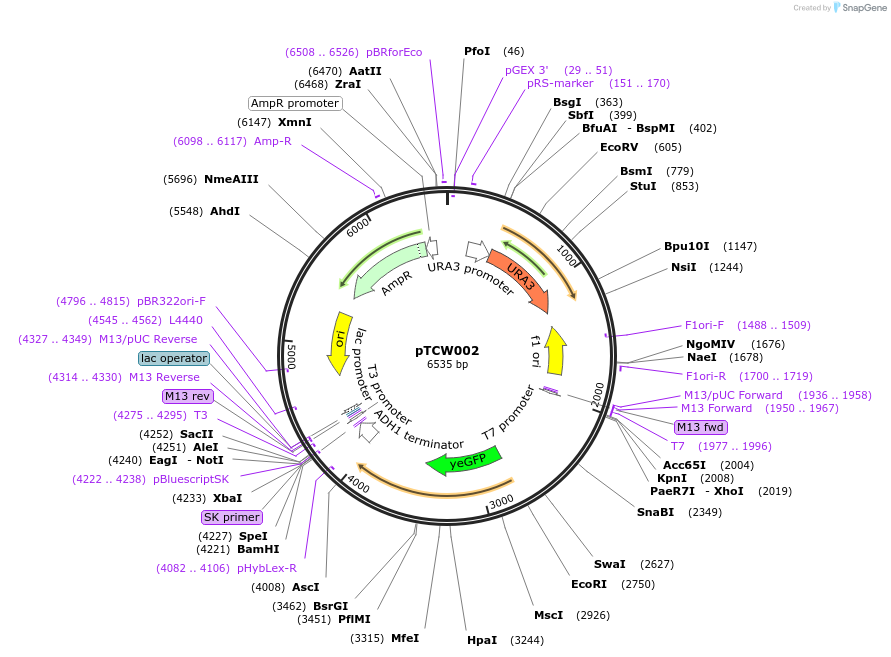
pTCW002
Plasmid#83522Purposepheromone mediated destablised-GFP expression in yeastDepositorInsertpFUS1-yEGFPCLN2PEST
ExpressionYeastPromoterpFUS1Available SinceDec. 16, 2016AvailabilityAcademic Institutions and Nonprofits only -
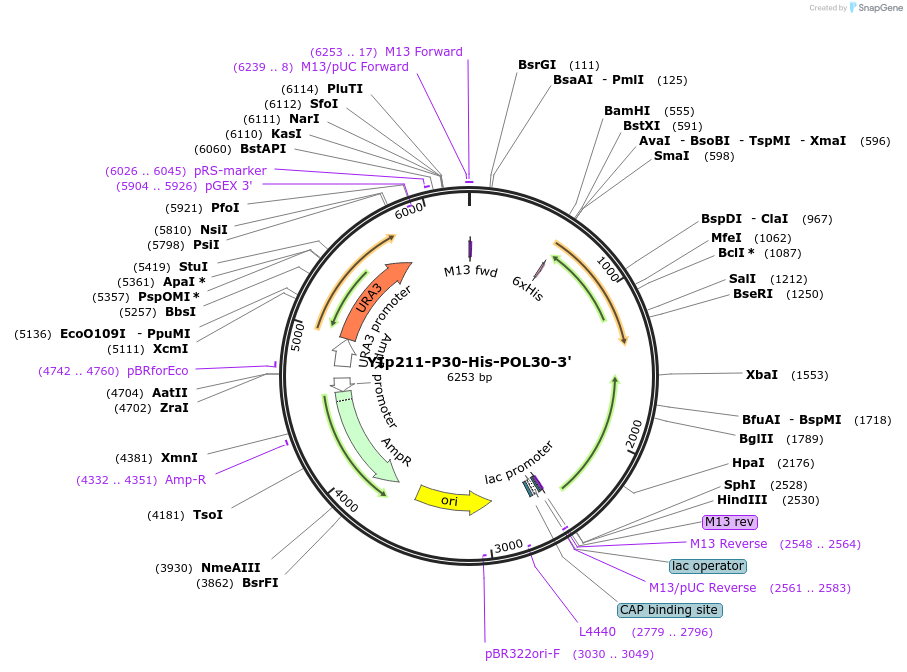
YIp211-P30-His-POL30-3'
Plasmid#99546PurposeYeast integrative vector for expression of His-tagged POL30 (PCNA); URA3 markerDepositorAvailable SinceSept. 14, 2017AvailabilityAcademic Institutions and Nonprofits only -
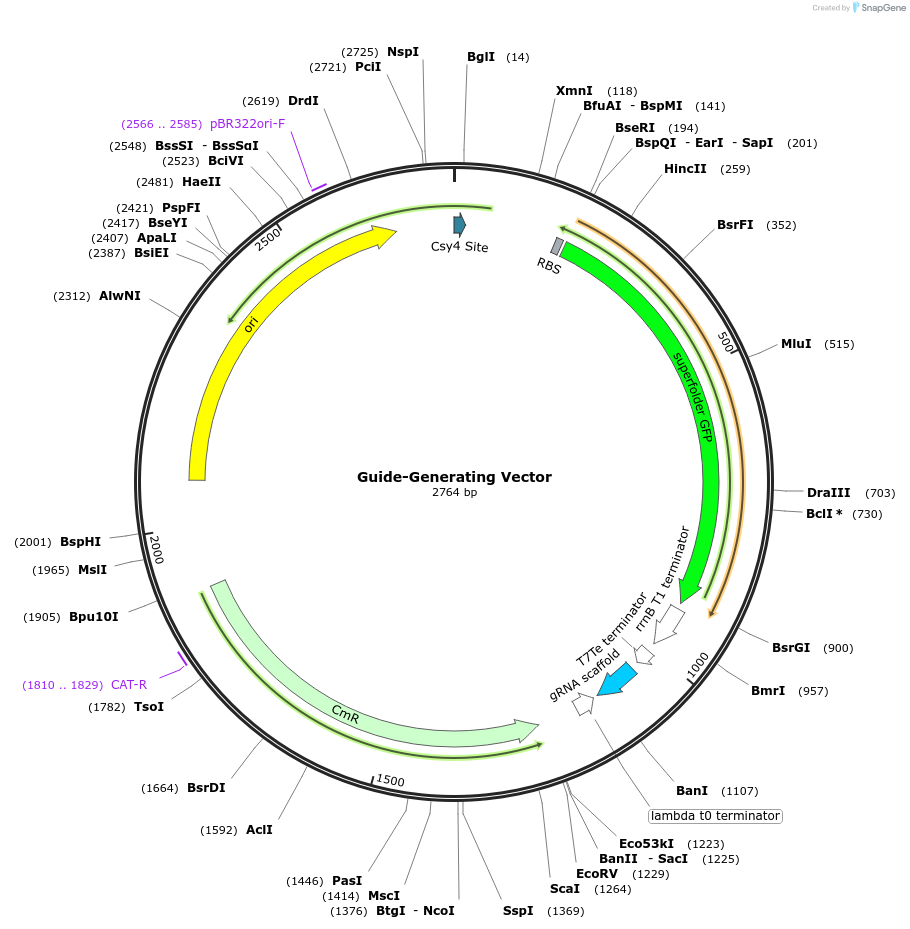
Guide-Generating Vector
Plasmid#125752PurposeGuide-Generating Vector for creating the Cys4-guide-scaffold structure in the first round of assemblyDepositorInsertCys4-GFPdropout-SpCas9scaffold
UseSynthetic BiologyAvailable SinceJuly 10, 2019AvailabilityAcademic Institutions and Nonprofits only -
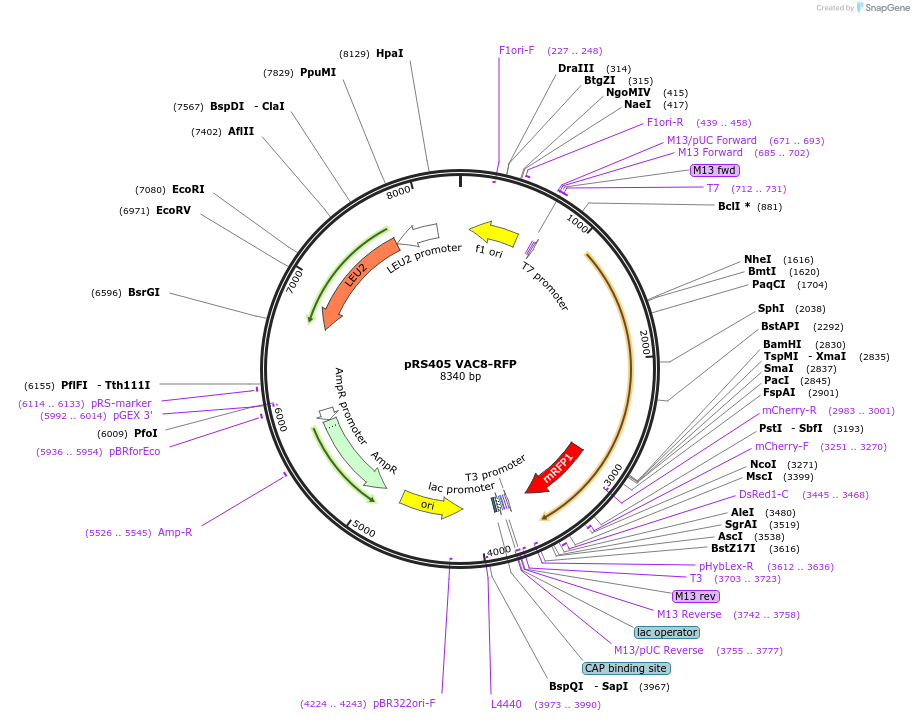
pRS405 VAC8-RFP
Plasmid#166841PurposeExpresses Vac8-RFP in yeastsDepositorAvailable SinceApril 2, 2021AvailabilityAcademic Institutions and Nonprofits only -
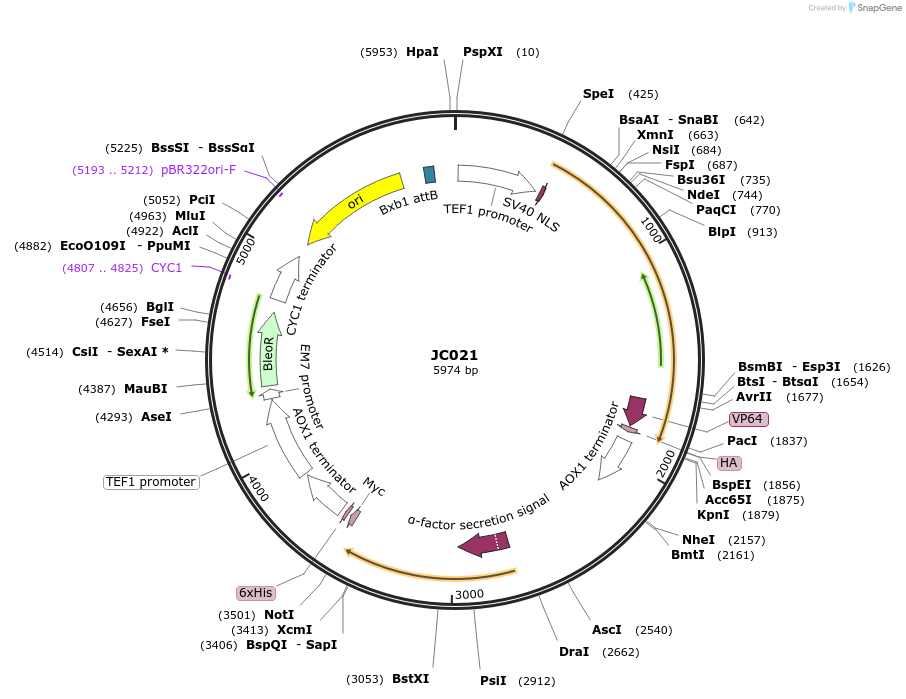
JC021
Plasmid#104944PurposeEstrogen inducing G-CSFDepositorInsertG-CSF
Available SinceJune 4, 2018AvailabilityAcademic Institutions and Nonprofits only -
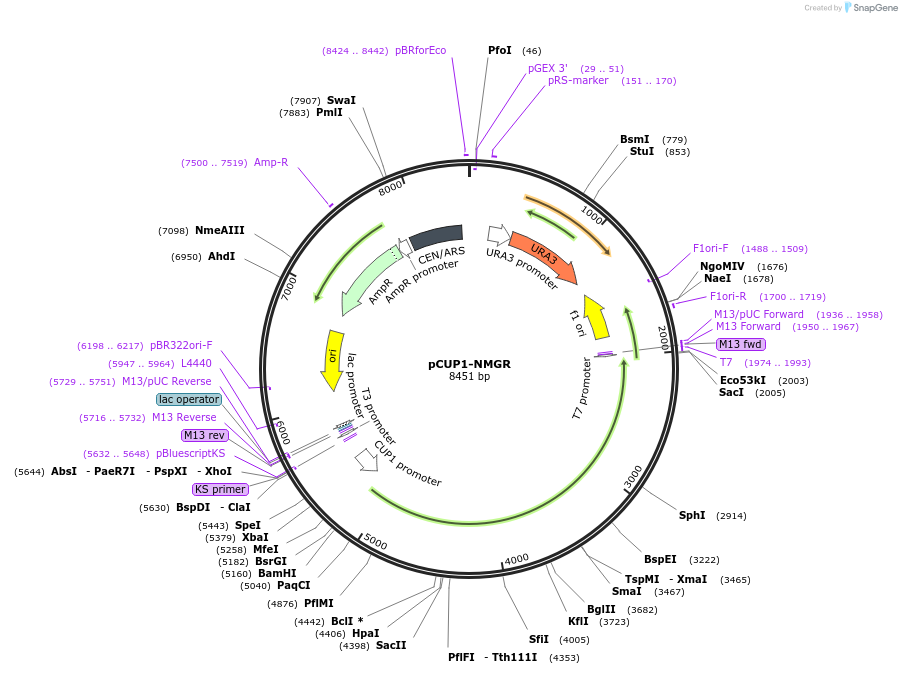
pCUP1-NMGR
Plasmid#1211DepositorAvailable SinceJan. 6, 2005AvailabilityAcademic Institutions and Nonprofits only -
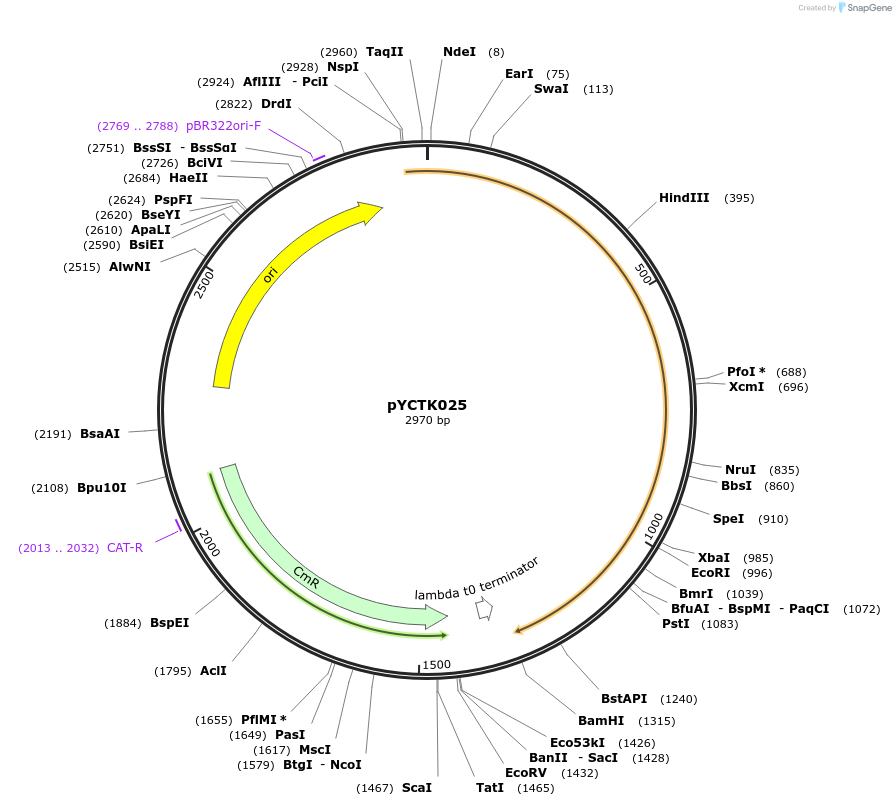
pYCTK025
Plasmid#176729PurposeThe plasmid is part of a GG toolkit for cell-cell communication in S. cerevisiae. This plasmid is an L0 part plasmid and contains the Ste2 gene of Lachancea mirantina (receiver part).DepositorArticleInsertste2Lm
UseSynthetic BiologyMutationCodon-optimized for expression in S. cerevisiaeAvailable SinceJan. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
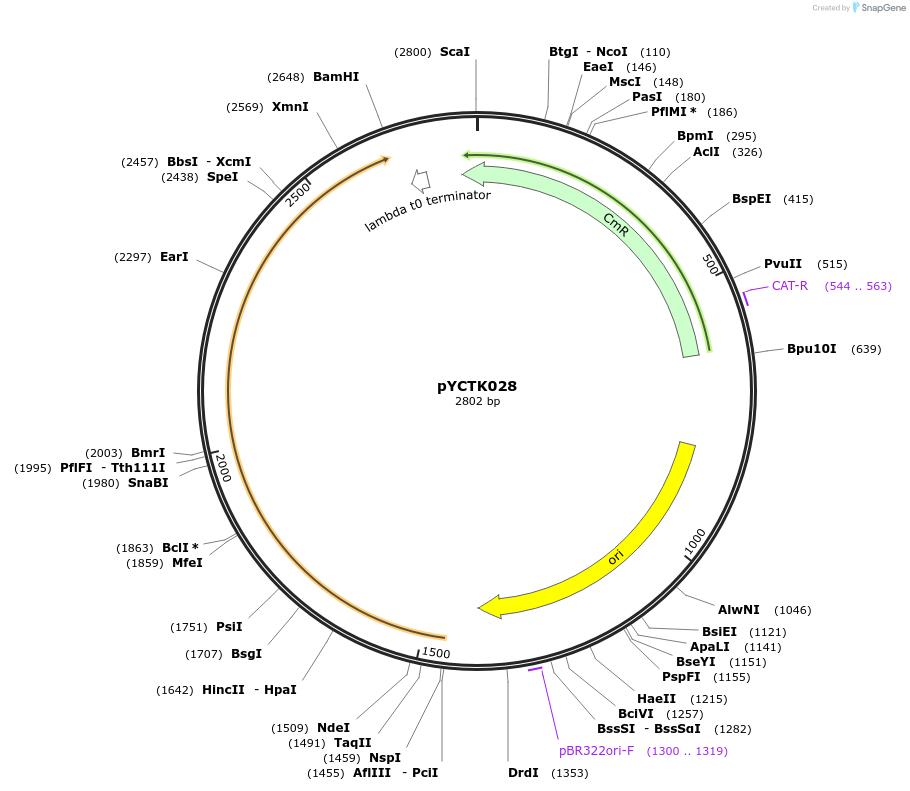
pYCTK028
Plasmid#176732PurposeThe plasmid is part of a GG toolkit for cell-cell communication in S. cerevisiae. This plasmid is an L0 part plasmid and contains the Ste2 gene of Tetrapisispora phaffii (receiver part).DepositorArticleInsertste2Tp
UseSynthetic BiologyMutationCodon-optimized for expression in S. cerevisiaeAvailable SinceJan. 6, 2022AvailabilityAcademic Institutions and Nonprofits only -
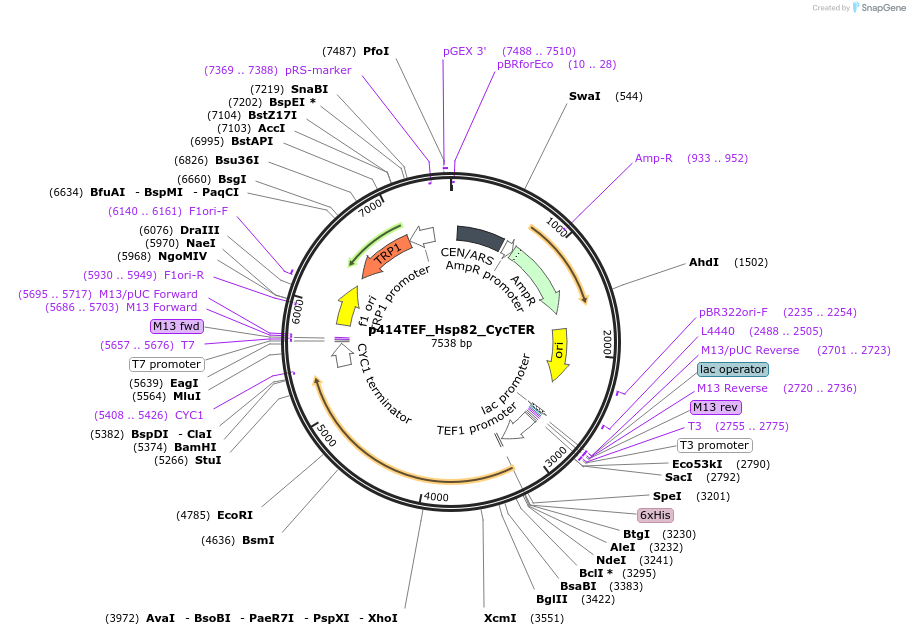
p414TEF_Hsp82_CycTER
Plasmid#61711PurposeYeast plasmid with Hsp82DepositorAvailable SinceFeb. 12, 2015AvailabilityAcademic Institutions and Nonprofits only -
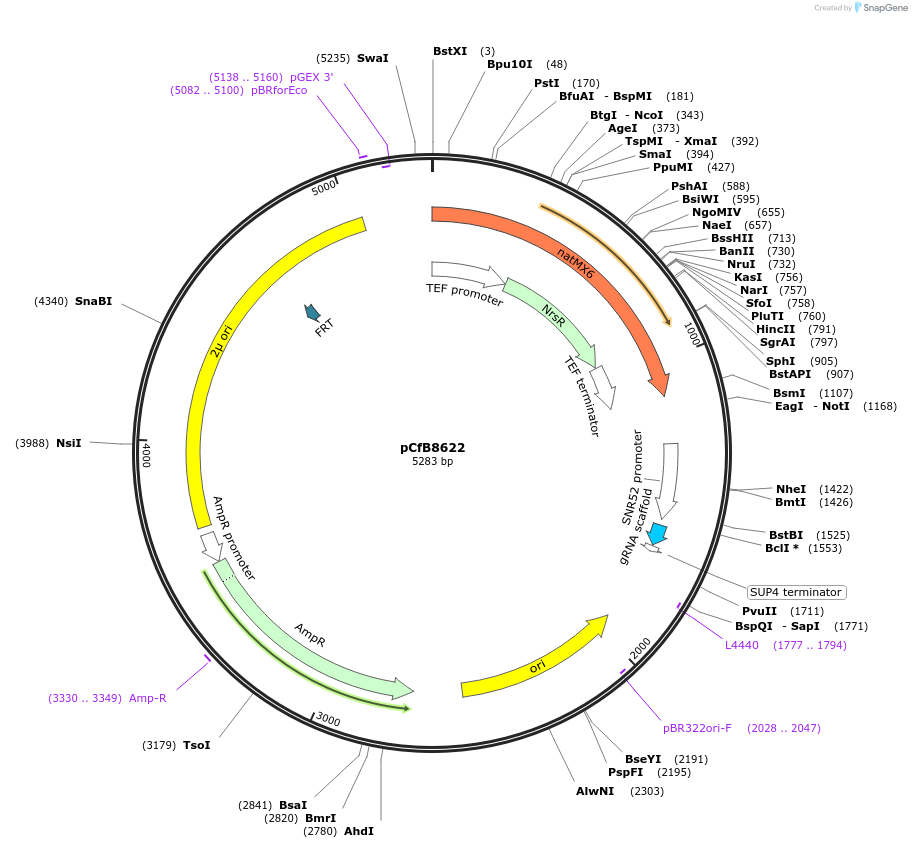
pCfB8622
Plasmid#126912PurposePlasmid containing gRNA expression cassette targeting ADE2 in Saccharomyces cerevisiaeDepositorInsertADE2 gRNA
UseCRISPRAvailable SinceDec. 17, 2019AvailabilityAcademic Institutions and Nonprofits only -
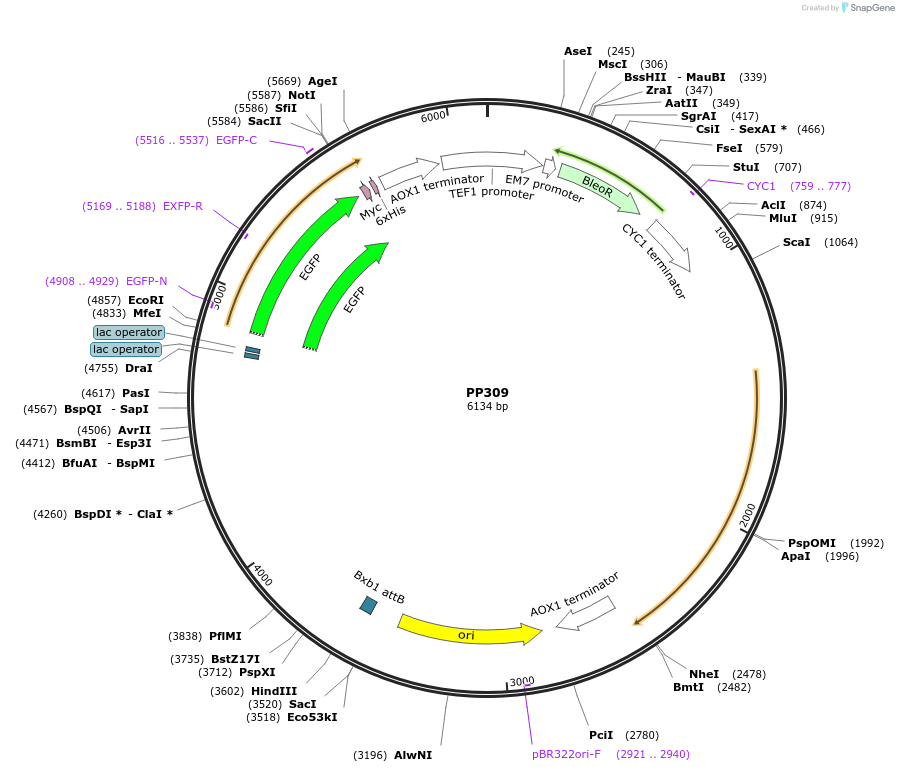
PP309
Plasmid#104951PurposeIPTG inducing GFPDepositorInsertanti-CTLA4
Available SinceSept. 17, 2018AvailabilityAcademic Institutions and Nonprofits only -
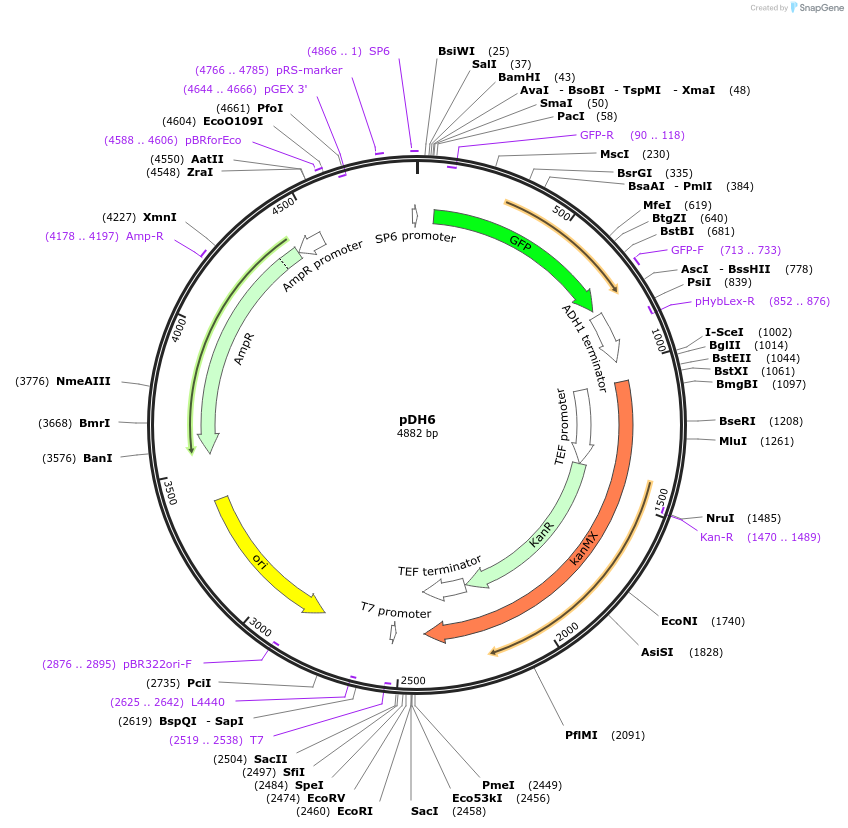
pDH6
Plasmid#83777PurposeUseful for tagging yeast proteins with C-terminal YFP tag; G418 selectionDepositorInsertYFP
TagsYFPExpressionYeastMutationS65G, V68L, Q69K, S72A, T203Y mutations added.Available SinceSept. 30, 2016AvailabilityAcademic Institutions and Nonprofits only -
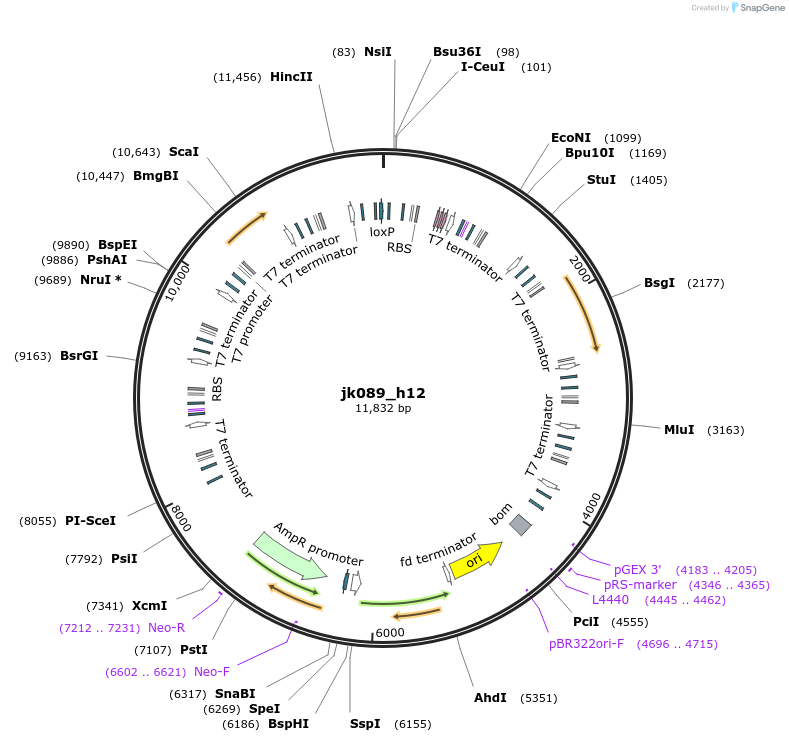
jk089_h12
Plasmid#112151PurposeC. thermophilum DASH/Dam1 complex (construct used for cryo-EM structure determination)DepositorInsertC. thermophilum DASH/Dam1 complex
Tagshis tag on Spc34 and strep tag on Dad1Available SinceSept. 20, 2018AvailabilityAcademic Institutions and Nonprofits only -
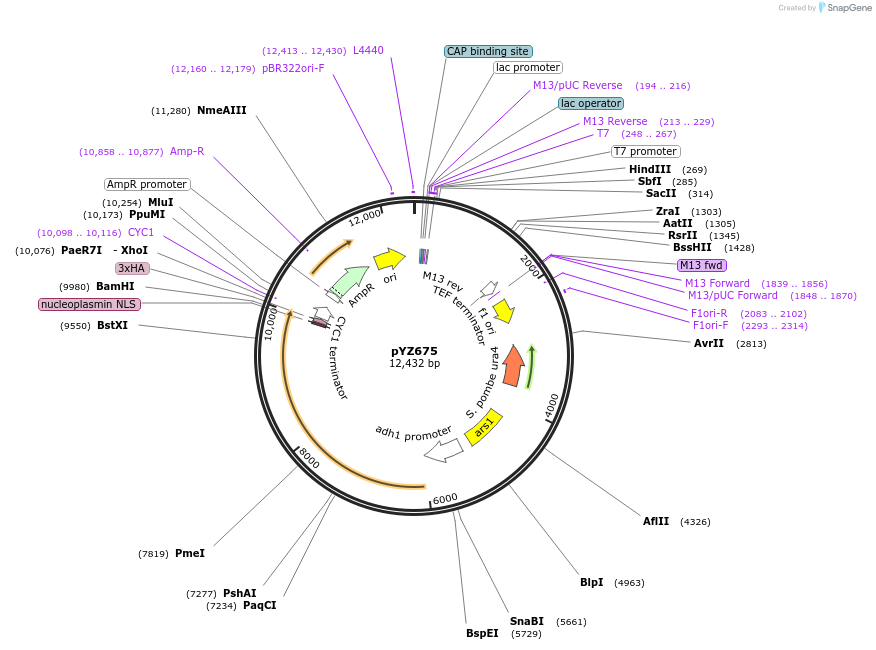
pYZ675
Plasmid#132954PurposecrRNA entry vector of Pfba1-BsaI pad-TEF1 Term with dFnCas12a Schizosaccharomyces pombeDepositorInsertdFnCas12a
UseCRISPR and Synthetic BiologyTagsnucleoplasmin NLSPromoteradh1Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
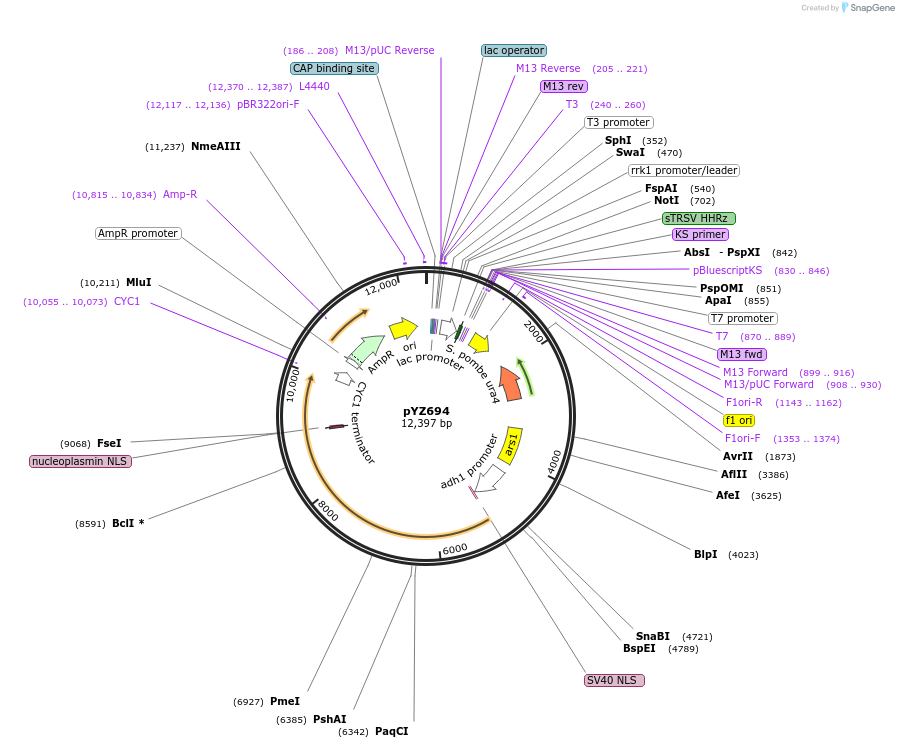
pYZ694
Plasmid#132956PurposecrRNA entry vector of Prrk1-Not1-HHR with dFnCas12a-Clr4 in Schizosaccharomyces pombeDepositorInsertdFnCas12a
UseCRISPR and Synthetic BiologyTagsSV40 NLS and nucleoplasmin NLSPromoteradh1Available SinceNov. 2, 2022AvailabilityAcademic Institutions and Nonprofits only -
p184.3
Plasmid#44521DepositorInsertHistone 3 N-terminus
TagsGSTExpressionBacterialMutationaa1-46Available SinceOct. 23, 2013AvailabilityAcademic Institutions and Nonprofits only -
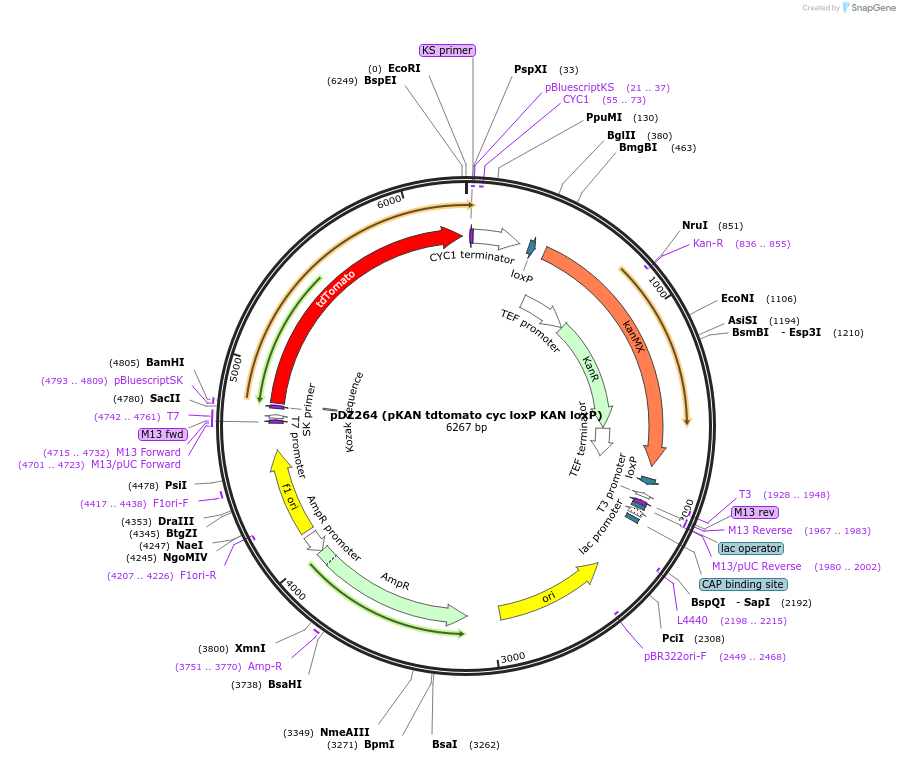
pDZ264 (pKAN tdtomato cyc loxP KAN loxP)
Plasmid#35193DepositorInserttdtomato cyc loxP KAN loxP
Available SinceMarch 5, 2012AvailabilityAcademic Institutions and Nonprofits only -
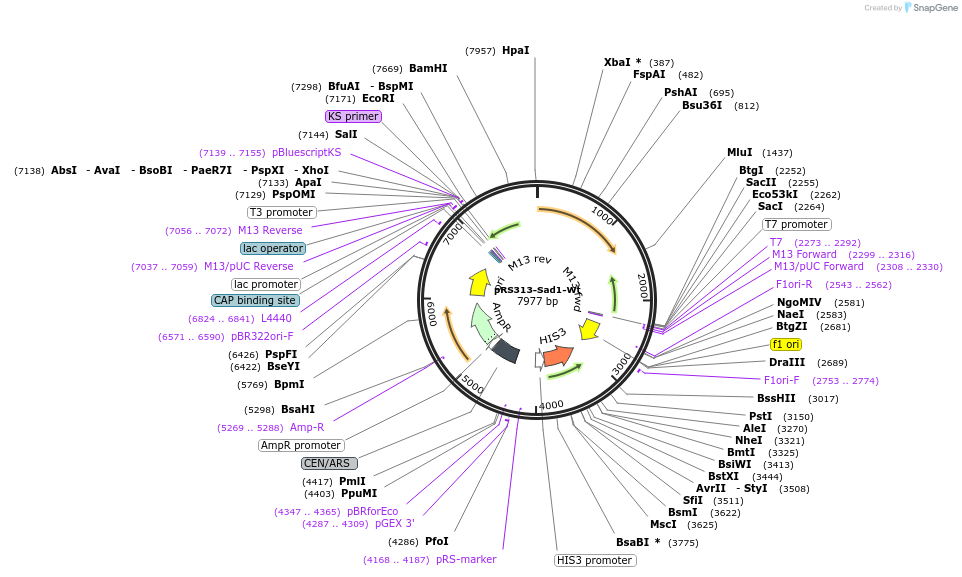
pRS313-Sad1-Wt
Plasmid#90308Purposeyeast expressionDepositorInsertSAD1
Tagsno tagExpressionYeastAvailable SinceJune 29, 2018AvailabilityAcademic Institutions and Nonprofits only -
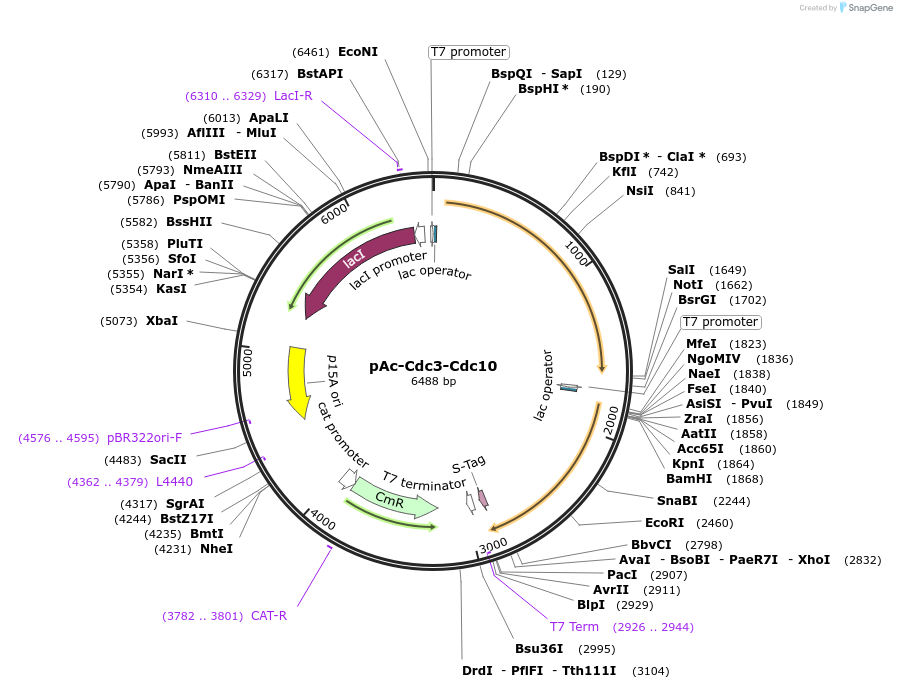
pAc-Cdc3-Cdc10
Plasmid#131160PurposeExpression of Septin rodsDepositorInsertCdc3 + Cdc10
UseBicistronicTagsCdc10: S TagExpressionBacterialPromoterT7Available SinceSept. 5, 2019AvailabilityAcademic Institutions and Nonprofits only